Abstract
Nucleocapsid serological assay sensitivity to identify severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections among vaccinees and for Omicron cases is unclear. In this prospective study, the Elecsys nucleocapsid assay was 89% sensitive in identifying SARS-CoV-2 infections 14–607 days pre–blood collection. Sensitivity was similar when comparing by vaccination status, and in Omicron (vs pre-Omicron) cases.
Keywords: SARS-CoV-2, COVID-19, vaccine, antibodies, nucleocapsid
Serological assays are essential to understanding community-level coronavirus disease 2019 (COVID-19) prevalence, accounting for both symptomatic and asymptomatic infections, allowing a wider window for detection of prior infection compared with polymerase chain reaction (PCR) and rapid antigen tests (RATs), and filling in gaps in infection rate knowledge due to testing shortages and changes in testing strategies [1, 2]. Given the rise of the Omicron variant, coinciding with decreased emphasis on case identification in many jurisdictions, antibody testing may play an increasingly important role in monitoring COVID-19 and vaccine effectiveness.
With widespread use of spike protein–targeted vaccines, prior severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection can be assessed using nucleocapsid antibody assays. Using samples collected 3–82 days post–positive PCR, the Elecsys nucleocapsid assay demonstrated 97.2% sensitivity and 99.8% specificity for classifying preceding SARS-CoV-2 infections [3]. However, performance was evaluated in unvaccinated individuals, with relatively short observation periods, and before the Omicron variant. Emerging data have questioned whether the sensitivity of nucleocapsid assays is compromised in vaccinated populations, potentially due to lower antibody responses [4]. Further, sensitivity in the Omicron era is unclear and may be affected by nucleocapsid mutations [5]. We sought to estimate the sensitivity of the Elecsys Nucleocapsid assay using a large prospectively enrolled cohort, many of whom were vaccinated.
METHODS
Study Setting and Design
We employed samples from the COVID-19 Occupational Risks, Seroprevalence and Immunity among Paramedics in Canada (CORSIP) study, an observational prospective cohort of adult Canadian paramedics [6]. Participants provided blood samples and questionnaires every 6 months and documented positive COVID-19 tests and vaccinations through an online portal. Omicron-variant SARS-CoV-2 was identified in Canada on November 28, 2021; by mid and late December 2021, the Omicron variant accounted for 85% and >95% of Canadian COVID-19 cases, respectively [7].
Patient Consent
The CORSIP study was approved by the Universities of British Columbia (H20-03620) and Toronto (#40435), and all participants provided written consent.
Selection of Participants
We included CORSIP participants in this analysis who reported having COVID-19 and who provided a blood sample after the diagnosis date. We excluded samples that were collected within 14 days of COVID-19 diagnosis, given that antinucleocapsid antibodies are only reliably detected after 2 weeks [6].
Serological Testing
All samples underwent Roche Elecsys Anti-SARS-Cov-2 N assay testing (Roche Diagnostics Corp., Indianapolis, IN, USA) (Supplementary Data).
Statistical Analyses
The reference standard was SARS-CoV-2 infection, defined as a positive PCR or RAT. We calculated sensitivity (with 95% CI) of the nucleocapsid assay using the manufacturer’s threshold for positivity of 1.0. We examined sensitivity overall and within subgroups, categorized by (1) vaccination status and (2) COVID-19 time period (“pre-Omicron” [before November 28, 2021] or “Omicron era” [December 15, 2021, and onward]). Given the temporal proximity of the Omicron wave and the study end date, we anticipated that Omicron-era cases would have shorter COVID-19-to-blood collection intervals (possibly affecting sensitivity); thus, in a sensitivity analysis we excluded pre-Omicron subgroup samples with COVID-19-to-blood collection intervals greater than the maximum Omicron-era COVID-19-to-blood collection interval. We did not calculate specificity given that we could not assume those not reporting positivity were truly negative, as our study population was not routinely and frequently tested for COVID-19. We used GraphPad Prism, version 9.2.0 (GraphPad Software, San Diego, CA, USA) for analyses.
RESULTS
As of April 8, 2022, the CORSIP study had 3241 participants; 420 (13%) reported having COVID-19, of whom 205 provided a blood sample ≥ 14 days postdiagnosis.
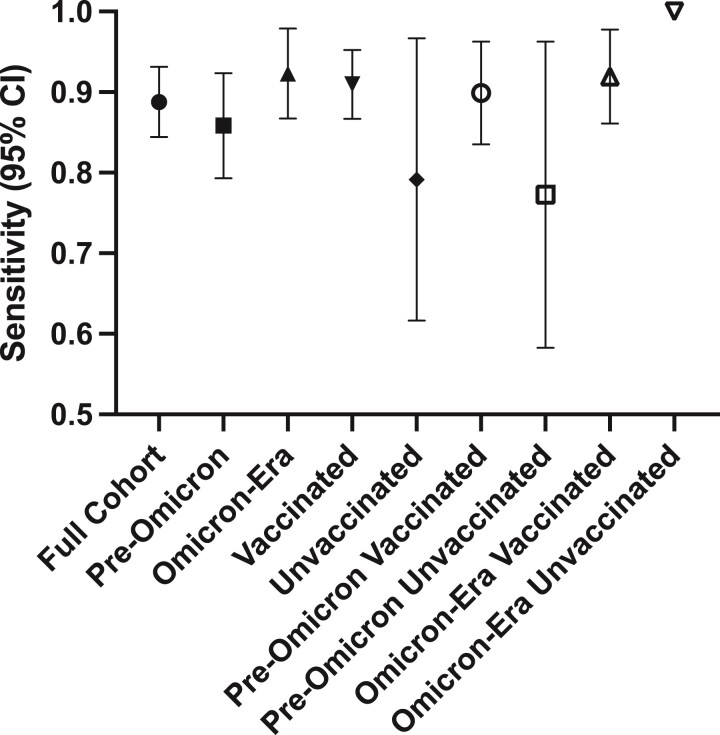
Of the study cohort (n = 205), the median age (interquartile range [IQR]) was 37 (32–46) years, 96 (47%) reported being of the female sex, and 177 (88%) were vaccinated (Table 1). The median date of positive COVID-19 test (IQR) was October 9, 2021 (December 26, 2020–January 1, 2022); 113 (55%) cases occurred pre-Omicron, 1 (0.49%) case occurred between the 2 time periods, and 91 (44%) occurred during the Omicron era. COVID-19 diagnosis was by RAT in 48 (23%) cases (14 of which were confirmed with PCR) and by PCR in 157 (77%). The median COVID-19-to-blood collection interval (IQR, range) was 63 (33–157, 14–607) days. For 182 participants, the nucleocapsid assay was reactive, for a sensitivity of 89% (95% CI, 84%–93%) (Figure 1). Of samples with reactive and unreactive tests, the median result (IQR) was 19 (5.8–57) and 0.088 (0.079–0.52), respectively. Of vaccinated participants, 161/177 had a reactive test (sensitivity, 91%; 95% CI, 87%–95%), and of unvaccinated participants 19/24 had a reactive test (sensitivity, 79%; 95% CI, 62%–97%).
Table 1.
Participant Characteristics, Overall and of Subgroups Classified by Time Period of COVID-19
| Participant Characteristics | Time Era Subgroups | ||
|---|---|---|---|
| Full Cohor (n = 205) | Pre-Omicron (n = 113) |
Omicron-Era (n = 91) |
|
| Age, median (IQR), y | 37 (32–46) | 38 (32–48) | 37 (32–43) |
| Female sex, No. (%) | 96 (47) | 53 (47) | 43 (47) |
| Missing | 10 (4.9) | 6 (5.3) | 4 (3.5) |
| Vaccinated, No. (%) | 176 (86) | 88 (78) | 88 (97) |
| 3 doses BNT162b2 | 65 (32) | 28 (25) | 37 (41) |
| 3 doses mRNA-1273 | 10 (4.9) | 4 (3.5) | 6 (6.6) |
| 3 doses combination of BNT162b2/mRNA-1273 | 34 (17) | 18 (16) | 16 (18) |
| 2 doses BNT162b2 | 50 (24) | 28 (25) | 22 (24) |
| 2 doses mRNA-1273 | 9 (5.1) | 4 (3.5) | 5 (5.5) |
| 2 doses ChAdOx1 | 1 (0.49) | 1 (0.88) | 0 |
| 2 doses combination of BNT162b2/mRNA-1273 | 2 (0.98) | 1 (0.88) | 1 (1.1) |
| 1 dose BNT162b2 | 4 (2.0) | 4 (3.5) | 0 |
| 1 dose mRNA-1273 | 1 (0.49) | 0 | 1 (1.1) |
| COVID-to-BC interval, median (IQR), d | 63 (33–157) | 147 (86–244) | 31 (21–46) |
| Medical history,a No. (%) | |||
| Hypertension | 15 (7.3) | 9 (7.8) | 6 (6.6) |
| Diabetes | 4 (2.0) | 3 (2.7) | 1 (1.1) |
| Asthma | 19 (9.3) | 13 (12) | 6 (6.6) |
| Lung disease | 0 | 0 | 0 |
| Heart disease | 1 (0.49) | 0 | 1 (1.1) |
| Kidney disease | 0 | 0 | 0 |
| Liver disease | 0 | 0 | 0 |
| Cancer | 4 (2.0) | 4 (3.5) | 0 |
| Hematologic disease | 2 (0.98) | 1 (0.88) | 1 (1.1) |
| Autoimmune disease | 6 (2.9) | 4 (3.5) | 2 (2.2) |
| Neurological disease | 0 | 0 | 0 |
| Missing data | 11 (5.4) | 6 (5.3) | 5 (5.5) |
| Race/ethnicity,b No. (%) | |||
| Arab | 1 (0.49) | 1 (0.88) | 0 |
| Black | 2 (0.98) | 1 (0.88) | 1 (1.1) |
| Chinese | 6 (2.9) | 2 (1.8) | 4 |
| Filipino | 2 (0.98) | 0 | 2 (2.2) |
| Japanese | 0 | 0 | 0 |
| Korean | 1 (0.49) | 0 | 1 (1.1) |
| Latin American | 2 (0.98) | 1 (0.88) | 1 (1.1) |
| South Asian | 1 (0.49) | 1 (0.88) | 0 |
| Southeast Asian | 2 (0.98) | 0 | 2 (2.2) |
| West Asian | 1 (0.49) | 1 (0.88) | 0 |
| White, prefer to self-describe | 178 (87) | 101 (89) | 76 (84) |
| Prefer not to describe | 1 (0.49) | 1 (0.88) | 0 |
| Missing data | 11 (5.4) | 6 (5.3) | 5 (5.5) |
Abbreviations: BC, blood collection; COVID-19, coronavirus disease 2019; COVID-to-BC interval, the interval (in days) between the COVID-19 diagnosis and the blood collection date; IQR, interquartile range.
Participants answered the question: “Have you been diagnosed by a physician with any of the following chronic medical conditions? (Select all that apply.)”
Participants answered the question: “How would you describe your ethnicity or race? (Check all that apply.)” Three participants identified with more than 1 race/ethnicity.
Figure 1.
Sensitivity (with 95% CI) of the Elecsys nucleocapsid assay for classifying preceding SARS-CoV-2 infections, overall and within subgroups. Error bars represent 95% CIs (not shown for the “Omicron-Era Unvaccinated” subgroup due to low sample size). Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Of the 91 cases that occurred in the Omicron era, the median COVID-19-to-blood collection interval (IQR, range) was 31 (21–45, 14–72) days. For 84 participants, the nucleocapsid assay was reactive, for a sensitivity of 92% (95% CI, 87%–98%). Of samples with reactive and unreactive tests, the median result (IQR) was 11 (2.9–28) and 0.20 (0.078–0.54), respectively. Of vaccinated participants, 80/87 had a reactive test (sensitivity, 92%; 95% CI, 86%–98%), and of unvaccinated participants 2/2 had a reactive test (sensitivity, 100%).
Of the 113 cases that occurred pre-Omicron, the median COVID-19-to-blood collection interval (IQR, range) was 147 (86–244, 19–607) days. For 97 participants, the nucleocapsid assay was reactive, for a sensitivity of 86% (95% CI, 79%–92%). After excluding cases with a COVID-19-to-blood collection interval of >72 days, 19/22 cases had reactive tests (sensitivity, 86%; 95% CI, 71%–100%). Of samples with reactive and unreactive tests, the median result (IQR) was 36 (10–113) and 0.086 (0.080–0.40), respectively. Of vaccinated participants, 80/89 had a reactive test (sensitivity, 90%; 95% CI, 84%–96%), and of unvaccinated participants 17/22 had a reactive test (sensitivity, 77%; 95% CI, 58%–96%).
DISCUSSION
From an observational prospective study of paramedics in Canada, the sensitivity of a nucleocapsid commercially available chemiluminescent serological assay to identify preceding SARS-CoV-2 infections was 89%. Contrary to concerns regarding performance among vaccinated or Omicron-era cases, we did not find evidence of decreased sensitivity.
While subgroups tested in our study varied in sample size (and thus confidence intervals), all yielded similar performance. We specifically examined whether test performance would vary for samples from vaccinated participants. Although confidence intervals overlapped, the point estimate of sensitivity for vaccinated cases was higher than unvaccinated, suggesting that vaccination does not impair sensitivity. We also examined whether test performance worsened in the Omicron era. The point estimate for Omicron-era sensitivity was higher in pre-Omicron samples, suggesting that Omicron did not decrease sensitivity.
We postulated that test performance may be related to the COVID-19-to-blood collection interval. We did not have prolonged periods of observation among Omicron-era samples to test this hypothesis. For pre-Omicron samples, we removed samples with COVID-19-to-blood collection intervals >72 days; however, we did not see a change in sensitivity.
Although the Elecsys nucleocapsid assay is not quantitative, we examined results to determine if Omicron or vaccinated results tended to be closer to the threshold for positivity. While the majority of results remained distinct from the threshold value, Omicron-era reactive and unreactive samples did tend to move closer to the threshold.
Previous studies examining Elecsys nucleocapsid assay performance have yielded mixed results. In the prevaccination and pre-Omicron eras, a longitudinal study comparing Elecsys sensitivity with other commercial assays reported sensitivity of 97.6% (95% CI, 87%–100%) in tests performed >81 days postdiagnosis, similar to performance at 21–80 days [8]. Alen and colleagues reported similar results (sensitivity, 95%; 95% CI, 92%–96%), with sensitivity maintained over the 33-week study [9]. Among patients with moderate/severe COVID-19 and a positive RT-PCR test, Chamkhi et al. reported the Elecsys assay to be less sensitive when testing samples collected within 1 week of symptom onset (sensitivity, 50%; 95% CI, 42%–58%), in comparison with 82% (95% CI, 74%–91%) in samples collected >21 days post–symptom onset [10]. Another study reported 90% sensitivity for samples collected ≤54 days post–PCR positivity [11].
Few previous studies have examined the performance of the Elecsys assay in vaccinated subjects, and none in the Omicron era. Allen and colleagues tested 23 vaccinated health care workers with breakthrough infections ≥14 days postvaccination (median second vaccine-to-COVID-19 interval [IQR], 30 [25–50] days), reporting that 6 were reactive (sensitivity, 26%; 95% CI, 11%–49%), a finding contradictory to our data. These cases occurred shortly after a second mRNA vaccine (when individuals have their highest concentration of neutralizing antibodies), which may in part explain why these individuals were less likely to mount a detectable nucleocapsid antibody response when infected. In a previous study examining the V-PLEX nucleocapsid assay to identify preceding SARS-CoV-2 infections, Asamoah-Boaheng found significant differences in specificity between samples from vaccinated and unvaccinated individuals, reporting that different thresholds were required for optimal test performance [12].
Limitations
We relied on participant reporting of COVID-19-positive results, which may have introduced error; however, while participants may have neglected to report all COVID-19 diagnoses, entering fictitious diagnoses would be unlikely. COVID-19 diagnoses were made by PCR and/or RAT (RAT used alone in 17% of cases), which may not be 100% specific. It is likely that many SARS-CoV-2 infections were not diagnosed; thus, it would be difficult to define a true negative case, and therefore this study could not comment on specificity. Our study participants included paramedics, who were predominantly healthy middle-aged individuals; results may differ in other populations.
CONCLUSIONS
The Elecsys nucleocapsid assay demonstrated 89% sensitivity for identifying preceding SARS-CoV-2 infections in vaccinated and unvaccinated participants over an observation period of 602 days, which spanned the SARS-CoV-2 wild-type to Omicron time periods.
Supplementary Material
Acknowledgments
We would like to acknowledge the contributions of all participating paramedics, Tracy Kirkham, Paul Demers, Laura Zunino, Tara Martin, David O'Neill, Dong Vo, Yann-Charles Lafontant, Richard Armour, Scott Haig, Jeff Maxim, Cheryl Cameron, Troy Clifford and the Ambulance Paramedics of BC, Dave Deines and the National Paramedics Association of Canada, Justin Yap, Ryan Sneath, Jennifer Bolster, Nechelle Wall, Tali Romero, Ali Scott, Carol Shen, Ashley Curtis, Chelsie Osmond, Veronica Chow and the BC Children’s Hospital Biobank, and other participating paramedic services and unions.
Financial support. This work was supported by the COVID-19 Immunity Task Force, Government of Canada.
Potential conflicts of interest. S.D. has acted as a content expert for respiratory viruses for Johnson & Johnson (Janssen). All other authors: no reported conflicts of interest. The authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Brian Grunau, Centre for Health Evaluation & Outcome Sciences, St. Paul’s Hospital, Vancouver, British Columbia, Canada; Department of Emergency Medicine, University of British Columbia, Vancouver, British Columbia, Canada; British Columbia Emergency Health Services, Vancouver, British Columbia, Canada.
Janessa Tom, British Columbia Emergency Health Services, Vancouver, British Columbia, Canada.
Michael Asamoah-Boaheng, Department of Emergency Medicine, University of British Columbia, Vancouver, British Columbia, Canada; Faculty of Medicine, Memorial University of Newfoundland, St. John’s, Newfoundland and Labrador, Canada.
Sheila F O’Brien, Canadian Blood Services, Ottawa, Ontario, Canada.
Steven J Drews, Canadian Blood Services, Ottawa, Ontario, Canada; Laboratory Medicine and Pathology, University of Alberta, Edmonton, Alberta, Canada.
Sadaf Sediqi, Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, British Columbia, Canada.
Pascal M Lavoie, Department of Pediatrics, University of British Columbia, Vancouver, British Columbia, Canada.
Vilte Barakauskas, Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, British Columbia, Canada.
David M Goldfarb, Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, British Columbia, Canada.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. Whitman JD, Hiatt J, Mowery CT, et al. . Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat Biotechnol 2020; 38:1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Byambasuren O, Cardona M, Bell K, Clark J, McLaws M-L, Glasziou P. Estimating the extent of asymptomatic COVID-19 and its potential for community transmission: systematic review and meta-analysis. Off J Assoc Med Microbiol Infect Dis Canada 2020; 5:223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ainsworth M, Andersson M, Auckland K, et al. . Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis 2020; 20:1390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allen N, Brady M, Carrion Martin AI, et al. . Serological markers of SARS-CoV-2 infection; anti-nucleocapsid antibody positivity may not be the ideal marker of natural infection in vaccinated individuals. J Infect 2021; 83:e9–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shah M, Woo HG. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front Immunol 2021; 12:830527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grunau B, O’Brien SF, Kirkham TL, et al. . A prospective observational cohort comparison of SARS-CoV-2 seroprevalence between paramedics and matched blood donors in Canada during the COVID-19 pandemic. Ann Emerg Med 2022; 80:38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. The Government of Canada . COVID-19 daily epidemiology update. Available at: https://health-infobase.canada.ca/covid-19/epidemiological-summary-covid-19-cases.html. Accessed April 25, 2022.
- 8. Muecksch F, Wise H, Batchelor B, et al. . Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J Infect Dis 2021; 223:389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Allen N, Brady M, Carrion Martin AI, et al. . SARS-CoV-2 antibody testing in health care workers: a comparison of the clinical performance of three commercially available antibody assays. Microbiol Spectr 2021; 9:e0039121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chamkhi S, Dhaouadi T, Sfar I, et al. . Comparative study of six SARS-CoV-2 serology assays: diagnostic performance and antibody dynamics in a cohort of hospitalized patients for moderate to critical COVID-19. Int J Immunopathol Pharmacol 2022; 36:205873842110732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poore B, Nerenz RD, Brodis D, Brown CI, Cervinski MA, Hubbard JA. A comparison of SARS-CoV-2 nucleocapsid and spike antibody detection using three commercially available automated immunoassays. Clin Biochem 2021; 95:77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Asamoah-Boaheng M, Goldfarb DM, Barakauskas V, et al. . Evaluation of the performance of a multiplexed serological assay in the detection of SARS-CoV-2 infections in a predominantly vaccinated population. Microbiol Spectr 2022; 10:e0145421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.