Abstract
Background:
High-risk human papillomavirus (hrHPV) testing is utilized in primary cervical cancer screening, generally along with cytology, to triage abnormalities to colposcopy. Most screening-based hrHPV testing involves pooled detection of any hrHPV or of HPV16/18. Cervical neoplasia progression risks based on extended hrHPV genotyping—particularly non-16/18 hrHPV types—are not well characterized. HPV genotype-specific incidence of high-grade cervical intraepithelial neoplasia or more severe (CIN2+) following an abnormal screening result was examined.
Methods:
We assessed a US-based prospective, multiracial, clinical cohort of 343 colposcopy patients with normal histology (n=226) or CIN1 (n=117). Baseline cervical samples underwent HPV DNA genotyping, and participants were followed up to five years. Genotype-specific CIN2+ incidence rates (IR) were estimated with accelerated failure time models. Five-year CIN2+ risks were estimated non-parametrically for hierarchical hrHPV risk groups (HPV16; else HPV18/45; else HPV31/33/35/52/58; else HPV39/51/56/59/68).
Results:
At enrollment, median participant age was 30.1 years; most (63%) were hrHPV-positive. Over follow-up, 24 participants progressed to CIN2+ (7.0%). CIN2+ IR among hrHPV-positive participants was 3.4/1,000 person-months. CIN2+ IRs were highest for HPV16 (8.3), HPV33 (7.8), and HPV58 (4.9). Five-year CIN2+ risk was higher for HPV16 (0.34) compared to HPV18/45 (0.12), HPV31/33/35/52/58 (0.12), and HPV39/51/56/59/68 (0.16) (p=0.05).
Conclusions:
Non-16/18 hrHPV types are associated with differential CIN2+ progression rates. HPV16, 33, and 58 exhibited the highest rates over five years. HPV risk groups warrant further investigation in diverse US populations.
Impact:
These novel data assessing extended HPV genotyping in a diverse clinical cohort can inform future directions to improve screening practices in the general population.
Keywords: cervical cancer, CIN, HPV, screening, prospective study
Introduction
Cervical cancer screening programs—historically Papanicolaou (Pap) cytology testing—have dramatically reduced morbidity and mortality from cervical cancer over the past several decades.(1) While cytology testing alone has a high specificity (95–99%) to rule out high-grade cervical intraepithelial neoplasia or more severe (CIN2/CIN3+), it has a relatively low sensitivity (55–80%) for CIN2+ detection.(2–4) In the United States (US), the American Society of Colposcopy and Cervical Pathology (ASCCP) screening guidelines have incorporated testing for high-risk human papillomavirus (hrHPV) to improve risk stratification for Pap cytology tests collected during routine screening for those aged 30 and older.(5,6) Recent recommendations have evolved to include not only overall hrHPV positivity (due to the wide availability of “pooled” hrHPV tests), but also partial genotyping for HPV 16/18 (due to the distinctly elevated risk associated with HPV 16/18).(6–8) These current management recommendations are “risk-based”—based on estimates of risk of CIN3+. Testing for hrHPV has allowed for more tailored risk-based management for millions of cervical cancer screening abnormalities detected every year. Importantly, HPV testing in the primary screening setting has allowed providers to decrease colposcopy referrals and lengthen follow-up from one year between screenings to as long as five years for certain lower-risk abnormalities.(6) Extended genotyping—to detect individual hrHPV types beyond 16 and 18—can further improve risk stratification of screen-detected abnormalities, both by triaging individuals at higher progression risk to immediate colposcopy, and by preventing unnecessary biopsies, cost, and psychological distress for those at relatively lower risk.(9,10)
Extended HPV genotyping has not yet been incorporated into national guidelines, despite evidence that non-16/18 hrHPV types confer differential risks of future CIN2+/CIN3+.(11,12) Indeed, some hrHPV genotypes may confer progression risks more similar to HPV 16/18, while others types may confer lower risks. Thus, under the “equal management for equal risk” principles of US guidelines,(6) certain hrHPV genotypes can potentially be managed more conservatively than others. Despite this potential to improve clinical management, many prospective studies investigating type-specific hrHPV infection progression to CIN2+/CIN3+ were performed outside of the US, in populations with different hrHPV genotype distributions and socio-demographics. Additionally, most US-based studies have assessed only pooled or partial HPV genotyping, which does not provide data on genotype-specific progression risks.(7,8) Further, it is especially important to ensure diverse study populations—with adequate representation of black, indigenous, and people of color (BIPOC)—in screening studies to optimally inform clinical decision-making, as these groups bear a disproportionate burden of cervical cancer morbidity and mortality.(13,14) Thus, studies of HPV genotype-specific CIN2+ progression among diverse US populations are necessary to continue to inform risk stratification and improve patient care.
The purpose of this study is to understand the incidence of CIN2+ associated with specific hrHPV genotypes—particularly non-16/18 genotypes—in the Cervical Intraepithelial Neoplasia Cohort Study (CINCS). CINCS is a US-based prospective, multi-racial cohort of women presenting to colposcopy following abnormal cervical cancer screening results. Our first aim was to describe the distribution of HPV genotypes and associated genotype-specific incidence rates of CIN2+ over five years. Our second aim was to assess the association between a priori HPV “risk groups” and their five-year cumulative risk of progression to CIN2+; we hypothesized that hrHPV risk groups would exhibit differential five-year risks of progression to CIN2+.
Materials and Methods
Study population
This is analysis of secondary data from the Cervical Intraepithelial Neoplasia Cohort Study (CINCS). The CINCS is a prospective clinical cohort of 1,372 women with abnormal cervical cancer screening results who presented for follow-up referral colposcopy in Durham, North Carolina, between September 2010 and March 2016. All those presenting for colposcopy had a previously abnormal cervical cancer screening test—by cytology or cytology/HPV co-testing—that triggered referral to colposcopy in accordance with U.S. national guidelines.(5) Figure S1 depicts the general screening guidelines in place during the study period. Colposcopy clinic attendees at 10 Duke University clinics were invited to participate, as previously described.(15) Participants were study-eligible if they were 21–79 years old, English or Spanish speakers, new visitors to the clinic, and provided written consent. Patients were excluded if they had previous treatment for cervical lesions [i.e., cold knife conization (CKC), loop electrosurgical excision procedure (LEEP), cryotherapy], had a hysterectomy, had moved out of the study area, or did not intend to receive follow-up care at a participating clinic.
At enrollment, all participants underwent a physician-directed pelvic exam, which included collection of exfoliated cervical cells (for cytology and HPV DNA genotyping) and a colposcopy examination with biopsies (for histology). An endocervical component (ECC) was collected on anyone with insufficient transformation zone, or a Pap cytology result of atypical glandular cells (AGC), adenocarcinoma in situ (AIS), or high-grade squamous intraepithelial lesion (HSIL). Cervical cytology was also collected at follow-up visits for up to 5 years; colposcopy-directed biopsies and ECC were collected at follow-up visits if the colposcopist found it necessary, per conservative clinical practice. All colposcopies were performed by experienced colposcopists affiliated with Duke University. Abnormal cytology and histology results during follow-up were managed per U.S. national clinical guidelines.(5) Study staff administered study questionnaires to participants at enrollment and each follow-up visit to collect socio-demographic, behavioral, and clinical characteristics.
This study was conducted in accordance with ethical guidelines and approval was granted by the Institutional Review Boards at Duke University (Durham, NC, US; IRB Pro00022943), North Carolina State University (Raleigh, NC, US; IRB 3565) and University of North Carolina (Chapel Hill, NC, US; IRB 15–2364).
Ascertainment of Cervical Cytology and Histology
Cervical cytology was ascertained from exfoliated cervical specimens collected at each study visit via ThinPrep® liquid-based cytology (Hologic Corporation, Marlborough, MA, US). Cervical exfoliated specimens were suspended in a ThinPrep® vial containing proprietary fluid with at least 50% methanol (Cytyc®, Marlborough, MA, US). Cytology was evaluated according to Bethesda criteria.(16) Residual specimens were stored at 4°C prior to HPV DNA testing.
Cervical histology was ascertained from colposcopy-directed biopsy specimens at enrollment for all participants and at follow-up per clinical indication. Biopsy results were reviewed and graded for severity by Duke-affiliated pathologists, and specimens were tested for adequacy per 2012 ASCCP guidelines.(5) Cytology and histology information were abstracted from patient medical records.
Exposure assessment: HPV Typology
HPV DNA was detected from exfoliated cervical cells collected at enrollment.(15) Following DNA extraction, PGMY09/PGMY11 primers were used in PCR to target a 450-bp region of the HPV L1 genome. Amplification of the human β-globin gene was included as an internal control to ensure sample sufficiency. HPV-positive specimens were subsequently genotyped using the HPV Linear Array® (Roche Diagnostics, Branchburg, NJ, US). This assay detects 13 hrHPV types (16,18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68) and 24 low-risk HPV (lrHPV) types (6, 11, 26, 40, 42, 53, 54, 61, 62, 64, 66, 67, 69, 70, 71, 72, 73, 81, 82, 83, 84, Is39, and cp6108). Multiple-type infections were defined as HPV positivity with two or more hrHPV or lrHPV types. HPV genotypes were evaluated individually, stratified by hrHPV and lrHPV status, and by single- and multiple-type infections. For multiple type infections, person-time and progression events count towards all individual HPV types present at enrollment. We also examined four hrHPV “risk stratification groups”: HPV 16, HPV 18/45, HPV 31/33/35/52/58, and HPV 39/51/56/59/68. These risk groups were originally proposed by Demarco et al. based on risks of CIN3+ progression in the NCI-Kaiser Permanente Northern California Persistence and Progression (PaP) Study.(17)
Outcome assessment: Incident CIN2+
The outcome of interest was a new (first) diagnosis of CIN2+ (“progression”) at any point during the follow-up period. Incident CIN2+ was defined as a histologic diagnosis of CIN2+ (CIN2, CIN2/3, CIN3, or invasive cervical cancer). In the absence of a histologic diagnosis, a cytologic diagnosis of HSIL, LSIL-H (LSIL, cannot exclude HSIL), or ASC-H (atypical squamous cells, cannot exclude HSIL) at follow-up were considered progression events. Outcome status was determined on the earliest date of the progression event, and participants were right-censored from further follow-up thereafter. For participants who received treatment during follow-up (LEEP, CKC, cryotherapy, or hysterectomy), the more severe histologic result between the colposcopic biopsy and the excisional treatment specimen was used for the final follow-up diagnosis. Those receiving treatment during follow-up were right-censored from further follow-up on the date of treatment.
Time-to-progression was measured in person-months from the date of study enrollment to the date of progression. Participants contributed person-months up to the time of progression, to the date of treatment, or to the date of their last attended clinical study visit, whichever occurred first. Progression events were considered interval-censored events, since we knew they occurred at some point in the interval between the previous visit and the visit at which progression was detected.
Analytic sample
This analysis included CINCS participants with normal or CIN1 histology at study enrollment who had HPV genotyping data, were not pregnant or HIV-positive at enrollment, reported no history of HPV vaccination, and returned for at least one follow-up visit (Figure 1). Of 1,372 enrolled CINCS participants, 803 had HPV DNA laboratory results. Of these, 62 women had inconclusive enrollment histology and were excluded. An additional 105 with CIN2+ at enrollment were excluded. Of the remaining 636 participants, 128 with a history of HPV vaccination were excluded; an additional 8 were excluded due to pregnancy, and 135 were excluded who did not return for a follow-up visit. Of the 365 remaining participants, 18 who received immediate treatment and 4 with an inconclusive or missing follow-up diagnosis were excluded, leaving a final analytic sample of 343 participants.
Figure 1. Flowchart for 343 CINCS participants included in analytic sample.
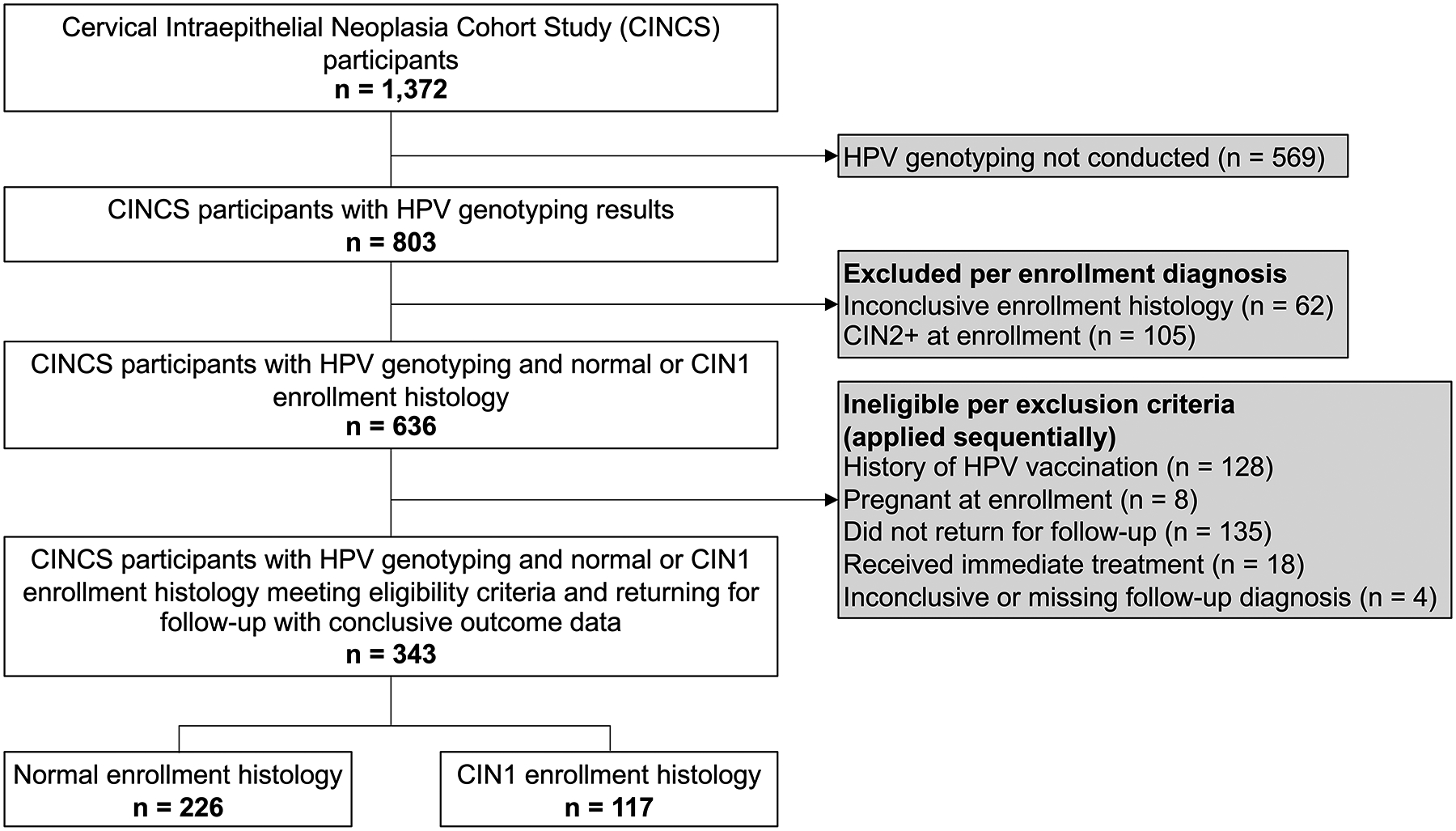
Eligibility and inclusion criteria for a secondary data analysis of the Cervical Intraepithelial Neoplasia Cohort Study (CINCS), based in Durham, North Carolina, US.
Statistical analysis
Descriptive statistics were generated to summarize the baseline distribution of socio-demographic characteristics and HPV genotypes. Pearson’s chi-square test was used to compare a.) characteristics stratified by enrollment histology (normal vs. CIN1), and b.) characteristics of those retained in the study versus those who were lost to follow-up to assess potential bias due to attrition.
Incidence rates (IRs) of CIN2+ and 95% confidence intervals (CIs) were estimated with exponential accelerated failure time (AFT) models for interval-censored data.(18–20) Parametric AFT models allow statistically efficient estimations of incidence rates that account for interval censoring. An exponential model assumes that survival times take on an exponential distribution with respect to the exposure, thus constraining the shape of the survival curve accordingly. To evaluate the appropriateness of this model, we graphically compared the shape of the exponential curve estimated from the data to a non-parametric Kaplan-Meier curve that imposed no constraints. If no events were observed in a group after stratifying by HPV type, the Poisson exact method was used to estimate 95% CIs. Rates were estimated among a) pooled hrHPV-positive and pooled lrHPV-positive participants, b) the four pre-defined hrHPV risk groups (HPV 16, HPV 18/45, HPV 31/33/35/52/58, and HPV 39/51/56/59/68), and c) each individual hrHPV genotype. These rates were estimated overall, stratified by enrollment histologic diagnosis (normal vs. CIN1), and stratified by single- versus multiple-type HPV infections.
Cumulative risk of CIN2+ was estimated using Turnbull’s nonparametric estimator for interval-censored failure time data.(21,22) The log-rank test was used to compare CIN2+ risk among the three hrHPV risk categories, where participants with multiple-type infections were categorized hierarchically according to their highest-risk HPV type (HPV 16 vs. else HPV 18/45 vs. else HPV 31/33/35/52/58 vs. else HPV 39/51/56/59/68). In addition, the log-rank test was used to directly compare CIN2+ risk among age groups (ages 21–24, 25–29, and 30+).
Three sensitivity analyses were conducted. For all sensitivity analyses, we re-estimated HPV genotype-specific prevalence, CIN2+ IRs, and five-year risks. First, we restricted the analytic sample to participants who initially had low-grade cytologic abnormalities [LSIL or atypical squamous cells of undetermined significance (ASCUS)] on their screening Pap test and excluded those with more severe screening cytology. Second, we assessed the impact of determining outcome status using only histologically confirmed CIN2+ cases. Third, we conducted a sensitivity analysis for age comparisons, using a binary age variable (30+ years vs. <30 years) rather than a three-category age variable.
All statistical analyses were conducted using R version 4.0.1 (Vienna, Austria) and the survival(22), interval(23), and epitools(24) packages.
Data availability
Study data are not publicly available due to participant privacy but are available upon reasonable request from the corresponding author.
Results
Enrollment characteristics
The distributions of socio-demographic and clinical characteristics among the 343 participants are presented in Table 1. Over half of participants identified as a race/ethnicity other than non-Hispanic White. At enrollment, 226 participants had normal histology (65.9%) and 117 had CIN1 (34.1%). Comparing those with normal histology to those with CIN1, distributions of race/ethnicity, smoking status, and parity were similar. However, CIN1 cases were more likely to be hrHPV-positive (71.8% vs. 58.8%; p=0.02), report hormonal contraception use (35.0% vs. 22.6%; p=0.01), and were younger (median 27.3 vs. 33.1 years) compared to those with normal histology. No differences in enrollment characteristics were observed between participants who did not return for any follow-up visits and those retained in the study (Table S1).
Table 1.
Characteristics of 343 CINCS colposcopy referral participants with normal or CIN1 enrollment histology
| Enrollment Histology | ||||
|---|---|---|---|---|
| Enrollment characteristic | Overall N (%)a |
Normal Histology N (%) |
CIN1 N (%) |
p-valueb |
| Total | 343 | 226 (65.9) | 117 (34.1) | |
| Age (years) | ||||
| Median (Range) | 30.1 (21.0–72.2) | 33.1 (21.3–72.2) | 27.3 (21.0–64.4) | |
| 21–24 | 75 (21.9) | 31 (13.7) | 44 (37.6) | <0.001 |
| 25–29 | 95 (27.7) | 61 (27.0) | 34 (29.1) | |
| 30+ | 173 (50.4) | 134 (59.3) | 39 (33.3) | |
| High-risk HPV | ||||
| Positive | 217 (63.3) | 133 (58.8) | 84 (71.8) | 0.02 |
| Negative | 126 (36.7) | 93 (41.2) | 33 (28.2) | |
| Referral cytology | ||||
| LSIL | 195 (57.0) | 119 (52.7) | 76 (65.5) | 0.05 |
| ASCUS | 92 (26.9) | 63 (27.9) | 29 (25.0) | |
| ASC-H | 30 (8.8) | 27 (11.9) | 3 (2.6) | |
| LSIL-H | 12 (3.5) | 7 (3.1) | 5 (4.3) | |
| HSIL | 7 (2.0) | 5 (2.2) | 2 (1.7) | |
| Normal or Otherc | 6 (1.8) | 5 (2.2) | 1 (0.9) | |
| Race/Ethnicity | ||||
| Black or African-American | 157 (45.9) | 104 (46.2) | 53 (45.3) | 0.13 |
| Non-Hispanic White | 148 (43.3) | 102 (45.3) | 46 (39.3) | |
| Otherd | 37 (10.8) | 19 (8.4) | 18 (15.4) | |
| Current smoking | ||||
| No | 277 (81.0) | 178 (79.1) | 99 (84.6) | 0.22 |
| Yes | 65 (19.0) | 47 (20.9) | 18 (15.4) | |
| Current hormonal contraceptive use e | ||||
| No | 251 (73.2) | 175 (77.4) | 76 (65.0) | 0.01 |
| Yes | 92 (26.8) | 51 (22.6) | 41 (35.0) | |
| Parity | ||||
| Nulliparous | 152 (44.7) | 99 (44.2) | 53 (45.7) | 0.91 |
| Primiparous (1) | 84 (24.7) | 57 (25.4) | 27 (23.3) | |
| Multiparous (2+) | 104 (30.6) | 68 (30.4) | 36 (31.0) | |
Numbers may not add up to total sample size due to missing data (referral cytology n=1; race/ethnicity n=1; current smoking: n=1; parity: n=3). Percentages calculated as percent of non-missing for each characteristic.
Chi-square test p-value, comparing normal histology to CIN1 at enrollment.
Other referral cytology includes unknown or inconclusive results.
Participants self-classified their race and ethnicity in a multiple-choice questionnaire at enrollment; “Other” includes self-classifications of “Asian/Pacific Islander” (n=19 overall, 9 normal histology, 10 CIN1) and “Hispanic White” (n=18 overall, 10 normal histology, 8 CIN1).
Hormonal contraceptives include oral, patch, injectable, and implant contraceptives.
Table 2 displays the distribution of HPV genotypes at enrollment. Most participants were positive for HPV (87.8%) and hrHPV (63.3%). Multiple-type HPV infections were more frequent among participants with CIN1 (62.4%) than normal histology (37.2%) (Table S2). Overall, the most frequently detected hrHPV types at enrollment were 16 (10.5%), 51 (9.6%), and 39 (8.7%).
Table 2.
Distribution of single- and multiple-type HPV infections at study enrollment in 343 CINCS participants
| HPV genotype | Total N (%)a |
Single-type infection N (%) |
Multiple-type infection N (%) |
|---|---|---|---|
| HPV-positive | 301 (87.8) | 144 (42.0) | 157 (45.8) |
| High-risk HPV | |||
| Any type | 217 (63.3) | 95 (27.7) | 129 (37.6) |
| 16 | 36 (10.5)b | 16 (4.7) | 20 (5.8) |
| 18 | 20 (5.8) | 6 (1.7) | 14 (4.1) |
| 31 | 19 (5.5) | 7 (2.0) | 12 (3.5) |
| 33 | 8 (2.3) | 1 (0.3) | 7 (2.0) |
| 35 | 17 (5.0) | 10 (2.9) | 7 (2.0) |
| 39 | 30 (8.7)b | 7 (2.0) | 23 (6.7) |
| 45 | 13 (3.8) | 6 (1.7) | 7 (2.0) |
| 51 | 33 (9.6)b | 8 (2.3) | 25 (7.3) |
| 52 | 29 (8.5) | 13 (3.8) | 16 (4.7) |
| 56 | 19 (5.5) | 3 (0.9) | 16 (4.7) |
| 58 | 18 (5.2) | 6 (1.7) | 12 (3.5) |
| 59 | 26 (7.6) | 8 (2.3) | 18 (5.2) |
| 68 | 16 (4.7) | 4 (1.2) | 12 (3.5) |
| Low-risk HPV | |||
| Any type | 184 (53.6) | 46 (13.4) | 138 (40.2) |
| HPV-negative | 42 (12.2) | -- | -- |
Percentages are out of total number of participants.
Top 3 most prevalent high-risk HPV genotypes overall
Incidence rates of CIN2+
Median study follow-up time was 24.3 person-months (range 3.8–62.3). There were 24 events of progression to CIN2+ (7.0% overall; 6.2% of normal histology and 8.5% of CIN1) (Table 3). CIN2+ IRs among all single- and multiple-type infections, stratified by HPV genotype, are displayed in Table 4. The highest rates overall were observed for HPV 16 (IR 8.3; 95% CI 4.1, 16.6), HPV 33 (IR 7.8; 95% CI 2.0, 31.3), and HPV 58 (IR 4.9; 95% CI 1.2, 19.6). The CIN2+ IR for single-type hrHPV infections was 1.9 per 1,000 person-months (95% CI 0.8, 4.6) and for multiple-type hrHPV infections was 4.4 (95% CI 2.6, 7.3). Among single-type HPV infections, the highest IRs were observed for HPV 16, 51, and 35, though these estimates were imprecise due to a relatively small number of participants infected with each HPV genotype after stratification. CIN2+ rates for all infections, stratified by normal histology vs. CIN1, are presented in Table S3. Single-type infections only, stratified by enrollment histology, are presented in Table S4.
Table 3.
Outcomes of 343 CINCS participants followed for up to 5 years, overall and stratified by enrollment histology
| Enrollment histology | |||
|---|---|---|---|
| Outcomea | Overall N (%)a |
Normal Histology N (%)a |
CIN1 N (%)a |
| Total | 343 | 226 | 117 |
| Progression to high-grade a | 24 (7.0) | 14 (6.2) | 10 (8.5) |
| CIN2 | 9 (2.6) | 5 (2.2) | 4 (3.4) |
| CIN2–3 | 4 (1.2) | 1 (0.4) | 3 (2.6) |
| CIN3 | 3 (0.9) | 3 (1.3) | 0 |
| HSIL | 3 (0.9) | 1 (0.4) | 2 (1.7) |
| ASC-H | 4 (1.2) | 3 (1.3) | 1 (0.9) |
| LSIL-H | 1 (0.3) | 1 (0.4) | 0 |
| No progression to high-grade | 319 (93.0) | 212 (93.8) | 107 (91.5) |
| Normal | 245 (71.4) | 172 (76.1) | 73 (62.4) |
| CIN1 | 11 (3.2) | 3 (1.3) | 8 (6.8) |
| ASCUS | 40 (11.7) | 19 (8.4) | 21 (17.9) |
| LSIL | 23 (6.7) | 18 (8.0) | 5 (4.3) |
Percentages are column percentages.
Participants counted as “progressed” on first instance of a progression event, Histology was preferred over cytology to categorize progression events, but cytology alone was used in the absence of confirmatory histology. Of the progression events, 6 eventually progressed to CIN3 later in follow-up (3 CIN2, 2 ASC-H, and 1 HSIL); however, all were counted as “progression” events earlier in their disease course per study protocol.
Table 4.
Incidence rates of CIN2+ among 343 participants followed for up to 5 years, overall and stratified by single- and multiple-type HPV infection status
| Overall (n=343)a | Single-type infections (n=144) | Multiple-type infections (n=157) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HPV genotype | N | CIN2+ events | Person-monthsb | Incidence rate (95% CI)c |
CIN2+ events | Person-monthsb | Incidence rate (95% CI)c |
CIN2+ events | Person-monthsb | Incidence rate (95% CI)c |
| HPV-positive | 301 | 22 | 8210 | 2.7 (1.8, 4.1) | 7 | 3882 | 1.8 (0.9, 3.8) | 15 | 4329 | 3.5 (2.1, 5.9) |
| High-risk HPV | ||||||||||
| Any type | 217 | 20 | 5972 | 3.4 (2.2, 5.3) | 5 | 2645 | 1.9 (0.8, 4.6) | 15 | 3521 | 4.4 (2.6, 7.3) |
| 16d | 36 | 8 | 1027 | 8.3 (4.1, 16.6)e | 2 | 433 | 4.9 (1.2, 19.6)e | 6 | 594 | 10.8 (4.8, 24.0)e |
| 18/45d | 33 | 2 | 1020 | 2.0 (0.5, 7.9) | 0 | 376 | 0.0 (0.0, 9.8) | 2 | 644 | 3.2 (0.8, 12.6) |
| 31/33/35/52/58d | 86 | 8 | 2257 | 3.6 (1.8, 7.2) | 2 | 998 | 2.0 (0.5, 8.1) | 6 | 1259 | 4.9 (2.2, 10.8) |
| 39/51/56/59/68d | 106 | 8 | 2901 | 2.8 (1.4, 5.6) | 1 | 838 | 1.2 (0.2, 8.5) | 7 | 2062 | 3.5 (1.6, 7.2) |
| 18 | 20 | 2 | 633 | 3.2 (0.8, 12.9) | 0 | 175 | 0.0 (0.0, 21.0) | 2 | 458 | 4.5 (1.1, 17.9) |
| 31 | 19 | 1 | 437 | 2.3 (0.3, 16.4) | 0 | 150 | 0.0 (0.0, 24.6) | 1 | 287 | 3.5 (0.5, 25.1) |
| 33 | 8 | 2 | 265 | 7.8 (2.0, 31.3)e | 1 | 30 | --f | 1 | 234 | 4.3 (0.6, 30.7) |
| 35 | 17 | 1 | 401 | 2.5 (0.4, 17.8) | 1 | 211 | 4.8 (0.7, 34.0)e | 0 | 190 | 0.0 (0.0, 19.4) |
| 39 | 30 | 3 | 712 | 4.3 (1.4, 13.3) | 0 | 125 | 0.0 (0.0, 29.4) | 3 | 586 | 5.2 (1.7, 16.2) |
| 45 | 13 | 0 | 387 | 0.0 (0.0, 9.5) | 0 | 201 | 0.0 (0.0, 18.4) | 0 | 187 | 0.0 (0.0, 19.8) |
| 51 | 33 | 3 | 918 | 3.3 (1.1, 10.3) | 1 | 206 | 4.9 (0.7, 34.5)e | 2 | 712 | 2.9 (0.7, 11.5) |
| 52 | 29 | 3 | 840 | 3.6 (1.2, 11.2) | 0 | 453 | 0.0 (0.0, 8.1) | 3 | 387 | 7.9 (2.6, 24.6)e |
| 56 | 19 | 1 | 578 | 1.7 (0.2, 12.3) | 0 | 76 | 0.0 (0.0, 48.7) | 1 | 502 | 2.0 (0.3, 14.2) |
| 58 | 18 | 2 | 420 | 4.9 (1.2, 19.6)e | 0 | 154 | 0.0 (0.0, 24.0) | 2 | 266 | 7.8 (2.0, 31.4)e |
| 59 | 26 | 1 | 763 | 1.3 (0.2, 9.4) | 0 | 231 | 0.0 (0.0, 16.0) | 1 | 532 | 1.9 (0.3, 13.5) |
| 68 | 16 | 1 | 466 | 2.2 (0.3, 15.3) | 0 | 200 | 0.0 (0.0, 18.5) | 1 | 266 | 3.8 (0.5, 26.8) |
| Low-risk HPV only | 81 | 2 | 2210 | 0.9 (0.2, 3.6) | 2 | 1208 | 1.7 (0.4, 6.7) | 0 | 1002 | 0.0 (0.0, 3.7) |
| HPV-negative | 42 | 2 | 1106 | 1.8 (0.5, 7.3) | -- | -- | -- | -- | -- | -- |
Includes both single- and multiple-type HPV infections (ie, for multiple types, progression events count towards all individual types present at enrollment)
Sum of person-months for all participants in group infected with noted genotype at enrollment; overall median follow-up time per person: 24.3 months
Incidence rate per 1000 person-months (95% CI = 95% confidence interval); estimated with exponential accelerated failure time models for interval-censored data; for genotypes with zero events, estimated with Poisson exact method
HPV risk stratification groups from Demarco et al. (17)
Highest three incidence rates by individual HPV type
Insufficient data to calculate point estimate and confidence interval (only one participant in HPV group)
CIN2+ risk by HPV risk group
Figure 2 shows that the five-year cumulative probability (risk) of CIN2+ differed by HPV risk group (HPV 16 vs. else HPV 18/45 vs. else HPV 31/33/35/52/58 vs. else HPV 39/51/56/59/68). The HPV 16 conferred the greatest risk of CIN2+ during follow-up: the five-year risk of CIN2+ was 0.34 for HPV 16, 0.12 for HPV 18/45, 0.12 for HPV 31/33/35/52/58, and 0.16 for 39/51/56/59/68. (log-rank p=0.05).
Figure 2. Five-year risk of progression to CIN2+ adapted by HPV risk group in CINCS participants with normal or CIN1 enrollment histology.
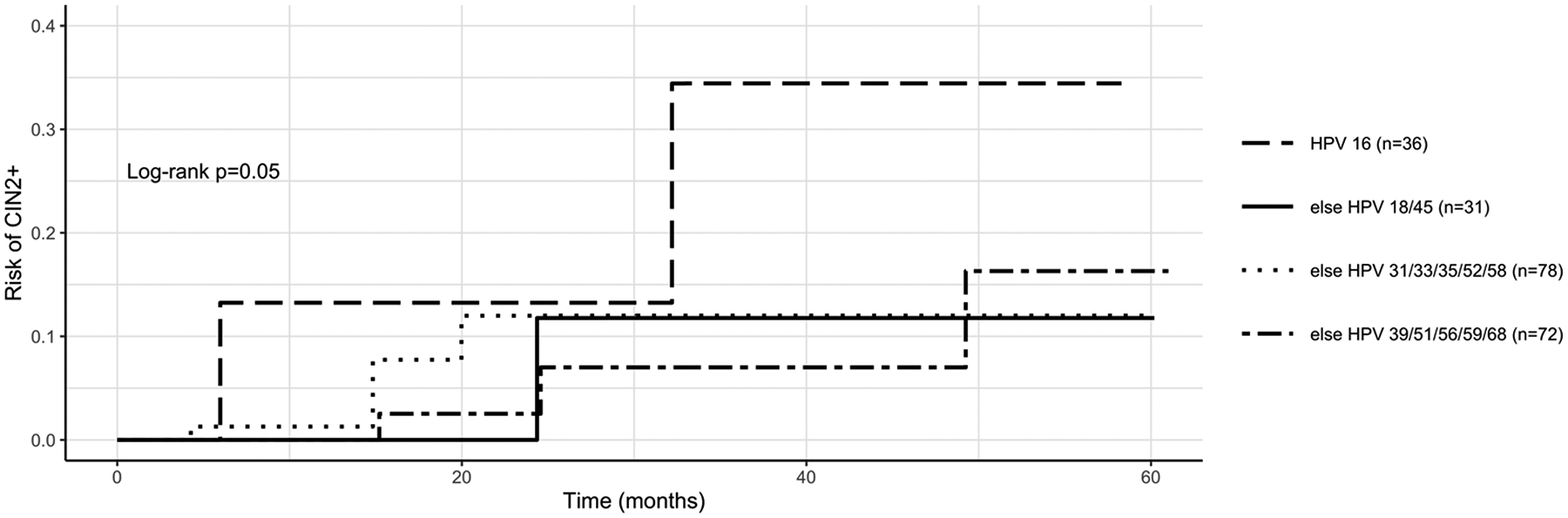
HPV risk stratification groups from Demarco et al. (17) Five-year risks of CIN2+ for each HPV risk group: 0.34 for “HPV 16”, 0.12 for “else HPV 18/45”, 0.12 for “else HPV 31/33/35/52/58”, and 0.16 for “else HPV 39/51/56/59/68”.
CIN2+ rates and risk by age
CIN2+ IR among hrHPV-positive participants was 3.4 among those aged 21+ (95% CI 2.2, 5.3), 3.3 among those 25+ (95% CI 2.0, 5.5), and 4.5 among those 30+ (95% CI 2.5, 7.8) (Table S5). In the full 21+ cohort, rates were highest for HPV 16, 33, and 58. Comparatively, rates were highest for HPV 33, 16, and 58 among those 25+ and for HPV 68, 16, and 58 among those 30+. Five-year risks by HPV risk group and age group are shown in Figure S2. Similar findings were observed when alternatively using a binary age variable (ages <30 vs. 30+).
Sensitivity analyses of low-grade enrollment cytology and CIN2+ outcome histology
After restricting to those who had LSIL or ASCUS on their enrollment screening Pap test prior to presenting to colposcopy (n=287), trends were similar to those observed in the main analytic sample (Table S6). After restricting to only histologically confirmed CIN2+ cases, trends were also similar. In both sensitivity analyses, the HPV types that exhibited the highest IRs (HPV 16, 58, and 33) were similar to those in the main analytic cohort. When comparing five-year risks among HPV risk groups, findings were similar, and p-values comparing the groups became smaller (p=0.05 in main cohort; p=0.04 in enrollment cytology sensitivity analysis; and p=0.02 in outcome histology sensitivity analysis.)
Discussion
This study is among the few prospective studies of extended HPV genotyping to characterize HPV genotype-specific risk of cervical disease progression in the US. This five-year prospective study investigated CIN2+ incidence by HPV genotype in a high-risk colposcopy referral population and found that CIN2+ incidence rates were highest among hrHPV types 16, 33, and 58. Thus, in addition to the well-established hrHPV types 16 and 18, hrHPV types 33 and 58 were associated with relatively higher risk of progression to CIN2+ over a five-year period compared to other hrHPV types. Our study utilized HPV risk stratification groups proposed by Demarco et al.: HPV 16, HPV 18/45, HPV 31/33/35/52/58, and HPV 39/51/56/59/68.(17) Consistent with Demarco et al., we found a distinctly elevated five-year risk of progression associated with HPV 16, while HPV 18/45 and HPV 31/33/35/52/58 had lower risks that remained similar to each other over follow-up. Demarco et al. found HPV 39/51/56/59/68 to have a lower risk (<5%) compared to the other non-16 groups (>5%), but we found this HPV 39/51/56/59/68 group to exhibit similar five-year risks of CIN2+ HPV 18/45 and HPV 31/33/35/52/58 (0.16, 0.12, and 0.12, respectively).
The current findings support evidence that additional non-16/18 hrHPV types may be considered in future screening practices to inform clinical management of screening abnormalities. Demarco et al. found that three-year-risks of CIN3+ were highest for HPV 16 (21.9%) HPV 18 (11.5%), HPV 33/58 (8.6%), HPV 31 (8.1%), HPV 52 (5.6%), and HPV 45 (5.4%) in the NCI-Kaiser Permanente PaP study.(17) In our study, HPV 45 did not yield any progression events, which may explain why the HPV 18/45 group appeared to have a relatively low risk of progression that was similar to the other “lower risk” groups. Otherwise, our findings support the relatively elevated progression risk associated with HPV 33 and 58. Our study showed that the HPV 39/51/56/59/68 group conferred a similar five-year risk to the HPV 18/45 group and HPV 31/33/35/52/58 groups, while the Demarco study found the HPV 39/51/56/59/68 group to have a lower risk. This difference may be driven by our relatively small sample size, and the higher progression rates observed in our study for HPV 39 (4.3 per 1,000 person-months) and HPV 51 (3.3 per 1,000 person-months), relative to the other types in their group. A notable difference between the Demarco study and our CINCS study is that Demarco et al. studied CIN3+ in a general screening population, while we assessed CIN2+ in a colposcopy referral population; additionally, the patient population in northern California (the setting of Kaiser Permanente) differs demographically from that in Durham, NC. For another study comparison, the Onclarity trial assessed CIN2+ and CIN3+ risk among individuals from a general screening population with negative and ASCUS/LSIL cytology. Among negative cytology, HPV 16 and 31 carried the highest progression risks, while an “intermediate risk” group was comprised of HPV types 18, 33, 58, and 52.(25) Among ASCUS/LSIL cases, HPV 16 carried the highest risk for CIN2+, while HPV types 31, 18, 33, and 58 comprised an “intermediate” risk group.(26) Our findings are also consistent with this, given the high CIN2+ incidence rates observed in the CINCS cohort for HPV types 16, 33, and 58 overall, and for types 33, 31, and 58 among CIN1 cases. In contrast, we observed a relatively lower rate of progression associated with HPV 31 compared to the Onclarity trial, potentially due to a relatively small number of participants infected with HPV 31 at enrollment in our study.
Variations in the incidence of high-grade lesions attributed to specific hrHPV types in different studies may be a function of geographic location and population. For example, a study among Japanese women with LSIL cytology and CIN1 histology found that hazard ratios for progression to CIN3+ were highest for HPV 31, 33, 35, and 52, compared to women with lrHPV or no infection.(27) Additionally, in the placebo arm of the VIVIANE phase III trial, which recruited from multiple centers internationally, HPV 33 had the highest two-year risk of progression to CIN2+, followed by types 16, 31, and 45.(28) The higher rates of progression that we observed for HPV 33 are concordant with these findings. The CINCS cohort included a diverse population with over 50% BIPOC representation in the southeastern US, and genotype-specific progression observed here may be more specific to this patient population. HPV type distributions have been shown to vary by geography and patient characteristics, such as race. For example, studies have shown that black, Hispanic, and Asian women with CIN2/CIN3 are significantly less likely to be HPV 16/18-positive compared to their white counterparts.(29,30) Variations in HPV type-specific progression risks may be related to these distributional differences. Given this, these reported findings contribute important knowledge about US-based HPV type-specific risk profiles and highlight the importance of continuing to study the shifting landscape of HPV and cervical disease, especially in understudied populations.
As new methods for screening are developed, the impact of single- versus multiple-type HPV infections must be considered in risk assessment for cervical cancer screening. Over half of HPV infections in the CINCS cohort were multiple-type infections, and participants with multiple-type infections exhibited higher CIN2+ rates than those with single-type infections. The presence of multiple-type infections may be a marker of elevated individual risk, potentially due to synergistic effects of multiple HPV types or to individual inability to clear HPV, leading to concomitant persistent infections. There is no consensus about whether infection with multiple hrHPV types confers a greater risk of progression to CIN2+. A small number of prospective studies in general screening populations and among women with low-grade abnormalities have observed no evidence of elevated risk of CIN2+ over two to three years in multiple-type infections, compared to single-type infections.(31),(32) However, some cross-sectional studies have shown a higher prevalence of multiple-type hrHPV infections with increasing CIN lesion severity.(33–36) Our prospective findings lend support to the potential use of extended HPV genotyping to identify patients with multiple infections and triage them for closer follow-up based on this elevated risk profile.
Study strengths include the use of longitudinal data over five years to assess genotype-specific CIN2+ risk; this prospective study design is advantageous to quantify risk over time and is critical to inform clinical practice guidelines. Additionally, most studies assessing HPV-related progression risk have examined pooled hrHPV genotypes; the current study leverages extended HPV genotyping for 37 individual HPV types, including identification of multiple-type infections. Finally, this study utilizes a unique clinical cohort in the US with most participants identifying as a non-white race or ethnicity—patient subpopulations that are historically underrepresented in many large studies. Inclusion of BIPOC participants in clinical research is critically important to elucidating a more representative landscape of HPV-related cervical disease and creating appropriate clinical guidelines. Leveraging these unique strengths, our findings support the clinical utility of non-16/18 hrHPV extended genotyping to improve risk stratification and triage screening-detected cervical abnormalities in a multiracial US population.
A primary limitation of this study is that HPV genotyping at study follow-up was not collected. The only true way to ascertain HPV progression rates is to examine time since incident infection. Since this was a clinical cohort, HPV genotyping was only available at time of enrollment, and thus this was not possible to assess with this dataset. Second, the relatively small sample size, especially after stratification by genotype and histology, impacted the ability to make strong inferences about some individual HPV genotypes. We additionally reported many stratified results with accompanying 95% CIs; this may have induced a higher risk of type II error. Third, there was potential for selection bias in this study population. Approximately one-quarter of participants potentially eligible for this analysis did not return for a follow-up visit; we found no significant differences in characteristics between those retained and those lost, including enrollment histology. Fourth, this study investigated a colposcopy referral population, so results may not generalize to a general screening population. In particular, colposcopy referral populations have higher baseline prevalence of disease as compared to general screening populations. Fifth, there is risk of detection bias and outcome misclassification since this study was performed in a clinical cohort. Namely, per conservative clinical practice, biopsies were not performed with each colposcopy exam, which may have led to under-detection of disease. Additionally, there was no external expert pathology review of biopsy specimens, which may have led to some disease misclassification. Sixth, while nearly half of our study sample identified as Black or African-American, far fewer individuals identified as Asian, or as Hispanic or Latina; improving representation of these and other BIPOC groups is an important priority for future screening studies. Finally, the analytic endpoint was defined as CIN2+, but CIN3+ is more proximal to invasive cancer and thus would have strengthened our study. However, there is important clinical value in determining risk stratification at earlier timepoints—such as CIN2+—since clinical decision-making to undergo treatment or more frequent testing occurs at these earlier points. Indeed, CIN2 is often treated in clinical practice, thus precluding observation of many CIN3+ cases.
In conclusion, the current data highlight the potential utility of non-16/18 hrHPV types—particularly 33 and 58—for predicting progression to CIN2+. Extended HPV genotyping beyond HPV 16/18 may be useful to identify patients who exhibit different risks of progression to CIN2+ to further improve HPV-based risk stratification. For example, identifying higher- and lower-risk non-16/18 genotypes can help triage patients to more intensive or more conservative management, respectively, and thereby support the current “equal management for equal risk” US guidelines and improve the efficiency of cervical cancer screening cascades. This study in a diverse clinical cohort in the southeastern US contributes to the literature assessing risk attribution of hrHPV genotypes to progression to CIN2+ for cervical cancer screening. Further investigation of the applicability of these results to a general screening population is warranted.
Supplementary Material
Acknowledgements
NIH NCI F30CA257181 (A. Bukowski), NIH NCI T32 5T32CA57726-29 (A. Bukowski), NIH NCI R01CA142983 (C. Hoyo, F. Valea, R.C. Bentley, R.L. Maguire, J.W. Schmitt, S.K. Murphy, J.S. Smith), and NIH NCI R01CA142983-02S1 (A.C. Vidal). Ayodele Gomih (UNC Department of Epidemiology). Chris Wiesen (Odum Institute).
Conflicts of interest:
J.S.S. has received research grants and consultancies from Hologic and Becton Dickinson for the past 5 years. Remaining authors A.B., C.H., M.G.H., W.R.B., F.V., R.C.B., A.C.V, R.L.M., J.W.S., S.K.M., and K.E.N. have no conflicts to disclose.
References
- 1.Yang DX, Soulos PR, Davis B, Gross CP, Yu JB. Impact of Widespread Cervical Cancer Screening. American Journal of Clinical Oncology: Cancer Clinical Trials. Lippincott Williams and Wilkins; 2018. page 289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayrand M-H, Duarte-Franco E, Rodrigues I, Walter SD, Hanley J, Ferenczy A, et al. Human Papillomavirus DNA versus Papanicolaou Screening Tests for Cervical Cancer. New England Journal of Medicine. Massachusetts Medical Society; 2007;357:1579–88. [DOI] [PubMed] [Google Scholar]
- 3.Nanda K, McCrory DC, Myers ER, Bastian LA, Hasselblad V, Hickey JD, et al. Accuracy of the papanicolaou test in screening for and follow-up of cervical cytologic abnormalities: A systematic review. Annals of Internal Medicine. American College of Physicians; 2000;132:810–9. [DOI] [PubMed] [Google Scholar]
- 4.Soost HJ, Lange HJ, Lehmacher WR-KB. The validation of cervical cytology. Sensitivity, specificity and predictive values. Acta Cytol. 1991;35:8–14. [PubMed] [Google Scholar]
- 5.Massad LS, Einstein MH, Huh WK, Katki HA, Kinney WK, Schiffman M, et al. 2012 Updated Consensus Guidelines for the Management of Abnormal Cervical Cancer Screening Tests and Cancer Precursors (ASCCP). Journal of Lower Genital Tract Disease. 2013;17:1–27. [DOI] [PubMed] [Google Scholar]
- 6.Perkins RB, Guido RS, Castle PE, Chelmow D, Einstein MH, Garcia F, et al. 2019 ASCCP Risk-Based Management Consensus Guidelines for Abnormal Cervical Cancer Screening Tests and Cancer Precursors. Journal of Lower Genital Tract Disease [Internet]. Lippincott Williams and Wilkins; 2020. [cited 2020 Oct 10];24:102–31. Available from: https://www-ncbi-nlm-nih-gov.libproxy.lib.unc.edu/pmc/articles/PMC7147428/ [Google Scholar]
- 7.Egemen D, Cheung LC, Chen X, Demarco M, Perkins RB, Kinney W, et al. Risk Estimates Supporting the 2019 ASCCP Risk-Based Management Consensus Guidelines. Journal of Lower Genital Tract Disease. Lippincott Williams and Wilkins; 2020;24:132–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demarco M, Egemen D, Raine-Bennett TR, Cheung LC, Befano B, Poitras NE, et al. A Study of Partial Human Papillomavirus Genotyping in Support of the 2019 ASCCP Risk-Based Management Consensus Guidelines. Journal of Lower Genital Tract Disease. Lippincott Williams and Wilkins; 2020;24:144–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsui J, Llanos AAM, Doose M, Rotter D, Stroup A. Determinants of abnormal cervical cancer screening follow-up and invasive cervical cancer among uninsured and underinsured women in New Jersey. Journal of Health Care for the Poor and Underserved. Johns Hopkins University Press; 2019;30:680–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kupets R, Paszat LF. Follow-up of abnormal pap smear results: A population-based study. Journal of Clinical Oncology. American Society of Clinical Oncology (ASCO); 2010;28:6076–6076. [Google Scholar]
- 11.Bonde JH, Sandri MT, Gary DS, Andrews JC. Clinical Utility of Human Papillomavirus Genotyping in Cervical Cancer Screening: A Systematic Review. Journal of Lower Genital Tract Disease. Lippincott Williams and Wilkins; 2020. page 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonde J, Bottari F, Parvu V, Pedersen H, Yanson K, Iacobone AD, et al. Bayesian analysis of baseline risk of CIN2 and ≥CIN3 by HPV genotype in a European referral cohort. International Journal of Cancer. Wiley-Liss Inc; 2019;145:1033–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olusola P, Banerjee HN, Philley JV., Dasgupta S. Human Papilloma Virus-Associated Cervical Cancer and Health Disparities. Cells. Multidisciplinary Digital Publishing Institute (MDPI); 2019;8:622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo W, Kim S, Huh WK, Dilley S, Coughlin SS, Partridge EE, et al. Recent trends in racial and regional disparities in cervical cancer incidence and mortality in United States. PLoS ONE. Public Library of Science; 2017;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vidal AC, Smith JS, Valea F, Bentley R, Gradison M, Yarnall KSH, et al. HPV genotypes and cervical intraepithelial neoplasia in a multiethnic cohort in the southeastern USA. Cancer Causes and Control. Kluwer Academic Publishers; 2014;25:1055–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solomon D, Davey D, Kurman R, Moriarty A, O’Connor D, Prey M, et al. The 2001 Bethesda System: Terminology for reporting results of cervical cytology. J Am Med Assoc. American Medical Association; 2002;287:2114–9. [DOI] [PubMed] [Google Scholar]
- 17.Demarco M, Hyun N, Carter-Pokras O, Raine-Bennett TR, Cheung L, Chen X, et al. A study of type-specific HPV natural history and implications for contemporary cervical cancer screening programs. EClinicalMedicine [Internet]. Lancet Publishing Group; 2020. [cited 2021 Jan 12];22. Available from: https://pubmed-ncbi-nlm-nih-gov.libproxy.lib.unc.edu/32510043/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collett D Modelling Survival Data in Medical Research. 3rd ed. CRC Press; 2015. [Google Scholar]
- 19.Lindsey J, Ryan L. Methods for interval-censored data. Statistics in Medicine. 1998;17:219–38. [DOI] [PubMed] [Google Scholar]
- 20.Gómez G, Calle ML, Oller R, Langohr K. Tutorial on methods for interval-censored data and their implementation in R: https://doi.org/101177/1471082X0900900402. SAGE PublicationsSage India: New Delhi, India; 2009;9:259–97. [Google Scholar]
- 21.Turnbull B The empirical distribution function with arbitrarily grouped, censored and truncated data. Journal of the Royal Statistical Society: Series B (Methodological). 1976;38:290–5. [Google Scholar]
- 22.Therneau T. A Package for Survival Analysis in R. R package version 32–13. 2021; [Google Scholar]
- 23.Fay M, Shaw P. Exact and Asymptotic Weighted Logrank Tests for Interval Censored Data: The interval R Package. Journal of Statistical Software. 2010;36:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aragon TJ. epitools: Epidemiology Tools. R package version 05–101. 2020; [Google Scholar]
- 25.Stoler MH, Wright TC, Parvu V, Yanson K, Cooper CK, Andrews J. Stratified risk of high-grade cervical disease using onclarity HPV extended genotyping in women, ≥25 years of age, with NILM cytology. Gynecol Oncol. Gynecol Oncol; 2019;153:26–33. [DOI] [PubMed] [Google Scholar]
- 26.Wright TC, Stoler MH, Parvu V, Yanson K, Cooper C, Andrews J. Risk detection for high-grade cervical disease using Onclarity HPV extended genotyping in women, ≥21 years of age, with ASC-US or LSIL cytology. Gynecologic Oncology. Academic Press Inc; 2019;154:360–7. [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto K, Oki A, Furuta R, Maeda H, Yasugi T, Takatsuka N, et al. Predicting the progression of cervical precursor lesions by human papillomavirus genotyping: A prospective cohort study. International Journal of Cancer. Int J Cancer; 2011;128:2898–910. [DOI] [PubMed] [Google Scholar]
- 28.Rachel Skinner S, Wheeler CM, Romanowski B, Castellsagué X, Lazcano-Ponce E, Rowena Del Rosario-Raymundo M, et al. Progression of HPV infection to detectable cervical lesions or clearance in adult women: Analysis of the control arm of the VIVIANE study. International Journal of Cancer. Wiley-Liss Inc; 2016;138:2428–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saadeh K, Park I, Gargano JW, Whitney E, Querec TD, Hurley L, et al. Prevalence of human papillomavirus (HPV)-vaccine types by race/ethnicity and sociodemographic factors in women with high-grade cervical intraepithelial neoplasia (CIN2/3/AIS), Alameda County, California, United States. Vaccine. Elsevier Ltd; 2020;38:39–45. [DOI] [PubMed] [Google Scholar]
- 30.Niccolai LM, Russ C, Julian PJ, Hariri S, Sinard J, Meek JI, et al. Individual and geographic disparities in human papillomavirus types 16/18 in high-grade cervical lesions. Cancer. John Wiley and Sons Inc; 2013;119:3052–8. [DOI] [PubMed] [Google Scholar]
- 31.Wheeler CM, Hunt WC, Schiffman M, Castle PE. Human Papillomavirus Genotypes and the Cumulative 2-Year Risk of Cervical Precancer. The Journal of Infectious Diseases. Oxford Academic; 2006;194:1291–9. [DOI] [PubMed] [Google Scholar]
- 32.Adcock R, Cuzick J, Hunt WC, McDonald RM, Wheeler CM. Role of HPV genotype, multiple infections, and viral load on the risk of high-grade cervical neoplasia. Cancer Epidemiology Biomarkers and Prevention. American Association for Cancer Research Inc; 2019;28:1816–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spinillo A, Dal Bello B, Gardella B, Roccio M, Dacco’ MD, Silini EM. Multiple human papillomavirus infection and high grade cervical intraepithelial neoplasia among women with cytological diagnosis of atypical squamous cells of undetermined significance or low grade squamous intraepithelial lesions. Gynecologic Oncology. Gynecol Oncol; 2009;113:115–9. [DOI] [PubMed] [Google Scholar]
- 34.Pista A, Oliveira A, Verdasca N, Ribeiro F. Single and multiple human papillomavirus infections in cervical abnormalities in Portuguese women. Clinical Microbiology and Infection. Blackwell Publishing Ltd; 2011;17:941–6. [DOI] [PubMed] [Google Scholar]
- 35.Trottier H, Mahmud S, Costa MC, Sobrinho JP, Duarte-Franco E, Rohan TE, et al. Human papillomavirus infections with multiple types and risk of cervical neoplasia. Cancer Epidemiology Biomarkers and Prevention. Cancer Epidemiol Biomarkers Prev; 2006;15:1274–80. [DOI] [PubMed] [Google Scholar]
- 36.Patel SJ, Mugo NR, Cohen CR, Ting J, Nguti R, Kwatampora J, et al. Multiple human papillomavirus infections and HIV seropositivity as risk factors for abnormal cervical cytology among female sex workers in Nairobi. International Journal of STD and AIDS. SAGE Publications Ltd; 2013;24:221–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Study data are not publicly available due to participant privacy but are available upon reasonable request from the corresponding author.


