Conspectus

This Account summarizes the progress in protein–calixarene complexation, tracing the developments from binary recognition to the glue activity of calixarenes and beyond to macrocycle-mediated frameworks. During the past 10 years, we have been tackling the question of protein–calixarene complexation in several ways, mainly by cocrystallization and X-ray structure determination as well as by solution state methods, NMR spectroscopy, isothermal titration calorimetry (ITC), and light scattering. Much of this work benefitted from collaboration, highlighted here. Our first breakthrough was the cocrystallization of cationic cytochrome c with sulfonato-calix[4]arene leading to a crystal structure defining three binding sites. Together with NMR studies, a dynamic complexation was deduced in which the calixarene explores the protein surface. Other cationic proteins were similarly amenable to cocrystallization with sulfonato-calix[4]arene, confirming calixarene–arginine/lysine encapsulation and consequent protein assembly. Calixarenes bearing anionic substituents such as sulfonate or phosphonate, but not carboxylate, have proven useful.
Studies with larger calix[n]arenes (n = 6, 8) demonstrated the bigger better binder phenomenon with increased affinities and more interesting assemblies, including solution-state oligomerization and porous frameworks. While the calix[4]arene cavity accommodates a single cationic side chain, the larger macrocycles adopt different conformations, molding to the protein surface and accommodating several residues (hydrophobic, polar, and/or charged) in small cavities. In addition to accommodating protein features, the calixarene can bind exogenous components such as polyethylene glycol (PEG), metal ions, buffer, and additives. Ternary cocrystallization of cytochrome c, sulfonato-calix[8]arene, and spermine resulted in altered framework fabrication due to calixarene encapsulation of the tetraamine. Besides host–guest chemistry with exogenous components, the calixarene can also self-assemble, with numerous instances of macrocycle dimers.
Calixarene complexation enables protein encapsulation, not merely side chain encapsulation. Cocrystal structures of sulfonato-calix[8]arene with cytochrome c or Ralstonia solanacearum lectin (RSL) provide evidence of encapsulation, with multiple calixarenes masking the same protein. NMR studies of cytochrome c and sulfonato-calix[8]arene are also consistent with multisite binding. In the case of RSL, a C3 symmetric trimer, up to six calixarenes bind the protein yielding a cubic framework mediated by calixarene dimers. Biomolecular calixarene complexation has evolved from molecular recognition to framework construction. This latter development contributes to the challenge in design and preparation of porous molecular materials. Cytochrome c and sulfonato-calix[8]arene form frameworks with >60% solvent in which the degree of porosity depends on the protein:calixarene ratio and the crystallization conditions. Recent developments with RSL led to three frameworks with varying porosity depending on the crystallization conditions, particularly the pH. NMR studies indicate a pH-triggered assembly in which two acidic residues appear to play key roles. The field of supramolecular protein chemistry is growing, and this Account aims to encourage new developments at the interface between biomolecular and synthetic/supramolecular chemistry.
Key References
Rennie M. L.; Fox G. C.; Pérez J.; Crowley P. B.. Auto-regulated Protein Assembly on a Supramolecular Scaffold. Angew. Chem., Int. Ed. 2018, 57, 13764–13769.1Switch on/switch off oligomerization as a function of protein–calixarene ratio. The first cocrystal structures of a protein and sulfonato-calix[8]arene. Three types of porous frameworks occur, one of which is assembled exclusively by calixarene-mediated interfaces.
Alex J. M.; Rennie M. L.; Engilberge S.; Lehoczki G.; Dorottya H.; Fizil Á.; Batta G.; Crowley P. B.. Calixarene-mediated Assembly of a Small Antifungal Protein. IUCrJ 2019, 6, 238–247.2Complexation of a ∼ 6 kDa cationic protein with the sulfonato-calix[n]arene (n = 4,6,8) series studied by solution and solid state methods. All three calixarenes bind the same solvent exposed site comprising lysine and hydrophobic side chains. Evidence of dimerization only with n = 8.
Engilberge S.; Rennie M. L.; Dumont E.; Crowley P. B.. Tuning Protein Frameworks via Auxiliary Supramolecular Interactions. ACS Nano 2019, 13, 10343–10350.3Ternary mixtures of protein, sulfonato-calix[8]arene and spermine result in framework duplication. The tetracationic additive is encapsulated by the calixarene and enables new calixarene-mediated junctions.
Ramberg K. O.; Engilberge S.; Skorek T.; Crowley P. B.. Facile Fabrication of Protein-Macrocycle Frameworks. J. Am. Chem. Soc. 2021, 143, 1896–1907.4The first cocrystal structures of a calixarene and a neutral protein. Two types of porous frameworks are mediated exclusively by sulfonato-calix[8]arene in a pH triggered process.
1. Background
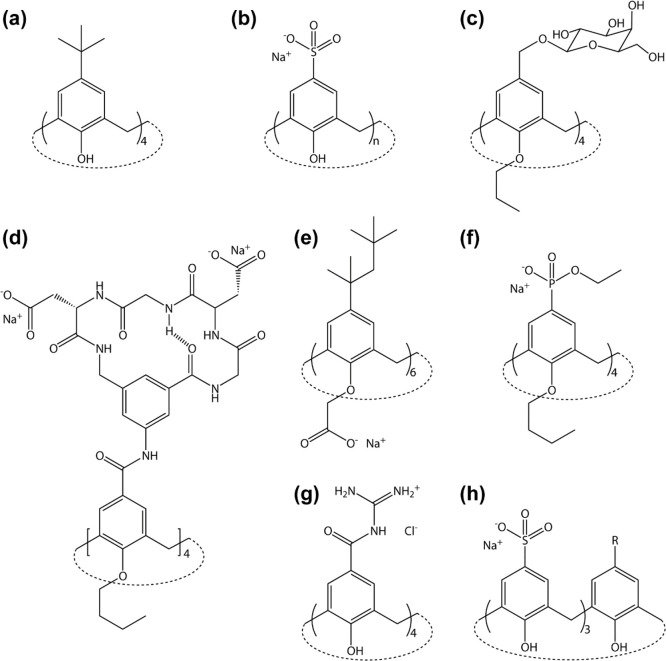
Calix[n]arenes (Figures 1 and 2), cyclic host molecules available in an array of sizes with variable conformations and cavity volumes, were investigated originally as synthetic enzyme mimics.5 Molecular recognition, required for substrate binding and possible catalysis, was central to this research. In 1984, Shinkai and co-workers produced a sulfonic acid derivative (Figure 1b) yielding a highly water-soluble calixarene, and demonstrated presently that calixarene cavities were capable of binding guests in water.6−8 The mid-1990s onward saw the development of bioinspired calixarenes bearing glyco or peptido features (Figure 1c,d). Here, calix[4]arene served as a rigid scaffold supporting biologic units.9−12 These sophisticated receptors were designed for biomolecule recognition, including protein complexation. Hamilton and co-workers reported the first example in which a calixarene bearing four peptide loops bound selectively the lysine-rich cytochrome c.10 Subsequently, Aoyama and co-workers described calixarene-based saccharide clusters that agglutinated lectins.11
Figure 1.
Schematic structures of (a) Gutsche’s canonical calix[4]arene with t-butyl groups on the upper rim5 and (b–h) water-soluble, protein-binding calixarenes. (b) Shinkai’s sulfonato-calix[n]arenes.6−8 (c) Representative glyco-calixarene, the tetra-galactoside from Parma.9 (d) Hamilton’s peptido-calixarene, containing glycine and aspartate.10 (e) Goto’s amphipathic calix[6]arene with lower rim carboxylates.20 (f) Schrader’s phosphonate-containing calix[4]arene.21 (g) de Mendoza’s guanidinio-calix[4]arene.24 (h) Hof’s asymmetric trisulfonato-calix[4]arene.33
Figure 2.
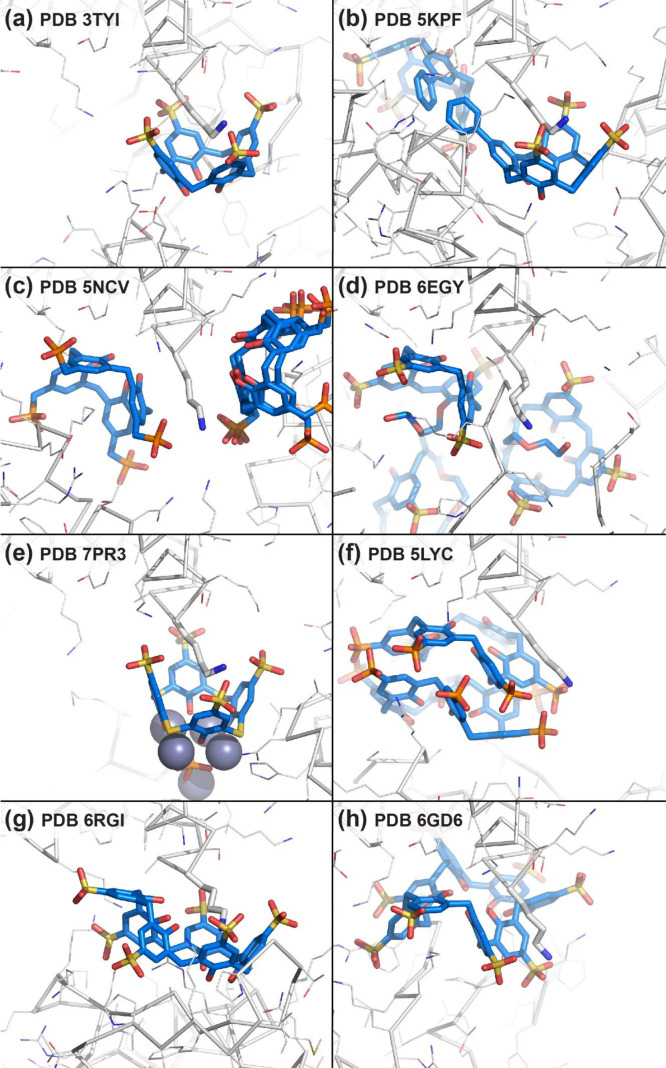
Binding site details from cocrystal structures of cytochrome c with (a) sulfonato-calix[4]arene, (b) phenyl-sulfonato-calix[4]arene, (c) methylphosphonato-calix[4]arene, (d) mono-PEGylated sulfonato-calix[4]arene, (e) sulfonato-thiacalix[4]arene with zinc, (f) phosphonato-calix[6]arene, (g) sulfonato-calix[6]arene, and (h) sulfonato-calix[8]arene. Lys4, shown as sticks, is encapsulated or bound exo depending on the calixarene.
Fifteen years after Shinkai’s seminal work, there appeared the first reports on sulfonato-calix[4]arene complexation of amino acids including arginine and lysine or short peptides thereof.13−18 These studies affirmed the potential of simple calixarenes as protein receptors. Small molecule X-ray crystal structures were particularly instructive, revealing partial encapsulation of the amino acid side chain in the calix[4]arene cavity.14,16,17 During the 2000s, multisite binding between sulfonato-calix[n]arenes and bovine serum albumin (BSA) was reported, leading to protein precipitation in salt-free solutions.19 Multisite calixarene–protein binding was suggested also by elegant experiments from Goto and co-workers who transferred cytochrome c from water into chloroform by complexation with an amphipathic calix[6]arene (Figure 1e).20 Schrader and co-workers used amphipathic calix[4]arenes for protein sensing.21,22 Lower rim butoxy groups enabled membrane-embedment while the upper rims were functionalized with amino or phosphonate groups (Figure 1f) to complement the charged properties of the target protein. In 2008, protein–calixarene complexation was given a twist. Rather than avail of the macrocycle cavity for side chain encapsulation, de Mendoza and co-workers plugged the cavities of the p53 tetramerization domain with a calix[4]arene.23 Guanidinium functionalities on the calixarene rescued an arginine to histidine mutation and stabilized the protein. The plug concept was taken further with complete matching between the C4 symmetric potassium channel and guanidinio-calix[4]arenes (e.g., Figure 1g).24 Meanwhile there were further developments with multivalent glyco-calixarenes25 and calixarene-based protein inhibitors.26−29
In 2010, Hof and co-workers revisited sulfonato-calix[4]arene complexation of the cationic amino acids.14,15,17,30,31 The focus was on arginine/lysine methylation. Modification of the lysine-ζNH3+ primary amine to the mono-, di-, and trimethylated forms increased the binding affinity for sulfonato-calix[4]arene (e.g., ∼70-fold tighter for trimethyllysine).27 A tetrapeptide snippet from the disordered N-terminus of histone H3 had ∼18-fold increased affinity for sulfonato-calix[4]arene when the lysine side chain was trimethylated. In a subsequent study, longer H3 peptides had micromolar affinities for the calixarene, acting akin to the aromatic cage motif of histone reader proteins.32 Improved affinity and selectivity toward trimethyllysine were obtained using trisulfonated calix[4]arenes (Figure 1h).33
With this overview of the key stepping stones, we turn now to our investigation of calixarene complexation, in particular X-ray cocrystal structures with model proteins (Tables 1 and 2). Numerous reviews are available for further insights to the past developments and the current biological applications of calixarenes.34−36
Table 1. Model Protein Characteristics.
| protein, organism | oligomer | fold | MW (kDa) | #Lys | #Arg | pIcalc |
|---|---|---|---|---|---|---|
| cytochrome c, S. cerevisiae | monomer | all alpha heme core | 12.8 | 16 | 3 | 9.5 |
| Lysozyme, G. gallus | monomer | alpha and beta two domain | 14.3 | 6 | 11 | 9.3 |
| PAF, P. chrysogenum | monomer | small protein disulfide-rich | 6.2 | 13 | 0 | 8.9 |
| PAFB, P. chrysogenum | monomer | small protein disulfide-rich | 6.5 | 8 | 2 | 8.8 |
| RSL, R. solanacearum | trimer | beta-propeller 6-blades | 29.1 | 9 | 9 | 6.8 |
Table 2. Protein–Calixarene Crystal Structures in the Protein Data Bank.
| PDB ID | space group | protein | ligand |
|---|---|---|---|
| 3TYI | P212121 | cyt c | sulfonato-calix[4]arene |
| 5LFT | P22121 | cyt c | bromo-sulfonato-calix[4]arene |
| 5KPF | C2221 | cyt c | phenyl-sulfonato-calix[4]arene |
| 5NCV | P1211 | cyt c | methylphosphonato-calix[4]arene |
| 6EGY | I4132 | cyt c | sulfonato-calix[4]arene monoPEG |
| 6EGZ | I4132 | cyt c | sulfonato-calix[4]arene diPEG |
| 6SUV | P43 | cyt ca | octa-anionic-calix[4]arene |
| 6SUY | P3221 | cyt c | octa-anionic-calix[4]arene |
| 7PR3 | P212121 | cyt c | sulfonato-thiacalix[4]arene + Zn |
| 5LYC | P43212 | cyt c | phosphonato-calix[6]arene |
| 6RGI | P3221 | cyt c | sulfonato-calix[6]arene |
| 6GD6 | H3 | cyt c | sulfonato-calix[8]arene |
| 6GD8 | P31 | cyt c | sulfonato-calix[8]arene |
| 6GD9 | P43212 | cyt c | sulfonato-calix[8]arene |
| 6RSK | P43212 | cyt c | sulfonato-calix[8]arene + spermine |
| 6Y0J | P61 | cyt c | calix[6]arene and calix[8]arene |
| 7BBT | C121 | cyt c | extended arm calix[8]arene |
| 4PRQ | P1211 | lysozyme | sulfonato-calix[4]arene |
| 4N0J | P1211 | lysozymeb | sulfonato-calix[4]arene |
| 6HA4 | P1211 | PAF | sulfonato-calix[4]arene |
| 6HAH | P1211 | PAF | sulfonato-calix[6]arene |
| 6HAJ | P61 | PAF | sulfonato-calix[8]arene |
| 7BAD | P31 | PAFB | sulfonato-calix[8]arene |
| 7PR5 | P212121 | RSL | sulfonato-thiacalix[4]arene + Zn |
| 6Z5X | P213 | RSL | sulfonato-calix[8]arene |
| 6Z5G | I23 | RSL | sulfonato-calix[8]arene |
| 6Z5Q | P3 | RSL | sulfonato-calix[8]arene |
| 6Z5P | P3 | RSL-R8 | sulfonato-calix[8]arene |
Equus caballus cytochrome c.
Dimethylated protein with R–NH3+ converted to R–NH(CH3)2+.
2. First Steps with Sulfonato-calix[4]arene
2.1. The Complex of Cytochrome c and Sulfonato-calix[4]arene
One strategy for protein surface recognition involves synthetic receptor molecules with a hydrophobic core enabling a water-occluded interface, and a polar/charged periphery complementing the charged features of the protein.10,12,21,37−39 This concept was illustrated beautifully by Aya and Hamilton (Figure 1 in ref (37)) who reported anionic porphyrins with nanomolar affinity for cytochrome c. Other proteins with different surface attributes could be targeted using porphyrins bearing the appropriately charged functionality.37,38 Following this line of research we obtained NMR data suggesting nonspecific binding between two anionic porphyrins and Saccharomyces cerevisiae cytochrome c.39 Attempts to cocrystallize these complexes were fruitless. Replacing the planar porphyrin with the bowl-shaped calix[4]arene proved to be a game-changer.
In Autumn 2010, we began cocrystallization trials of cytochrome c and sulfonato-calix[4]arene (745 Da), the latter provided by colleague Nicholas Power. Our idea was to maximize the protein–calixarene attraction by maintaining a low ionic strength. Therefore, PEG 8000 was used as a precipitant in the absence of buffer or salt. That year, the International Conference on Crystallization of Biological Macromolecules (ICCBM13) was held in Dublin. On returning from the conference, PhD student Róise McGovern emerged excitedly from the laboratory. Her first trial had yielded crystals! The X-ray diffraction images, obtained in collaboration with Amir Khan, contained smeared and overlapping spots, ugly but promising. Optimization involved adjusting the salt composition and preparing homogeneous protein–calixarene mixtures. Eventually, high-quality diffraction data were obtained at the European Synchrotron Radiation Facility (Grenoble). Meanwhile, we had collected multiple NMR data sets. HSQC-monitored titrations of 15N-labeled protein indicated a lysine-rich binding patch that accommodated at least two calixarenes with millimolar affinities.
The crystal structure of the cytochrome c–sulfonato-calix[4]arene complex was informative for three reasons.40 (1) The structure proved unambiguously that calix[4]arene was capable of protein complexation by encapsulation (endo binding) of one lysine side chain (Figure 2a). Of the three crystallographic sites, Lys4 and Lys89 were consistent with the NMR data while Lys22 was not, suggesting that it arose via crystal packing. (2) The occurrence of three binding sites confirmed the concept of multisite protein–calixarene complexation.19,20 We suggested that the calixarene could hop between lysine side chains and camouflage the protein surface. (3) In the crystal, each calixarene occurred as a junction between two or more proteins. Exo interactions with lysines and other side chains resulted in this glue activity, altering the protein solubility in favor of assembly/crystallization. Apparently, calix[4]arene complexation of lysine, with interfaces of ∼200 Å2, is an example of surface-entropy reduction facilitating protein crystallization.41 In addition to clamping down a lysine, the calixarene converts a heterogeneous protein surface to a C4 symmetric cap. Prior to publication, I presented this work at the 2011 Bürgenstock Conference, where Ivan Huc and Tom Fyles offered great encouragement.
Unknown to us at the time, Falson, Coleman, and co-workers had earlier reported an asymmetric carboxylato-calixarene bearing a lipid for the extraction and purification of membrane proteins.42 This surfactant calixarene was cocrystallized with a Bacillus ABC transporter, but the diffraction data were insufficient to detect the macrocycle. Subsequently, a calixarene-containing crystallization kit was commercialized by CALIXAR (section 3.2).
Cytochrome c–sulfonato-calix[4]arene crystals are robust, an attribute which makes them attractive for applications. Through an EU Cost Action, I met Fred Lisdat who was constructing multilayer electrodes of cytochrome c and polyaniline sulfonate.43 The possibility of replacing polyaniline sulfonate with sulfonato-calix[4]arene was immanent. In collaboration, we grew cocrystals of cytochrome c and sulfonato-calix[4]arene on modified gold chip electrodes and obtained direct electrochemical characterization by cyclic voltammetry.44
2.2. Cocrystals of Lysozyme and Sulfonato-calix[4]arene
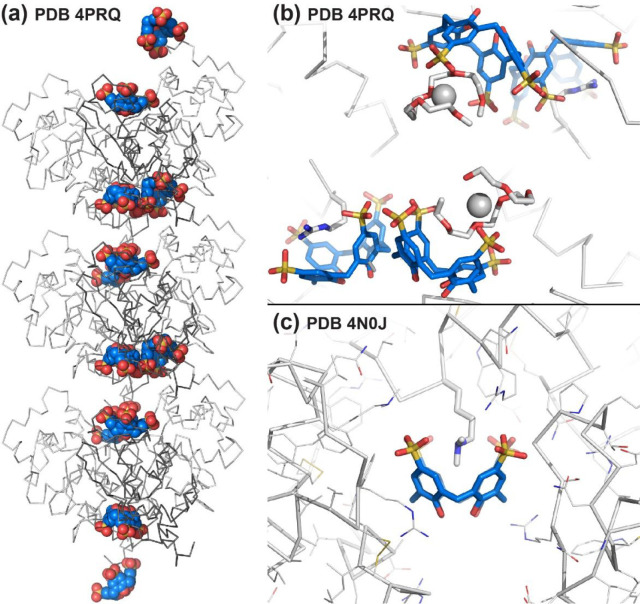
The easily crystallizable hen egg white lysozyme was an obvious target for sulfonato-calix[n]arene complexation. Similar to cytochrome c, lysozyme has an isoelectric point (pI) of ∼9 but it is arginine-rich rather than lysine-rich (Table 1). The combination of lysozyme and sulfonato-calix[4]arene in water resulted in instantaneous precipitation.45 Similar crystallization conditions to those used for cytochrome c(40) formed heavy precipitates that eventually yielded small cubic crystals. X-ray diffraction was performed at SOLEIL synchrotron (Gif-sur-Yvette, France) in collaboration with Andrew McCarthy. The lysozyme–sulfonato-calix[4]arene cocrystal structure comprised a D2-symmetric tetramer arranged in filaments (Figure 3).45 The lysozyme tetramer had a central channel (∼10 Å diameter) plugged at either end by a pair of calixarenes with their cavities projected outward, reminiscent of earlier work on calix[4]arenes with tetrameric channels.23,24 Interestingly, N-terminal Lys1 of lysozyme was bound exo to the calixarene dimer. One calixarene encapsulated the side chain of Arg128 from a neighboring molecule. Located in the C-terminus, Arg128 is the most sterically accessible of the 11 arginines in lysozyme. The other calixarene complexed a magnesium ion and a fragment of PEG, behaving like a crown ether (Figure 3b). This entity was supported by data from Raston and co-workers who had described structures of sulfonato-calix[6]arene, 18-crown-6 and lanthanides.46
Figure 3.
(a) Filament of lysozyme tetramers (three shown) mediated by dimers of sulfonato-calix[4]arene and (b) detail of the calixarene dimer with encapsulation of Arg128 and a complex of Mg2+ and PEG. (c) Cocrystal structure of dimethylated lysozyme and sulfonato-calix[4]arene showing encapsulation of Lys116*.
The same year that we reported calix[4]arene-mediated assembly of lysozyme, Yang and co-workers reported that sulfonato-calix[4]arene inhibits pentamer formation by capsid protein L1 (pI ∼ 9) from human papillomavirus.47 In 2016, supramolecular assembly of sulfonato-calix[4]arene and cationic protamine was described by Liu and co-workers,48 while Mohanty and co-workers used sulfonato-calix[4]arene to inhibit insulin amyloidogenesis.49
2.3. Lysine Methylation and Sulfonato-calix[4]arene
Considering the enhanced affinity for dimethyllysine over lysine,31 we cocrystallized sulfonato-calix[4]arene with dimethylated lysozyme (lysozyme*). The cocrystal structure revealed calixarene complexation of Lys116*, the most sterically accessible such group (Figure 3c).50 The binding mode, with a pronounced cation-pi contribution, contrasted to that of unmodified lysine. The selectivity for Lys116* was supported by an NMR study performed by Fraser Hof and co-workers. In addition to Lys116* complexation, the crystal structure revealed binding to Arg14. This result emphasized that crystal packing can produce binding sites that do not occur in solution. For example, two lysozyme molecules interacted exo to the calixarene at Arg14, with salt bridges formed between the lower rim phenols and Arg21.
Meanwhile, calixarene-functionalized agarose resin was developed for peptide purification based on lysine methylation.51 Zhong and co-workers devised a related strategy by host-assisted capillary electrophoresis.52 In addition to sulfonato-calix[n]arenes, related macrocycles were developed as Lys(Me)n receptors.53,54
3. Cytochrome c Complexation with Other Calix[4]arenes
3.1. Asymmetric Trisulfonato-calixarenes
Working with trisulfonato-calixarenes from the Hof laboratory,33 MSc student Aishling Doohan tested if host asymmetry altered the binding specificity. The bromo derivative (Figure 1h, R = Br, 744 Da) cocrystallized readily with cytochrome c under conditions similar to those reported previously.40,55 The crystal structure yielded evidence for selective binding, as only Lys86 was encapsulated. The bromo substituent made van der Waals contact with the Lys86 backbone carbonyl, raising the possibility of halogen bonding. On the other hand, there was evidence also of nonspecific binding.55 A calixarene dimer,56 with encapsulated bromo substituents, occurred at 70% occupancy wedged between two protein chains. One protein used Lys5 and Lys89, while the other protein used Lys4 and Lys100 to bind this calixarene dimer via cation−π bonds.
Cocrystals of the phenyl derivative (Figure 1h, R = C6H5, 741 Da) and cytochrome c were obtained by microseeding with cytochrome c–sulfonato-calix[4]arene cocrystal seeds.55 The crystal structure revealed a single site with Lys4 bound endo to the calixarene. Interestingly, the phenyl substituent made van der Waals contact with Ala3 and a calixarene bound to a symmetry mate in the crystal packing (Figure 2b). This weak calixarene dimerization at a protein–protein interface suggested a mechanism for protein–calixarene aggregation observed in buffered solutions. While the X-ray data revealed variations in specificity of the trisulfonato-calix[4]arenes, the NMR data suggested a broad binding patch, though not as extensive as for sulfonato-calix[4]arene. Thermodynamic analysis, by ITC, yielded two site binding and apparent dissociation constants of 0.02 and 0.03 mM for the bromo derivative and for sulfonato-calix[4]arene, respectively. The phenyl derivative resisted ITC analysis due to aggregation.
3.2. Anionic Calix[4]arenes from Parma
PhD student Jimi Alex tested the CALIXAR kit42 containing calix[4]arenes variously functionalized with carboxylato or phosphonato substituents at the upper or lower rims. No cocrystals were obtained with the carboxylato derivatives, a result borne out by other carboxylato-macrocycles57,58 that have resisted cocrystallization. Only the upper rim methylphosphonato-calix[4]arene yielded cocrystals with cytochrome c. This compound, synthesized originally by Ungaro and co-workers added impetus to our ongoing collaboration with Alessandro Casnati.81 Cocrystallization occurred at 2 equiv of this ligand, compared to the 10 equiv required for sulfonato-calix[4]arene. A crystal structure of methylphosphonato-calix[4]arene (800 Da) in complex with cytochrome c was instructive for several reasons.59 Lys86 was selected as the binding site, similar to the complex with the bromo derivative. One of the methylphosphonato substituents rotated into the cavity affording new interactions between the encapsulated cation and the upper rim anion. A second binding site at Lys54, was likely a result of crystal packing as it was not evident in NMR experiments. Within the crystal packing, key residue Lys4, was sandwiched exo between two calixarenes (Figure 2c).
Jimi Alex investigated another calix[4]arene from Parma, with upper rim sulfonato- and lower rim carboxylato- groups. This octa-anionic calixarene (977 Da) is locked in the cone conformation by lower rim coordination of a Na+ ion. Silvano Geremia and co-workers obtained cocrystals with horse cytochrome c making for a comparison with our data on the yeast protein.60 In both cases the calixarene bound to charge rich patches and yielded porous assemblies (section 7). Curiously, in the yeast case, the calixarene did not encapsulate any side chain.
3.3. PEGylated Calix[4]arenes
Postdoc Srinu synthesized sulfonato-calix[4]arenes bearing one, two, or four PEG chains. Our goal was supramolecular PEGylation of proteins, using the calixarene to tether the PEG to the protein. As noted in 2016, a related concept was developed originally with PEGylated triazine dyes to improve enzyme solubility.61 While our work was in progress, Isaacs, Langer, Anderson, and co-workers reported a system based on PEGylated cucurbit[7]uril.62 Working with cytochrome c and the mono- or di-PEGylated calix[4]arene (1.5 and 1.9 kDa, respectively), Srinu obtained crystallographic proof of supramolecular PEGylation.63 The protein–calixarene binding sites were familiar but the PEG appendages gave rise to new features. In the mono-PEG case, the calixarene occurred in either the cone or partial cone conformation (Figure 2d). Self-encapsulation of a PEG fragment together with Mg2+ occurred also, similar to earlier observations.45
3.4. Thiacalix[4]arene
Recently, we demonstrated the capacity of sulfonato-thiacalix[4]arene (816 Da) for macrocycle- and metal-mediated protein assembly.64 PhD student Ronan Flood obtained cocrystals of two model proteins and sulfonato-thiacalix[4]arene in combination with zinc. In cocrystals with cytochrome c, the thiacalixarene supported penta-nuclear zinc clusters that acted as nodes for protein complexation (Figure 2e). Due to space limitations, we direct the reader to the paper for full details.64
4. Bigger Better Binders
4.1. Protein Dimerization with Phosphonato-calix[6]arene
Shortly after the 12th International Conference on Calixarenes, I received a package of phosphonato-calix[n]arenes from Colin Raston.65 A cocrystal structure of phosphonato-calix[6]arene (1117 Da) and cytochrome c, together with solution state studies by postdoc Martin Rennie, marked another turning point in our research.66 The crystal structure contained a calix[6]arene dimer. Each calixarene bound one protein by encapsulating Lys4, Lys100, and a small hydrophobic patch to form an ∼350 Å2 interface (Figure 2f). A porous assembly occurred with an ∼60% solvent content (compared to the 30–40% in typical protein crystals). The interface area of protein–protein contacts was approximately equal to that of protein–calixarene and calixarene–calixarene contacts, suggesting a pivotal role for the calixarene within the assembly. Importantly, the calixarene-mediated protein dimer was not merely a consequence of crystal packing. Three different solution state techniques, NMR spectroscopy, size exclusion chromatography coupled with multiangle light scattering (SECMALS), and ITC, pointed to the existence of dimers and higher order oligomers. Furthermore, the ITC data indicated a binding affinity in the low micromolar range. This improved affinity, with respect to calix[4]arene, may be attributed in part to the larger interface size.66
Curiously, sulfonato-calix[6]arene (1117 Da) did not exert the same effects on cytochrome c. In this case, crystals were obtained in the presence of imidazole, which displaced the Met80 heme ligand resulting in a partially unfolded cytochrome supported by calix[6]arene complexation (Figure 2g).67
4.2. Protein Assembly and Encapsulation with Sulfonato-calix[8]arene
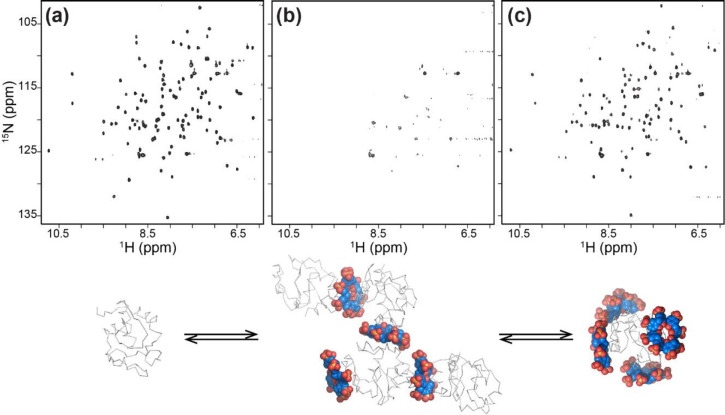
The bigger better binder theme comes to the fore with sulfonato-calix[8]arene (1489 Da). Long before a crystal structure was available, we had NMR data pointing to unusual behavior of cytochrome c and sulfonato-calix[8]arene (Figure 4). The 1H–15N HSQC spectrum of cytochrome c was obliterated by ∼1 equiv of the macrocycle.1 In itself, this result is not unusual as ligand-mediated aggregation may result in NMR-invisible high molecular weight species. However, further additions of macrocycle resulted in spectral improvement, and at ∼3 equiv the spectrum was restored albeit with multiple chemical shift perturbations. The existence of a high molecular weight species, specifically a tetramer, was evident from light scattering measurements including small-angle X-ray scattering (SAXS) data obtained in collaboration with Javier Pérez (SWING beamline, SOLEIL synchrotron). Biphasic ITC data that could not be fitted to a bidentate ligand model were also consistent with a multispecies process.68 Therefore, it appeared that the calix[8]arene could switch on and off protein assembly. Such autoregulated assembly is characteristic of cytokine–heparin interactions as well as the assembly of cationic proteins by polyphosphates.1
Figure 4.
NMR and X-ray data suggest autoregulated assembly of cytochrome c by sulfonato-calix[8]arene. 1H–15N HSQC spectra of (a) pure protein and protein plus (b) ∼1 and (c) ∼3 equiv of calixarene. Spectral obliteration may be due to the formation of high molecular weight species such as a tetramer (based on PDB 6GD8). Higher equivalents of ligand result in masking/encapsulation (based on PDB 6GD9), disassembly, and spectral recovery.
Cocrystallization trials with cytochrome c and sulfonato-calix[8]arene initially proved ineffective but revealed the counterintuitive observation that the degree of precipitation was inversely proportional to the calixarene concentration. Martin Rennie made the breakthrough, and as if seed crystals flew(69) cocrystallization with sulfonato-calix[8]arene is now straightforward. Three crystal forms of cytochrome c in complex with sulfonato-calix[8]arene lent support for a tetramer assembly (Figures 2h and 4) and revealed three essential properties: (1) promiscuity of binding, where five distinct calix[8]arene-binding patches were evident, involving interface areas up to ∼500 Å2 and varying degrees of macrocycle molding to the surface of cytochrome c; (2) protein masking/encapsulation, where each structure included two or more sulfonato-calix[8]arene binding sites with up to ∼35% surface coverage suggesting a mechanism for disassembly of high molecular weight species as deduced from the NMR experiments (protein encapsulation, Figure 4), and (3) porous framework fabrication, where all three structures were highly porous, with solvent contents > 60%. These frameworks were elaborated by the addition of spermine (section 7).3
4.3. Alternative Assembly by an “Extended Arm” Calix[8]arene
Aspiring to affect even greater protein surface coverage (encapsulation), we worked with an “extended arm” calix[8]arene from Colin Raston’s laboratory. PhD student Niamh Mockler cocrystallized this 2.2 kDa macrocycle bearing benzyl extensions with cytochrome c. Although improved encapsulation beyond the capacity of sulfonato-calix[8]arene was hypothesized, we obtained a novel binding mode by a supramolecular entity.70 The crystal included a trimeric stack of the extended arm calixarene. Remarkably, the benzyl extensions projected from this stack forming four grooves, each of which accommodated the N-terminal α helix of cytochrome c. Apparently, bigger is not always better. The structure was further surprising in that the calixarene stack was threaded by a PEG fragment yielding a pseudorotaxane.
4.4. Mixed Calixarenes for Protein Crystallization
To add a final layer of complexity, consider the composite structure of cytochrome c with phosphonato-calix[6]arene and sulfonato-calix[8]arene. Ternary cocrystals, obtained at an approximately 1:1:1 ratio of the components, involved two types of calixarene-mediated interfaces.71 Phosphonato-calix[6]arene formed a dimer and bound two molecules of cytochrome c via the Lys4 and Lys100 pair. Interestingly, the direction of rotation within this dimeric assembly was opposite to that in the original structure.66 Sulfonato-calix[8]arene also bound a known site (Lys72, Lys73, and Lys86)1 and again mediated a cytochrome c dimer. The crystal packing was a highly porous (70% solvent content), dendrite-like assembly of supramolecular copolymers with alternating phosphonato-calix[6]arene or sulfonato-calix[8]arene junctions as well as one protein–protein interface.71
5. Antifungal Proteins
At the 2016 Chianti Workshop, Gyula Batta presented his NMR studies of Penicillium antifungal protein (PAF).72 This small and highly cationic protein immediately caught our attention as a candidate for calixarene complexation. Jimi Alex took on the challenge and her work proved seminal (Figure 5). Simple conditions, comprising only PEG and a buffer,40,59 resulted in cocrystallization of PAF with the sulfonato-calix[n]arene series, while PAF alone did not yield diffraction-quality crystals.2 The cocrystal structures of PAF with sulfonato-calix[4]arene or sulfonato-calix[6]arene were solved in the same space group (P1211) with similar unit cell parameters and one calixarene per polypeptide. In contrast, the cocrystal with sulfonato-calix[8]arene was hexagonal with one calixarene per two proteins. The same highly exposed feature (Pro29, Lys30, and Phe31) bound the calix[n]arene in each structure. Adopting the double cone conformation, sulfonato-calix[8]arene interacted exo to the Pro29/Lys30/Phe31 patch on two PAF molecules. In the crystal packing, each calix[n]arene bound to at least four proteins, substantiating the glue activity (Figure 5). A fragment of PEG completed the protein–calixarene binding sites in the calix[6]arene and calix[8]arene structures. The latter was threaded with a heptaethylene glycol unit forming extensive interactions with the calixarene as well as crown-ether-like complexes with Lys9 residues. The PAF–sulfonato-calix[n]arene binding sites identified by X-ray crystallography were confirmed in solution by NMR spectroscopy. Micromolar binding affinities were determined by ITC and the tightest binding sulfonato-calix[8]arene acted as a bidentate ligand, consistent with the X-ray model of ligand-mediated dimerization.2
Figure 5.
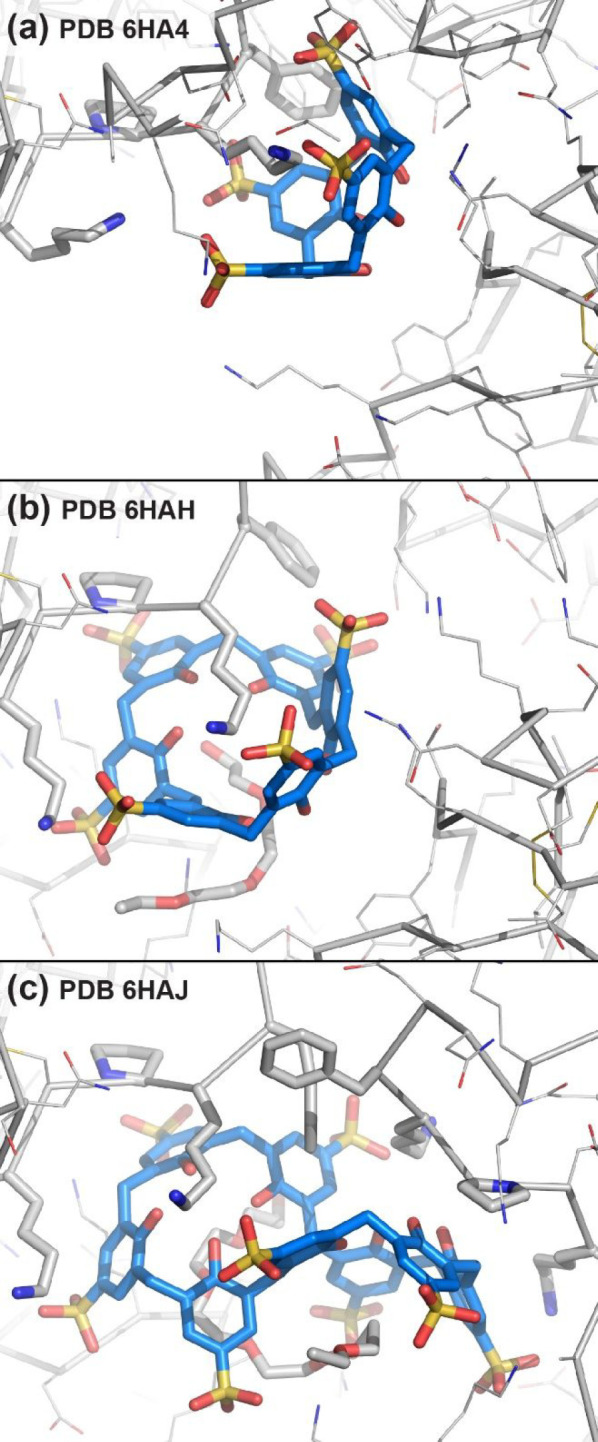
Binding sites in cocrystal structures of PAF with (a) sulfonato-calix[4]arene, (b) sulfonato-calix[6]arene, and (c) sulfonato-calix[8]arene. Lys27, Pro29, Lys30, and Phe31 shown as sticks. Note PEG fragments in (b) and (c).
In a follow-up study, we collaborated with Florentine Marx and co-workers to study the histidine-rich PAFB with ∼35% sequence identity to PAF. Postdoc Francesca Guagnini obtained diffraction-quality crystals of PAFB with sulfonato-calix[8]arene in the presence of zinc. The cocrystal structure (space group P31) was highly porous and included a trinuclear Zn cluster, assembled by three proteins each contributing two histidines.73 The adjacent calix[8]arene binding site, masked ∼400 Å2 of the protein surface.
6. A Symmetric, Neutral Target—Toward Encapsulation
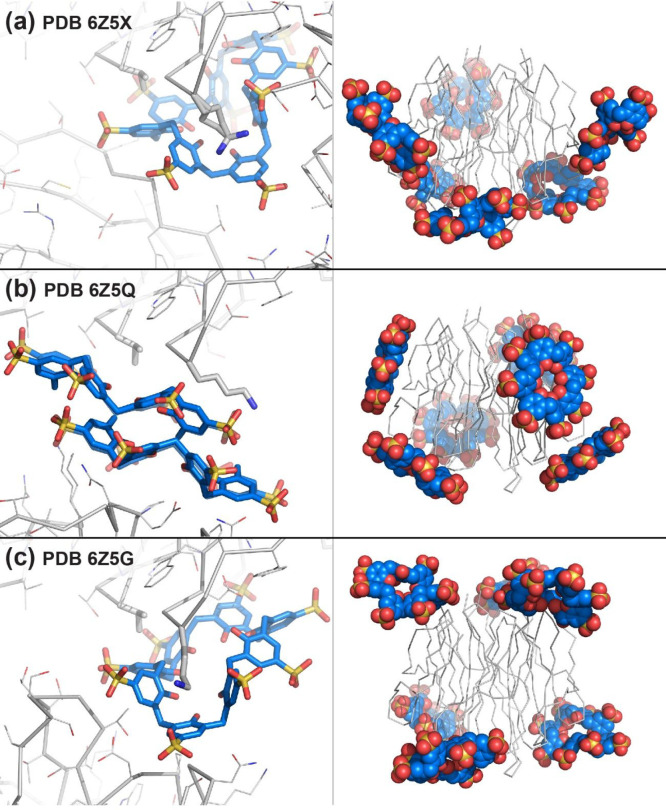
Thus far, we have discussed cationic targets.1,2,45,50 Until recently, there was limited evidence for calixarene complexation of acidic proteins.19,21,49 NMR studies revealed sulfonato-calix[4]arene complexation of arginine and/or lysine side chains in human ubiquitin74 and the WW domain of peptidyl-prolyl isomerase Pin1, each with a pI < 7.75 What about a neutral protein? RSL is a highly stable C3 trimer with a 6-bladed β-propeller topology and a pI close to 7. The absence of a cationic patch and the high symmetry made RSL an interesting target for calixarene complexation. Postdoc Sylvain Engilberge made the first progress obtaining cocrystals at >30 equiv of sulfonato-calix[8]arene in high concentrations of ammonium sulfate across a wide pH range.4 These results suggested that charge–charge interactions were minimal, and assembly was driven by the hydrophobic effect. Cocrystals were obtained also at ≤1 M ammonium sulfate with ≤10 equiv of calixarene, but only at pH ≤ 4 where the protein is cationic. PhD student Kiefer Ramberg discovered a third cocrystal form in a low pH NMR sample after overnight incubation in the fridge. Each of the three crystal forms involved calixarene complexation of adjacent residues Val13 and Lys34 (Figure 6). The calixarene adopted conformations ranging from highly puckered to fully extended (pleated loop). The puckered conformation was molded neatly while the extended conformation was bound tangentially to the protein surface. Furthermore, the extended conformation occurred within a calixarene dimer (Figure 6b). Interestingly, the crystals obtained at high ammonium sulfate were densely packed, while the low pH crystals were porous (section 7).4 NMR studies revealed negligible calixarene binding at pH 6 but significant binding at pH 4. Two of the six acidic residues in RSL had elevated pKa values in the presence of sulfonato-calix[8]arene suggesting pH-triggered assembly. The crystals provided supporting evidence, as the two acidic residues participated in calixarene complexation. Remarkably, one of the crystal forms accommodated different mutants, including an arginine-enriched and highly cationic variant (RSL-R8) that did not require low pH for cocrystallization.
Figure 6.
Binding sites and surface masking/encapsulation in three cocrystal structures of RSL with sulfonato-calix[8]arene, space groups (a) P213, (b) I23 and (c) P3. The recurring epitope, Val13 and Lys34, shown as sticks. Note the disorder Lys34 in (a).
With C3 symmetry, RSL presents three equivalent surfaces with interesting consequences for protein encapsulation by calix[8]arene. Each structure comprised the trimer masked to varying degrees with at least 6 calixarenes (Figure 6). Recently, large quaterphen[n]arenes, synthetically more challenging than calixarenes, have been used to encapsulate antimicrobial peptides.76
7. Protein–Calixarene Frameworks
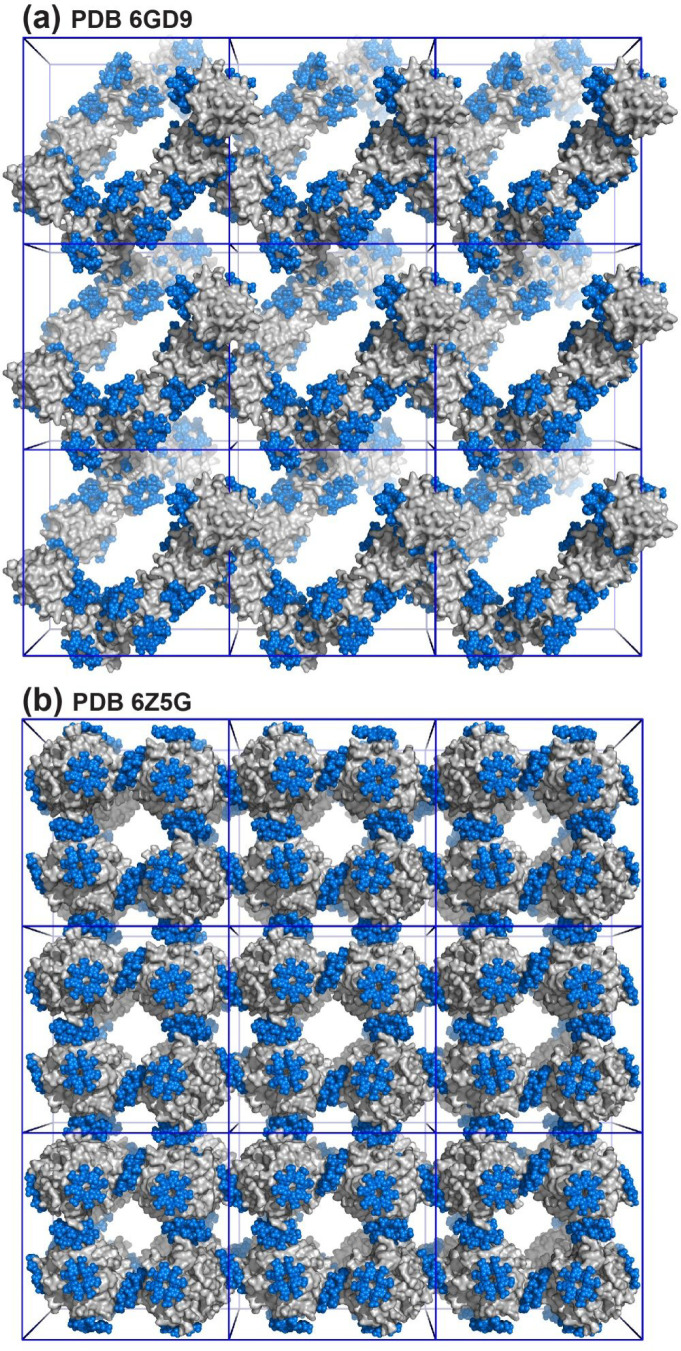
With the developments from supramolecular to materials chemistry, the focus of molecular recognition has shifted from discrete complexes to frameworks, especially porous crystals. A similar shift is evident here. Although originally investigated as enzyme mimics and later as receptors for protein surfaces, it transpires that calixarenes are versatile scaffolds for constructing protein frameworks.1,3,4 Porous frameworks with solvent contents exceeding 50% were obtained by cocrystallization of cytochrome c with phosphonato-calix[6]arene,61 sulfonato-calix[6]arene,62 sulfonato-calix[8]arene,1,3 or the octa-anionic calix[4]arene.55 RSL and sulfonato-calix[8]arene also yielded highly porous structures.4 Here, I compare two such structures to illustrate framework fabrication by calixarenes (Figure 7).
Figure 7.
Porous frameworks of (a) cytochrome c (P43212) and (b) RSL (I23) with unit cell axes a = b ≈ 10 nm. All interfaces are mediated by sulfonato-calix[8]arene.
In the presence of ≥3 equiv of sulfonato-calix[8]arene, cytochrome c forms an exceptionally porous framework with 85% solvent content.1,3 This diamondoid structure is held together exclusively by protein–calixarene–protein junctions and contains 3–10 nm wide cavities, suggesting the possibility of accommodating additional protein. The framework has no protein–protein contacts as each protein is substantially masked by the calixarene. Interestingly, one calixarene is highly solvent exposed, pointing into the crystal void. We wondered if this site could accommodate additional guests. Sylvain Engilberge tested ternary cocrystallization of protein, calixarene, and tetracationic spermine (structurally analogous to the dilysine motif), yielding two new structures with framework duplication and decreased porosity.3 RSL and sulfonato-calix[8]arene, at pH ≤ 4, also yielded two porous crystal forms devoid of protein–protein interfaces. A particularly aesthetic structure in the high symmetry (and rare), cubic space group I23 had 66% solvent content.4 Crystals in space group P3 were similarly porous and were obtained under trivial conditions in the absence of precipitant. This crystal form accommodated the cationic arginine-enriched RSL-R8, with only minor changes to the assembly despite an altered calixarene binding site. It remains to be seen what functionality can be achieved in these crystals, but efforts in other laboratories are promising.77
Porous protein assemblies provide a basis for new types of biocompatible and sustainable materials with broad applications. Different preparation strategies are in development,77 including designed protein assembly78 and the repurposing of natural protein cages.79 Macrocycle-mediated protein assembly confers advantages such as ease of fabrication and enhanced functionality via host–guest chemistry.3,4
8. Concluding Remarks
Almost 50 years ago, Cram and Cram wrote “the host molecule is the larger, and the guest molecule is the smaller of the two.”80 Host–guest chemistry of sulfonato-calix[n]arenes began with the trimethylanilinium cation6−8 and progressed to amino acids13−18,30,31 and later to proteins.1−4,19,40 Individual side chain encapsulation has led to protein encapsulation by multiple calixarenes, in which the “guest” is >10-fold larger than the “host.” What began as investigations of protein surface recognition (binary interactions) evolved to protein assembly (oligomerization) and crystallization (extended frameworks).1,10,40 Simple conditions containing PEG and a buffer/salt are sufficient to obtain cocrystals of cationic proteins with variously functionalized anionic calix[4]arenes.2,40,45,59 To date, success has been achieved with calixarenes bearing sulfonate or phosphonate substituents, but not carboxylates. Cationic calixarenes, although established protein binders,23,24 have yet to be cocrystallized with a protein. For example, guanidinio-containing calix[4]arenes have resisted cocrystallization, despite valiant efforts by PhD student Marta Giuliani. It remains to be seen what other functional groups may prove favorable. For example, the benzyl-sulfonate “extended arm” calix[8]arene formed a trimeric macrocycle stack leading to a new protein binding mode.70 And framework fabrication has taken a new direction with thiacalixarene, which enables dual macrocycle-/metal-mediated assembly and great scope for generating metal clusters in combination with proteins.64 The relatively simple and commercially available sulfonato-calix[8]arene has proven to be a versatile scaffold, mediating oligomerization, such as tetramerization of cytochrome c,1 enabling framework fabrication, such as the highly porous cubic assembly of RSL,4 or encapsulating individual proteins. Much remains to be discovered in protein assembly and encapsulation by macrocycles. Increased collaboration between biochemists and supramolecular chemists will lead to valuable advances, especially in the area of biomaterials.
Acknowledgments
Thanks to all past and present co-workers and collaborators. NUI, Science Foundation Ireland, Irish Research Council, Royal Society of Chemistry, EU Cost Action and SOLEIL Synchrotron are acknowledged for their support.
Biography
Peter B. Crowley graduated with a BSc in chemistry from University College Dublin and a PhD in biophysics from Leiden University. Since 2017, he is professor of protein chemistry at Galway.
The author declares no competing financial interest.
References
- Rennie M. L.; Fox G. C.; Pérez J.; Crowley P. B. Auto-regulated Protein Assembly on a Supramolecular Scaffold. Angew. Chem., Int. Ed. 2018, 57, 13764–13769. 10.1002/anie.201807490. [DOI] [PubMed] [Google Scholar]
- Alex J. M.; Rennie M. L.; Engilberge S.; Lehoczki G.; Dorottya H.; Fizil Á.; Batta G.; Crowley P. B. Calixarene-mediated Assembly of a Small Antifungal Protein. IUCrJ. 2019, 6, 238–247. 10.1107/S2052252519000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engilberge S.; Rennie M. L.; Dumont E.; Crowley P. B. Tuning Protein Frameworks via Auxiliary Supramolecular Interactions. ACS Nano 2019, 13, 10343–10350. 10.1021/acsnano.9b04115. [DOI] [PubMed] [Google Scholar]
- Ramberg K. O.; Engilberge S.; Skorek T.; Crowley P. B. Facile Fabrication of Protein-Macrocycle Frameworks. J. Am. Chem. Soc. 2021, 143, 1896–1907. 10.1021/jacs.0c10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsche C. D. Calixarenes. Acc. Chem. Res. 1983, 16, 161–170. 10.1021/ar00089a003. [DOI] [Google Scholar]
- Shinkai S.; Mori S.; Tsubaki T.; Sone T.; Manabe O. New Water-soluble Host Molecules Derived from Calix[6]arene. Tetrahedron Lett. 1984, 25, 5315–5318. 10.1016/S0040-4039(01)81592-6. [DOI] [Google Scholar]
- Shinkai S.; Araki K.; Manabe O. NMR Determination of Association Constants for Calixarene Complexes. Evidence for the Formation of a 1:2 Complex with Calix[8]arene. J. Am. Chem. Soc. 1988, 110, 7214–7215. 10.1021/ja00229a046. [DOI] [Google Scholar]
- Shinkai S.; Araki K.; Matsuda T.; Nishiyama N.; Ikeda H.; Takasu I.; Iwamoto M. NMR and Crystallographic Studies of a p-sulfonatocalix[4]arene-guest Complex. J. Am. Chem. Soc. 1990, 112, 9053–9058. 10.1021/ja00181a004. [DOI] [Google Scholar]
- Marra A.; Scherrmann M.-C.; Dondoni A.; Ungaro R.; Casnati A.; Minari P. Sugar Calixarenes: Preparation of Calix[4]arenes Substituted at the Lower and Upper Rims with O-Glycosyl Groups. Angew. Chem., Int. Ed. 1995, 33, 2479–2481. 10.1002/anie.199424791. [DOI] [Google Scholar]
- Hamuro Y.; Calama M. C.; Park H. S.; Hamilton A. D. A Calixarene with Four Peptide Loops: An Antibody Mimic for Recognition of Protein. Surfaces. Angew. Chem. Int. Ed. 1997, 36, 2680–2683. 10.1002/anie.199726801. [DOI] [Google Scholar]
- Fujimoto T.; Shimizu C.; Hayashida O.; Aoyama Y. Ternary Complexation Involving Protein. Molecular Transport to Saccharide-binding Proteins Using Macrocyclic Saccharide Cluster as Specific Transporter. J. Am. Chem. Soc. 1998, 120, 601–602. 10.1021/ja972966p. [DOI] [Google Scholar]
- Park H. S.; Lin Q.; Hamilton A. D. Protein Surface Recognition by Synthetic Receptors: A Route to Novel Submicromolar Inhibitors for α-chymotrypsin. J. Am. Chem. Soc. 1999, 121, 8–13. 10.1021/ja981504o. [DOI] [Google Scholar]
- Douteau-Guével N.; Coleman A. W.; Morel J.-P.; Morel-Desrosiers N. Complexation of the Basic Amino Acids Lysine and Arginine by Three Sulfonatocalix[n]arenes (n = 4, 6 and 8) in Water: Microcalorimetric Determination of the Gibbs Energies, Enthalpies and Entropies of Complexation. J. Chem. Soc., Perkin Trans. 1999, 2, 629–634. 10.1039/a806855k. [DOI] [Google Scholar]
- Selkti M.; Coleman A. W.; Nicolis I.; Douteau-Guével N.; Villain F.; Tomas A.; De Rango C. The First Example of a Substrate Spanning the Calix[4]arene Bilayer: The Solid State Complex of p-sulfonatocalix[4]arene with L-lysine. Chem. Commun. 2000, 161–162. 10.1039/a906546f. [DOI] [Google Scholar]
- Douteau-Guével N.; Perret F.; Coleman A. W.; Morel J.-P.; Morel-Desrosiers N. Binding of Dipeptides and Tripeptides Containing Lysine or Arginine by p-sulfonatocalixarenes in Water: NMR and Microcalorimetric Studies. J. Chem. Soc., Perkin Trans. 2002, 2, 524–532. 10.1039/b109553f. [DOI] [Google Scholar]
- Atwood J. L.; Ness T.; Nichols P. J.; Raston C. L. Confinement of Amino Acids in Tetra-p-Sulfonated Calix[4]arene Bilayers. Cryst. Growth Des. 2002, 2, 171–176. 10.1021/cg0200053. [DOI] [Google Scholar]
- Lazar A.; Da Silva E.; Navaza A.; Barbey C.; Coleman A. W. A New Packing Motif for para-sulfonatocalix[4]arene: The Solid State Structure of the para-sulfonatocalix[4]arene D-arginine Complex. Chem. Commun. 2004, 2162–2163. 10.1039/b408863h. [DOI] [PubMed] [Google Scholar]
- Arena G.; Casnati A.; Contino A.; Magrì A.; Sansone F.; Sciotto D.; Ungaro R. Inclusion of Naturally Occurring Amino Acids in Water Soluble Calix[4]arenes: a Microcalorimetric and 1H NMR Investigation Supported by Molecular Modeling. Org. Biomol. Chem. 2006, 4, 243–249. 10.1039/B514896K. [DOI] [PubMed] [Google Scholar]
- Memmi L.; Lazar A.; Brioude A.; Ball V.; Coleman A. W. Protein-calixarene Interactions: Complexation of Bovine Serum Albumin by Sulfonatocalix[n]arenes. Chem. Commun. 2001, 2474–2475. 10.1039/b109190p. [DOI] [PubMed] [Google Scholar]
- Oshima T.; Goto M.; Furusaki S. Complex Formation of Cytochrome c with a Calixarene Carboxylic Acid Derivative: A Novel Solubilization Method for Biomolecules in Organic Media. Biomacromolecules 2002, 3, 438–444. 10.1021/bm010148q. [DOI] [PubMed] [Google Scholar]
- Zadmard R.; Schrader T. Nanomolar Protein Sensing with Embedded Receptor Molecules. J. Am. Chem. Soc. 2005, 127, 904–915. 10.1021/ja045785d. [DOI] [PubMed] [Google Scholar]
- Kolusheva S.; Zadmard R.; Schrader T.; Jelinek R. Color Fingerprinting of Proteins by Calixarenes Embedded in Lipid/Polydiacetylene Vesicles. J. Am. Chem. Soc. 2006, 128, 13592–13598. 10.1021/ja064957z. [DOI] [PubMed] [Google Scholar]
- Gordo S.; Martos V.; Santos E.; Menéndez M.; Bo C.; Giralt E.; de Mendoza J. Stability and Structural Recovery of the Tetramerization Domain of p53-R337H Mutant Induced by a Designed Templating Ligand. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 16426–16431. 10.1073/pnas.0805658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martos V.; Bell S. C.; Santos E.; Isacoff E. Y.; Trauner D.; de Mendoza J. Calix[4]arene-based Conical-shaped Ligands for Voltage-dependent Potassium Channels. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 10482–10486. 10.1073/pnas.0813396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecioni S.; Lalor R.; Blanchard B.; Praly J. P.; Imberty A.; Matthews S. E.; Vidal S. Achieving High Affinity Towards a Bacterial Lectin Through Multivalent Topological Isomers of Calix[4]arene Glycoconjugates. Chem.—Eur. J. 2009, 15, 13232–13240. 10.1002/chem.200901799. [DOI] [PubMed] [Google Scholar]
- Chini M. G.; Terracciano S.; Riccio R.; Bifulco G.; Ciao R.; Gaeta C.; Troisi F.; Neri P. Conformationally Locked Calixarene-based Histone Deacetylase Inhibitors. Org. Lett. 2010, 12, 5382–5385. 10.1021/ol102420b. [DOI] [PubMed] [Google Scholar]
- Vovk A. I.; Kononets L. A.; Tanchuk V. Y.; Cherenok S. O.; Drapailo A. B.; Kalchenko V. I.; Kukhar V. P. Inhibition of Yersinia Protein Yyrosine Phosphatase by Phosphonate Derivatives of Calixarenes. Bioorg. Med. Chem. Lett. 2010, 20, 483–487. 10.1016/j.bmcl.2009.11.126. [DOI] [PubMed] [Google Scholar]
- Sayin S.; Yilmaz E.; Yilmaz M. Improvement of Catalytic Properties of Candida rugosa Lipase by Sol-gel Encapsulation in the Presence of Magnetic Calix[4]arene Nanoparticles. Org. Biomol. Chem. 2011, 9, 4021–4024. 10.1039/c1ob05115f. [DOI] [PubMed] [Google Scholar]
- Dings R. P. M.; Miller M. C.; Nesmelova I.; Astorgues-Xerri L.; Kumar N.; Serova M.; Chen X.; Raymond E.; Hoye T. R.; Mayo K. H. Antitumor Agent Calixarene 0118 Targets Human Galectin-1 as an Allosteric Inhibitor of Carbohydrate Binding. J. Med. Chem. 2012, 55, 5121–5129. 10.1021/jm300014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G.; Menon S. Recognition of Lysine, Arginine and Histidine by Novel p-sulfonatocalix[4]arene Thiol Functionalized Gold Nanoparticles in Aqueous Solution. Chem. Commun. 2009, 24, 3563–3565. 10.1039/b905141d. [DOI] [PubMed] [Google Scholar]
- Beshara C. S.; Jones C. E.; Daze K. D.; Lilgert B. J.; Hof F. A Simple Calixarene Recognizes Post–translationally Methylated Lysine. ChemBioChem. 2010, 11, 63–66. 10.1002/cbic.200900633. [DOI] [PubMed] [Google Scholar]
- Daze K. D.; Pinter T.; Beshara C. S.; Ibraheem A.; Minaker S. A.; Ma M. C. F.; Courtemanche R. J. M.; Campbell R. E.; Hof F. Supramolecular Hosts that Recognize Methyllysines and Disrupt the Interaction Between a Modified Histone Tail and its Epigenetic Reader Protein. Chem. Sci. 2012, 3, 2695–2699. 10.1039/c2sc20583a. [DOI] [Google Scholar]
- Daze K. D.; Ma M. C. F.; Pineux F.; Hof F. Synthesis of New Trisulfonated Calix[4]arenes Functionalized at the Upper Rim, and Their Complexation with the Trimethyllysine Epigenetic Mark. Org. Lett. 2012, 14, 1512–1515. 10.1021/ol300243b. [DOI] [PubMed] [Google Scholar]
- Perret F.; Coleman A. W. Biochemistry of Anionic Calix[n]arenes. Chem. Commun. 2011, 47, 7303–7319. 10.1039/c1cc11541c. [DOI] [PubMed] [Google Scholar]
- Guo D. S.; Liu Y. Supramolecular Chemistry of p-sulfonatocalix[n]arenes and its Biological Applications. Acc. Chem. Res. 2014, 47, 1925–1934. 10.1021/ar500009g. [DOI] [PubMed] [Google Scholar]
- Giuliani M.; Morbioli I.; Sansone F.; Casnati A. Moulding Calixarenes for Biomacromolecule Targeting. Chem. Commun. 2015, 51, 14140–14159. 10.1039/C5CC05204A. [DOI] [PubMed] [Google Scholar]
- Aya T.; Hamilton A. D. Tetrabiphenylporphyrin-based Receptors for Protein Surfaces Show Sub–nanomolar Affinity and Enhance Unfolding. Bioorg. Med. Chem. Lett. 2003, 13, 2651–2654. 10.1016/S0960-894X(03)00551-1. [DOI] [PubMed] [Google Scholar]
- Zhou H.; Baldini L.; Hong J.; Wilson A. J.; Hamilton A. D. Pattern Recognition of Proteins Based on an Array of Functionalized Porphyrins. J. Am. Chem. Soc. 2006, 128, 2421–2425. 10.1021/ja056833c. [DOI] [PubMed] [Google Scholar]
- Crowley P. B.; Ganji P.; Ibrahim H. Protein surface recognition: Structural Characterisation of Cytochrome c - Porphyrin Complexes. ChemBioChem. 2008, 9, 1029–1033. 10.1002/cbic.200700736. [DOI] [PubMed] [Google Scholar]
- McGovern R. E.; Fernandes H.; Khan A. R.; Power N. P.; Crowley P. B. Protein Camouflage in Cytochrome c - Calixarene complexes. Nat. Chem. 2012, 4, 527–533. 10.1038/nchem.1342. [DOI] [PubMed] [Google Scholar]
- Derewenda Z. S.; Vekilov P. G. Entropy and Surface Engineering in Protein Crystallization. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 116–124. 10.1107/S0907444905035237. [DOI] [PubMed] [Google Scholar]
- Matar-Merheb R.; Rhimi M.; Leydier A.; Huché F.; Galián C.; Desuzinges-Mandon E.; Ficheux D.; Flot D.; Aghajari N.; Kahn R.; Di Pietro A.; Jault J. M.; Coleman A. W.; Falson P. Structuring Detergents for Extracting and Stabilizing Functional Membrane Proteins. PLoS One 2011, 6, e18036. 10.1371/journal.pone.0018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarauli D.; Tanne J.; Xu C.; Schulz B.; Trnkova L.; Lisdat F. Insights into the Formation and Operation of Polyaniline Sulfonate/Cytochrome c Multilayer Electrodes: Contributions of Polyelectrolytes’ Properties. Phys. Chem. Chem. Phys. 2010, 12, 14271–14277. 10.1039/c0cp00793e. [DOI] [PubMed] [Google Scholar]
- McGovern R. E.; Feifel S. C.; Lisdat F.; Crowley P. B. Microscale Crystals of Cytochrome c and Calixarene on Electrodes: Interprotein Electron Transfer between Defined Sites. Angew. Chem., Int. Ed. 2015, 54, 6356–6359. 10.1002/anie.201500191. [DOI] [PubMed] [Google Scholar]
- McGovern R. E.; McCarthy A. A.; Crowley P. B. Protein Assembly Mediated by Sulfonatocalix[4]arene. Chem. Commun. 2014, 50, 10412–10415. 10.1039/C4CC04897K. [DOI] [PubMed] [Google Scholar]
- Dalgarno S. J.; Hardie M. J.; Makha M.; Raston C. L. Controlling the Conformation and Interplay of p-sulfonatocalix[6]arene as Lanthanide Crown Ether Complexes. Chem.—Eur. J. 2003, 9, 2834–2839. 10.1002/chem.200304963. [DOI] [PubMed] [Google Scholar]
- Zheng D. D.; Fu D. Y.; Wu Y.; Sun Y. L.; Tan L. L.; Zhou T.; Ma S. Q.; Zha X.; Yang Y. W. Efficient Inhibition of Human Papillomavirus 16 L1 Pentamer Formation by a Carboxylatopillarene and a p–sulfonatocalixarene. Chem. Commun. 2014, 50, 3201–3203. 10.1039/c3cc49789e. [DOI] [PubMed] [Google Scholar]
- Wang K.; Guo D.-S.; Zhao M.-Y.; Liu Y. A Supramolecular Vesicle Based on the Complexation of p-sulfonatocalixarene with Protamine and its Trypsin-triggered Controllable-release Properties. Chem.—Eur. J. 2016, 22, 1475–1483. 10.1002/chem.201303963. [DOI] [PubMed] [Google Scholar]
- Shinde M. N.; Barooah N.; Bhasikuttan A. C.; Mohanty J. Inhibition and Disintegration of Insulin Amyloid Fibrils: A Facile Supramolecular Strategy with p-sulfonatocalixarenes. Chem. Commun. 2016, 52, 2992–2995. 10.1039/C5CC10159J. [DOI] [PubMed] [Google Scholar]
- McGovern R. E.; Snarr B. D.; Lyons J. A.; McFarlane J.; Whiting A. L.; Paci I.; Hof F.; Crowley P. B. Structural Study of a Small Molecule Receptor Bound to Dimethyllysine in Lysozyme. Chem. Sci. 2015, 6, 442–449. 10.1039/C4SC02383H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett G. A.; Starke M. J.; Shaurya A.; Li J.; Hof F. Supramolecular Affinity Chromatography for Methylation-Targeted Proteomics. Anal. Chem. 2016, 88, 3697–703. 10.1021/acs.analchem.5b04508. [DOI] [PubMed] [Google Scholar]
- Lee J.; Perez L.; Liu Y.; Wang H.; Hooley R. J.; Zhong W. Separation of Methylated Histone Peptides via Host-Assisted Capillary Electrophoresis. Anal. Chem. 2018, 90, 1881–1888. 10.1021/acs.analchem.7b03969. [DOI] [PubMed] [Google Scholar]
- Pinalli R.; Brancatelli G.; Pedrini A.; Menozzi D.; Hernández D.; Ballester P.; Geremia S.; Dalcanale E. The Origin of Selectivity in the Complexation of N-Methyl Amino Acids by Tetraphosphonate Cavitands. J. Am. Chem. Soc. 2016, 138, 8569–8580. 10.1021/jacs.6b04372. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Perez L.; Mettry M.; Gill A. D.; Byers S. R.; Easley C. J.; Bardeen C. J.; Zhong W.; Hooley R. J. Site Selective Reading of Epigenetic Markers by a Dual-mode Synthetic Receptor Array. Chem. Sci. 2017, 8, 3960–3970. 10.1039/C7SC00865A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolan A. M.; Rennie M. L.; Crowley P. B. Protein Recognition by Functionalized Sulfonatocalix[4]arenes. Chem.—Eur. J. 2018, 24, 984–991. 10.1002/chem.201704931. [DOI] [PubMed] [Google Scholar]
- Garnett G. A. E.; Daze K. D.; Peña Diaz J. A.; Fagen N.; Shaurya A.; Ma M. C.; Collins M. S.; Johnson D. W.; Zakharov L. N.; Hof F. Attraction by Repulsion: Compounds with Like Charges Undergo Self-assembly in Water that Improves in High Salt and Persists in Real Biological Fluids. Chem. Commun. 2016, 52, 2768–2771. 10.1039/C5CC10527G. [DOI] [PubMed] [Google Scholar]
- Li C.; Ma J.; Zhao L.; Zhang Y.; Yu Y.; Shu X.; Li J.; Jia X. Molecular Selective Binding of Basic Amino Acids by a Water-soluble Pillar[5]arene. Chem. Commun. 2013, 49, 1924–1926. 10.1039/c3cc38622h. [DOI] [PubMed] [Google Scholar]
- Cholewa P. P.; Beavers C. M.; Teat S. J.; Dalgarno S. J. Directed Assembly via Selectively Positioned Host Functionality. Chem. Commun. 2013, 49, 3203–3205. 10.1039/c3cc40564h. [DOI] [PubMed] [Google Scholar]
- Almi M.; Arduini A.; Casnati A.; Pochini A.; Ungaro R. Chloromethylation of calixarenes and synthesis of new water soluble macrocyclic hosts. Tetrahedron 1989, 45, 2177–2182. 10.1016/S0040-4020(01)80077-6. [DOI] [Google Scholar]
- Alex J. M.; Rennie M. L.; Volpi S.; Sansone F.; Casnati A.; Crowley P. B. Phosphonated Calixarene as a ″Molecular Glue″ for Protein Crystallization. Cryst. Growth Des. 2018, 18, 2467–2473. 10.1021/acs.cgd.8b00092. [DOI] [Google Scholar]
- Alex J. M.; Brancatelli G.; Volpi S.; Bonaccorso C.; Casnati A.; Geremia S.; Crowley P. B. Probing the Determinants of Porosity in Protein Frameworks: Co-crystals of Cytochrome c and an Octa-anionic Calix[4]arene. Org. Biomol. Chem. 2020, 18, 211–214. 10.1039/C9OB02275A. [DOI] [PubMed] [Google Scholar]
- Antonik P. M.; Eissa A. M.; Round A. R.; Cameron N. R.; Crowley P. B. Noncovalent PEGylation via Lectin-Glycopolymer Interactions. Biomacromolecules 2016, 17, 2719–2725. 10.1021/acs.biomac.6b00766. [DOI] [PubMed] [Google Scholar]
- Webber M. J.; Appel E. A.; Vinciguerra B.; Cortinas A. B.; Thapa L. S.; Jhunjhunwala S.; Isaacs L.; Langer R.; Anderson D. G. Supramolecular PEGylation of Biopharmaceuticals. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 14189–14194. 10.1073/pnas.1616639113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummidivarapu V. V. S.; Rennie M. L.; Doolan A. M.; Crowley P. B. Noncovalent PEGylation via Sulfonatocalix[4]arene - A Crystallographic Proof. Bioconj. Chem. 2018, 29, 3999–4003. 10.1021/acs.bioconjchem.8b00769. [DOI] [PubMed] [Google Scholar]
- Flood R. J.; Ramberg K. O.; Mengel D. B.; Guagnini F.; Crowley P. B. Protein Frameworks with Thiacalixarene and Zinc. Cryst. Growth Des. 2022, 22, 3271–3276. 10.1021/acs.cgd.2c00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A. D.; Raston C. L. Multifunctional p-phosphonated Calixarenes. Chem. Commun. 2011, 47, 9764–9772. 10.1039/c1cc12048d. [DOI] [PubMed] [Google Scholar]
- Rennie M. L.; Doolan A. M.; Raston C. L.; Crowley P. B. Protein Dimerization on a Phosphonated Calix[6]arene Disc. Angew. Chem., Int. Ed. 2017, 56, 5517–5521. 10.1002/anie.201701500. [DOI] [PubMed] [Google Scholar]
- Engilberge S.; Rennie M. L.; Crowley P. B. Calixarene Capture of Partially Unfolded Cytochrome c. FEBS Lett. 2019, 593, 2112–2117. 10.1002/1873-3468.13512. [DOI] [PubMed] [Google Scholar]
- Rennie M. L.; Crowley P. B. A Thermodynamic Model of Auto-regulated Protein Assembly by a Supramolecular Scaffold. ChemPhysChem 2019, 20, 1011–1017. 10.1002/cphc.201900153. [DOI] [PubMed] [Google Scholar]
- Hoffmann R. At first sight. Cryst. Growth Des. 2001, 1, 3. 10.1021/cg0000058. [DOI] [Google Scholar]
- Mockler N. M.; Ramberg K. O.; Guagnini F.; Raston C. L.; Crowley P. B. Noncovalent Protein-Pseudorotaxane Assembly Incorporating an Extended Arm Calix[8]arene with α-Helical Recognition Properties. Cryst. Growth Des. 2021, 21, 1424–1427. 10.1021/acs.cgd.0c01717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler N. M.; Engilberge S.; Rennie M. L.; Raston C. L.; Crowley P. B. Protein-macrocycle Framework Engineering: Supramolecular Copolymerisation with Two Disparate Calixarenes. Supramol. Chem. 2021, 33, 122–128. 10.1080/10610278.2021.1935946. [DOI] [Google Scholar]
- Fizil Á.; Gáspári Z.; Barna T.; Marx F.; Batta G. ″Invisible″ Conformers of an Antifungal Disulfide Protein Revealed by Constrained Cold and Heat Unfolding, CEST-NMR Experiments, and Molecular Dynamics Calculations. Chem.—Eur. J. 2015, 21, 5136–5144. 10.1002/chem.201404879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guagnini F.; Huber A.; Alex J. M.; Marx F.; Crowley P. B. Porous Assembly of an Antifungal Protein Mediated by Zinc and Sulfonato-calix[8]arene. J. Struct. Biol. 2021, 213, 107711. 10.1016/j.jsb.2021.107711. [DOI] [PubMed] [Google Scholar]
- Mallon M.; Dutt S.; Schrader T.; Crowley P. B. Protein Camouflage: Supramolecular Anion Recognition by Ubiquitin. ChemBioChem. 2016, 17, 774–783. 10.1002/cbic.201500477. [DOI] [PubMed] [Google Scholar]
- Hogeweg A.; Sowislok A.; Schrader T.; Beuck C. An NMR Method to Pinpoint Supramolecular Ligand Binding to Basic Residues on Proteins. Angew. Chem., Int. Ed. 2017, 56, 14758–14762. 10.1002/anie.201707950. [DOI] [PubMed] [Google Scholar]
- Chen J.; Meng Q.; Zhang Y.; Dong M.; Zhao L.; Zhang Y.; Chen L.; Chai Y.; Meng Z.; Wang C.; Jia X.; Li C. Complexation of an Antimicrobial Peptide by Large-Sized Macrocycles for Decreasing Hemolysis and Improving Stability. Angew. Chem., Int. Ed. 2021, 60, 11288–11293. 10.1002/anie.202102706. [DOI] [PubMed] [Google Scholar]
- Kojima M.; Abe S.; Ueno T. Engineering of Protein Crystals for use as Solid Biomaterials. Biomater Sci. 2022, 10, 354–367. 10.1039/D1BM01752G. [DOI] [PubMed] [Google Scholar]
- Lai Y. T.; Reading E.; Hura G. L.; Tsai K. L.; Laganowsky A.; Asturias F. J.; Tainer J. A.; Robinson C. V.; Yeates T. O. Structure of a Designed Protein Cage that Self-assembles into a Highly Porous Cube. Nat. Chem. 2014, 6, 1065–1071. 10.1038/nchem.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwardson T. G. W.; Levasseur M. D.; Tetter S.; Steinauer A.; Hori M.; Hilvert D. Protein Cages: From Fundamentals to Advanced Applications. Chem. Rev. 2022, 122, 9145–9197. 10.1021/acs.chemrev.1c00877. [DOI] [PubMed] [Google Scholar]
- Cram D. J.; Cram J. M. Host-Guest Chemistry: Complexes Between Organic Compounds Simulate the Substrate Selectivity of Enzymes. Science 1974, 183, 803–809. 10.1126/science.183.4127.803. [DOI] [PubMed] [Google Scholar]