ABSTRACT
A number of genomic mutations that are thought to be strongly involved in the development of schizophrenia (SCZ) and autism spectrum disorder (ASD) have been identified. Abnormalities involving oligodendrocytes have been reported in SCZ, and as a related gene, oligodendrocyte lineage transcription factor 2 (OLIG2) has been reported to be strongly associated with SCZ. In this study, based on the common disease–rare variant hypothesis, target sequencing of candidate genes was performed to identify rare mutations with a high effect size and the possibility that the identified mutations may increase the risks of SCZ and ASD in the Japanese population. In this study, the exon region of OLIG2 was targeted; 370 patients with SCZ and 192 with ASD were subjected to next-generation sequencing. As a result, one rare missense mutation (A33T) was detected. We used the Sanger method to validate this missense mutation with a low frequency (<1%), and then carried out a genetic association analysis involving 3299 unrelated individuals (1447 with SCZ, 380 with ASD, and 1472 healthy controls) to clarify whether A33T was associated with SCZ or ASD. A33T was not found in either case group, and in only one control. We did not find evidence that p.A33T is involved in the onset of ASD or SCZ; however, associations with this variant need to be evaluated in larger samples to confirm our results.
Key Words: OLIG2, rare variant, schizophrenia, autism spectrum disorder
INTRODUCTION
Schizophrenia (SCZ) is a mental disorder with symptoms characterized by disturbances in thought, perception, emotion, language, sense of self, and behavior. Risk factors include birth complications, birth timing, maternal undernourishment, maternal influenza in pregnancy, family history, childhood trauma, social isolation, cannabis abuse, minority ethnicity, and urbanization.1 Although the cause is unknown, it is well understood that genetic factors make a significant contribution.2 Genome wide association studies (GWAS) have shown that common variants may be important factors contributing to an increased risk for SCZ,3-6 while epidemiological twin studies have estimated the heritability of SCZ to be 81%.7
Autism spectrum disorder (ASD) is a developmental disorder characterized by disturbances in social interaction and communication, and by restricted and repetitive behaviors.8 ASD is caused by a combination of genetic and environmental factors.9 A large number of studies have shown that SCZ and ASD are closely related, especially in terms of the undelaying genetic risk factors.10
Oligodendrocytes are a type of glial cells in the central nervous system that are responsible for the formation of myelin (the myelin sheath). The primary function of oligodendrocytes is to provide support and insulation to axons in the central nervous system, which they accomplish by creating the myelin sheath. Candidate gene association studies or genome-wide approaches have suggested that oligodendrocyte/myelin-related (OMR) abnormalities are a primary contributor to SCZ.11OLIG2 is a basic helix-loop-helix (bHLH) transcription factor essential for oligodendrocyte and motoneuron development in the spinal cord.12,13 In addition, OLIG2, as one of the OMR genes, is an excellent candidate gene for SCZ. Simultaneously, OLIG2 might affect the expression of many other OMR genes because it influences precursor as well as fully matured oligodendrocytes and is both necessary and sufficient for the genesis of oligodendrocytes and myelination. OLIG2 abnormalities have been observed in several postmortem brain studies of patients with SCZ.14-17 Although the relationship between ASD pathology and oligodendrocytes remains unclear, an analysis of autism model animals focusing on abnormalities in oligodendrocyte development is ongoing.18 Therefore, the main focus of the present study is to explore the association between OLIG2 and SCZ or ASD.
Recently, large-scale whole-exome sequencing19,20 and copy number variation (CNV) analysis using SCZ and ASD samples have indicated that deleterious rare genetic variants such as single-nucleotide variants (SNVs) and CNVs are also important genetic risk factors that increase the risk of ASD and SCZ.21,22 Furthermore, rare SNVs discovered from the sequencing of susceptibility genes may have a large effect size and account for a portion of the heritability of SCZ and ASD, and could contribute to our understanding of the etiopathology of neurodevelopmental disorders from a mechanistic point of view.23-25 Therefore, in the present study, we focused on the identification of rare SNVs within OLIG2 that may increase the risk for ASD or SCZ in the Japanese population.
MATERIALS AND METHODS
Participants
In this study, two independent groups of samples were used: one included 370 SCZ (196 men and 174 women; mean age ± standard deviation (SD), 49.7±14.8 years) and 192 ASD (149 men and 43 women; mean age±SD, 16.3±8.4 years) samples, which were re-sequenced for mutation discovery, and the other included 1447 SCZ (836 men and 611 women; mean age±SD, 50.8±15.3 years) samples, 380 ASD (292 men and 88 women; mean age±SD, 19.2±10.2 years) samples, and 1472 controls (667 men and 805 women; mean age±SD, 41.2±16.1 years) for genotyping and association analysis; these were used for an association analysis of specific variants detected in the first group.
All participants in this study were unrelated citizens of Japan, self-proclaimed as Japanese, and lived on the Japanese mainland. All participants in the control group were recruited from Nagoya University Hospital. All patients had been diagnosed with SCZ or ASD according to the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition.8 Healthy controls were selected from the general population and had no personal or family history of mental disorders (based on responses to a self-report questionnaire or evaluation through an unstructured interview to ensure that they had no relevant mental disorders). We explained the study purpose and methods to all participants (or their parents/guardians in the case they were minors) both verbally and in writing before obtaining consent. This study was approved by the Ethics Committees of the Nagoya University Graduate School of Medicine.
Preparation and resequencing
We used the QIAamp DNA blood kit or tissue kit (QIAGEN Ltd., Hilden, Germany) to extract genomic DNA from whole blood or saliva. We then used next-generation sequencing (NSC) technology on the Ion Torrent PGMTM to re-sequence the OLIG2 coding regions (Ensembl Transcript ID: ENST00000333337.3; Chr 21: 34,398,243-34,401,504;323 amino acids) according to protocols identified in the Ion AmpliSeqTM Library Preparation User Guide (Thermo Fisher Scientific, Waltham, MA, USA) and the Ion PGMTM Sequencing 200 Kit (Thermo Fisher Scientific). We first targeted specific PCR amplification and then prepared libraries to obtain 200-bp PCR fragments flanked by adaptor and barcode sequences; these sequences allowed sequencing and sample identification, respectively. The concentration of each library was determined using the Ion Library TaqMan Quantitation Assay Kit (Thermo Fisher Scientific). Amplified libraries were subjected to emulsion PCR and subsequent enrichment for template-positive Ion SphereTM particles (ISPs) using the Ion OneTouchTM system (Life Technologies, Carlsbad, CA, USA). ISPs were enriched and sequenced in a 200-bp configuration run using 318 chips (Life Technologies).
Data collection and analysis
Sequence reads were run through a data analysis using the Ion ReporterTM Software, Torrent SuiteTM Software 4.4, to generate sequence reads filtered according to the pipeline software quality controls and remove those that were low quality. Raw data output from the sequencer with the default parameters call quality ≥20 and read depth ≥10 were uploaded to the Torrent Server (Life Technologies) for variant calling with NCBI GRCh37 as a reference. All data were analyzed using Ingenuity Variant AnalysisTM (http://www.ingenuity.com/variants) for annotation and visualization.
The candidate variants we selected were defined as exonic or splice-site mutations with allele frequencies of <1% in following public exome databases: dbSNP Build 153 (https://www.ncbi.nlm.nih.gov/snp/), the Genome Aggregation Database (gnomAD) (https://gnomad.broadinstitute.org), and two databases as a reference for Japanese: the Human Genetic Variation Database (HGVD) (http://www.hgvd.genome.med.kyoto-u.ac.jp) and the Japanese Multi Omics Reference Panel (jMorp) (https://jmorp.megabank.tohoku.ac.jp/ijgvd/).
Validation
The filtered variants were validated using the Sanger method with the ABI 3130XL Genetic Analyzer (Life Technologies) and Sequence analysis software, version 6.0 (Applied Biosystems, Foster City, CA, USA) to analyze all sequence data.
Genetic association analysis
Each variant was subjected to a custom TaqMan® SNP Genotyping assay (Applied Biosystems). DNA samples were prepared on 384-well microtiter plates with four non-template controls as negative controls and samples with the variant as positive controls. The Genotyping Master Mix and Sequence Detection System (Applied Biosystems) was used to perform the data analysis and reactions according to the standard protocols. Differences in genotype distributions between cases and controls were tested using one-tailed Fisher’s exact tests.
We computed the statistical power and effective sample sizes using the Genetic Association Study Power Calculator (http://csg.sph.umich.edu/abecasis/gas_power_calculator).26
RESULTS
Mutation screening and data analysis
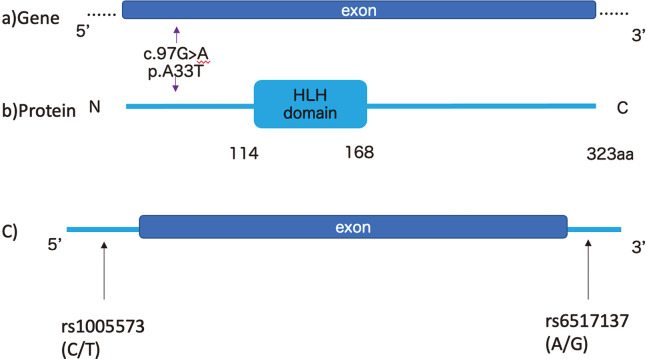
Our sequence data are available with the accession No. DRA004490DNA in the Data Bank of Japan (DDBJ) database (http://www.ddbj.nig.ac.jp). Re-sequencing of the OLIG2 coding regions using the Ion PGM platform identified one rare (minor allele frequency [MAF] <1%) missense, heterozygous variation within the OLIG2 exons (Table 1). A33T was not located in the bHLH domain according to UniProt (Figure 1). We searched the genetic databases dbSNP build153, gnmoAD, HGVD, and jMorp, which are frequently used to determine if this missense mutation is new variant. A33T had already been registered in gnmoAD and jMorp, but was not found in the other databases.
Table 1.
Details of discovered rare missense mutations and in silico analyses
| Chr | Gene
Region |
Positiona | Transcript
variant |
Protein
variant |
Translation
Impact |
SCZ
(n=370) |
ASD
(n=192) |
our
cohort MAFb |
dsSNPIDc | 1000
Genomes Frequency |
gnomADb | jMorp
(4.7KJPN)b |
HGVD
Frequency |
| 21 | Exonic | 34399267 | c.97G>A | p.A33T | missense | 0 | 2 | 0.0017 | rs1375013666 | — | 0.000004 | 0.0001 | — |
Chr: chromosome
SCZ: schizophrenia
ASD: autism spectrum disorders
MAF: minor allele frequency
gnomAD: The Genome Aggregation Database Exome Aggregation Consortium
jMorp: Japanese Multi Omics Reference Panel
HGVD: Human Genetic Variation Database
a Genomic position based on NCBI build GRCh 37 (Transcript ID ENST00000333337.3)
b minor allele count/total allele count
c dbSNP Build 153
Fig. 1.
Locations of novel rare variant in OLIG2
Fig. 1a:OLIG2 structure is based on ENST00000333337.3(GRCh37.p13).
Fig. 1b: The protein structure of OLIG2 is based on the Human Protein Reference Database. Arrows indicate the locations of novel rare variant in OLIG2. A33T is located in the N-terminal region.
Fig. 1c: SNPs which are reported to be associated with ASD and/or schizophrenia.
In silico analysis
We conducted in silico analysis of A33T to predict the possible impact of an amino acid substitution on the structure and function by using silico tools SIFT and PolyPhen-2. SIFT showed that it was tolerated and PolyPhen-2 showed that it was benign.
Genetic association analysis
We selected a large sample set for A33T in OLIG2 with 1447 SCZ cases, 380 ASD cases, and 1472 controls for the association analysis. We found no mutations in the SCZ and ASD cases, and only one in the controls. The results of the genetic association analysis of A33T are shown in Table 2. We then used the Genetic Power Calculator.27
Table 2.
Association analysis of rare missense variant
| variant | Genome data | Case (SCZ+ASD) | Control | |||||||
| p.A33T | positiona | Transcript
variant |
Genotype
countb |
MAF | P-valuec | Odds
radio |
Genotype
count |
MAF | ||
| 21:34399267 | c.97G>A | 0/0/1827 | 0 | N/A | N/A | 0/1/1472 | 0.0003 | |||
SCZ: schizophrenia
ASD: autism spectrum disorders
MAF: minor allele frequency
a Genomic position based on NCBI build GRCh 37.p13 (Transcript ID ENST00000371236.2).
b Genotype count; homozygote of minor allele/heterozygote/homozygote of major allele.
c P-values were calculated with Fisher’s exact test (2 × 2 contingency table, one-tail).
The total number of cases was 1827, disease prevalence of 0.01, significance level of 0.05, and MAF 0.0003 (observed in our sample). We determined that our sample had sufficient statistical power (beta >80%), but only with a genotype relative risk (GRR) >7.
DISCUSSION
Several studies on common variants as susceptibility genes for SCZ and ASD have been reported,24-25 but to the best of our knowledge, few studies have investigated rare variants for those illnesses. We performed a systematic study by sequencing the coding regions of OLIG2 genes in SCZ and ASD. We performed mutation screening of exons in OLIG2 for 562 Japanese patients and detected one rare nonsynonymous mutation (SNV). One SNV (A33T) was present at a very low frequency in some databases. Association analysis was then performed with a cohort comprising 1827 (1447 SCZ and 380 ASD) cases and 1472 controls. Although several similar studies have identified a statistical association between rare SNVs and neurodevelopmental disorders, we found no statistically significant associations between the rare variant we investigated in this study and SCZ or ASD.
LIMITATIONS
This study has several limitations. First, our mutation screening sample was relatively small, so we cannot exclude the possibility that there may be other more promising rare missense variants within OLIG2. Furthermore, regarding the association analysis and samples used in the current study, our statistical power calculation showed that the beta level was >80% only in the case that the GRR was >7. This relatively high GRR indicates that our study may be underpowered, considering that the rare variant associated with neurodevelopmental disorders may have a GRR of 2.28
CONCLUSIONS
We sequenced the exons of OLIG2 in Japanese patients with SCZ or ASD and identified one rare missense variant (A33T) that may contribute to increased susceptibility to SCZ and ASD. However, we could not confirm or deny the association between A33T and ASD or SCZ in the follow-up analysis because of the variant’s low frequency. Therefore, to assess the relevance of this variant unequivocally, further studies involving much larger sample sizes are needed. In addition to evaluating comprehensively the contribution of rare SNVs located within OMR genes, detailed sequencing studies of genes other than OLIG2 (ie, OLIG1, MOG, and SOX) may be warranted.
ACKNOWLEDGMENTS
This study was supported by research grants from the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant No. Scientific Research (A)(18H04040) and AMED under grant Nos. JP20dm0107087, P20dm0207075, JP20dk0307081, and JP20ek0109488.
FUNDING/SUPPORT
Dr N Ozaki has received research support or speakers’ honoraria from, or has served as a consultant to, Sumitomo Dainippon, Eisai, Otsuka, KAITEKI, Mitsubishi Tanabe, Shionogi, Eli Lilly, Mochida, DAIICHI SANKYO, Nihon Medi-Physics, Takeda, Meiji Seika Pharma, EA Pharma, Pfizer, MSD, Lundbeck Japan, Taisho Pharma, outside the submitted work.
CONFLICT OF INTEREST
All authors have no conflicts of interest to declare.
Abbreviations
- SCZ
schizophrenia
- ASD
autism spectrum disorder
- OLIG2
oligodendrocyte lineage transcription factor 2
- SNVs
single-nucleotide variants
REFERENCES
- 1.Hany M, Rehman B, Azhar Y, Chapman J. Schizophrenia. In: StatPearls. Treasure Island, FL: StatPearls Publishing; 2022.
- 2.Riley B. Linkage studies of schizophrenia. Neurotox Res. 2004;6(1):17–34. doi: 10.1007/BF03033293. [DOI] [PubMed]
- 3.Pulver AE, Karayiorgou M, Wolyniec PS, et al. Sequential strategy to identify a susceptibility gene for schizophrenia: report of potential linkage on chromosome 22q12–q13.1: Part 1. Am J Med Genet. 1994;54(1):36–43. doi: 10.1002/ajmg.1320540108. [DOI] [PubMed]
- 4.Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71(4):877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed]
- 5.Petryshen TL, Middleton FA, Kirby A, et al. Support for involvement of neuregulin 1 in schizophrenia pathophysiology. Mol Psychiatry. 2005;10(4):366–374,328. doi: 10.1038/sj.mp.4001608. [DOI] [PubMed]
- 6.Seshadri S, Kamiya A, Yokota Y, et al. Disrupted-in-Schizophrenia-1 expression is regulated by beta-site amyloid precursor protein cleaving enzyme-1-neuregulin cascade. Proc Natl Acad Sci U S A. 2010;107(12):5622–5627. doi: 10.1073/pnas.0909284107. [DOI] [PMC free article] [PubMed]
- 7.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed]
- 8.American Psychiatric Association. Diagnostic & Statistical Manual of Mental Disorders. 5th ed. Washington, DC: American Psychiatric Association; 2013.
- 9.Chaste P, Leboyer M. Autism risk factors: genes, environment, and gene-environment interactions. Dialogues Clin Neurosci. 2012;14(3):281–292. doi: 10.31887/DCNS.2012.14.3/pchaste. [DOI] [PMC free article] [PubMed]
- 10.Chisholm K, Lin A, Abu-Akel A, Wood SJ. The association between autism and schizophrenia spectrum disorders: a review of eight alternate models of co-occurrence. Neurosci Biobehav Rev. 2015;55:173–183. doi: 10.1016/j.neubiorev.2015.04.012. [DOI] [PubMed]
- 11.Roussos P, Haroutunian V. Schizophrenia: susceptibility genes and oligodendroglial and myelin related abnormalities. Front Cell Neurosci. 2014;8:5. doi: 10.3389/fncel.2014.00005. [DOI] [PMC free article] [PubMed]
- 12.Ono K, Takebayashi H, Ikeda K, et al. Regional- and temporal-dependent changes in the differentiation of Olig2 progenitors in the forebrain, and the impact on astrocyte development in the dorsal pallium. Dev Biol. 2008;320(2):456–468. doi: 10.1016/j.ydbio.2008.06.001. [DOI] [PubMed]
- 13.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109(1):61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed]
- 14.Tkachev D, Mimmack ML, Ryan MM, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362(9386):798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed]
- 15.Katsel P, Davis KL, Haroutunian V. Variations in myelin and oligodendrocyte-related gene expression across multiple brain regions in schizophrenia: a gene ontology study. Schizophr Res. 2005;79(2–3):157–173. doi: 10.1016/j.schres.2005.06.007. [DOI] [PubMed]
- 16.Iwamoto K, Bundo M, Yamada K, et al. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005;25(22):5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed]
- 17.Mauney SA, Pietersen CY, Sonntag KC, Woo TW. Differentiation of oligodendrocyte precursors is impaired in the prefrontal cortex in schizophrenia. Schizophr Res. 2015;169(1–3):374–380. doi: 10.1016/j.schres.2015.10.042. [DOI] [PMC free article] [PubMed]
- 18.Bronzuoli MR, Facchinetti R, Ingrassia D, et al. Neuroglia in the autistic brain: evidence from a preclinical model. Mol Autism. 2018;9:66. doi: 10.1186/s13229-018-0254-0. [DOI] [PMC free article] [PubMed]
- 19.Hashimoto R, Nakazawa T, Tsurusaki Y, et al. Whole-exome sequencing and neurite outgrowth analysis in autism spectrum disorder. J Hum Genet. 2016;61(3):199–206. doi: 10.1038/jhg.2015.141. [DOI] [PMC free article] [PubMed]
- 20.Hornig T, Grüning B, Kundu K, et al. GRIN3B missense mutation as an inherited risk factor for schizophrenia: whole-exome sequencing in a family with a familiar history of psychotic disorders. Genet Res (Camb). 2017;99:e1. doi: 10.1017/S0016672316000148. [DOI] [PMC free article] [PubMed]
- 21.Kushima I, Aleksic B, Nakatochi M, et al. High-resolution nucleotide number variation analysis of schizophrenia in Japan. Mol Psychiatry. 2017;22(3):430–440. doi: 10.1038/mp.2016.88. [DOI] [PubMed]
- 22.Li Z, Chen J, Xu Y, et al. Genome-wide analysis of the role of copy number variation in schizophrenia risk in Chinese. Biol Psychiatry. 2016;80(4):331–337. doi: 10.1016/j.biopsych.2015.11.012. [DOI] [PubMed]
- 23.Kim MJ, Biag J, Fass DM, et al. Functional analysis of rare variants found in schizophrenia implicates a critical role for GIT1-PAK3 signaling in neuroplasticity. Mol Psychiatry. 2017;22(3):417–429. doi: 10.1038/mp.2016.98. [DOI] [PMC free article] [PubMed]
- 24.Georgieva L, Moskvina V, Peirce T, et al. Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proc Natl Acad Sci U S A. 2006;103(33):12469–12474. doi: 10.1073/pnas.0603029103. [DOI] [PMC free article] [PubMed]
- 25.Usui H, Takahashi N, Saito S, et al. The 2',3'-cyclic nucleotide 3'-phosphodiesterase and oligodendrocyte lineage transcription factor 2 genes do not appear to be associated with schizophrenia in the Japanese population. Schizophr Res. 2006;88(1–3):245–250. doi: 10.1016/j.schres.2006.07.019. [DOI] [PubMed]
- 26.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed]
- 27.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed]
- 28.Rees E, O’Donovan MC, Owen MJ. Genetics of schizophrenia. Curr Opin Behav Sci. 2015;2:8–14. doi: 10.1016/j.cobeha.2014.07.001. [DOI]