Dear Editor,
Introducing beneficial genes/alleles from wild relatives into the cultivated tomato has been an important approach for tomato breeding. Solanum habrochaites and S. galapagense have been widely used as germplasm donors in modern breeding to improve biotic and abiotic stress tolerance of tomato. S. habrochaites grows in the Peruvian Andes at altitudes up to 3300 m and is notable for its tolerance of chilling and drought and resistance to many diseases and pests. S. galapagense is endemic to the Galápagos Islands, has extraordinary salt tolerance and insect resistance, and appears even more closely related to the cultivated tomato (Solanum lycopersicum) than Solanum pimpinellifolium, the wild progenitor of cultivated tomato [1]. Due to their importance, draft genomes of these two species have been assembled using Illumina short-read sequencing [2] or PacBio long-read sequencing [3]. However, high levels of fragmentation and/or the lack of chromosome-scale assemblies have limited their applications in tomato breeding and research.
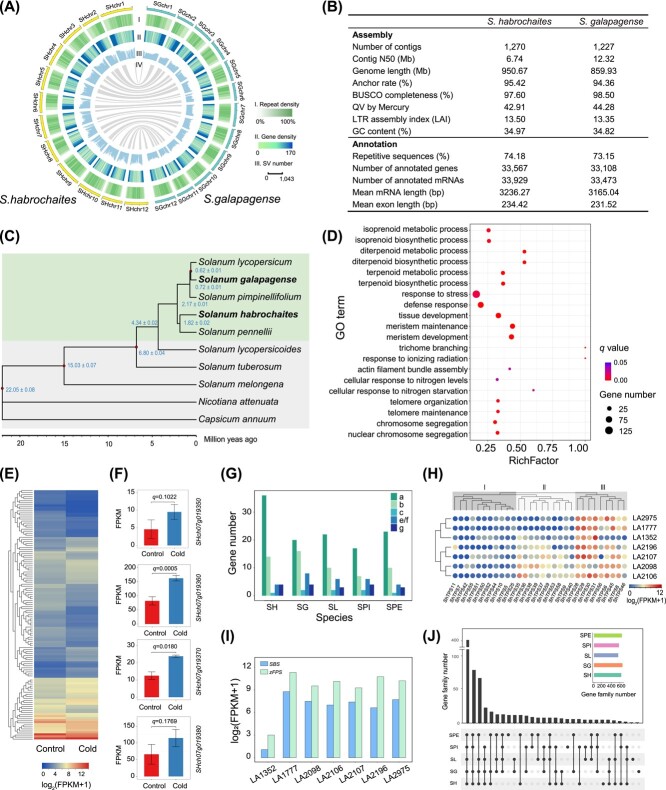
In this study, chromosome-scale assemblies of S. habrochaites (accession LA0407) and S. galapagense (accession LA0317) were developed using PacBio HiFi reads and chromatin interaction maps generated with Hi-C technology. The final assemblies of S. habrochaites and S. galapagense had total contig sizes of 950.7 and 859.9 Mb, respectively, and contig N50 sizes of 6.74 and 12.32 Mb, with 95.4 and 94.4% of the contigs anchored and ordered on the 12 chromosomes (Fig. 1A, Supplementary Data Fig. S1). The S. habrochaites and S. galapagense assemblies captured 97.6 and 98.5% of the 1614 Embryophyta conserved genes, respectively, and had LTR (long terminal repeat) assembly index (LAI) scores of 13.50 and 13.35. Moreover, the consensus quality values (QVs) of S. habrochaites and S. galapagense assemblies were 42.91 and 44.28, respectively, corresponding to a base accuracy of 99.995 and 99.996%. Taken together, the results indicated the high degree of contiguity, completeness, and base accuracy of these two genome assemblies.
Figure 1.
Solanum habrochaites and Solanum galapagense genomes. (A) Genomic landscape of S. habrochaites and S. galapagense. Densities of repeat sequences (I), genes (II), and SVs compared with Solanum lycopersicum (III) in 1-Mb windows, and syntenic blocks between S. habrochaites and S. galapagense (IV) are shown. (B) Statistics of the S. habrochaites and S. galapagense genome assemblies and annotations. (C) Phylogeny of 10 Solanaceae species with estimated divergence times. Red dots on the tree node indicate divergence times obtained from the TimeTree database (http://www.timetree.org/) that were used for calibration. (D) Gene Ontogeny (GO) terms enriched in genes overlapping with insertions and expansions in S. habrochaites. (E) Expression heat map of significantly up- or downregulated genes (q < .05) under cold stress in S. habrochaites with coding regions overlapping with insertions and expansions. (F) Expression of four tandemly duplicated ShRCI3s under cold stress. Error bars represent the standard deviation of three independent replicates. (G) Number of different TPS subfamily genes detected in five tomato species. (H, I) Expression of TPS genes (H) and zFPS (SHch08g004680) and SBS (ShTPS45, SHch08g004730) (I) in stem/petiole trichomes of seven different S. habrochaites accessions. (J) UpSet plot of RGA gene families among five tomato species. SH, S. habrochaites; SG, S. galapagense; SL, S. lycopersicum; SPI, S. pimpinellifolium; SPE, S. pennellii.
The S. habrochaites and S. galapagense genomes harbored 74.2% (705.2 Mb) and 73.2% (632.2 Mb) repetitive sequences, respectively, of which LTR retrotransposons accounted for 58.8 and 59.0% (Fig. 1B, Supplementary Data Table S1). A total of 33 567 and 33 108 protein-coding genes were predicted from the S. habrochaites and S. galapagense genome assemblies, respectively, and around 98% of the predicted genes could be annotated in public databases.
The phylogenetic tree constructed for S. habrochaites, S. galapagense, and eight other Solanaceae species using 3011 single-copy orthologous genes revealed that S. habrochaites was close to Solanum pennellii and that S. galapagense appeared close to S. lycopersicum (Fig. 1C), consistent with the previous phylogenomic study [4]. Molecular dating suggested that S. habrochaites diverged from S. pennellii around 1.82 million years ago (Mya), and S. galapagense and S. lycopersicum diverged around 0.62 Mya. A total of 16 666 gene families were shared among S. habrochaites, S. galapagense, S. lycopersicum, S. pimpinellifolium, and S. pennellii, and 366 and 190 were unique to S. habrochaites and S. galapagense, respectively (Supplementary Data Fig. S2). KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis suggested that these S. habrochaites-specific genes were significantly enriched with those involved in plant–pathogen interaction and in the MAPK signaling pathway, while the S. galapagense-specific genes were significantly enriched with those involved in zeatin and terpenoid biosynthesis.
To identify structure variants (SVs) relative to cultivated tomato, genome sequences of S. habrochaites and S. galapagense were compared with the S. lycopersicum genome (version SL4.0), according to the pipeline described in our previous study [5]. A total of 336 319 SVs with a total length of 257.9 Mb between S. habrochaites and S. lycopersicum and 98 443 SVs with a total length of 62.2 Mb between S. galapagense and S. lycopersicum were identified (Supplementary Data Fig. S3, Supplementary Data Table S2). The insertion and expansion regions in S. habrochaites, representing the S. habrochaites-specific sequences, overlapped with the coding regions of 5250 genes, which were significantly enriched with those involved in response to stress, defense response, terpenoid biosynthetic, and metabolic processes etc. (Fig. 1D). Coding regions of 1336 genes were found overlapping with the insertion and expansion regions of S. galapagense, and these genes were significantly enriched with those associated with defense response, pyrimidine nucleotide metabolism, and lipid metabolism etc. (Supplementary Data Fig. S4). These results suggested that the inserted and expanded genome regions in S. habrochaites and S. galapagense might contribute to the higher stress tolerance of the two wild tomato species. We found that the expression of 122 of these genes in S. habrochaites was significantly changed after cold treatment (Fig. 1E), including two of four tandem duplicates (Shch07g019350–Shch07g019380) homologous to Arabidopsis rare cold-inducible protein 3 (RIC3) [6], which corresponded to only one copy (Solyc07g049240) in SL4.0 (Fig. 1F, Supplementary Data Fig. S5). The upregulation of ShRIC3 genes by cold treatment suggested their potential roles in cold stress responses.
As mentioned above, the inserted/expanded genes in S. habrochaites were enriched in the terpenoid biosynthetic process. Terpenoids play roles in plant defense against pathogens and pests. Terpene synthases (TPSs) are key enzymes in generating terpenoids. A total of 59, 50, 43, 36, and 41 TPS genes were identified in genomes of S. habrochaites, S. galapagense, S. lycopersicum, S. pimpinellifolium (LA2093), and S. pennellii (LA0716), respectively (Fig. 1G, Supplementary Data Tables S3 and S4). Five TPS subfamilies, including TPS-a, -b, -c, -e/f and -g, were identified, and TPS-a was the most abundant (Supplementary Data Fig. S6). Since TPS-a members mainly encode sesquiterpene synthases, a remarkable expansion of this subfamily in S. habrochaites suggested potentially diverse or unique sesquiterpene synthesis in this species. Eighteen ShTPSs were not expressed in any of the investigated tissues, including leaf, stem, root, flower, and fruit, while the remainder were mainly expressed in a tissue-specific manner (Supplementary Data Fig. S7). Trichomes play roles in plant defense by providing specialized metabolites, including terpenes. Nearly half of the ShTPSs were expressed in stem/petiole trichomes of seven S. habrochaites accessions, and these ShTPSs were further divided into three groups based on their expression patterns (Fig. 1H). The various TPS expression patterns probably contributed to the diversity of terpene composition in these accessions [7]. A novel sesquiterpene biosynthesis pathway involving SBS (santalene and bergamotene synthase, a TPS-e/f member) and zFPS (Z-isoprenyl pyrophosphate synthase) has been proposed in S. habrochaites [8]. Our results showed that SBS and zFPS had similar expression levels in the seven S. habrochaites accessions, except LA1352, suggesting both conserved and diverged sesquiterpene biosynthesis in these accessions (Fig. 1I).
The wild relatives of tomato are the main gene source for tomato resistance breeding [9]. To explore the reservoir of resistance genes in tomato species, resistance gene analogs (RGAs) were identified in genomes of S. habrochaites, S. galapagense, S. lycopersicum, S. pimpinellifolium, and S. pennellii. In total, 4668 RGAs were detected in these five species, including 2482 receptor-like protein kinases (RLKs), 831 nucleotide binding site (NBS)-encoding proteins and 391 receptor-like proteins (RLPs) (Supplementary Data Table S5). Gene family analysis indicated that 401 gene families (2685 genes) were shared in all five tomato species, while 187 gene families (919 genes) were not found in S. lycopersicum (Fig. 1J). In addition, 163 and 36 RGAs were found in the insertion/expansion regions of S. habrochaites and S. galapagense, respectively. These extra RGAs might contribute to the high disease resistance of the two species.
In summary, the high-quality genome assemblies of S. habrochaites and S. galapagense provide robust references, in particular, new gene sources of stress tolerance and terpene biosynthesis for functional genomic research and genetic improvement in tomato.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32170395), the US National Science Foundation (IOS-1855585), the Foundations of Hubei Hongshan Laboratory (2021hszd017), and the Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences.
Author contributions
L.G., Z.F., X.Y., and S.G conceived the project. Y.S., M.Q., S.G., and C.H. performed the experiments. X.Y. and M.Q. analyzed the data and wrote the manuscript. L.G., Z.F., and S.G revised the manuscript.
Data availability
Raw sequencing data and genome assemblies have been deposited in the Genome Sequence Archive (GSA) under the BioProject accession number PRJCA008297.
Conflict of interest
The authors declare no conflicts of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
Supplementary Material
Contributor Information
Xiaofen Yu, CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Innovative Academy of Seed Design, Chinese Academy of Sciences, Wuhan 430074, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
Minghao Qu, CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Innovative Academy of Seed Design, Chinese Academy of Sciences, Wuhan 430074, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yanna Shi, Zhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Zhejiang University, Hangzhou 310058, China.
Chenlu Hao, CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Innovative Academy of Seed Design, Chinese Academy of Sciences, Wuhan 430074, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Sumin Guo, CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Innovative Academy of Seed Design, Chinese Academy of Sciences, Wuhan 430074, China.
Zhangjun Fei, Boyce Thompson Institute, Cornell University, Ithaca, NY 14853, USA; US Department of Agriculture-Agricultural Research Service, Robert W. Holley Center for Agriculture and Health, Ithaca, NY 14853, USA.
Lei Gao, CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Innovative Academy of Seed Design, Chinese Academy of Sciences, Wuhan 430074, China; Hubei Hongshan Laboratory, Wuhan 430070, China.
References
- 1. Strickler SR, Bombarely A, Munkvold JDet al. Comparative genomics and phylogenetic discordance of cultivated tomato and close wild relatives. PeerJ. 2015;3:e793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aflitos S, Schijlen E, Jong Het al. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 2014;80:136–48. [DOI] [PubMed] [Google Scholar]
- 3. Seong K, Shaw CL, Seo Eet al. A draft genome assembly for the heterozygous wild tomato Solanum habrochaites highlights haplotypic structural variations of intracellular immune receptors. bioRxiv. 2022: 2022.2001.2021.477156. [Google Scholar]
- 4. Pease JB, Haak DC, Hahn MWet al. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 2016;14:e1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang X, Gao L, Jiao Cet al. Genome of Solanum pimpinellifolium provides insights into structural variants during tomato breeding. Nat Commun. 2020;11:5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Llorente F, López-Cobollo RM, Catalá Ret al. A novel cold-inducible gene from Arabidopsis, RCI3, encodes a peroxidase that constitutes a component for stress tolerance. Plant J. 2002;32:13–24. [DOI] [PubMed] [Google Scholar]
- 7. Gonzales-Vigil E, Hufnagel DE, Kim Jet al. Evolution of TPS20-related terpene synthases influences chemical diversity in the glandular trichomes of the wild tomato relative Solanum habrochaites. Plant J. 2012;71:921–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sallaud C, Rontein D, Onillon Set al. A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell. 2009;21:301–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seong K, Seo E, Witek Ket al. Evolution of NLR resistance genes with noncanonical N-terminal domains in wild tomato species. New Phytol. 2020;227:1530–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data and genome assemblies have been deposited in the Genome Sequence Archive (GSA) under the BioProject accession number PRJCA008297.