Abstract
Escherichia coli Fis is a small DNA binding and bending protein that has been implicated in a variety of biological processes. A minimal promoter sequence consisting of 43 bp is sufficient to generate its characteristic growth phase-dependent expression pattern and is also subject to negative regulation by stringent control. However, information about the precise identification of nucleotides contributing to basal promoter activity and its regulation has been scant. In this work, 72 independent mutations were generated in the fis promoter (fis P) region from −108 to +78 using both random and site-directed PCR mutagenesis. β-Galactosidase activities from mutant promoters fused to the (trp-lac)W200 fusion on a plasmid were used to conclusively identify the sequences TTTCAT and TAATAT as the −35 and −10 regions, respectively, which are optimally separated by 17 bp. We found that four consecutive substitutions within the GC-rich sequence just upstream of +1 and mutations in the −35 region, but not in the −10 region, significantly reduced the response to stringent control. Analysis of the effects of mutations on growth phase-dependent regulation showed that replacing the predominant transcription initiation nucleotide +1C with a preferred nucleotide (A or G) profoundly altered expression such that high levels of fis P mRNA were detected during late logarithmic and early stationary phases. A less dramatic effect was seen with improvements in the −10 and −35 consensus sequences. These results suggest that the acute growth phase-dependent regulation pattern observed with this promoter requires an inefficient transcription initiation process that is achieved with promoter sequences deviating from the −10 and −35 consensus sequences and, more importantly, a dependence upon the availability of the least favored transcription initiation nucleotide, CTP.
Fis is a small nucleoid-associated protein found in several enteric bacteria, including Escherichia coli, Salmonella typhimurium, Klebsiella pneumoniae, Serratia marcescens, Erwinia carotovora, Yersinia pestis, Proteus vulgaris, and three nonenteric bacteria (6, 22, 27, 37). Its gene, located at 73.4 min on the E. coli chromosome, is part of a two-gene operon transcribed from a single promoter (4, 36). Between the promoter and fis lies a well-conserved open reading frame (ORF1), whose function has not been described. Fis, however, has been shown to be involved in a variety of cellular processes. For example, it stimulates certain site-specific DNA recombination reactions (2, 3, 16, 23, 26, 47), stimulates stable RNA transcription (34, 42), regulates initiation of DNA replication at oriC (11, 14, 51), and modulates DNA topology (44).
Fis exhibits a distinct expression pattern (4, 35, 36, 47). When stationary-phase cells are batch cultured in rich medium such as Luria-Bertani (LB) medium, Fis protein levels rapidly increase from less than 250 to over 25,000 dimers per cell in the early logarithmic growth phase. Protein levels then decrease continually to less than 1% peak levels by early stationary phase (4). Transcription control is most likely the primary determinant of this peculiar expression pattern. mRNA and protein expression patterns are similar, and mRNA decay rates do not contribute to this pattern (4, 40). However, little is known about the regions of the fis promoter (fis P) that mediate this unique expression pattern.
Several observations demonstrated that fis P is controlled by negative autoregulation. Maximal fis mRNA levels were found to be sixfold higher in fis mutant cells than in otherwise isogenic fis+ cells (4). Likewise, β-galactosidase activities from fis P fused to lacZ are over fourfold higher in fis mutant cells than in fis+ cells (36). Six Fis binding sites have been identified in the promoter region, and binding of RNA polymerase to fis P in vitro was prevented if Fis was also present (4). So far, Fis sites I (centered at +24) and II (centered at −44) have been shown to be critical for autoregulation, with site II playing the more important role (36, 40). On the other hand, fis P transcription is stimulated in vivo three- to fourfold by integration host factor (IHF) when bound to a site centered at −116 (40). However, while Fis and IHF have opposing effects on the magnitude of fis P expression, neither is responsible for the growth phase-dependent regulation (4, 40). In fact, this expression pattern can be generated by the fis P sequences from −38 to +5, which lack the IHF and Fis binding sites (36, 40). Thus, this 43-bp region must contain the minimal elements required for basal transcription activity, as well as growth phase-dependent regulation.
Stringent control was also shown to negatively regulate expression from minimal fis P (36). When cells were starved for isoleucine, mRNA synthesized from fis P quickly subsided but was restored upon addition of chloramphenicol. A sequence of seven consecutive G · C base pairs starting 2 bp downstream from the putative −10 region is a likely candidate for a discriminator, which has been implicated in the stringent control of many promoters (9). Replacement of these sequences with the corresponding region from the β-lactamase (bla) promoter resulted in loss of both stringent and growth phase-dependent control (36). This was taken as evidence that the GC-rich motif was required for both regulatory processes and implied that growth phase-dependent regulation could be explained in terms of a sensitivity to variations in intracellular ppGpp levels. However, fis P maintains its unusual expression pattern in a relA spoT strain, indicating that growth phase-dependent regulation is not dependent on ppGpp levels (4).
Much uncertainty remains regarding the function and regulation of fis P, and this warrants a more detailed characterization of the nucleotide sequences comprising this promoter. Thus, we generated a comprehensive collection of mutations within the fis P region and tested their effects on transcription in vivo, stringent control, and growth phase-dependent regulation. We show that three DNA elements that affect growth phase-dependent regulation include the suboptimal −10 and −35 regions and the nucleotide at or near the transcriptional start site. We also found that single-point mutations in the GC-rich sequence in fis P do not affect the stringent control, but changing four sequential G · C base pairs to A · T base pairs significantly reduces susceptibility to this regulation. Moreover, we found that promoter mutants altered in growth phase-dependent regulation are still subject to stringent control and vice versa. Finally, mutations obtained in this work provided a functional basis for a reliable identification of the fis P recognition sequences.
MATERIALS AND METHODS
Chemicals, enzymes, and growth media.
Most chemicals were purchased from Sigma Chemical Co., Fisher Scientific Co., Life Technologies Inc. (Gibco BRL), Pharmacia, or VWR Scientific. Enzymes were from New England Biolabs, Promega Corp., or Boehringer Mannheim Corp. Radioisotopes ([γ-32P]ATP and [α-32P]dATP) were from Amersham Life Sciences. Most oligonucleotides were generated by a Perkin-Elmer automated DNA synthesizer operated within the Department of Biological Sciences, State University of New York at Albany; some were purchased from Ransom Hill Bioscience, Inc., Ramona Calif., or Biosynthesis, Inc., Lewisville, Tex.
Bacterial culture media were from Difco Laboratories. Cultures were grown at 37°C in LB medium or plated on MacConkey agar medium supplemented with 5% lactose (MacConkey-lactose) (43). For the stringent-control assay, cells were cultured in M9 salts (43) supplemented with 0.2% glucose, 4-μg/ml thiamine, 40-μg/ml thymine, and 18 amino acids (no valine or isoleucine) at 100 μg/ml each. Plasmid-containing cells were selected by adding ampicillin at 100 μg/ml to the growth medium.
Bacterial strains and plasmids.
RZ211 [F− Δ(lac pro) thi ara str recA56 srl] (24) and RJ1561 (RZ211 fis::767) (21) were used for the β-galactosidase assays, while RJ1561 was used in primer extension and stringent-control assays. RJ1882 [MG1655 (relAΔ1251 spoTΔ1207 fis::985Ω)] (R. C. Johnson, University of California at Los Angeles) was transformed with pTP127 and used in the stringent-control assay.
Plasmid pRJ800 is a pBR322-based plasmid containing the pUC18 polylinker region, followed by the (trp-lac)W200 fusion (4). All fis P regions were cloned into this plasmid such that transcription of the trp-lac fusion was under the control of fis P. pRJ1028 carries wild-type fis P sequences from −375 to +78 within the HincII site of pRJ800, and pTP127 contains the wild-type fis P sequences from −108 to +105 cloned into the KpnI and XbaI sites of pRJ800 (40). A number of fis P mutations were generated in the region from −108 to +78 as described below. These plasmids are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmida | Description or mutation(s)b | Plasmida | Mutation(s)b | |
|---|---|---|---|---|
| pRJ800 | pBR322-based vector | pKW241 | Δ-36T | |
| pRJ1028 | Wild-type fis P from −375 to +78 | pKW242 | +39A→G | |
| pTP127 | Wild-type fis P −108 to +105 | pKW243 | +3T→A | |
| pKW189 | −20A→G | pKW244 | −7A→G | |
| pKW197 | 13 bp deleted in region downstream of +56 (Δ13) | pKW245 | −59A→T | |
| pKW198 | −59A→G | pKW247 | −72T, −11A→G | |
| pKW199 | −20A→G, +27T→A | pKW248 | −57A→T | |
| pKW200 | −33C→A | pKW249 | −75T→C | |
| pKW201 | −57A→G | pKW250 | +27→C, +39A→G | |
| pKW202 | Δ13, −34T→G, +21A→T | pCA274c | −1C→T | |
| pKW203 | −54A→G, +37G→A | pCA275c | −2G→A | |
| pKW204 | −57A→C, −72T→A | pCA176c | −3C→T | |
| pKW205 | −37C→A, +52A→C | pCA277c | −4C→T | |
| pKW206 | −12A→T | pCA278c | −5G→T | |
| pKW207 | +5C→T | pCA279c | −5G→T | |
| pKW208 | −70T→C | pCA282c | +1C→A | |
| pKW211 | +21A→G | pCA286c | +3T→C | |
| pKW212 | −61T→C, +42G→T | pCA287c | +4G→T | |
| pKW213 | −35T→A | pKW288cd | −10T→A, −11A→T | |
| pKW214 | −19A→G | pKW289c | A inserted at −22 | |
| pKW215 | Δ−70T | pKW290c | C inserted at −22 | |
| pKW216 | −71T→G | pKW292 | −10T→A | |
| pKW217 | −69G→A | pKW293 | −34T→G | |
| pKW218 | −35T→G | pKW294 | −12A→G | |
| pKW219 | Δ−42T, Δ−43T, −10T→G | pKW295 | +2T→C | |
| pKW220 | Δ+17G | pKW296 | −36T→G | |
| pKW221 | −11A→G | pKW297 | −6C→T | |
| pKW222 | −75T→C, +22T→C | pKW298 | −8T→C | |
| pKW223 | −59A→G, −87T→A, +34T→A | pKW299 | −34T→G | |
| pKW224 | +2T→A | pKW300 | −37C→A | |
| pKW225 | −38C→T | pKW301ce | −33C→G, −31T→C, −30C→A | |
| pKW226 | −14G→C | pKW302c | −33C→G | |
| pKW227 | −21A→C | pKW303c | Δ−25G | |
| pKW228 | +17T→G | pKW304 | −10T→C | |
| pKW229 | −61T→A | pCA324c | +1C→G | |
| pKW230 | −34T→A | pCA325c | −2G→T | |
| pKW240 | −32A→T | pKW331c | GCCG→ATTT at −5 to −2 |
Plasmids pRJ800 and pTP127 are described in references 4 and 40, respectively. pRJ1028 was from R. C. Johnson. pKW292 to pKW298 were derived from mutated fis P on pRJ1028-based plasmids made by B. Hu and R. C. Johnson. All of the other plasmids listed were generated in this work. pCA274 to pCA279, pCA282, pCA286, pCA287, pCA324, and PCA325 and pKW331 contain fis P sequences from −108 to +105. All of the others listed contain fis P sequences from −108 to +78.
Nucleotide changes in the fis P region are given for the sense strand and are numbered relative to the predominant transcriptional start site. Δ13 is an aberrant mutation in which nucleotides +56, +60, +66, +67, +72, and +77 and the proximal 7 bp of the vector sequence are deleted. In addition, four substitutions are found in this region: +61T→G, +65G→A, +72C→T, and +76G→A.
Mutations were made by site-directed PCR mutagenesis.
These mutations create a consensus −10 region.
These mutations create a consensus −35 region 16 bp upstream from the −10 region.
Random PCR mutagenesis was performed as described below using pRJ1028 or pTP127 as the template. The regions from −108 to +78 in the resulting mutated pRJ1028-derived plasmids were subsequently amplified using oligonucleotides oRO165 and oRO150 (Fig. 1) and cloned into the KpnI and SphI sites of pRJ800. pKW292 to pKW298 were similarly constructed from mutated pRJ1028 derivatives originally isolated in the laboratory of R. C. Johnson and resequenced in our laboratory for verification. pCA274 to pCA279, pCA282, pCA286, pCA287, pCA324, and pCA325 all contain the fis P sequences from −108 to +105 and a single-point mutation created by site-directed PCR mutagenesis using pTP127 as the template; pKW331 is a similar construct with four consecutive base pairs substituted (Table 1). They were each digested and cloned into the KpnI and XbaI sites of pRJ800.
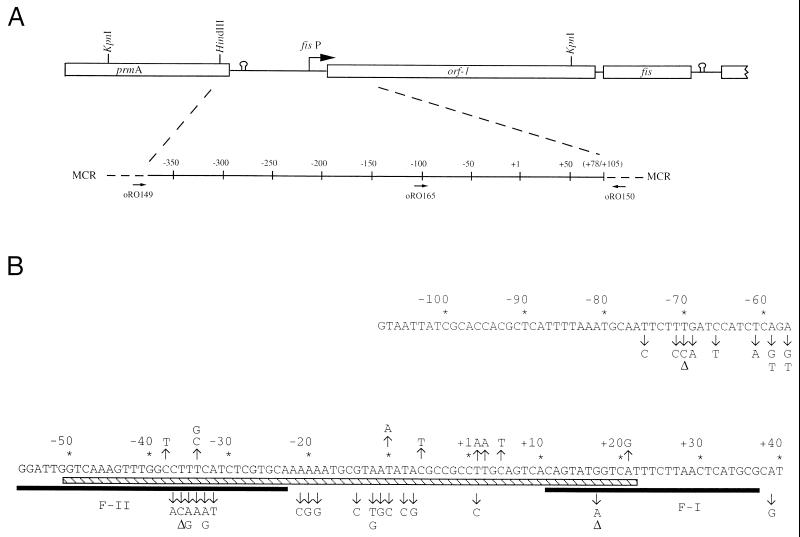
FIG. 1.
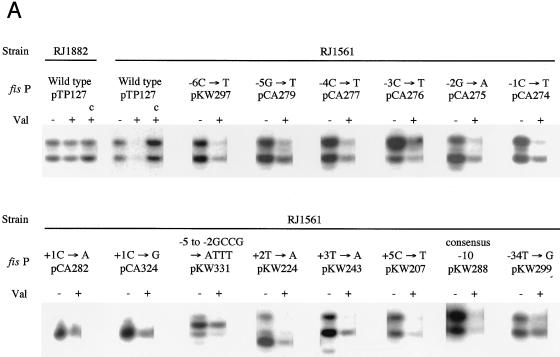
Random point mutations within the E. coli fis P region. (A) Schematic representation of the fis operon, which consists of fis, an open reading frame (ORF1), and fis P. The gene encoding L11 methyltransferase (prmA) is located upstream of the fis operon (49). Open boxes denote genes, the arrow represents the transcription initiation site, and the stem-loops represent presumed regions of transcription termination. The bottom portion shows the region of fis P that was subject to PCR mutagenesis and the oligonucleotides used in the PCRs (arrows). MCR is the pUC18-derived multicloning region within pRJ800, and numbering is relative to the predominant transcription start site. (B) fis P region containing the single point mutations. The nucleotide sequence is that of the nontemplate strand. The predominant transcription start site is identified as +1. Positions substituted are indicated by arrows with the nucleotide change(s); those that gave a more intense red colony color on MacConkey-lactose agar (compared to pTP127) are shown above the sequence, and those that gave a white or a less intense red color are shown below the sequence. ▵, single-base-pair deletion;  , Fis binding sites F-I and F-II; ▧ς70 RNA polymerase binding site (4).
, Fis binding sites F-I and F-II; ▧ς70 RNA polymerase binding site (4).
Copy number for various pRJ800-based plasmids carrying wild-type or mutant fis P was found not to vary by more than 6%. Triplicate samples of saturated overnight cultures of RJ1561 cells containing pTP127, pKW288, pKW299, pKW296, and pKW298 were diluted 1:75 into fresh LB medium and grown for 90 min. Plasmid DNA was extracted by the alkaline lysis method (43) from cell pellets containing equivalent amounts of RJ1561 cells carrying one of the plasmids examined. In addition, the cell pellets contained equivalent amounts of RJ1561 cells carrying pUC18 as an internal control. DNA from 50% of this preparation was linearized with HindIII and separated by electrophoresis on a 0.8% agarose gel in TBE buffer (43). The gel was photographed, and the negative was scanned with a UMAX S-6E scanner. The DNA bands were quantified by using NIH Image, version 1.61 (National Institutes of Health; available at http://rsb.info.nih.gov/nih-image). The intensities of the pUC18 bands were used as an internal control to correct for differences in handling and loading.
PCR mutagenesis.
Random PCR mutagenesis was performed by using a modified procedure (12). Reactions were performed in a 100-μl volume containing 50 ng of a plasmid (pRJ1028 or pTP127) template, 250 ng of each primer (oRO150 and either oRO165 or oRO149), 16.6 mM (NH4)2SO4, 67 mM Tris (pH 8.0), 6.7 mM Na2EDTA, 0.17-mg/ml bovine serum albumin, 10 mM β-mercaptoethanol, 10% dimethyl sulfoxide, 1 mM each deoxynucleoside triphosphate, 6.6 mM MgCl2, and 5 U of Taq polymerase from Promega Corp. The PCR products were purified by polyacrylamide gel electrophoresis, eluted by the crush-and-soak method (43), digested with KpnI and SphI (for pTP127-based fragments) or EcoRI and SphI (for pRJ1028-based fragments), and then cloned into the same sites in pRJ800. The resulting plasmids were transformed into RJ1561 and plated on MacConkey-lactose agar. Mutant phenotypes were screened by comparing colony color with that of cells containing either pTP127 or pRJ1028, as appropriate. Colonies carrying plasmids with up-promoter mutations were identified by a more intense red color. Conversely, colonies containing down-promoter mutations were identified by either a white or a more slowly developing red color. Plasmids from isolated colonies giving an altered phenotype were extracted, and the respective fis P regions were sequenced.
Site-directed mutagenesis was performed by a two-step megaprimer method (5) using Taq polymerase from Boehringer Mannheim under conditions specified by the manufacturer. In the first reaction, an upstream primer containing the desired mutation(s) was used together with the downstream primer, oRO150 (Fig. 1). One-half of this amplified product was then used as the downstream megaprimer in a second reaction with oRO165 as the upstream primer and 500 ng of template DNA. All PCR products were purified by polyacrylamide gel electrophoresis, eluted by the crush-and-soak method (43), digested with KpnI and XbaI or KpnI and SphI, as appropriate, and cloned into the same sites in pRJ800. All fis P regions were sequenced to verify the presence of the mutation and to ensure that no other mutations occurred during amplification.
DNA sequencing reactions.
Dideoxynucleotide sequencing was performed on alkali-denatured double-stranded plasmid DNA using Sequenase version 2.0 (U.S. Biochemicals) under conditions specified by the supplier.
β-Galactosidase assays.
β-Galactosidase assays were performed essentially as previously described (32). Saturated bacterial cultures were diluted 1:75 in fresh LB medium and grown at 37°C with constant shaking for 90 min, at which time fis P expression is near peak levels. The values reported are averages of at least three independent assays.
Primer extension analysis.
Primer extension reactions were performed essentially as previously described (18). For analysis of growth phase-dependent regulation, saturated cultures of RJ1561 containing the various fis P constructs were diluted 1:20 in fresh LB medium and grown at 37°C with constant shaking. At various times after subculturing, samples were removed and total RNA was extracted as previously described (8). For the data in Fig. 3, cells were harvested after 90 min of growth at 37°C. Primer extension reactions were performed with 10 μg of total RNA and 2 pmol of a DNA primer (oRO109) that hybridizes to the fis P mRNA from +56 to +40. The products were separated on 8% polyacrylamide–8 M urea gels and autoradiographed. Transcripts generated from chromosomal fis P were virtually undetectable under our exposure conditions. In some cases, primer-extended products were quantitated by using a Storm 860 PhosphorImager and ImageQuaNT software (Molecular Dynamics, Inc., Sunnyvale Calif.). Based on quantitation of extended products and free primer, the primer concentration in these reactions was in excess of fis P transcripts by greater than 75-fold, indicating that the primer concentration was not limiting.
FIG. 3.
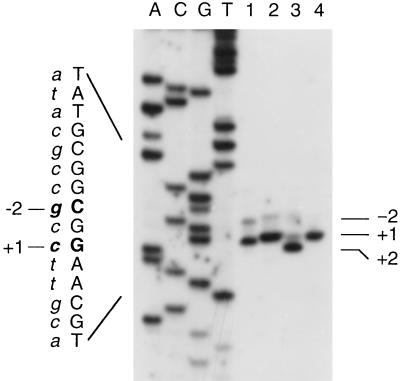
Transcription initiation sites in wild-type and mutant fis P. Primer extension reactions were performed with 32P-labeled oRO109 and 10 μg of total RNA obtained from RJ1561 carrying pTP127-based plasmids containing wild-type fis P (lane 1), or fis P with the mutation +3T→A (lane 2), +2T→A (lane 3), or +1C→A (lane 4). DNA sequencing reactions were performed with the same labeled primer and electrophoresed in parallel in 8% polyacrylamide–8 M urea gels. These reactions are indicated above the gel by A, C, G, and T. The sequence of part of the template DNA strand directly read from the autoradiograph is indicated on the left with capital letters, while the complementary strand is in lowercase italics. The positions of transcription initiation sites are indicated.
For analysis of stringent control, amino acid starvation was induced by adding valine to a cell culture lacking valine and isoleucine. Excess valine inhibits the biosynthesis of isoleucine and valine, thereby starving the cells for isoleucine (29). Initially, we used modified Hershey’s medium, which has been successfully applied in these kinds of experiments (15, 36), but later switched to a simpler, M9 salts-based medium that gave identical results. Saturated cultures of RJ1561 carrying various fis P-containing plasmids were diluted in supplemented M9 medium to an OD600 of about 0.1 and grown for 2 doublings. At this point, 10 ml was harvested and 500-μg/ml valine was added to the remaining culture. Ten minutes later, a second 10-ml sample was harvested. To verify that chloramphenicol could restore transcription of fis P, 200 μg/ml was added to cultures of RJ1561 or RJ1882 carrying pTP127 10 min after addition of valine. A final 10-ml sample was harvested 10 min after chloramphenicol addition. Total RNA was extracted from each sample, and primer extensions were performed as described above.
Nucleotide numbering reassignment.
When cells are grown in LB medium, transcription from fis P initiates primarily with CTP and less efficiently with GTP 2 bases upstream from the predominant start (4) (see Fig. 3). However, transcription was originally reported to initiate with the GTP, and the corresponding G in the DNA sequence was designated +1 (36). Since the present work characterizes the fis P region in detail, including the region around the start of transcription, we adjusted its nucleotide numbering to reflect the preference for initiation with CTP. All of the nucleotides in fis P are therefore numbered with the predominant start site C as +1 (Fig. 1).
RESULTS
Generation of mutations in the fis P region.
To identify DNA sequences downstream of the ihf site involved in regulating fis expression, we generated point mutations within the fis P region from −108 to +78 by PCR mutagenesis. This region allows transcription to proceed without stimulation by IHF but is still subject to autoregulation and growth phase-dependent control (36, 40). Mutations were screened for the ability to increase or decrease transcription in vivo. This was done by fusing the mutagenized fis P regions to the (trp-lac)W200 fusion in plasmid pRJ800, transforming the plasmid into E. coli RJ1561 (RZ211 fis::767), and plating the bacteria on MacConkey agar containing lactose and ampicillin. Colony color was compared to that of cells carrying the wild-type fis P region in pTP127. From this screen, we obtained 54 different mutations, of which 42 consisted of single substitutions or deletions (Fig. 1), 8 consisted of double mutations, and 4 consisted of 3 or more mutations (Table 1). About 10% of these mutations were obtained two or three times, suggesting that salient regulatory regions were likely to have been targeted by our mutagenesis procedure. The single mutations were clustered in the regions from −75 to −57, −38 to −32, −21 to −19, −14 to −6, and +2 to +5, suggesting that these may represent regulatory regions. Point mutations were also seen at +17, +21, and +39. In addition, 15 single-point mutations and 3 multiple mutations were specifically targeted, for a total of 72 mutations within this promoter region (Table 1). Mutations located downstream of +10 or upstream of −40 fell outside the core region known to contain the sequence required for growth phase-dependent regulation and stringent control (36, 40) and were not further analyzed.
The presence or absence of a functional fis gene has been found not to alter the copy number of pRJ800-based plasmids by more than 8% (40). We also showed that copy numbers for various pRJ800-based plasmids containing mutated fis P regions giving rise to notable changes in β-galactosidase activity were within 6% (<1.1-fold). Thus, variations in plasmid copy number had no appreciable influence on our analysis of effects of mutations on fis P transcription from these plasmids.
The fis P −10 region.
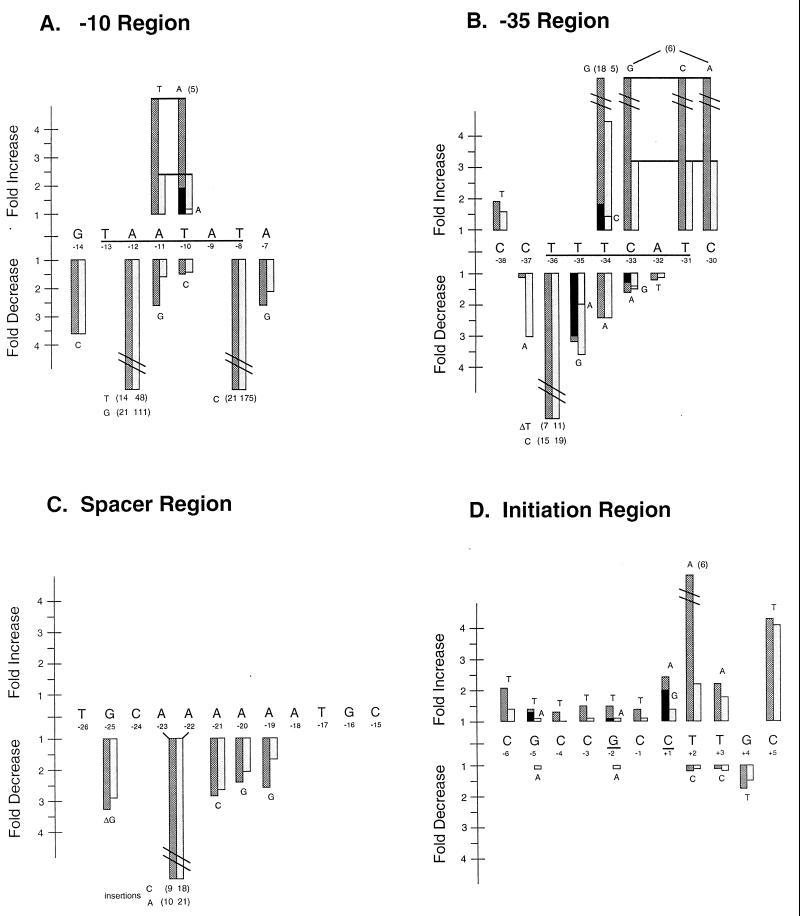
An AT-rich sequence located from −13 to −8 relative to the primary transcription start site (+1) matches the ς70 promoter −10 consensus sequence (17, 19) in four of its six nucleotides (TAATAT; matches to the consensus are underlined) and had been suggested to function as the −10 region for fis P (4, 36). Only one mutation, −8T→G, has been shown to affect fis P by substantially decreasing its activity (36). We now have seven mutations in this region, most of which decrease fis P transcription (Fig. 2A). Two of these changed the highly conserved −12A to either T or G, each of which severely reduced fis P transcription. The −12A→T mutation caused 14- and 47-fold reductions in fis P transcription in RZ211 (fis+) and RJ1561 (fis) cells, respectively; −12A→G caused 21- and 111-fold decreases in fis P transcription in RZ211 and RJ1561, respectively. Another mutation also changed the highly conserved −8T to C, resulting in 21- and 175-fold reductions in β-galactosidase activity in RZ211 and RJ1561 cells, respectively. The two nonconsensus nucleotides, −11A and −10T, are also the least conserved positions in the consensus. Mutations in each of these positions (−11A→G and −10T→C) caused relatively small decreases in transcription (about 1.5- to 2.6-fold). When −10T was replaced with the more conserved A, transcription increased about twofold in RZ211 and slightly in RJ1561 cells. However, when −11A and −10T were simultaneously replaced with T and A, such that all six nucleotides in this region matched the consensus, fis P transcription increased 5.1-fold in RZ211 and 2.4-fold in RJ1561. Primer extension analysis confirmed that this mutation results in increased levels of transcripts initiating at fis P start sites +1 and −2 (Fig. 3). These results validate the sequence TAATAT from −13 to −8 as the −10 promoter region.
FIG. 2.
Relative β-galactosidase activities of various fis P mutants. Saturated RZ211 or RJ1561 cultures carrying pTP127-based plasmids with mutated fis P were diluted 75-fold in LB medium and grown at 37°C for 90 min. The wild-type promoter gave 29 and 332 U of β-galactosidase activity in RZ211 and RJ1561, respectively. The fold change relative to pTP127 in RZ211 ( and
and  ) or RJ1561 (
) or RJ1561 ( and □) is indicated for mutations located in regions containing the −10 (A), the −35 (B), the spacer (C), or the transcription initiation (D) region. Nucleotide substitutions are generally indicated above or below the bars; a nucleotide deletion is indicated by Δ. Where more than one substitution occurred, the additional mutation is to the right of the bars, with the fold change in RZ211 indicated as
and □) is indicated for mutations located in regions containing the −10 (A), the −35 (B), the spacer (C), or the transcription initiation (D) region. Nucleotide substitutions are generally indicated above or below the bars; a nucleotide deletion is indicated by Δ. Where more than one substitution occurred, the additional mutation is to the right of the bars, with the fold change in RZ211 indicated as  and that in RJ1561 indicated as □. Bars connected with a line at the top are multiple mutations that were site directed to generate the consensus −10 or −35 region. Where the fold change exceeded the scale, the bars have slashes and the fold changes are shown in parentheses. Results were based on averages of at least three independent assays. Nucleotide positions are numbered below the sequence; underlined nucleotides represent sequences comprising the −10 region (A), the −35 region (B), or the transcription initiation sites (D).
and that in RJ1561 indicated as □. Bars connected with a line at the top are multiple mutations that were site directed to generate the consensus −10 or −35 region. Where the fold change exceeded the scale, the bars have slashes and the fold changes are shown in parentheses. Results were based on averages of at least three independent assays. Nucleotide positions are numbered below the sequence; underlined nucleotides represent sequences comprising the −10 region (A), the −35 region (B), or the transcription initiation sites (D).
The G at −14 also plays an important role, as its replacement with C resulted in a 3.6-fold decrease in β-galactosidase activity. Likewise, replacement of −7A with G resulted in an over-twofold reduction in activity. Thus, it appears that important nucleotides in the −10 region for fis P can be extended in both directions to include −14G and −7A.
The fis P −35 region.
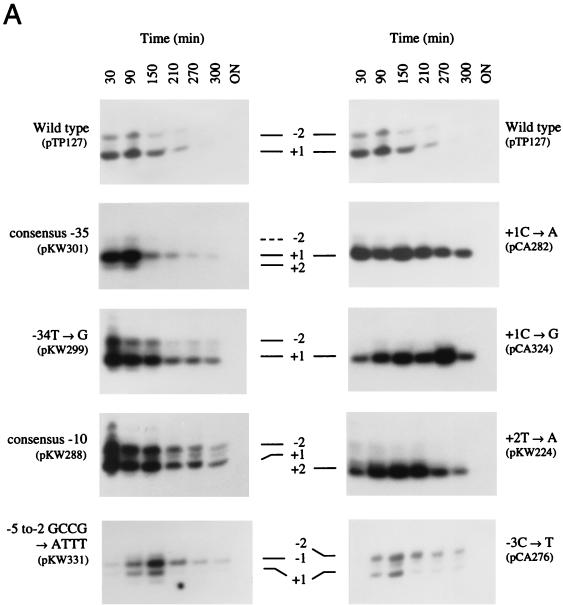
The nucleotide sequence expected to contain the fis P −35 region was difficult to assess correctly because matches to the consensus in this region are scarce. Since deletion of the region from −38 to −28 was shown to abolish fis P activity (40), we were certain that nucleotide sequences in this region were essential for promoter function. The sequence TTCATC (matches to the consensus are underlined) from −35 to −30 had been suggested to function as the −35 region because it contained three matches to the consensus (4, 36). However, the suboptimal 16-bp spacing between this sequence and the −10 region would be expected to limit the activity of this already weak promoter sequence. Nevertheless, when three mutations were introduced within this sequence so that it completely matched the −35 consensus (TTGACA), we observed 5.9- and 3.2-fold increases in β-galactosidase activity in RZ211 and RJ1561, respectively (Fig. 2B). This is consistent with previous observations (36) and demonstrates that an optimal −35 promoter sequence can function with a 16-bp spacer. Unexpectedly, primer extension analysis showed that this mutation caused an increase in transcription initiating at +1 and +2 but not at −2 (see Fig. 5A).
FIG. 5.
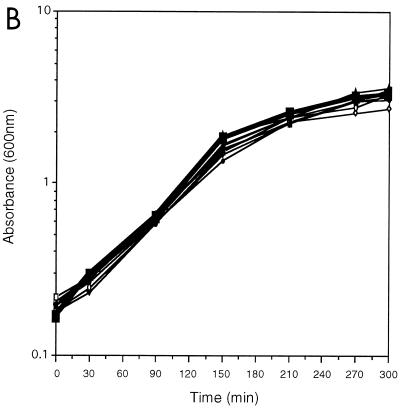
Effects of fis P mutations on growth phase regulation. Saturated cultures of RJ1561 carrying the indicated plasmids were diluted 20-fold into LB medium, grown at 37°C, and harvested at various times thereafter for total RNA preparation. (A) Primer extension reactions were performed with 10 μg of total RNA as described in the legend to Fig. 3. The promoter mutation(s) and host plasmids are described at the outer edges, and the positions of major transcription initiation sites are indicated at the center. (B) Growth of RJ1561 cultures carrying the plasmids shown in panel A. Symbols: ■, wild type; ◊, consensus −35; +, −34T→G; ⧫, consensus −10; □, GCCG→ATTT at −5 to −2; ○, +1C→A; ×, +1C→G; ●, +2T→A; ▵, −3C→T.
The single mutation −34T→G resulted in a 18-fold transcription increase in RZ211 and a 5-fold increase in RJ1561 (Fig. 2B). This mutation improved another poor match to the −35 sequence (TTTCAT to TTGCAT) positioned at the preferred 17-bp spacing from the −10 region. Primer extension reactions from this single up-promoter mutation showed an increase in transcription initiation at both the +1 and −2 sites (see Fig. 5A), as would be expected from an improvement of the wild-type promoter sequence. A comparable point mutation (−33C→G) improving the −35 sequence at the 16-bp spacing from TTCATC to TTGATC resulted in a slight decrease in transcription. Even the triple mutation that created a perfect match to the −35 region with the 16-bp spacing did not increase transcription as much as the single-point mutation −34T→G. These results, taken together, suggest that the wild-type sequence TTTCAT positioned 17 bp from the −10 region is the most likely one to serve as the −35 promoter region.
Ten other point mutations fell within this region, eight of which decreased transcription from fis P (Fig. 2B). Replacement of −35T with A or G resulted in a 2- to 3.5-fold reduction in transcription. The most severe effects on transcription were caused by point mutations at −36T (−36T→C or deletion of −36T), demonstrating that this nucleotide is essential for promoter function. This is consistent with the idea that the sequence TTTCAT serves as the −35 region. Although the T at −34 does not contribute to this consensus sequence, it is favored over A but less favored than C or G. These results further discredit the sequence TTCATC, since T at −34 would be predicted to be the preferred nucleotide. Curiously, −33C is preferred over G or A, even when these changes would improve the match to the consensus for one of the two −35 regions considered. Apparently, a C at this position is more favorable within the context of this promoter sequence and may represent a peculiarity of fis P. Finally, replacement of −32A with T does not significantly affect transcription, suggesting that it does not play an important role, which further invalidates TTCATC as the −35 promoter sequence.
Our data do not support a potential −35 region 18 bp from the −10 region. However, replacement of −37C with A resulted in a threefold decrease in transcription in RJ1561 cells, indicating that it contributes to fis P transcription. On the other hand, this mutation showed almost no effect in RZ211, suggesting that it may have affected Fis binding at site II, causing a reduction in autoregulation efficiency. The mutation −38C→T causes a modest (less-than-twofold) increase in transcription in both RZ211 and RJ1561. Therefore, the nucleotide sequence immediately upstream of the −35 region can positively or negatively influence transcription from fis P.
The fis P spacer region.
To determine if the distance of the fis P spacer region was somehow imposing a limitation on the efficiency of transcription, we examined the effect of altering it by 1 bp on promoter activity. When C was inserted between residues −22 and −23, transcription decreased 9-fold in RZ211 and 18-fold in RJ1561 (Fig. 2C). To rule out the possibility that this effect was due to an interruption in the sequence of A nucleotides in this region, we inserted an A in the same location to preserve this sequence while still increasing the spacing by 1 bp. Transcription from this mutant promoter was similarly reduced by about 10-fold in RZ211 cells and 21-fold in RJ1561 cells. Thus, increasing the spacer region by 1 bp is detrimental to fis P function. Shortening the spacer region by 1 bp by deleting −25G reduced transcription 3.3-fold in RZ211 and 2.9-fold in RJ1561. Therefore, the wild-type distance between the −10 and −35 regions is optimal for transcription.
Three other single-point mutations were obtained by random mutagenesis in the spacer region: −21A→C, −20A→G, and −19A→G. They caused transcription to decrease from about 2.4- to 2.9-fold in RZ211 and about 1.7- to 2.9-fold in RJ1561, suggesting that the A nucleotides at these positions assist in generating wild-type levels of transcription. Since these three nucleotides are part of an A tract that extends from −18 to −23, it is possible that the structure of this region contributes to basal transcriptional activity from fis P.
The fis P transcription initiation region.
Five single point mutations in the region around the transcription initiation site were obtained by random mutagenesis: −6C→T, +2T→A, +2T→C, +3T→A, and +5C→T (Fig. 1B). With the exception of +2T→C, these mutations caused transcription to increase. To more completely analyze this region, including the GC-rich sequence from −6 to +1, we generated 12 additional mutations (Fig. 2D). In general, replacement of individual nucleotides within the GC-rich region with T or A caused small increases in transcription in RZ211 and showed little or no effect in RJ1561. Replacement of four consecutive GC base pairs (in pKW331) similarly increased β-galactosidase activity twofold in RZ211 and not in RJ1561 (data not shown). It appears that increasing the AT richness in this region leads to a moderate reduction in autoregulation efficiency.
Of all of the mutations in this region, those affecting nucleotides downstream of −1 produced the largest increases in transcription. When +2T is replaced with A, transcription increases 6.2-fold in RZ211 and 2.2-fold in RJ1561. Replacement of +1C with A or G causes an over-2-fold increase in transcription in RZ211 and an about 1.5-fold increase in RJ1561. The +3T→A mutation increased transcription 2.2-fold in RZ211 and 1.8-fold in RJ1561. Primer extension analysis of mRNA synthesized from these mutant promoters revealed certain alterations in the selection of transcription initiation sites (Fig. 3; see Fig. 5A). In the +1C→A and +1C→G mutants, transcription initiated exclusively at +1. In the +2T→A mutant, transcription initiated almost exclusively at +2. This most likely reflects the strong preference of RNA polymerase for initiation of transcription with ATP or GTP over UTP or CTP (17, 31). Thus, increases in activity caused by these mutations may be attributed to more-efficient transcription initiation with ATP or GTP.
In the +3T→A mutant, transcription initiated at +1 and −2 much as in the wild type, and initiation at +3A was barely detectable upon overexposure (data not shown). This indicates that the outer limit of efficient transcription initiation from this promoter is +2 (9 bp from the −10 region). Yet this and the mutations +4G→T and +5C→T alter promoter activity, suggesting that a change in the initiating nucleotide is not the only manner in which this region affects transcription. A simple correlation between AT richness in the region from +1 to +5 and elevated levels of transcription cannot be established. The +4G→T mutant decreases transcription, +2T→C and +3T→C have negligible effects, and +3T→A, +2T→A, and +1C→G increase transcription while preserving the base pair composition.
Effects of mutations on stringent control.
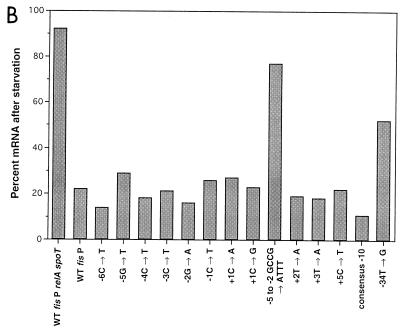
Since fis has been shown to be subject to stringent control, it was of interest to examine the effect of promoter mutations on this form of regulation. Particular focus was given to the GC-rich motif from −6 to +1. Since it has been shown that stringent control could be observed on promoters while in multicopy plasmids (15), we used the same fis P-containing plasmids from which β-galactosidase activities were determined. Cultures of plasmid-containing RJ1561 were grown to mid-logarithmic phase in media having all of the amino acids except valine and isoleucine. Valine was added to induce isoleucine starvation and, hence, the stringent response (29). Primer extension analysis was then conducted on RNA samples obtained before and after addition of valine. The decrease in fis P mRNA levels resulting from induced starvation was used to assess the effect of stringent control on various promoters. The results showed that transcription from the wild-type promoter decreased nearly fivefold to 22% in starved cells (Fig. 4A and B). Treatment with chloramphenicol, which relaxes the stringent response (9), restored transcription (Fig. 4A). In starved RJ1882 (relA spoT fis), fis P mRNA levels decrease only to 93%, consistent with the notion that these decreases are largely attributable to negative regulation by ppGpp. Stringent control of single-point mutations affecting the GC richness in the region from −6C to +1C gave relative mRNA levels as low as 14% (with −6C→T) and as high as 29% (with −5G→T), all of which were roughly comparable to the 22% relative mRNA levels observed with the wild-type promoter. However, the 4-bp replacement at −5 to −2 (GCCG→ATTT) caused significant resistance to stringent control, as 78% of the fis P mRNA remained after starvation.
FIG. 4.
Effects of fis P mutations on stringent control. (A) Primer extension reactions of several fis P from starved or nonstarved cells. Saturated cultures of RJ1561 carrying pTP127-based plasmids were diluted to an OD600 of about 0.1 in defined medium lacking valine or isoleucine and grown for 2 doublings at 37°C. A sample was taken just prior to (−) or 10 min after (+) addition of 500-μg/ml valine. For the wild-type promoter in RJ1882 (fis relA spoT) or RJ1561 (fis), 200-μg/ml chloramphenicol was added immediately after the second sample was removed, and a third sample was taken 10 min later (c). Total RNA was extracted from these cells, and 10 μg was used for primer extensions as described in the legend to Fig. 3. The fis P mutation, the plasmid name, and the strain that carried them are denoted above. Results are a compilation of several independent experiments, and band intensities from different promoters should not be compared. (B) Percent fis P mRNA remaining after starvation. Primer-extended products were quantitated from the data in panel A by PhosphorImaging. For each set, mRNA in the nonstarved lane was considered 100%, and mRNA in the starved lane was expressed relative to this one.
We also examined the effects of several up-promoter mutations near the transcriptional start site and in the −10 and −35 promoter regions. Promoter mutants +2T→A, +3T→A, and +5C→T still responded to stringent control with efficiencies comparable to that of the wild type (Fig. 4A and B). Moreover, the perfect match to the −10 promoter region responded similarly to the wild type. However, in the case of the −34T→G mutation, 53% mRNA levels remained after induced starvation, indicating a partial loss of negative regulation by stringent control. Thus, both the discriminator sequence and the −35 region in fis P appear to affect the response to stringent control.
Effects of mutations on growth phase-dependent regulation.
To determine if any of the mutations obtained affected the growth phase-dependent expression pattern, we performed primer extension analysis using total RNA from RJ1561 carrying plasmids with wild-type or mutant fis P regions grown in LB medium for various lengths of time (Fig. 5A). Transcripts from wild-type fis P were not detected in cells in stationary phase. However, upon reinitiation of growth in batch cultures, transcripts quickly accumulated, reaching the highest measured levels after about 90 min, when cells were in logarithmic growth phase. Thereafter, mRNA levels decreased, becoming barely detectable after about 210 min when cells had re-entered stationary phase (Fig. 5B).
Many down-promoter mutants could not be confidently analyzed in this fashion, as little or no transcription from them could be detected. Nevertheless, 31 promoter mutations were examined. The majority of these retained normal growth phase-dependent regulation. A notable example is shown with plasmid pKW301, containing the fis P region with a consensus −35 sequence 16 bp upstream of the −10 region (Fig. 5A). Peak mRNA levels originating from this promoter were significantly higher than those from the wild-type promoter, but transcription remained tightly growth phase regulated, emulating the pattern obtained with wild-type fis P. This shows that elevated peak levels of mRNA do not necessarily result in alteration of the growth phase regulation pattern.
The mutation −34T→G (pKW299) also results in a notable increase in transcription, but in this case, the growth phase regulation pattern is appreciably altered (Fig. 5A). mRNA levels from this promoter peak earlier than those from the wild type and then decrease more slowly, such that considerable levels are still observed by 300 min of growth. However, mRNA from this promoter could not be detected after 24 h of culturing. A similar pattern of expression was observed with the promoter containing the consensus −10 region (pKW288). Such alterations in regulation pattern cannot be attributed to differences in cell growth, since growth rates were very similar in all of the cultures examined (Fig. 5B). Therefore, the growth phase regulation pattern was affected by improvements in the −35 and −10 regions.
pKW331, carrying the 4-bp mutation that alters the discriminatory sequence and the response to stringent control, retains much of the growth phase regulation pattern (Fig. 5A). The accumulation of transcripts is somewhat slower from this promoter than from the wild type, such that they peak at some time after 90 min of growth. Nevertheless, after 150 min, mRNA levels decrease rapidly and become barely detectable by 300 min. Thus, while the expression pattern from pKW331 is not identical to that from the wild type (pTP127), it is still tightly growth phase dependently regulated. Plasmid pCA276 (−3C→T), which also alters the discriminator but does not have an effect on stringent control, similarly showed a slight deviation from the typical growth phase regulation pattern. However, for unknown reasons, this mutant plasmid caused a shift in the predominant start site to −2G, and the slight effect on growth phase regulation is seen only at this initiation site. Therefore, small changes in growth phase-dependent regulation can occur through processes other than the stringent response.
The most dramatic alterations in the growth phase regulation pattern were observed with the mutations +1C→A (pCA282), and +1C→G (pCA324). These promoters also responded to a nutritional upshift with a rapid increase in mRNA levels but maintained substantial levels of mRNA after about 300 min of growth (Fig. 5A). In the case of pCA282, maximum mRNA levels were observed as early as 30 min of growth and very gradually declined only after about 150 min. Overexposure allowed visualization of transcripts from a 24-h culture (data not shown). Elevated levels were also observed from pCA324 during logarithmic and early stationary phases (e.g., 30 to 300 min). mRNA levels from pKW224 (+2T→A) peaked at about 90 min of growth and remained elevated until after 210 min. By 300 min, mRNA levels were reduced but mRNA was still abundant. Thus, each of the single mutations +1C→A, +1C→G, and +2T→A significantly altered the normal growth phase-dependent regulation pattern of fis P.
DISCUSSION
The fis P promoter.
Results presented in this work have advanced the characterization of E. coli fis P to a finer detail, as well as our understanding of its function and regulation. Its sequence resembles that of a ς70 promoter, and indeed, ς70 RNA polymerase protects this promoter from DNase I cleavage in the region from −50 to +26 (4) and is able to transcribe this promoter in vitro (48). The −10 region was readily deciphered by our mutation analysis. Mutations within the sequence TAATAT from −13 to −8 that improved the match to the consensus sequence increased transcription, while those that deviated further from the consensus decreased transcription. Two additional nucleotides (−14G and −7A) might form part of an extended −10 promoter region, as they appear to contribute to promoter function. However, we cannot rule out the possibility that the negative effect of −7A→G is due to an increase in the GC richness of this region. Two possible sequences for the −35 region were considered: TTCATC from −35 to −30 with a 16-bp spacer region and TTTCAT from −36 to −31 with a 17-bp spacer region. Both represent poor matches to the −35 consensus (matches underlined) and lack the highly conserved G at the third position (17, 19). However, results from our mutation analysis led us to conclude that TTTCAT serves as the −35 promoter sequence. We see no reason to propose two or more overlapping promoters. Multiple transcription initiation sites 6 to 10 bp downstream from the −10 region can be reconciled with the activity of a single promoter. Factors such as the sequence around the transcription initiation site and the available nucleotide pools can influence the selection of start sites (7, 31).
In the spacer region, three point mutations replaced an A with C or G and caused moderate reductions in transcription. These three nucleotides are part of an A6 tract (from −23 to −18) that may contribute to a bend (28). It is possible that an intrinsic bend or flexibility in the spacer region might facilitate contacts with RNA polymerase (38). Indeed, divergence from the consensus −35 and −10 sequences can be compensated for, in part, with a spacer sequence having properly oriented curvature (10). While the GC-rich sequence separating the −10 and initiation regions contributes to modest improvements in the efficiency of Fis repression, it does not appear to restrict fis P basal transcriptional activity in fis mutant cells under conditions of rapid cell growth. This suggests that the free-energy requirement for DNA strand separation in this region is not limiting under these conditions. An effect of this GC-rich sequence in slower-growing cultures has not been tested. Nonetheless, under conditions of amino acid starvation (see below), this sequence plays an important role in mediating regulation by stringent control.
Stringent control.
The observation had been made that promoters negatively regulated by stringent control often exhibit two of the following three characteristics: (i) a second cryptic RNA polymerase binding site upstream of the target promoter, (ii) a weak −35 region, and (iii) a GC-rich discriminator sequence between the −10 region and the transcription start site (30). All three of these hold true for fis P. An upstream RNA polymerase binding site was previously identified in the region from −68 to −126, but transcripts initiating in this region have not been detected (4, 39).
Our results point to the −35 (TTTCAT) and GC-rich discriminator sequences as contributors to the regulation by stringent control (Fig. 6). An improved version of the −35 sequence (TTGCAT) partially reduced the effect of stringent control (Fig. 4). However, a perfect match to neither the −10 consensus (Fig. 4) nor the −35 consensus with a 16-bp spacer (36; data not shown) altered regulation by stringent control. These observations suggest that certain interactions between RNA polymerase and the fis P −35 region can limit the strength of negative regulation by stringent control.
FIG. 6.
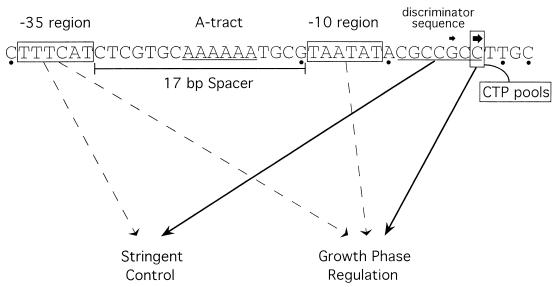
Sequences involved in basal transcription and regulation of fis P. Boxed nucleotides contain the −35 region, the −10 region, or the predominant transcription initiation site. The two filled arrows above the nucleotide sequence denote the relative strengths of the start sites. The discriminator sequence and A tract are underlined. Bases with a dot underneath are not formally part of the −35 and −10 hexameric regions or a transcription initiation site but are important for promoter activity. Solid arrows drawn from the sequence indicate regions shown to have a substantial effect on the response to stringent control or growth phase regulation, and dashed arrows indicate regions that have moderate effects on one or both of these regulatory processes. The line connecting CTP pools to +1C indicates that the role of the start site in growth phase regulation may rely on CTP concentrations.
While single-point mutations within the discriminator region showed little or no effect on the response to stringent control, the 4-bp mutation in pKW331 (GCCG→ATTT at −5 to −2) resulted in notable resistance. Similar observations have been made for rnpB (25). Three or four consecutive mutations in the rnpB GC-rich region were generally required to diminish the stringent response in this promoter. A 4-bp substitution of G · C for A · T base pairs also reduced the efficiency of stringent control at the tyrT promoter (29). However, in the case of the tufBp, certain single-point mutations within the GC-rich discriminator motif can completely diminish its response to stringent control (33), even though its GC-rich discriminator sequence is very similar to that of rnpB. This suggests that contributions of individual nucleotides within the discriminator region to the response to stringent control depend on the promoter context.
Growth phase-dependent regulation.
The fis P region from −38 to +5 contains sufficient information to generate growth phase-dependent regulation (36, 40). Many of the mutations obtained in this region had little or no effect on this regulation pattern. However, mutations in three distinct regions resulted in discernible deviations from the wild-type expression pattern: the −35 region, the −10 region, and the transcription initiation site (Fig. 6). While these promoters were still subject to growth phase-dependent regulation, the pattern was substantially altered compared to that of the wild type. Presumably, these mutations improved interactions with RNA polymerase to confer partial resistance to transcriptional shutdown during late-logarithmic and early stationary phases. However, the −35 up-promoter mutation in pKW301 (with a 16-bp spacer) did not cause a deviation from the wild-type regulation pattern, suggesting that this mutation might have affected a step during transcription initiation that is not associated with the mechanism of growth phase-dependent regulation.
The mutations +1C→A and +1C→G produced a more severe change in the regulation pattern (Fig. 5). A largely similar expression pattern was observed for +2T→A. In these promoters, transcription initiated almost exclusively from either +1 or +2 with ATP or GTP. These are more highly preferred initiation nucleotides than CTP (17, 31), most likely because RNA polymerase has a lower Km for purines as transcription initiation nucleotides (1, 52). Thus, we assume that an ability to use the kinetically favored ATP or GTP facilitates the process of transcription initiation at fis P such that high levels of expression are observed during late logarithmic and stationary phases. On the other hand, use of the least-preferred CTP nucleotide as the predominant transcription initiation nucleotide (31) could impose a severe restriction on transcription. We have noticed that high concentrations of CTP (but not ATP or GTP) significantly increased the proportion of heparin-resistant fis P-RNA polymerase complexes in runoff transcription assays (48), suggesting that the stability of open complexes in fis P may be enhanced by sufficient concentrations of CTP.
While our data showed that the GC-rich discriminator is important for stringent control, no prominent role was found for this sequence in growth phase regulation. Yet, it had been previously shown that replacing the fis P region from −6 to +3 with corresponding sequences from bla promoter significantly altered both stringent control and growth phase regulation, implying that the discriminator sequence played a central role in both processes (36). We note, however, that the hybrid promoter also contained G and A nucleotides positioned 7 and 8 bp downstream from the −10 region, respectively. As we have shown, this facilitates transcription initiation and alters the growth phase-dependent regulation but not stringent control. Thus, the dual effect of the hybrid promoter can be explained by the alteration of both the discriminator sequence and the transcription initiation nucleotide.
We envision that wild-type fis P is exquisitely sensitive to changes in available concentrations of CTP. A rapid intracellular accumulation of nucleoside triphosphates (NTPs) could result from an abrupt increase in metabolism under conditions of nutrient upshift. Transcription initiation from fis P could increase, at least in part, as a result of increasing CTP availability. Then, as transcription from highly active promoters (such as those of stable RNAs) steadily increases and DNA synthesis progresses, the available NTP pool can be expected to decline. CTP concentrations may readily fall to lower steady-state levels with a consequent decrease in transcription from fis P. Mutations affecting the −10 and −35 regions in pKW288 and pKW299 might help stabilize open complexes long enough to allow transcription initiation with suboptimal intracellular CTP pools during late logarithmic and early stationary phases. On the other hand, because of their lower Km as transcription initiation nucleotides, ATP and GTP continue to support transcription from most other promoters, as well as the +1C→A and +1C→G fis promoters, even if NTP pools decrease during late-logarithmic and early stationary phases. Thus, in this model, CTP could serve as a key transcriptional regulator of the fis expression pattern (Fig. 6).
Transcription initiation with purine nucleotides will change the fis P regulation pattern significantly only if the initiation site is at a proper distance from the −10 region. For instance, initiation with GTP at −2 (6 bp from the −10 region) in the wild-type promoter is both inefficient and growth phase regulated. The mutation −3C→T somehow caused initiation at −2 to be both more efficient and more resistant to repression during late logarithmic phase relative to initiation with CTP at +1 (Fig. 5). Efficient initiation with GTP or ATP at +1 had a substantial effect on the regulation pattern, more so than initiation with ATP at +2. Thus, it appears that the position of the preferred initiating nucleotide affects the efficiency of initiation and, consequently, growth phase-dependent regulation.
The regulation pattern of fis P might then arise from a combination of unfavorable transcription initiation conditions: (i) if transcription initiates with the least preferred initiation nucleotide (CTP), (ii) if transcription initiates at an unfavorable distance from the −10 region, and (iii) if the −10 and −35 regions deviate from their consensus sequences. These conditions may be conducive to nonproductive RNA polymerase-promoter complexes that may become productive with sufficiently high intracellular initiating NTP concentrations.
Sensitivity to pyrimidine NTP pools has been observed for other promoters. For instance, the E. coli pyrC (31, 50) and the Salmonella typhimurium pyrC and pyrD (45, 46) promoters initiate transcription with CTP when intracellular CTP levels are high or with GTP several bases downstream when CTP levels are low. Initiation with CTP results in synthesis of a 5′ mRNA sequence capable of forming a stem-loop structure that hinders translation, thus accounting for negative feedback. In other promoters, such as those for pyrBI and codBA, high UTP concentrations give rise to reiterative RNA synthesis whereby RNA products are made that contain long stretches of U residues near their 5′ termini, causing transcription to abort (20, 41).
Regulation of transcription by intracellular NTP pools has been shown to play a key role in growth rate-dependent regulation of rrnBp1 and rrnDp1 (13). In both cases, the initiating purine NTP increases the stability of RNA polymerase-promoter complexes. In the case of rrnBp1, a strong correlation was made among increasing cell growth rates, increasing intracellular levels of ATP, and increasing transcription activity in vivo. In a car mutant strain, in which intracellular ATP levels decreased with increasing growth rates, rrnBp1 transcription also decreased with increasing growth rates. These observations led to a persuasive model that explicates growth rate-dependent regulation of rRNA promoters in terms of promoter sensitivity to varying purine NTP pools under varying nutrient availability. It is possible that the varying levels of fis expression observed under different nutritional conditions (4) are also related to varying NTP pools during different cell growth rates.
ACKNOWLEDGMENTS
We thank B. Hu and R. C. Johnson (University of California, Los Angeles) for the mutant pRJ1028-based plasmids used in the construction of plasmids pKW292 to -298 and T. S. Pratt for the construction of pTP127. We also thank R. C. Johnson, M. B. Beach, and T. S. Pratt for useful comments.
This work was supported by funds from Public Health Service grant GM52051.
REFERENCES
- 1.Anthony D D, Goldthwait D A, Wu C W. Studies with the ribonucleic acid polymerase. II. Kinetic aspects of initiation and polymerization. Biochemistry. 1969;8:246–256. doi: 10.1021/bi00829a035. [DOI] [PubMed] [Google Scholar]
- 2.Ball C A, Johnson R C. Efficient excision of phage lambda from the Escherichia coli chromosome requires the Fis protein. J Bacteriol. 1991;173:4027–4031. doi: 10.1128/jb.173.13.4027-4031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball C A, Johnson R C. Multiple effects of Fis on integration and the control of lysogeny in phage lambda. J Bacteriol. 1991;173:4032–4038. doi: 10.1128/jb.173.13.4032-4038.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball C A, Osuna R, Ferguson K C, Johnson R C. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barik S, Galinski M. “Megaprimer” method of PCR: increased template concentration improves yield. BioTechniques. 1991;10:489–490. [PubMed] [Google Scholar]
- 6.Beach M B, Osuna R. Identification and characterization of the fis operon in enteric bacteria. J Bacteriol. 1998;180:5932–5946. doi: 10.1128/jb.180.22.5932-5946.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpousis A J, Stefano J E, Gralla J D. 5′ nucleotide heterogeneity and altered initiation of transcription at mutant lac promoters. J Mol Biol. 1982;157:619–633. doi: 10.1016/0022-2836(82)90502-2. [DOI] [PubMed] [Google Scholar]
- 8.Case C C, Roels S M, Gonzalez J E, Simons E, Simons R. Analysis of the promoters and transcripts involved in IS10 anti-sense transcriptional RNA control. Gene. 1988;72:219–236. doi: 10.1016/0378-1119(88)90147-3. [DOI] [PubMed] [Google Scholar]
- 9.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 10.Collis C M, Molloy P L, Both G W, Drew H R. Influence of the sequence-dependent flexure of DNA on transcription in E. coli. Nucleic Acids Res. 1989;17:9447–9468. doi: 10.1093/nar/17.22.9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filutowicz M, Ross W, Wild J, Gourse R L. Involvement of Fis protein in replication of the Escherichia coli chromosome. J Bacteriol. 1992;174:398–407. doi: 10.1128/jb.174.2.398-407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finkel S E. Ph.D. thesis. Los Angeles: University of California; 1994. [Google Scholar]
- 13.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 14.Gille H, Egan J B, Roth A, Messer W. The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 1991;19:4167–4172. doi: 10.1093/nar/19.15.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gourse R L, Stark M J, Dahlberg A E. Regions of DNA involved in the stringent control of plasmid-encoded rRNA in vivo. Cell. 1983;32:1347–1354. doi: 10.1016/0092-8674(83)90315-x. [DOI] [PubMed] [Google Scholar]
- 16.Haffter P, Bickle T A. Purification and DNA-binding properties of FIS and Cin, two proteins required for the bacteriophage P1 site-specific recombination system, cin. J Mol Biol. 1987;198:579–587. doi: 10.1016/0022-2836(87)90201-4. [DOI] [PubMed] [Google Scholar]
- 17.Harley C B, Reynolds R P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartz D, McPheeters D S, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 19.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin D J, Turnbough C L., Jr An Escherichia coli RNA polymerase defective in transcription due to its overproduction of abortive initiation products. J Mol Biol. 1994;236:72–80. doi: 10.1006/jmbi.1994.1119. [DOI] [PubMed] [Google Scholar]
- 21.Johnson R C, Ball C A, Pfeffer D, Simon M I. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci USA. 1988;85:3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson R C, Bruist M F, Simon M I. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986;46:531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- 23.Johnson R C, Simon M I. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985;41:781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- 24.Johnson R C, Yin J C, Reznikoff W S. Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell. 1982;30:873–882. doi: 10.1016/0092-8674(82)90292-6. [DOI] [PubMed] [Google Scholar]
- 25.Jung Y H, Lee Y. Escherichia coli rnpB promoter mutants altered in stringent response. Biochem Biophys Res Commun. 1997;230:582–586. doi: 10.1006/bbrc.1996.6005. [DOI] [PubMed] [Google Scholar]
- 26.Kahmann R, Rudt F, Koch C, Mertens G. G inversion in bacteriophage Mu DNA is stimulated by a site within the invertase gene and a host factor. Cell. 1985;41:771–780. doi: 10.1016/s0092-8674(85)80058-1. [DOI] [PubMed] [Google Scholar]
- 27.Koch C, Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem. 1986;261:15673–15678. [PubMed] [Google Scholar]
- 28.Koo H S, Drak J, Rice J A, Crothers D M. Determination of the extent of DNA bending by an adenine-thymine tract. Biochemistry. 1990;29:4227–4234. doi: 10.1021/bi00469a027. [DOI] [PubMed] [Google Scholar]
- 29.Lamond A I, Travers A A. Genetically separable functional elements mediate the optimal expression and stringent regulation of a bacterial tRNA gene. Cell. 1985;40:319–326. doi: 10.1016/0092-8674(85)90146-1. [DOI] [PubMed] [Google Scholar]
- 30.Lamond A I, Travers A A. Stringent control of bacterial transcription. Cell. 1985;41:6–8. doi: 10.1016/0092-8674(85)90050-9. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Turnbough C L., Jr Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J Bacteriol. 1994;176:2938–2945. doi: 10.1128/jb.176.10.2938-2945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 33.Mizushima-Sugano J, Kaziro Y. Regulation of the expression of the tufB operon: DNA sequences directly involved in the stringent control. EMBO J. 1985;4:1053–1058. doi: 10.1002/j.1460-2075.1985.tb03738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsson L, Vanet A, Vijgenboom E, Bosch L. The role of FIS in trans activation of stable RNA operons of E. coli. EMBO J. 1990;9:727–734. doi: 10.1002/j.1460-2075.1990.tb08166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsson L, Verbeek H, Vijgenboom E, van Drunen C, Vanet A, Bosch L. FIS-dependent trans activation of stable RNA operons of Escherichia coli under various growth conditions. J Bacteriol. 1992;174:921–929. doi: 10.1128/jb.174.3.921-929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ninnemann O, Koch C, Kahmann R. The E. coli fis promoter is subject to stringent control and autoregulation. EMBO J. 1992;11:1075–1083. doi: 10.1002/j.1460-2075.1992.tb05146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osuna R, Lienau D, Hughes K T, Johnson R C. Sequence, regulation, and functions of fis in Salmonella typhimurium. J Bacteriol. 1995;177:2021–2032. doi: 10.1128/jb.177.8.2021-2032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Martin J, Rojo F, de Lorenzo V. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pratt, T. S., and R. Osuna. Unpublished results.
- 40.Pratt T S, Steiner T, Feldman L S, Walker K A, Osuna R. Deletion analysis of the fis promoter region in Escherichia coli: antagonistic effects of integration host factor and Fis. J Bacteriol. 1997;179:6367–6377. doi: 10.1128/jb.179.20.6367-6377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi F, Turnbough C L., Jr Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J Mol Biol. 1995;254:552–565. doi: 10.1006/jmbi.1995.0638. [DOI] [PubMed] [Google Scholar]
- 42.Ross W, Thompson J F, Newlands J T, Gourse R L. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Schneider R, Travers A, Muskhelishvili G. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol Microbiol. 1997;26:519–530. doi: 10.1046/j.1365-2958.1997.5951971.x. [DOI] [PubMed] [Google Scholar]
- 45.Sorensen K I, Baker K E, Kelln R A, Neuhard J. Nucleotide pool-sensitive selection of the transcriptional start site in vivo at the Salmonella typhimurium pyrC and pyrD promoters. J Bacteriol. 1993;175:4137–4144. doi: 10.1128/jb.175.13.4137-4144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorensen K I, Neuhard J. Dual transcriptional initiation sites from the pyrC promoter control expression of the gene in Salmonella typhimurium. Mol Gen Genet. 1991;225:249–256. doi: 10.1007/BF00269856. [DOI] [PubMed] [Google Scholar]
- 47.Thompson J F, Moitoso de Vargas L, Koch C, Kahmann R, Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987;50:901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- 48.Undamatla, J., T. S. Pratt, and R. Osuna. Unpublished results.
- 49.Vanet A, Plumbridge J A, Alix J H. Cotranscription of two genes necessary for ribosomal protein L11 methylation (prmA) and pantothenate transport (panF) in Escherichia coli K-12. J Bacteriol. 1993;175:7178–7188. doi: 10.1128/jb.175.22.7178-7188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson H R, Archer C D, Liu J K, Turnbough C L., Jr Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J Bacteriol. 1992;174:514–524. doi: 10.1128/jb.174.2.514-524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wold S, Crooke E, Skarstad K. The Escherichia coli Fis protein prevents initiation of DNA replication from oriC in vitro. Nucleic Acids Res. 1996;24:3527–3532. doi: 10.1093/nar/24.18.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu C W, Goldthwait D A. Studies of nucleotide binding to the ribonucleic acid polymerase by equilibrium dialysis. Biochemistry. 1969;8:4458–4464. doi: 10.1021/bi00839a035. [DOI] [PubMed] [Google Scholar]