Abstract
The inositol pyrophosphates (diphosphoinositol polyphosphates), which include 1-InsP7, 5-InsP7, and InsP8, are highly ‘energetic’ signalling molecules that play important roles in many cellular processes, particularly with regards to phosphate and bioenergetic homeostasis. Two classes of kinases synthesize the PP-InsPs: IP6Ks and PPIP5Ks. The significance of the IP6Ks - and their 5-InsP7 product - has been widely reported. However, relatively little is known about the biological significance of the PPIP5Ks. The purpose of this review is to provide an update on developments in our understanding of key features of the PPIP5Ks, which we believe strengthens the hypothesis that their catalytic activities serve important cellular functions. Central to this discussion is the recent discovery that the PPIP5K is a rare example of a single protein that catalyses a kinase/phosphatase futile cycle.
Introduction
Inositol pyrophosphates (PP-InsPs; Fig 1) are a specialized class of cell signaling molecules that boast a unique, functionally significant feature: the most crowded three-dimensional arrangement of phosphate groups that has been observed throughout Nature. Crammed around a six-carbon inositol scaffold are as many as seven or eight phosphates (for “InsP7” and “InsP8” respectively, see Fig 1), including diphosphate groups that are highly ‘energetic’ (Hand and Honek 2007; Shears 2015).
Figure 1. Inositol Pyrophosphate Turnover and the PPIP5Ks.
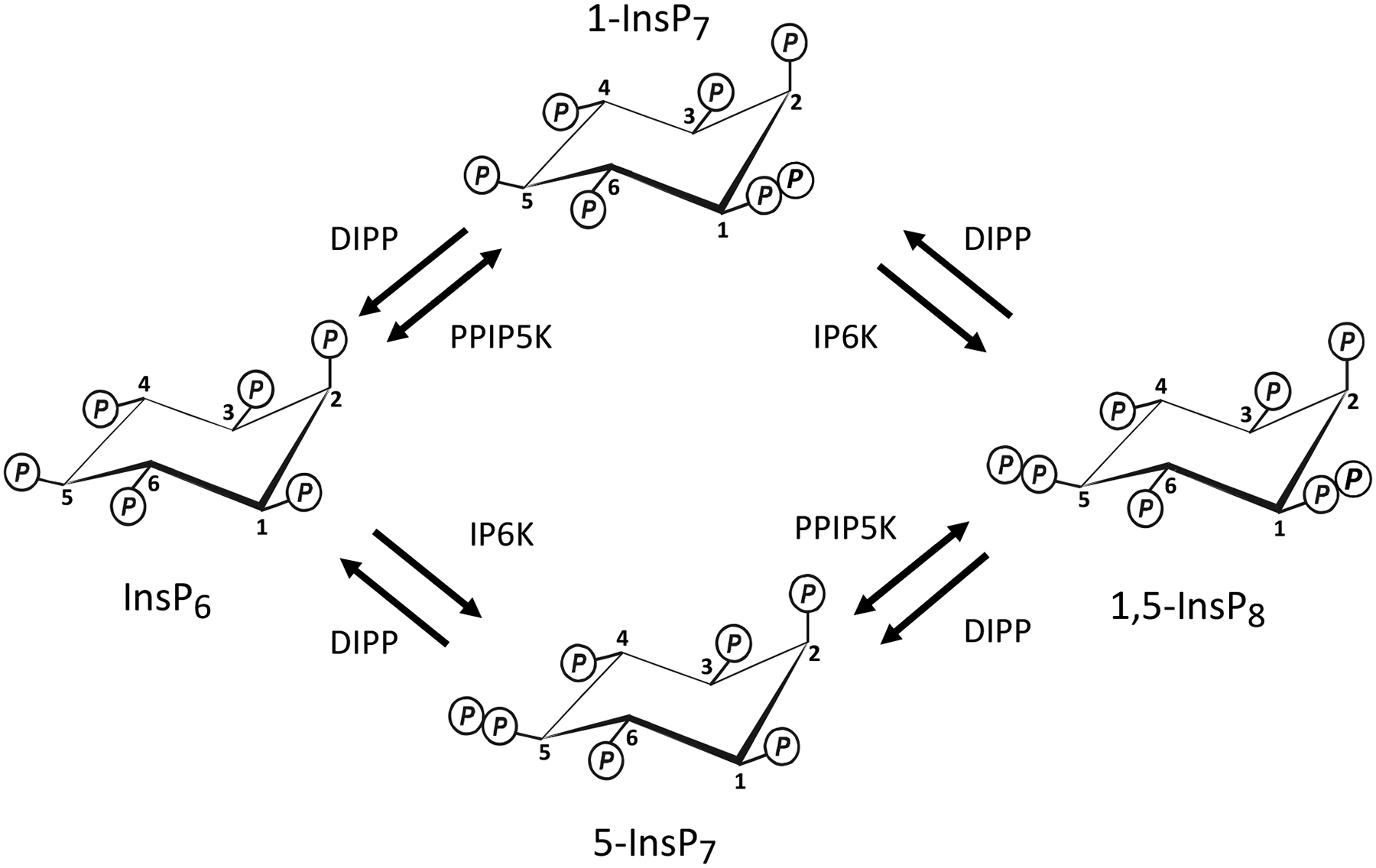
The schematic denotes the contributions to inositol pyrophosphate turnover of the 5-kinase activities of IP6Ks, the 1-kinase and 1-phosphatase activities of PPIP5Ks, and the DIPP phosphatases.
Yeasts and metazoan cells can phosphorylate InsP6 to InsP8 through two parallel pathways (Fig 1), which utilize two separate classes of enzymes to form diphosphate groups: the 5-kinases (the IP6Ks (E.C.2.7.4.21); Draskovic et al. 2008; Saiardi et al. 1999) and the 1-kinases (the PPIP5Ks (E.C. 2.7.4.24); Lin et al. 2009; Wang et al. 2012). Consequently, cells contain two InsP7 isomers, which are distinguished by whether the diphosphate is attached to either the 5- or 1-position on the inositol ring; InsP8 possesses both of these diphosphates (Fig 1). The IP6Ks can also add a diphosphate group to Ins(1,3,4,5,6)P5 (Draskovic et al. 2008; Saiardi et al. 2000), but with less positional specificity, since both the 1- and 5-positions can become diphosphorylated (Draskovic et al. 2008). These particular PP-InsPs are not well studied, and lie outside the main focus of this review which concerns the catalytic activities of the PPIP5Ks. A family of phosphatases - the DIPPs (E.C. 3.6.1.52) (Safrany et al. 1998) - hydrolyze both the 1- and 5-diphosphate groups.
In most studies that have analyzed cellular PP-InsP levels, the HPLC methodology that was used was unable to resolve 1-InsP7 from 5-InsP7, and so total InsP7 has been recorded. Its levels in mammalian cells have generally been reported to lie in the 1 – 2 μM range (see (Ingram, Safrany, and Barnes 2003; Wilson, Livermore, and Saiardi 2013)). As for InsP8, its concentration is typically around 10% that of total InsP7.
To gain insight into the relative contributions from the two InsP7 isomers, TNP (N2-(m-(trifluoromethyl)benzyl) N6-(p-nitrobenzyl)purine), a cell permeant pan-IP6K inhibitor, was added to intact cells to attenuate 5-InsP7 synthesis (Padmanabhan et al. 2009). This inhibitor reduced the size of the InsP7 peak by around 90% (Padmanabhan et al. 2009). Thus, it was concluded that 1-InsP7 accounts for no more than 10% of total InsP7. A more direct approach -the introduction of an HPLC technique which can resolve the two InsP7 isomers - has further reduced estimates of 1-InsP7 levels to less than 2% of total InsP7, at least in HCT116 cells (Gu et al. 2016).
The fact that 5-InsP7 is the most abundant of the PP-InsPs has translated into its preeminence in the PP-InsP literature. Thus, 5-InsP7 has been reported to regulate multiple, diverse biological processes: vesicle trafficking (Saiardi et al. 2000; Saiardi et al. 2002), apoptosis (Morrison et al. 2001; Nagata et al. 2005), autophagy (Nagata et al. 2010), repair of DNA repair by homologous recombination (Jadav et al. 2013; Luo et al. 2002), transcription of glycolytic genes (Szijgyarto et al. 2011), hemostasis (Ghosh et al. 2013), phagocytic and bactericidal activities of neutrophils (Prasad et al. 2011), craniofacial morphogenesis (Sarmah and Wente 2010), epigenetic modifications to chromatin (Burton et al. 2013), resistance to peroxide stress (Onnebo and Saiardi 2009), cell adhesion (Rao et al. 2015), dynein-driven vesicle transport (Chanduri et al. 2016) and exocytic insulin secretion (Illies et al. 2007). This extensive list of publications serves to underscore how little we understand concerning the specific significance of 1-InsP7 and InsP8. Against this backdrop, the purpose of this review is to draw attention to some important features of the PPIP5Ks, so as to make the case that their control over cellular levels of PP-InsPs must be functionally significant.
Structural Characterization of the PPIP5K kinase domain.
PPIP5Ks are distributed throughout eukaryotic kingdoms; the orthologs have various aliases: Asp1 in Schizosaccharomyces pombe, Vip1 in Saccharomyces cerevisiae, VIH in Arabidopsis thaliana. Yeasts and plants diverged from the animal kingdom 1.3 and 1.5 billion years ago, respectively (see www.timetree.org), so it is apparent that PPIP5Ks - and hence 1-InsP7 / InsP8 -are evolutionarily ancient. Two PPIP5K genes are expressed in both mammals and plants (Choi et al. 2007; Fridy et al. 2007; Laha et al. 2015; Mulugu et al. 2007). Both the type 1 and 2 forms of mammalian PPIP5Ks are relatively large proteins - approximately 160 and 140 kDa respectively; under some circumstances, they may even oligomerize into even larger complexes (Choi et al. 2007). The kinase domain is self-contained at the N-terminal one-third of these proteins (Fridy et al. 2007; Mulugu et al. 2007; Pohlmann et al. 2014; Wang et al. 2012). This certainly facilitated the expression and preparation of the kinase domain of human PPIP5K2 to the concentration and purity required to solve its structure (Wang et al. 2012).
This structural analysis was driven by the need to obtain an atomic level rationalization of some specialized features - for example, catalytic specificity: PPIP5Ks exhibit negligible activity towards 1-InsP7, Ins(1,3,4,5,6)P5 and 5-PP-Ins(1,3,4,6)P4 (Wang et al. 2012) which, among the naturally-occurring inositol phosphates, are those that are the most similar to 5-InsP7 and InsP6. Another catalytic accomplishment is that the active site accommodates sterically hindered and intensely electronegative substrates - and yet adds to those ligands another bulky and negatively charged phosphate group. Furthermore, the substrates are metabolized with high affinity: initial kinetic analyses determined that the Km values for InsP6 and 5-InsP7 are both in the 0.1 to 0.3 μM range (Choi et al. 2007; Fridy et al. 2007). Subsequent work derived an even lower Km value for 5-InsP7 (0.06 μM), perhaps in part reflecting technical improvements in the procedures for preparation of pure PP-InsPs (Weaver, Wang, and Shears 2013).
Our structural data derived from the PPIP5K2 kinase domain were obtained at a resolution of 1.8 Angstroms. The conclusions that we drew likely also apply to human PPIP5K1, since the two kinase domains share 84% sequence identity, and residues that are dissimilar are confined to the protein surface, distal to the active site (Wang et al. 2012). Our results established that ligand specificity is in part enforced by the geometry of the substrate-binding pocket, which comprises two near-parallel grooves that form a staggered “H”-shape. This architecture makes a perfectly-tailored aperture for accommodating six phosphate/pyrophosphate groups attached to an inositol ring in its chair conformation (Wang et al. 2012). Specificity within the catalytic pocket is additionally imposed by an array of Lys and Arg residues that make polar contacts with every phosphate group in both InsP6 and 5-InsP7; Mg2+ ions also contribute to specificity by electrostatic bridging between certain phosphate groups (Wang et al. 2012). In fact, both a Mg2+ ions and Lys214 of human PPIP5K2 specifically interact with the 5-β-phosphate in 5-InsP7, which of course is missing from InsP6, thereby accounting for the latter’s lower catalytic efficiency (kcat/Km = 7.6 × 104 M/s for InsP6; 2.2 × 106 M/s for 5-InsP7; Weaver, Wang, and Shears 2013). Indeed, most current evidence points to the phosphorylation of 5-InsP7 being the predominant activity of PPIP5Ks in vivo, while the phosphorylation of InsP6 is generally considered to be a relatively minor activity (Padmanabhan et al. 2009; Wang et al. 2012; Weaver, Wang, and Shears 2013). Bearing that in mind, and the relatively low levels of 1-InsP7 in cells, it is reasonable to propose that InsP8 may be the more physiologically-important product.
By comparing crystals with and without substrates, we established that their occupation of the catalytic pocket is accompanied by induced fit motion of side-chains of three active-site residues, namely, Arg262, Arg281 and Lys329; such conformational dynamics are recognized to help define ligand specificity, while also reducing the degree of free-energy of activation (Herschlag 1988). Were there instead to be backbone rearrangements, the energetic demand would have been higher (Wang et al. 2012).
As for understanding how the catalytic cycle moves from the substrate-bound ground state to the transition state, a crystal structure that was complexed with a MgF3− transition-state mimic revealed that conformational dynamics of both enzyme and substrate are involved (Wang et al. 2012). There are conformational changes in five amino-acid side chains in particular – Lys54, Arg213, Lys214, Arg262 and Lys329 – that advance the 1-alpha-phosphate of the substrate towards the gamma-phosphate of ATP. During this process, the 1-alpha-phosphate rotates almost 20°. That is, PPIP5Ks exploit a specific property of a phosphoester bond - its ability to rotate. Such a transition is not available for inositol phosphate kinases that phosphorylate the hydroxyl groups, which lack conformational flexibility.
Another feature of catalysis by PPIP5Ks is that an extensive negative charge balance by positively charged amino-acid residues facilitates a partly-associative, in-line phosphoryl transfer mechanism (Wang et al. 2012). That is, the new P-O bond can be formed by nucleophilic attack of the acceptor oxygen, before the original P-O bond with the donor oxygen is broken. Finally, in the active site of the kinase the inositol ring of InsP6 and 5-InsP7 is presented perpendicular to the plane of the nucleotide’s beta-phosphate, which avoids steric and electrostatic clashing between the nucleotide and the non-reacting oxygens on the 1-phosphate of the substrate (Wang et al. 2012). This innovative topology distinguishes PPIP5Ks from all of the other inositol phosphate kinases that phosphorylate hydroxyl groups (Wang et al. 2012).
Nevertheless, these structural data also begged some questions: ATP is enveloped between two sets of anti-parallel beta-sheets, so that less than 10% of the nucleotide is predicted to be solvent accessible. Thus, it can be anticipated that the enzyme may undergo considerable conformational changes when admitting ATP and releasing ADP. Unfortunately, the nature of these rearrangements is unknown, as the structure of the apo-enzyme has not yet been solved. A second issue is, perhaps, more subtle: considering how highly constraining is the structure of the substrate-binding pocket, that will only admit ligand presented in the appropriate orientation - i.e., a direct hit - could that limit the enzyme’s ability to attract ligand that rotates and diffuses randomly through the bulk phase? To illustrate the point about the need for a “direct hit”, consider the usual outcome of a classic carnival game as an analogy: the high failure rate when attempting to throw a table tennis ball into the narrow neck of a distant goldfish bowl.
Further work has uncovered a near-unique mechanism by which PPIP5Ks circumvent the possibility of such a “high failure rate”. Through X-ray analysis of several synthetic PP-InsP analogues in crystal complexes with the PPIP5K2 kinase domain, together with biochemical assays and mutagenesis, it has been shown that this class of enzyme utilizes a surface-mounted, ligand-binding site that is adjacent to the catalytic pocket (Wang et al. 2014). It has been concluded that this unusual ligand-binding site facilitates substrate capture from the bulk phase, prior to its transfer into the catalytic pocket; a “catch-and-pass” reaction mechanism (Wang et al. 2014).
Once again, a structural rationalization of one question raised another - and this one remains to be resolved: why does ligand occupation of the substrate-capture site stimulates an intrinsic, non-productive ATPase activity (Wang et al. 2014)? Intrinsic ATPase - reactivity of the gamma phosphate of ATP with water instead of substrate - appears to be an inherent property of most, if not all, kinases. However, the catalytic cycle of the kinase domain of PPIP5K is apparently unique in that the ATPase activity is stimulated by the substrate itself, specifically, during its occupation of the capture site (Wang et al. 2014; Weaver, Wang, and Shears 2013). It has been estimated that during the phosphorylation of 5-InsP7 by human PPIP5K2, an accompanying substrate-stimulated ATPase activity accounts for 17% of the total ATP that is consumed (Weaver, Wang, and Shears 2013). The proportion is much higher during InsP6 phosphorylation: 50% (Weaver, Wang, and Shears 2013). It seems reasonable to speculate that this ATP hydrolysis is not simply ‘wasted’, but instead has some significance for the catalytic cycle. For example, transfer of ligand from the capture site to the catalytic pocket involves not only a lateral migration of 7 Angstroms, but also a ring flip, and a 100 degree rotation (Wang et al. 2014). Such molecular gymnastics are an appropriate candidate for assistance from the free energy of ATP hydrolysis.
Beyond the Kinase Activities: The Phosphatase Domain.
Off-switches for cell-signaling are as vitally important as the on-switches. Until recently, the off-switches for the PP-InsPs were thought to be the prerogative of the DIPP family (Kilari et al. 2013; Safrany et al. 1998). Indeed, the DIPPS seem to have one feature that makes them especially well suited to this role: highly activate rates of PP-InsP dephosphorylation (Safrany et al. 1998). Nevertheless, their lack of specificity for a particular PP-InsP is not consistent with a need to regulate the levels of an individual PP-InsP that may have distinct functions.
Against that puzzling backdrop, one recent finding brings a new dimension to our understanding of the pathways of PP-InsP turnover: PPIP5Ks host a phosphatase domain (Fig. 2). This remarkable feature was originally recognized by John York’s laboratory (Fridy et al. 2007; Mulugu et al. 2007). The amino-acid sequence of the active site bears similarities to the relatively non-specific phytase subgroup of histidine acid phosphatases (Fridy et al. 2007; Mulugu et al. 2007). That observation belies the actual positional specificity of the PPIP5K phosphatase domain: only the 1-beta-phosphate from either 1-InsP7 or 1,5-InsP8 is hydrolyzed (Wang et al. 2015). In other words, the PPIP5K family catalyzes 1-kinase/1-phosphatase futile cycles. It has been shown that InsP8 is the preferred of the two substrates; InsP8 is dephosphorylated about 10-fold faster than is 1-InsP7 (Wang et al. 2015). Since it is also the case that the kinase domain prefers 5-InsP7 over InsP6 (see above), we can conclude that, in vivo, flux through the 5-InsP7 / InsP8 cycle should exceed that through the InsP6 / 1-InsP7 cycle (see Fig 1). It should be interesting to establish from a structural analysis how this exquisite specificity is enforced; data obtained from Asp1 are particularly illuminating - its phosphatase domain does not hydrolyze either InsP6, or the 2-, 3-, 4-, 5-, or 6-isomers of InsP7 (Wang et al. 2015).
Figure 2. Domain graphic for PPIP5K1 and PPIP5K2.

The figure depicts a domain graphic for human PPIP5K1 (BC057395.1) and human PPIP5K2 (XM_005271938). The so-called CPA (Contains Penta-Arginine) region in PPIP5K2 is rigorously conserved throughout metazoans; the eponymous pent-arginine is a functional nuclear localization sequence (Yong et al. 2015). However, transcripts containing this CPA domain (e.g., XM_005277536.1) are relatively poorly expressed in human cells (or at least those that we have examined), possibly as a consequence of rare, alternate splicing. In the figure, the amino acid residues defining each domain in PPIP5K1 are numbered as in a previous study, which also defined the intrinsically disordered domain (IDR) (Machkalyan et al. 2016). These boundaries were matched to those of the corresponding domains in PPIP5K2 by sequence alignments using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The aligned IDR boundaries in PPIP5K2 are consistent with those independently predicted from the PSIPRED Protein Sequence Analysis Workbench (http://bioinf.cs.ucl.ac.uk/psipred/).The % sequence identities across each of the three domains are also indicated.
It is very rare that a ‘futile’ kinase/phosphatase cycle can be catalyzed by a single protein at two separate catalytic sites. Understanding what are the particular advantages of such kinase/phosphatase bifunctionality is a topic of considerable general interest (Dasgupta et al. 2014). In the context of cell signaling, a substrate cycle that is driven by separate proteins is susceptible to incoherent output within a particular tissue, due to stochastic cell-to-cell variation in the expression of each of the individual proteins; such random variability in protein expression is even observed within populations of genetically identical cells cultured under highly similar conditions (Atay and Skotheim 2014). This potential for incoherent output is avoided when there is co-expression of the kinase and phosphatase - such as is inevitable when the two proteins are fused together (Dasgupta et al. 2014). Of course, for the maintenance of coherent output to be a benefit to arise from bifunctionality of the PPIP5Ks, their regulation of 1-InsP7 / InsP8 levels must fulfil some important biological functions.
Another potential advantage of bifunctionality could be its offering an opportunity for coordinated, reciprocal regulation of the competing kinase/phosphatase activities in PPIP5Ks; this could form the basis for a sensitive mechanism of control over 1-InsP7 / InsP8 synthesis and metabolism. However, as yet, this idea is purely speculative; no such regulatory process has been demonstrated for any metazoan PPIP5K. On the other hand, there has been a description of a mechanism for regulating the phosphatase activity of Asp1. This protein hosts an Fe-S cluster with a [2Fe-2S]2+ arrangement that inhibits phosphatase activity and thereby promotes net kinase activity (Wang et al. 2015). This is an unprecedented role for an Fe-S cluster: despite their being widely distributed throughout Nature, no others have previously been found to have a direct catalytic influence upon a phosphatase activity. Data obtained by electron paramagnetic resonance indicate the cluster is not redox active (Wang et al. 2015). It has been proposed that the role of the cluster in Asp1 may be to couple PP-InsP turnover to Fe availability (Wang et al. 2015). Indeed, it is intriguing that the environmentally controlled switch to an alternative morphology for S. pombe - pseudohyphal invasive growth - is influenced by both Fe (Prevorovsky et al. 2009) and the Asp1 phosphatase domain (Pohlmann et al. 2014). In any case, the regulatory impact of altered [2Fe-2S]2+ cluster assembly upon PP-InsP metabolism might be expected to be a relatively long-term adaptive process, since the mechanism by which the [2Fe-2S]2+ cluster inhibits phosphatase activity appears not to be readily reversible, at least in vitro (Wang et al. 2015).
It remains to be established whether or not there are Fe-S clusters in mammalian PPIP5Ks. Reliable predictions have not been obtained from multiple sequence alignments, in part because Cys-based consensus motifs for [2Fe-2S] centers are so diverse (Wang et al. 2015). Furthermore, towards the C-terminus of Asp1, where the Fe-S cluster is most likely to be hosted, the amino-acid sequence diverges substantially from those in PPIP5K1 and PPIP5K2, which in turn diverge considerably from each other at their own C-termini (Wang et al. 2015). In fact, even the identity of the Cys residues that co-ordinate the Fe-S cluster in Asp1 has not yet been unequivocally established (Wang et al. 2015). A systematic approach (individual mutation of the twelve candidate Cys residues to Ser) revealed six - residues 607, 663, 864, 868, 879 and 905 - that each led to a reduction in the Fe content of the Asp1 phosphatase domain (Wang et al. 2015). However, no more than four Cys residues are required to ligate a [2Fe-2S]2+ cluster (Beinert, Holm, and Munck 1997; Johnson and Smith 2005). Thus, these mutagenic data raise the possibility that two of these six mutations alter protein structure, and thereby indirectly impact Fe-S cluster assembly. Another possibility to consider is that there are alternate cluster ligation states. This lack of clarity does not help us predict which Cys residues in mammalian PPIP5Ks might co-ordinate any Fe-S cluster. A final confounding issue is that the biophysical techniques that can unequivocally detect Fe-S clusters require considerable (i.e. mg) quantities of highly-pure enzyme; it has not yet proved possible to satisfy that requirement, due to difficulties expressing and purifying such large proteins that are also rather labile (Choi et al. 2007).
Beyond the Kinase Activities: Other Motifs and Domains in the PPIP5Ks
Within the phosphatase domain of the PPIP5Ks there is a cryptic PH domain, and it has especially high affinity for PtdIns(3,4,5)P3 (Gokhale et al. 2013; Gokhale, Zaremba, and Shears 2011). In some cells, stimulus-dependent activation of PtdIns(3,4,5)P3 synthesis has been associated with a degree of relocalization of PPIP5K1 from the cytoplasm to the plasma membrane (Gokhale et al. 2013; Gokhale, Zaremba, and Shears 2011). We have proposed that the significance of PPIP5K translocation lies in it regulating the activity of the protein kinase AKT. The latter is a classic example of a PH-domain containing protein that is recruited to the plasma membrane and activated by binding to PtdIns(3,4,5)P3 (Cantley 2002). Both InsP6 and 5-InsP7 are competitive inhibitors of AKT activation, in large part by competing with PtdIns(3,4,5)P3 binding (Chakraborty et al. 2010; Luo et al. 2003). We (Gokhale et al. 2013) have suggested this to be a necessary process for preventing any non-sustained, stochastic increases in PtdIns(3,4,5)P3 from inappropriately recruiting and activating AKT and other signaling proteins with PH-domains. The aberrant PtdIns(3,4,5)P3-signaling that underlies much of cancer biology is a good illustration of why PH-domain recruitment is so carefully regulated (Cantley 2002).
We have proposed that translocation of PPIP5K1 to the plasma membrane will locally deplete levels of InsP6 and 5-InsP7; moreover, the products of this catalytic activity, 1-InsP7 and InsP8, have 10 to 20-fold lower affinity for AKT (Gokhale et al. 2013). That is, PPIP5K kinase activity will attenuate the inositolphosphate-based inhibition of PtdIns(3,4,5)P3 signaling through PH domains. Thus, optimum signaling by the PI3K cascade may depend upon coincidence detection: the translocation of both PPIP5K and a PH-domain protein. This raises a rather ironic conclusion (at odds with the tone of the rest of this review) that one aspect of PPIP5K functionality rests on the relative biological inactivity of InsP8 and 1-InsP7, at least as PH-domain ligands.
Another functionally significant region that is present in both PPIP5Ks is an unusually long, intrinsically disordered region (IRD) that lies at their C-termini (Machkalyan et al. 2016). Such domains are typically considered as molecular scaffolds for promoting binding of other proteins (Machkalyan et al. 2016). A proteomic-based search for protein partners of PPIP5K1 yielded many that participate in trafficking of intracellular vesicles, particularly those in the exocyst complex that tether post-Golgi vesicles to plasma membranes (Machkalyan et al. 2016). Additionally, there is evidence that PPIP5K1 interacts with proteins that participate in lipid metabolism and cytoskeletal organization (Machkalyan et al. 2016). Thus, it appears likely that further delving into scaffolding functions for PPIP5Ks will be a productive future research direction.
Within the IDR of PPIP5K2 (but not PPIP5K1) is a penta-arginine nuclear localization sequence (NLS) (Yong et al. 2015). The location of this NLS sequence is unlikely to be coincidental; such sequences typically reside in relatively unstructured regions of proteins (Bauer, Doetsch, and Corbett 2015). It is through interactions of the NLS with specialized molecular chaperones - karyopherins - that the host protein gains access to the nucleus (Bauer, Doetsch, and Corbett 2015). This NLS lies in a sequence of 63 amino-acid residues in PPIP5K2 that we have named as a CPA (Contains Penta-Arginine) region (Fig 2). This domain is rigorously conserved throughout metazoans, which presumably testifies to its importance (Yong et al. 2015). However, mRNA transcripts containing this CPA domain (e.g., XM_005277536.1) are relatively poorly expressed in human cells (or at least those that we have examined), possibly as a consequence of rare, alternate splicing. Nevertheless, this NLS is functional: we expressed a CPA-containing version of GFP-tagged, human PPIP5K2 in HEK cells, and a distinct nuclear pool of this protein was observed (at a concentration that was approximately 30% of that found in the cytoplasm (Yong et al. 2015)). Mutagenesis of the NLS (RRRRR to RAAAR) reduced the degree of nuclear localization almost 4-fold (Yong et al. 2015).
A Ser residue that lies immediately to the C-terminus of the penta-arginine is phosphorylated by an as yet unknown protein kinase (Yong et al. 2015); analysis of consensus motifs (Phosida database; http://141.61.102.18/phosida/index.aspx) indicate that candidates include PKA, CaMK2 and AKT. The mutation of this Ser residue to Ala increases the nuclear concentration of the PPIP5K2 by 60% (Yong et al. 2015). These data indicate that phosphorylation of the native Ser inhibits the functionality of the NLS. Such a regulatory process presumably supervises a specific, nuclear function for 1-InsP7 and/or InsP8. That conclusion indicates it would be valuable to systematically investigate which cell types might express isoform(s) of PPIP5K2 that contain the CPA domain.
Potential Signaling functions for 1-InsP7 and InsP8
While the PPIP5Ks synthesize 1-InsP7 and InsP8, the latter is at least 5 to 10-fold more abundant (see above). Despite 1-InsP7 earning a booby-prize among PP-InsPs for its low relative abundance, evidence of its biological significance accompanied the initial description of a protein responsible for 1-kinase activity - the Vip1 ortholog in S. cerevisiae (Lee et al. 2007; Mulugu et al. 2007); depriving this yeast of inorganic phosphate was reported to substantially activate 1-InsP7 synthesis, which was proposed to drive adaptive transcriptional responses (Lee et al. 2007). On the other hand, subsequent studies have described the opposite response of InsP7 levels to phosphate starvation: a decrease in its concentration (Lonetti et al. 2011; Wild et al. 2016). An explanation for these inconsistent results has not been forthcoming, but it has previously been noted that phosphate deprivation is associated with a decline in cellular ATP levels, upon which all PP-InsP synthesis depends (see Saiardi 2012; Shears 2015).
1-InsP7 has also been proposed to mediate certain innate immune responses - specifically, RIG-1 dependent transcription of interferon-β in response to viral invasion (Pulloor et al. 2014). Evidence for a mechanism underlying this response was obtained in vitro: 1-InsP7 enhances the degree of phosphorylation of IRF3, which promotes its dimerization and entry into the nucleus, where it stimulates interferon-β transcription (Pulloor et al. 2014). In those experiments, 5-InsP7 was found to be ineffective, while 1-InsP7 acted more potently than did InsP8. Again, interpreting whether 1-InsP7 over InsP8 is the more active molecule in vivo requires us to take their relative cellular levels into account.
Considering the apparent role of PPIP5Ks in innate immunity in mammalian cells (see above) it is intriguing that the VIH2 ortholog in A. thaliana also regulates innate immune responses (Laha et al. 2015) - and in this case it is InsP8 that is suggested to be the functional signal. In this particular plant, cellular levels of InsP8 are elevated by methyl-jasmonate, one of a group of plant hormones that helps protect plants from herbivorous insects and necrotrophic fungi (Laha et al. 2015). This protective response is impaired in lines of A. thaliana in which the kinase domain of VIH2 is disrupted by insertional mutagenesis (Laha et al. 2015). It has been speculated that InsP8 may augment transcriptional activation by methyl-jasmonate (Laha et al. 2015); we await further developments in this area with interest.
A typical characteristic of an intracellular signal is stimulus-dependent changes in its cellular levels. The only published report that 1-InsP7 levels are regulated is the aforementioned study with phosphate-deprived yeast, which others have failed to reproduce. On the other hand, stimulus-dependent regulation of InsP8 concentration has been demonstrated in several studies. For example, hyperosmotic stress substantially elevates InsP8 levels (Choi et al. 2008; Padmanabhan et al. 2009; Pesesse et al. 2004). This response has the hallmark of an adaptive response to an environmental stress, although as yet no functional significance has been attributed to it. Thermal challenges - both heat and cold - also stimulate InsP8 synthesis (Choi, Mollapour, and Shears 2005), although it is unclear if this is a regulated stress response, or merely differential effects of temperature changes upon the competing activities of PP-InsP kinases and phosphatases. Finally, InsP8 has been implicated as being a sensor of bioenergetic health in mammalian cells (Choi et al. 2008); levels of this PP-InsP decrease following relatively mild bioenergetic challenges, such as those known to elevate AMP, although AMP-dependent protein kinase appears not to play a role in regulating InsP8 turnover (Choi et al. 2008). To date, a mechanistic basis for this phenomenon has not been derived, neither has a functional consequence been established. Nevertheless, these data indicate that InsP8 serves some important homeostatic function(s).
Future Directions
Genetically-modified animal models have frequently illuminated biological functions of intracellular signaling cascades. At the time of writing, there are no published phenotypes for knock-out of either PPIP5K1 or PPIP5K2 in any animal model. Such information is likely to be available soon, since a PPIP5K2 knock-out mouse strain has been produced, according to the International Mouse Phenotyping Consortium (http://www.mousephenotype.org). The relative ease with which CRISPR can be used to genetically modify cells in culture can also accelerate molecular studies into PP-InsP biology.
Nevertheless, although it is incontestable that genetic approaches have substantially contributed to our understanding of the biological roles of inositol phosphate kinase (e.g. Chakraborty et al. 2010; Ghosh et al. 2013; Prasad et al. 2011; Scoumanne et al., 2016; Szijgyarto et al., 2011), such experiments do have certain drawbacks: the long term nature of such experiments introduces the possibility that the nature of a particular phenotype arising from gene deletion may be confounded by secondary genetic changes. Thus, pharmacological approaches are often a useful complement to genetic work. For example, it would be useful to have cell-permeable inhibitors of the kinase and phosphatase activities of the PPIP5Ks. To this end, we recently began a program to develop inhibitors of the kinase activity of PPIP5Ks. We have screened the human PPIP5K2 kinase domain against a library of almost 5000 molecules that have been curated because of their known - or predicted - actions as inhibitors of ATP-binding to protein kinases (Baughman et al. 2016). Some promising lead molecules have been identified (Baughman et al. 2016) which we intend to develop further.
It would be useful to be able to specifically modify levels of a particular PP-InsP in intact cells without genetic modification of their metabolic pathways. To this end, methods are also being introduced that circumvent the impermeability to the plasma membrane of highly charged molecules such as the PP-InsPs. For example, guanidinium-rich molecular transporters have been developed (Pavlovic et al. 2016). This method was recently used to deliver caged 5-InsP7 into HeLa cells; the 5-InsP7 was subsequently released upon cell exposure to UV-light. Furthermore, metabolically stable PP-InsP analogues are now available (Riley et al. 2012; Wu et al. 2013).
As we learn more about the unexpected and sometimes complex features of PPIP5Ks and their orthologs, it seems entirely reasonable to anticipate that the major product of these enzymes - InsP8 - is serving important cellular roles. Recent developments (see above) make us optimistic that the shroud of uncertainty covering the biological significance of InsP8 will shortly be removed.
Acknowledgment
Work in the authors’ laboratory was supported by the Intramural Research Program of the NIH / National Institute of Environmental Health Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Atay O, Skotheim JM. Modularity and predictability in cell signaling and decision making. Mol Biol Cell. 2014;25:3445–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer NC, Doetsch PW, Corbett AH. Mechanisms Regulating Protein Localization. Traffic. 2015;16:1039–61 [DOI] [PubMed] [Google Scholar]
- Baughman BM, Wang H, An Y, Kireev D, Stashko M, Jessen HJ et al. A High-Throughput Screening-Compatible Strategy for the Identification of Inositol Pyrophosphate Kinase Inhibitors. PLoS One. 2016; in press [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Beinert H, Holm RH, Munck E. Iron-sulfur clusters: nature’s modular, multipurpose structures. Science. 1997;277:653–9 [DOI] [PubMed] [Google Scholar]
- Burton A, Azevedo C, Andreassi C, Riccio A, Saiardi A. Inositol pyrophosphates regulate JMJD2C-dependent histone demethylation. Proc Natl Acad Sci U S A. 2013;110:18970–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7 [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR et al. Inositol pyrophosphates inhibit akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanduri M, Rai A, Malla AB, Wu M, Fiedler D, Mallik R et al. Inositol hexakisphosphate kinase 1 (IP6K1) activity is required for cytoplasmic dynein-driven transport. Biochem J. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Williams J, Cho J, Falck JR, Shears SB. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem. 2007;282:30763–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Mollapour E, Choi JH, Shears SB. Cellular Energetic Status Supervises the Synthesis of Bis-Diphosphoinositol Tetrakisphosphate Independently of AMP-Activated Protein Kinase. Mol Pharmacol. 2008;74:527–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Mollapour E, Shears SB. Signal transduction during environmental stress: InsP8 operates within highly restricted contexts. Cell Signal. 2005;17:1533–41 [DOI] [PubMed] [Google Scholar]
- Dasgupta T, Croll DH, Owen JA, Vander Heiden MG, Locasale JW, Alon U et al. A fundamental trade-off in covalent switching and its circumvention by enzyme bifunctionality in glucose homeostasis. J Biol Chem. 2014;289:13010–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draskovic P, Saiardi A, Bhandari R, Burton A, Ilc G, Kovacevic M et al. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem Biol. 2008;15:274–86 [DOI] [PubMed] [Google Scholar]
- Fridy PC, Otto JC, Dollins DE, York JD. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J Biol Chem. 2007;282:30754–62 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Shukla D, Suman K, Lakshmi BJ, Manorama R, Kumar S et al. Inositol hexakisphosphate kinase 1 maintains hemostasis in mice by regulating platelet polyphosphate levels. Blood. 2013;122:1478–86 [DOI] [PubMed] [Google Scholar]
- Gokhale NA, Zaremba A, Janoshazi AK, Weaver JD, Shears SB. PPIP5K1 Modulates Ligand Competition Between Diphosphoinositol Polyphosphates and PtdIns(3,4,5)P3 for Polyphosphoinositide-Binding Domains. Biochem J. 2013;453:413–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NA, Zaremba A, Shears SB. Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem J. 2011;434:415–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Wilson MSC, Jessen HJ, Saiardi A, Shears SB. Inositol Pyrophosphate Profiling of two HCT116 Cell Lines Uncovers Variation in InsP8 Levels. submitted. PLoS One 2016; in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand CE, Honek JF. Phosphate transfer from inositol pyrophosphates InsP5PP and InsP4(PP)2: a semi-empirical investigation. Bioorg Med Chem Lett. 2007;17:183–8 [DOI] [PubMed] [Google Scholar]
- Herschlag D The role of induced fit and conformational changes in specificity and catalysis. Bioorganic Chemistry. 1988;16:62–96 [Google Scholar]
- Illies C, Gromada J, Fiume R, Leibiger B, Yu J, Juhl K et al. Inositol pyrophosphates determine exocytic capacity. Science. 2007;318:1299–302 [DOI] [PubMed] [Google Scholar]
- Ingram SW, Safrany ST, Barnes LD. Disruption and overexpression of the Schizosaccharomyces pombe aps1 gene and the effects on growth rate, morphology, and intracellular diadenosine 5’, 5”’-P1, P5-pentaphosphate and diphosphoinositol polyphosphate concentrations. Biochem J. 2003;369:519–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadav RS, Chanduri MV, Sengupta S, Bhandari R. Inositol Pyrophosphate Synthesis by Inositol Hexakisphosphate Kinase 1 is Required for Homologous Recombination Repair. J Biol Chem. 2013;288:3312–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MK, Smith AD: Iron-sulphur proteins. In Encyclopedia of Inorganic Chemistry. Edited by King RB. Chichester: John Wiley & Sons; 2005:2589–2619. [Google Scholar]
- Kilari RS, Weaver JD, Shears SB, Safrany ST. Understanding inositol pyrophosphate metabolism and function: Kinetic characterization of the DIPPs. FEBS Lett. 2013;587:3464–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha D, Johnen P, Azevedo C, Dynowski M, Weiss M, Capolicchio S et al. VIH2 Regulates the Synthesis of Inositol Pyrophosphate InsP8 and Jasmonate-Dependent Defenses in Arabidopsis. Plant Cell. 2015;27:1082–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Fridy PC, Ribeiro AA, Choi JH, Barma DK, Vogel G et al. Structural analysis and detection of biological inositol pyrophosphates reveals that the VIP/PPIP5K family are 1/3-kinases. J Biol Chem. 2009;284:1863–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetti A, Szijgyarto Z, Bosch D, Loss O, Azevedo C, Saiardi A. Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J Biol Chem. 2011;286:31966–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K et al. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–72 [DOI] [PubMed] [Google Scholar]
- Luo HR, Saiardi A, Yu H, Nagata E, Ye K, Snyder SH. Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase C1 mutant yeast. Biochemistry. 2002;41:2509–15 [DOI] [PubMed] [Google Scholar]
- Machkalyan G, Trieu P, Petrin D, Hebert TE, Miller GJ. PPIP5K1 interacts with the exocyst complex through a C-terminal intrinsically disordered domain and regulates cell motility. Cell Signal. 2016;28:401–11 [DOI] [PubMed] [Google Scholar]
- Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-b in ovarian carcinoma cells. J Biol Chem. 2001;276:24965–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE et al. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316:106–9 [DOI] [PubMed] [Google Scholar]
- Nagata E, Luo HR, Saiardi A, Bae BI, Suzuki N, Snyder SH. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J Biol Chem. 2005;280:1634–40 [DOI] [PubMed] [Google Scholar]
- Nagata E, Saiardi A, Tsukamoto H, Satoh T, Itoh Y, Itoh J et al. Inositol hexakisphosphate kinases promote autophagy. Int J Biochem Cell Biol. 2010;42:2065–71 [DOI] [PubMed] [Google Scholar]
- Onnebo SM, Saiardi A. Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem J. 2009;423:109–18 [DOI] [PubMed] [Google Scholar]
- Padmanabhan U, Dollins DE, Fridy PC, York JD, Downes CP. Characterization of a selective inhibitor of inositol hexakisphosphate kinases: Use in defining biological roles and metabolic relationships of inositol pyrophosphates. J Biol Chem. 2009;284:10571–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic I, Thakor DT, Vargas JR, McKinlay CJ, Hauke S, Anstaett P et al. Cellular delivery and photochemical release of a caged inositol-pyrophosphate induces PH-domain translocation in cellulo. Nat Commun. 2016;7:10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesesse X, Choi K, Zhang T, Shears SB. Signalling by higher inositolpolyphosphates: Synthesis of bis-diphosphoinositol tetrakisphosphate (“InsP8”) is selectively activated by hyperosmotic stress. J Biol Chem. 2004;279:43378–81 [DOI] [PubMed] [Google Scholar]
- Pohlmann J, Risse C, Seidel C, Pohlmann T, Jakopec V, Walla E et al. The vip1 inositol polyphosphate kinase family regulates polarized growth and modulates the microtubule cytoskeleton in fungi. PLoS Genet. 2014;10:e1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Jia Y, Chakraborty A, Li Y, Jain SK, Zhong J et al. Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat Immunol. 2011;12:752–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevorovsky M, Stanurova J, Puta F, Folk P. High environmental iron concentrations stimulate adhesion and invasive growth of Schizosaccharomyces pombe. FEMS Microbiol Lett. 2009;293:130–4 [DOI] [PubMed] [Google Scholar]
- Pulloor NK, Nair S, Kostic AD, Bist P, Weaver JD, Tyagi R et al. Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response. PLoS Pathog. 2014;10:e1003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, Xu J, Fu C, Cha JY, Gadalla MM, Xu R et al. Inositol pyrophosphates promote tumor growth and metastasis by antagonizing liver kinase B1. Proc Natl Acad Sci U S A. 2015;112:1773–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley AM, Wang H, Weaver JD, Shears SB, Potter BVL. First synthetic analogues of diphosphoinositol polyphosphates: interaction with PPIP5 kinase. Chem Commun. 2012;48:11292–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safrany ST, Caffrey JJ, Yang X, Bembenek ME, Moyer MB, Burkhart WA et al. A novel context for the “MutT” module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998;17:6599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A How inositol pyrophosphates control cellular phosphate homeostasis? Adv Biol Regul. 2012;52:351–9 [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The Inositol Hexakisphosphate Kinase Family: Catalytic Flexibility, and Function in Yeast Vacuole Biogenesis. J Biol Chem. 2000;275:24686–92 [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman A, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–6 [DOI] [PubMed] [Google Scholar]
- Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Nat Acad Sci USA. 2002;99:14206–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah B, Wente SR. Inositol hexakisphosphate kinase-2 acts as an effector of the vertebrate Hedgehog pathway. Proc Natl Acad Sci U S A. 2010;107:19921–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoumanne A, Molina-Ortiz P, Monteyne D, Perez-Morga D, Erneux C, Schurmans S. Specific expression and function of inositol 1,4,5-trisphosphate 3-kinase C (ITPKC) in wild type and knock-out mice. Adv Biol Regul. 2016. 62:1–10. [DOI] [PubMed] [Google Scholar]
- Shears SB. Inositol pyrophosphates: why so many phosphates? Adv Biol Regul. 2015. 57:203–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334:802–5 [DOI] [PubMed] [Google Scholar]
- Wang H, Falck JR, Hall TM, Shears SB. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol. 2012;8:111–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Godage HY, Riley AM, Weaver JD, Shears SB, Potter BVL. Synthetic Inositol Phosphate Analogs Reveal that PPIP5K2 Has a Surface-Mounted Substrate Capture Site that Is a Target for Drug Discovery. Chemistry & Biology. 2014;21:689–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nair VS, Holland AA, Capolicchio S, Jessen HJ, Johnson MK et al. Asp1 from Schizosaccharomyces pombe Binds a [2Fe-2S](2+) Cluster Which Inhibits Inositol Pyrophosphate 1-Phosphatase Activity. Biochemistry. 2015;54:6462–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver JD, Wang H, Shears SB. The kinetic properties of a human PPIP5K reveal that its kinase activities are protected against the consequences of a deteriorating cellular bioenergetic environment. Biosci Rep. 2013;33:228–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild R, Gerasimaite R, Jung JY, Truffault V, Pavlovic I, Schmidt A et al. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science. 2016;352:986–90 [DOI] [PubMed] [Google Scholar]
- Wilson MS, Livermore TM, Saiardi A. Inositol pyrophosphates: between signalling and metabolism. Biochem J. 2013;452:369–79 [DOI] [PubMed] [Google Scholar]
- Wu M, Dul BE, Trevisan AJ, Fiedler D. Synthesis and characterization of non-hydrolysable diphosphoinositol polyphosphate second messengers. Chem Sci. 2013;4:405–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong ST, Nguyen HN, Choi JH, Bortner CD, Williams J, Pulloor NK et al. Identification of a functional nuclear translocation sequence in hPPIP5K2. BMC Cell Biol. 2015;16:17. [DOI] [PMC free article] [PubMed] [Google Scholar]


