Abstract
Objectives
To investigate the gene mutational profile of urachal carcinoma in correlation with its clinicopathologic features.
Methods
We analyzed genetic mutations in 30 cases of urachal carcinoma by next-generation sequencing (NGS) test. Histologic slides and clinical data were reviewed.
Results
The patients included 21 men and 9 women, with a mean age of 53 years (range, 24-75 years). The urachal carcinomas included mucinous (11), enteric (10), signet ring cell (8), and high-grade neuroendocrine (1) subtypes. Targeted NGS analysis demonstrated genetic mutations in all the urachal tumors (mean, 2; range, 1-4). TP53 was the most mutated gene (25), followed by KRAS (9) and GNAS (8) genes. TP53 mutations were more common in the signet ring cell subtype (7/8), and GNAS mutations were present only in the mucinous (5/11) and signet ring cell subtypes (3/8) but not in the enteric subtype (0/10). KRAS mutations were significantly associated with cancer stage IV (P = .02) and younger patient age (P = .046). Furthermore, the presence of KRAS mutations in urachal carcinoma portended a poorer overall survival (P = .006).
Conclusions
Urachal carcinoma demonstrates frequent gene mutations that are associated with distinct clinicopathologic features. Gene mutation may underlie the development and progression of this aggressive disease.
Keywords: Urachal carcinoma, Next-generation sequencing, Gene mutations, TP53, KRAS, GNAS
Key Points.
• Urachal carcinoma demonstrates frequent gene mutations, with the TP53, KRAS, and GNAS genes being the most mutated. Gene mutations are associated with distinct clinicopathologic features.
• TP53 mutations are most frequent in the signet ring cell subtype, while GNAS mutations are associated with mucinous and signet ring cell subtypes.
• KRAS mutations are more common in high-stage cancers and young patients. The presence of KRAS mutations in urachal carcinoma portends a poor clinical outcome.
Introduction
Urachal carcinoma is a rare malignant neoplasm that arises from the urachal remnant.1-7 The urachus is a vestigial structure that connects the urinary bladder to the allantois during embryonal development, facilitating liquid waste discharge and gas exchange.2,6 After birth, the urachus is usually obliterated and becomes a fibrous cord called the median umbilical ligament; however, approximately 30% of adults have a urachal remnant, a tubular or cystic structure in the bladder dome or elsewhere along the midline of the anterior wall.1,2 Although the urachal remnant is usually lined by urothelium, most urachal carcinomas are composed of adenocarcinoma, including mucinous, enteric, signet ring cell, and not otherwise specified (NOS) subtypes.7 Other carcinomas, such as urothelial carcinoma, squamous carcinoma, and neuroendocrine carcinoma, may also occur in rare instances.8,9 The histologic divergence between the urachal remnant and urachal carcinoma is likely due to the intestinal metaplastic change of the urachal epithelium.2,8 Other researchers think that primitive colonic remnants (cloacal) may also contribute to this morphologic discrepancy, since the cloaca is in close proximity to the urachus during embryonal development.8,10
Urachal carcinoma is an aggressive disease that frequently spreads to the bladder and other organs.1-7 Several staging systems have been proposed for urachal carcinoma, and the Sheldon system, which is endorsed by the World Health Organization, is the most widely used.1,2,4,7 Over 90% of urachal carcinomas are diagnosed at advanced stages (Sheldon stage III or IV), and about 21% of patients have developed distant metastasis at initial presentation.11 The overall prognosis is poor, and the average 5-year cancer-specific survival rate is 64.4%.11 At advanced stages, it is difficult, if not impossible, to eradicate the disease by surgery alone, and systemic therapy is often part of the treatment regimen. As only 30% to 40% of patients experience a response to conventional chemotherapy, targeted therapy based on the molecular signature of urachal carcinoma is needed for this aggressive disease.6,11-13 A better understanding of genetic mutations in urachal carcinoma may lead to more effective targeted therapy. To elucidate the mutational profile of urachal carcinoma, we performed a next-generation sequencing (NGS) analysis in a large cohort of urachal carcinomas from a single institution in correlation with the clinicopathologic features.
MATERIALS AND METHODS
This study was approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center (Houston, TX). We retrospectively searched the pathology files at MD Anderson from 2008 to 2020 and identified 30 patients with urachal carcinoma whose tumor samples had undergone NGS.
Patients’ H&E and immunohistochemical slides were reviewed to confirm the diagnosis in accordance with the histopathologic criteria set by the World Health Organization: the tumor was located at the dome or anterior wall of the bladder, the tumor predominantly involved the muscularis propria or perivesical soft tissue with the epicenter at the bladder wall, extensive cystitis cystica or cystitis glandularis was not present beyond the dome or anterior wall, and there was no known primary carcinoma of a similar histologic type at other anatomic locations.7 The tumor clinical stage was based on the Sheldon staging system for urachal carcinoma: I, tumors were limited to the urachal mucosa; II, tumors invaded into but not beyond the urachal muscular layer; III, tumors extended to the bladder, abdominal wall, and other adjacent organs; and IV, tumors metastasized to the lymph nodes or other distant organs.2,7 Data collected from the pathologic review and medical record included age, sex, and clinical presentation of symptoms; tumor size, location, stage, and histologic subtype; presence of signet ring cell features; and clinical outcome. Survival times were recorded from the date of initial diagnosis to the date of death or last follow-up.
NGS was performed on formalin-fixed, paraffin-embedded urachal carcinoma tissues and included a 50-Gene Somatic Mutation Analysis Panel (AmpliSeq Hotspot version 2) (n = 6), a 134-gene Solid Tumor Genomics Assay v1 (or Comprehensive Oncomine version 1) (n = 8), a 146-gene Solid Tumor Genomics Assay v3 (or Comprehensive Oncomine version 3) (n = 11), and a 409-Gene Somatic Mutation Analysis Panel (Ion AmpliSeq Comprehensive Cancer Panel) (n = 5) (Supplemental Tables 1-4; all supplemental materials can be found at American Journal of Clinical Pathology online).14-16 All four NGS panels were purchased from Thermo Fisher Scientific. Our analysis was focused on the initial 50 key oncogenes and tumor suppressor genes, which were included in all the four test panels.
Statistics were calculated using standard methods and IBM SPSS version 24 for Windows (SPSS). The Kaplan-Meier method was used to plot survival curves, and differences between these curves were analyzed for significance using the log-rank test. The χ 2 tests were performed to evaluate potential correlations between categorical groups. When categorical variables fell short of their expected count, the Fisher exact test was used. A P value of less than .05 was considered statistically significant.
RESULTS
Pathologic Findings
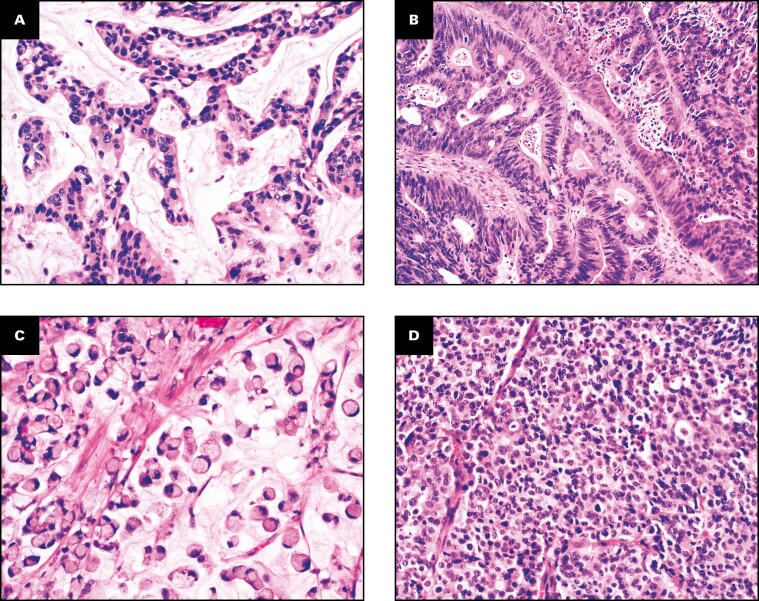
The patients with urachal carcinoma included 21 men and 9 women with a mean age of 53 years (range, 24-75 years). Twenty-seven patients had gross or microscopic hematuria, and three had abdominal pain. One patient had mucosuria accompanied by hematuria. The tumors were located at the bladder dome (n = 23) or anterior wall (n = 7). The mean tumor size was 5.7 cm (range, 2.5-17 cm). The urachal carcinomas included mucinous (n = 11), enteric (n = 10), signet ring cell (n = 8), and high-grade neuroendocrine carcinoma (n = 1) subtypes. The mucinous subtype produced abundant extracellular mucin with floating tumor cells Figure 1A. The enteric subtype consisted of pseudostratified columnar cells with marked nuclear atypia and necrosis Figure 1B. The signet ring cell subtype was characterized by infiltrative tumor cells with a prominent intracellular mucinous vacuole that pushed the indented nucleus to the periphery Figure 1C . High-grade neuroendocrine carcinoma subtype was composed of solid sheets of poorly differentiated carcinoma Figure 1D and showed diffuse immunoreactivity for synaptophysin and chromogranin. In six cases, the urachal tumor showed mixed histologic subtypes, and the tumor classification was based on the predominant subtype, from which tumor sample was taken for the NGS test. At initial presentation, 17 patients had metastatic disease at stage IV using the Sheldon system Table 1 . Thirteen patients had stage III, 10 of whom subsequently developed metastases in a mean of 2.8 years (range, 1.2-10.4 years). The metastatic sites included the lungs (n = 11), abdominal wall (not by direct extension) (n = 4), liver (n = 3), lymph nodes (n = 3), vertebra (n = 1), and vagina (n = 1).
Figure 1.
Urachal carcinoma. A, Mucinous subtype. B, Enteric subtype. C, Signet ring cell subtype. D, High-grade neuroendocrine carcinoma subtype. (H&E, x200)
Table 1.
Next-Generation Sequencing With Distinct Gene Mutations (Black) in Urachal Carcinomaa
| Case No. | Type | Age, y | Stage | TP53 | KRAS | GNAS | SMAD4 | KIT | PI3K3CA | NOTCH1 | ATM | FGFR1 | FGFR2 | BRAF | KDR | PDGFRA | RB1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MU | 31 | IV | ||||||||||||||
| 2 | MU | 50 | III | ||||||||||||||
| 3 | MU | 45 | III | ||||||||||||||
| 4 | MU | 54 | IV | ||||||||||||||
| 5 | MU | 62 | III | ||||||||||||||
| 6 | MU | 46 | IV | ||||||||||||||
| 7 | MU | 59 | III | ||||||||||||||
| 8 | MU | 58 | IV | ||||||||||||||
| 9 | MU | 50 | IV | ||||||||||||||
| 10 | MU | 52 | IV | ||||||||||||||
| 11 | MU | 56 | III | ||||||||||||||
| 12 | EN | 68 | IV | ||||||||||||||
| 13 | EN | 61 | IV | ||||||||||||||
| 14 | EN | 47 | IV | ||||||||||||||
| 15 | EN | 69 | III | ||||||||||||||
| 16 | EN | 75 | IV | ||||||||||||||
| 17 | EN | 52 | IV | ||||||||||||||
| 18 | EN | 53 | III | ||||||||||||||
| 19 | EN | 59 | III | ||||||||||||||
| 20 | EN | 51 | IV | ||||||||||||||
| 21 | EN | 36 | III | ||||||||||||||
| 22 | SR | 63 | III | ||||||||||||||
| 23 | SR | 71 | III | ||||||||||||||
| 24 | SR | 66 | III | ||||||||||||||
| 25 | SR | 50 | IV | ||||||||||||||
| 26 | SR | 42 | IV | ||||||||||||||
| 27 | SR | 33 | IV | ||||||||||||||
| 28 | SR | 52 | IV | ||||||||||||||
| 29 | SR | 67 | III | ||||||||||||||
| 30 | NE | 24 | IV |
EN, enteric subtype; MU, mucinous subtype; NE, high-grade neuroendocrine carcinoma subtype; SR, signet ring cell subtype.
aStage was based on the Sheldon staging system.
Molecular Data
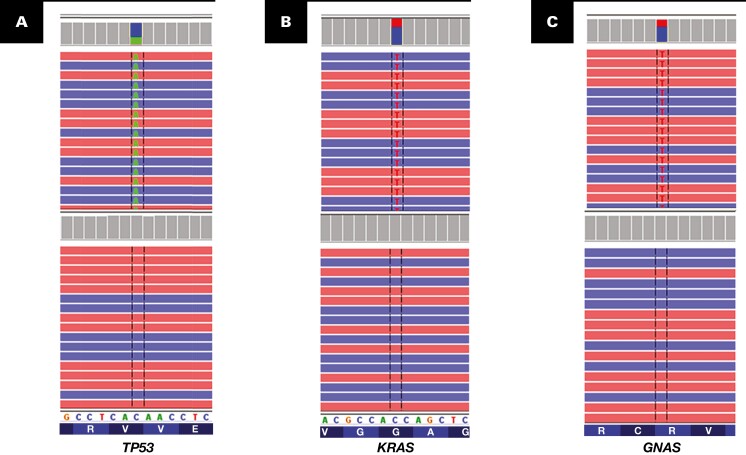
NGS was performed on primary (n = 13) and metastatic (n = 17) tumors. All except for one tumor showed gene mutations that were covered by the 50-gene panel (Supplemental Table 1). The number of mutated genes per case ranged from 1 to 4 with a mean of 2.2 Table 1 . The only tumor (case 11) with no gene mutations on the 50-gene panel was analyzed with a 409-gene panel, which demonstrated mutations in four other genes, including GUCY1A2, MAPK2K1, MAPK2K2, and PTPRD (Supplemental Table 4). Overall, TP53 was the most frequently mutated gene (n = 25) Figure 2A , and one tumor harbored two mutations at different exons of the TP53 gene (Supplemental Table 5). TP53 mutation was found more often in the signet ring cell tumor subtype (7/8) than in the non–signet ring cell subtype (P = .013). KRAS was the second most commonly mutated gene (n = 9) Figure 2B , followed by GNAS (n = 8) Figure 2C . Other commonly mutated genes included SMAD4 (n = 7), KIT (n = 3), MAP2K1 (n = 3), ARID1A (n = 3), PIK3CA (n = 3), NOTCH1 (n = 2), ATM (n = 2), and NF1 (n = 2). In addition, a number of other genes were also mutated Table 1 . GNAS mutations were present only in the mucinous (5/11) or signet ring cell subtypes (3/8) but not in the enteric subtype (0/10). This difference was statistically significant (P = .043). Interestingly, high-grade neuroendocrine carcinoma showed concurrent mutations in TP53 and RB1 genes, which were frequently present in lung and bladder small cell carcinomas.17
Figure 2.
Common genetic mutations (top) in urachal carcinomas vs normal (bottom). A, TP53 gene mutation. B, KRAS gene mutation. C, GNAS gene mutation.
Clinical Follow-up Findings
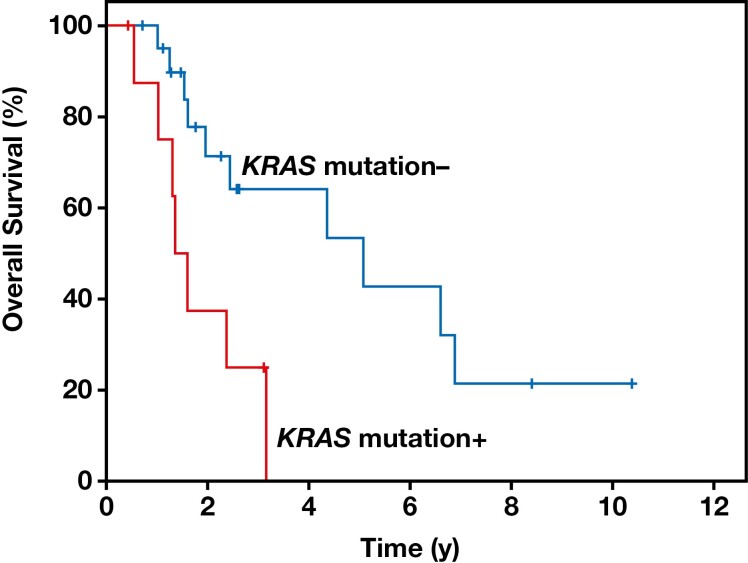
Follow-up information was available for all patients, with a mean follow-up time of 2.8 years (range, 0.4-10.4 years). Seventeen patients died of disease in a median of 1.6 years (range, 0.5-6.9 years). Ten patients were alive with disease (range, 0.4-10.4 years), and three patients were alive without evidence of disease (range, 0.7-3.1 years). Patients with stage IV disease had a significantly lower survival rate than did those with stage III disease (P = .005). KRAS mutations were significantly more common in high-stage tumors (P = .02), as eight of the nine tumors with KRAS mutations were stage IV. KRAS mutations were also more frequent in younger patients (P = .046), as seven of the nine patients with KRAS mutations were younger than 53 years (the mean age of our cohort). Furthermore, KRAS mutations were associated with a significantly poorer overall survival (P = .006) in patients with urachal carcinoma Figure 3 .
Figure 3.
Patients with KRAS mutations in urachal carcinoma showed a significantly poorer overall survival in the Kaplan-Meier curve (P = .006).
Discussion
There have been limited studies to investigate the molecular profile of urachal carcinoma, and most studies were conducted in a small series of cases due to the rarity of this malignant tumor.10,12,18-24 Maurer et al22 reported that urachal carcinoma expressed genomic alterations that were frequently present in bladder urothelial carcinoma and colorectal adenocarcinoma, suggesting that similar pathways might drive the tumorigenesis of these malignancies, despite the different tissue origins. However, Lee et al23 found that urachal adenocarcinoma showed a distinct molecular signature from bladder urothelial carcinoma and colorectal adenocarcinoma, indicating that urachal carcinoma might use different oncogenic pathways from those in bladder and colorectal carcinomas. Recently, Reis et al13 evaluated a large cohort of urachal carcinomas from multiple institutions, which demonstrated the prevalence of genetic mutations in urachal tumors. The most frequent mutations occurred in the TP53 gene, followed by KRAS, BRAF, and PIK3CA genes. Furthermore, a considerable number of urachal carcinomas showed genomic aberrations in the RAS/RAF/PI3K signal transduction pathway, suggesting a potential value of anti–epidermal growth factor receptor therapy in this aggressive disease. However, only a small number of genes (12-15 genes) were analyzed by the targeted NGS test in this multi-institutional study.13 In the current study, we conducted a mutational analysis of a cohort of urachal carcinomas from a single institution. Our NGS test included a large number of actionable or predictive genes (50-409), which demonstrated frequent mutations in TP53, KRAS, GNAS, and other genes. Importantly, our study reveals that several gene mutations are associated with distinct clinical and pathologic features in urachal carcinoma. GNAS mutations are present only in mucinous and signet ring cell subtypes, and KRAS mutations are more common in cancer stage IV and younger patients. Furthermore, the presence of the KRAS mutation in urachal carcinoma portends a significantly poorer overall survival. Nonetheless, due to the limited number of cases in our study, large independent cohort studies are needed to confirm the clinicopathologic significance of these gene mutations in urachal carcinoma.
Several studies have demonstrated that TP53 is the most mutated gene in urachal carcinoma.13,22-25 As a tumor suppressor, TP53 inhibits cell cycle, particularly G1-S progression, and regulates DNA damage repair, cell proliferation, and apoptosis.26 Mutations of the TP53 gene have been reported in nearly half of all human malignancies, from a wide range of tumors, and are associated with poor prognosis and treatment failure in some cancers.27 In the current study, our NGS analysis showed that the TP53 gene was mutated in 75% of urachal carcinomas, particularly those with signet ring cell features; however, we did not observe any significant clinical association with the TP53 gene mutations either. Reis et al13 also found urachal carcinoma had frequent mutations in the TP53 gene and expressed a high level of p53 protein, but neither TP53 gene mutations nor p53 overexpression was associated with patient survival.25 Nonetheless, targeted therapy against the TP53 gene may still be useful in human cancers with TP53 mutations, including urachal carcinoma.28,29
The KRAS gene is another frequently mutated gene in urachal carcinoma. This oncogene encodes a GTPase protein that plays a key role in the RAS/MAPK pathway.30 When the KRAS gene is constitutively activated by gene mutation, it increases cell proliferation, inhibits apoptosis, and regulates tumor microenvironment and immune response.30,31 Several studies have demonstrated that the KRAS mutation is often associated with a poor prognosis in adenocarcinomas of the lungs and gastrointestinal tract.32,33 Sirintrapun et al19 previously reported that the KRAS mutation was associated with a better overall survival in urachal carcinoma, but their cohort was small with only seven patients. In the current study, we studied a larger cohort of 30 patients and demonstrated that the KRAS mutation was significantly associated with high-stage disease and lower overall survival rate. The KRAS mutation also occurred more frequently in younger patients with urachal carcinoma in our cohort. Our findings are more in line with the previously known association between the KRAS mutation and poor prognosis at other anatomic sites.32,33 In addition, two patients showed KRASG12C mutants in our study, suggesting that KRASG12C inhibitors might be useful in the targeted therapy for urachal carcinoma.34
A considerable number of urachal carcinomas exhibited genetic mutations in the GNAS gene in the current study. The GNAS gene encodes the guanine nucleotide binding protein (G protein) α stimulating activity polypeptide 1 and regulates the G protein function in signaling pathways.35,36 Somatic mutations of GNAS have been found in several mucin-producing glandular neoplasms, such as intraductal papillary mucinous neoplasms of the pancreas and low-grade appendiceal mucinous neoplasms.37,38 Nishikawa et al37 reported that the GNAS mutation might regulate mucin production through the cyclic adenosine monophosphate–protein kinase A signaling pathway. Pietrantonio et al39 found that the GNAS mutation may serve as a prognostic biomarker in patients with relapsed peritoneal pseudomyxoma receiving metronomic capecitabine and bevacizumab. In the current study, GNAS mutations were present only in mucinous and signet ring cell subtypes, reaffirming the regulatory role of GNAS in mucinous production, although GNAS mutations are not significantly associated with the clinical outcome.
Our study demonstrated that distinct gene mutations are associated with clinicopathologic features in urachal carcinoma, but there are several limitations in our study. First, we used different NGS platforms, which consisted of 50, 134, 146, or 409 gene targets. However, our analysis was focused on the initial 50 actionable or predictive gene markers, such as TP53, KRAS, and GNAS, that were covered in all the platforms. In addition, these expanded NGS platforms identified novel gene mutations in urachal carcinoma that have not previously been reported. Second, the number of patients was limited in our study, and most patients presented at an advanced stage (Sheldon stage III or IV) in this study, probably because of the tertiary referral nature of our institution. Our patients had an overall survival duration of 1.6 years, which was compatible with the results of previous studies.1,6,8 Finally, our study was focused on pathologic features and clinical outcomes; the therapeutic implications of these gene mutations are beyond the realm of this study. Nonetheless, we identified a wide range of genetic mutations in urachal carcinoma, which may provide useful insight into potential therapeutic targets.
In conclusion, our NGS analysis of a large cohort of urachal carcinomas from a single institution demonstrates that gene mutations are highly frequent in this malignant disease. The most common mutations occur in the TP53, KRAS, and GNAS genes. Importantly, these gene mutations are associated with distinct clinicopathologic features. GNAS mutations are present only in urachal carcinomas with mucinous and signet ring cell features, and KRAS mutations are significantly associated with high-stage cancer and young patient age. Furthermore, the presence of the KRAS mutation portends a poor overall survival in patients with urachal carcinoma. Our results suggest that gene mutations may underlie the development of distinct clinicopathologic features in urachal carcinoma. Novel targeted therapies based on gene mutations may improve the treatment repertoire for this aggressive disease.
Supplementary Material
Acknowledgments:
The authors thank Ann M. Sutton, scientific editor, Research Medical Library at MD Anderson Cancer Center, for editing this article.
Contributor Information
Michael P Zaleski, Department Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Hui Chen, Department Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Sinchita Roy-Chowdhuri, Department Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Keyur P Patel, Department Hematopathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Rajyalakshmi Luthra, Department Hematopathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Mark J Routbort, Department Hematopathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Ashish M Kamat, Department Urology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Jianjun Gao, Genitourinary Medical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Arlene Siefker-Radtke, Genitourinary Medical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Bogdan Czerniak, Department Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Charles C Guo, Department Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Funding: This study was supported by the National Institutes of Health/National Cancer Institute under award number P30 CA016672.
References
- 1. Dhillon J, Liang Y, Kamat AM, et al. Urachal carcinoma: a pathologic and clinical study of 46 cases. Hum Pathol. 2015;46:1808-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sheldon CA, Clayman RV, Gonzalez R, et al. Malignant urachal lesions. J Urol. 1984;131:1-8. [DOI] [PubMed] [Google Scholar]
- 3. Gopalan A, Sharp DS, Fine SW, et al. Urachal carcinoma: a clinicopathologic analysis of 24 cases with outcome correlation. Am J Surg Pathol. 2009;33:659-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Molina JR, Quevedo JF, Furth AF, et al. Predictors of survival from urachal cancer: a Mayo Clinic study of 49 cases. Cancer. 2007;110:2434-2440. [DOI] [PubMed] [Google Scholar]
- 5. Bruins HM, Visser O, Ploeg M, et al. The clinical epidemiology of urachal carcinoma: results of a large population based study. J Urol. 2012;188:1102-1107. [DOI] [PubMed] [Google Scholar]
- 6. Siefker-Radtke A. Urachal adenocarcinoma: a clinician’s guide for treatment. Semin Oncol. 2012;39:619-624. [DOI] [PubMed] [Google Scholar]
- 7. Moch HHP, Ulbright TM, Retuer VE.. World Health Organization Classification of Tumours of the Urinary System and Male Genital Organs. 4th ed. Lyon, France: International Agency for Research on Cancer; 2016. [Google Scholar]
- 8. Paner GP, Lopez-Beltran A, Sirohi D, et al. Updates in the pathologic diagnosis and classification of epithelial neoplasms of urachal origin. Adv Anat Pathol. 2016;23:71-83. [DOI] [PubMed] [Google Scholar]
- 9. Wang G, Huang H, Kamat AM, et al. High-grade neuroendocrine carcinoma of the urachus-report of 3 cases. Hum Pathol. 2017;67:126-133. [DOI] [PubMed] [Google Scholar]
- 10. Kardos J, Wobker SE, Woods ME, et al. Comprehensive molecular characterization of urachal adenocarcinoma reveals commonalities with colorectal cancer, including a hypermutable phenotype. JCO Precis Oncol. 2017;1:PO.17.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szarvas T, Módos O, Niedworok C, et al. Clinical, prognostic, and therapeutic aspects of urachal carcinoma: a comprehensive review with meta-analysis of 1,010 cases. Urol Oncol. 2016;34:388-398. [DOI] [PubMed] [Google Scholar]
- 12. Loh KP, Mondo E, Hansen EA, et al. Targeted therapy based on tumor genomic analyses in metastatic urachal carcinoma. Clin Genitourin Cancer. 2016;14:e449-e452. [DOI] [PubMed] [Google Scholar]
- 13. Reis H, van der Vos KE, Niedworok C, et al. Pathogenic and targetable genetic alterations in 70 urachal adenocarcinomas. Int J Cancer. 2018;143:1764-1773. [DOI] [PubMed] [Google Scholar]
- 14. Luthra R, Patel KP, Routbort MJ, et al. A targeted high-throughput next-generation sequencing panel for clinical screening of mutations, gene amplifications, and fusions in solid tumors. J Mol Diagn. 2017;19:255-264. [DOI] [PubMed] [Google Scholar]
- 15. Singh RR, Patel KP, Routbort MJ, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607-622. [DOI] [PubMed] [Google Scholar]
- 16. Singh RR, Patel KP, Routbort MJ, et al. Clinical massively parallel next-generation sequencing analysis of 409 cancer-related genes for mutations and copy number variations in solid tumours. Br J Cancer. 2014;111:2014-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang MT, Penson A, Desai NB, et al. Small-cell carcinomas of the bladder and lung are characterized by a convergent but distinct pathogenesis. Clin Cancer Res. 2018;24:1965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collazo-Lorduy A, Castillo-Martin M, Wang L, et al. Urachal carcinoma shares genomic alterations with colorectal carcinoma and may respond to epidermal growth factor inhibition. Eur Urol. 2016;70:771-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sirintrapun SJ, Ward M, Woo J, et al. High-stage urachal adenocarcinoma can be associated with microsatellite instability and KRAS mutations. Hum Pathol. 2014;45:327-330. [DOI] [PubMed] [Google Scholar]
- 20. Singh H, Liu Y, Xiao X, et al. Whole exome sequencing of urachal adenocarcinoma reveals recurrent NF1 mutations. Oncotarget. 2016;7:29211-29215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hang JF, Pan CC. Absence of GNAS and BRAF mutations but presence of KRAS mutation in urachal adenocarcinoma. Pathology. 2017;49:316-317. [DOI] [PubMed] [Google Scholar]
- 22. Maurer A, Ortiz-Bruechle N, Guricova K, et al. Comparative genomic profiling of glandular bladder tumours. Virchows Arch. 2020;477:445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee S, Lee J, Sim SH, et al. Comprehensive somatic genome alterations of urachal carcinoma. J Med Genet. 2017;54:572-578. [DOI] [PubMed] [Google Scholar]
- 24. Cornejo KM, Cosar EF, Paner GP, et al. Mutational profile using next-generation sequencing may aid in the diagnosis and treatment of urachal adenocarcinoma. Int J Surg Pathol. 2020;28:51-59. [DOI] [PubMed] [Google Scholar]
- 25. Niedworok C, Panitz M, Szarvas T, et al. Urachal carcinoma of the bladder: impact of clinical and immunohistochemical parameters on prognosis. J Urol. 2016;195:1690-1696. [DOI] [PubMed] [Google Scholar]
- 26. Kirsch DG, Kastan MB. Tumor-suppressor p53: implications for tumor development and prognosis. J Clin Oncol. 1998;16:3158-3168. [DOI] [PubMed] [Google Scholar]
- 27. Soussi T, Legros Y, Lubin R, et al. Multifactorial analysis of p53 alteration in human cancer: a review. Int J Cancer. 1994;57:1-9. [DOI] [PubMed] [Google Scholar]
- 28. Blandino G, Di Agostino S. New therapeutic strategies to treat human cancers expressing mutant p53 proteins. J Exp Clin Cancer Res. 2018;37:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lane DP, Cheok CF, Lain S. p53-based cancer therapy. Cold Spring Harb Perspect Biol 2010;2:a001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bryant KL, Mancias JD, Kimmelman AC, et al. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahrendt SA, Decker PA, Alawi EA, et al. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001;92:1525-1530. [DOI] [PubMed] [Google Scholar]
- 33. Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992-3995. [DOI] [PubMed] [Google Scholar]
- 34. Hallin J, Engstrom LD, Hargis L, et al. The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao X, Sadana R, Dessauer CW, et al. Conditional stimulation of type V and VI adenylyl cyclases by G protein betagamma subunits. J Biol Chem. 2007;282:294-302. [DOI] [PubMed] [Google Scholar]
- 36. Brand CS, Sadana R, Malik S, et al. Adenylyl cyclase 5 regulation by Gβγ involves isoform-specific use of multiple interaction sites. Mol Pharmacol. 2015;88:758-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishikawa G, Sekine S, Ogawa R, et al. Frequent GNAS mutations in low-grade appendiceal mucinous neoplasms. Br J Cancer. 2013;108:951-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3:92ra-9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pietrantonio F, Berenato R, Maggi C, et al. GNAS mutations as prognostic biomarker in patients with relapsed peritoneal pseudomyxoma receiving metronomic capecitabine and bevacizumab: a clinical and translational study. J Transl Med. 2016;14:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.