Abstract
Background
The active metabolite of 5-Fluorouracil (5FU), used in the treatment of several types of cancer, acts by inhibiting the thymidylate synthase encoded by the TYMS gene, which catalyzes the rate-limiting step in DNA replication. The major failure of 5FU-based cancer therapy is the development of drug resistance. High levels of TYMS-encoded protein in cancerous tissues are predictive of poor response to 5FU treatment. Expression of TYMS is regulated by various mechanisms, including involving non-coding RNAs, both miRNAs and long non-coding RNAs (lncRNAs).
Aim
To delineate the miRNAs and lncRNAs network regulating the level of TYMS-encoded protein.
Main body
Several miRNAs targeting TYMS mRNA have been identified in colon cancers, the levels of which can be regulated to varying degrees by lncRNAs. Due to their regulation by the MALAT1 lncRNA, these miRNAs can be divided into three groups: (1) miR-197-3p, miR-203a-3p, miR-375-3p which are downregulated by MALAT1 as confirmed experimentally and the levels of these miRNAs are actually reduced in colon and gastric cancers; (2) miR-140-3p, miR-330-3p that could potentially interact with MALAT1, but not yet supported by experimental results; (3) miR-192-5p, miR-215-5p whose seed sequences do not recognize complementary response elements within MALAT1. Considering the putative MALAT1-miRNAs interaction network, attention is drawn to the potential positive feedback loop causing increased expression of MALAT1 in colon cancer and hepatocellular carcinoma, where YAP1 acts as a transcriptional co-factor which, by binding to the TCF4 transcription factor/ β-catenin complex, may increase the activation of the MALAT1 gene whereas the MALAT1 lncRNA can inhibit miR-375-3p which in turn targets YAP1 mRNA.
Conclusion
The network of non-coding RNAs may reduce the sensitivity of cancer cells to 5FU treatment by upregulating the level of thymidylate synthase.
Keywords: Micro RNA, miRNAs, ceRNA network, 5-FU resistance, Chemoresistance, Colorectal cancer, Capecitabine, Hippo-YAP, Wnt beta-catenin
Introduction
5-Fluorouracil (5FU) is a uracil analogue with a fluorine atom at the C-5 position of the pyrimidine (Fig. 1) and belongs to the class of chemotherapeutic agents known as the fluoropyrimidines. 5FU is used to treat several types of cancer. 5FU is used intravenously in palliative treatment of colorectal cancers (CRCs), breast cancers, gastric cancers, pancreatic cancers (Dean and Kane 2016a). Another fluoropyrimidine, namely capecitabine (trade name Xeloda), was rationally designed to mimic the continuous infusion of 5FU (Hoff et al. 2001) and is an oral 5FU prodrug used in the treatment of metastatic colon and breast cancers (Dean and Kane 2016b), including metastasis of HER2-positive breast cancer to the brain (Franchino et al. 2018). Tegafur, another oral prodrug of 5FU, is involved in chemotherapy for solid tumors including advanced non-small-cell lung cancer (NSCLC), CRC, gastric cancer, breast cancer, pancreatic cancer, cervical cancer (Okamoto and Fukuoka 2009). However, the therapeutic efficacy of tegafur is highly dependent on the variant of the CYP2A6 enzyme that bioactivates this prodrug (Tanner and Tyndale 2017). The anti-tumor activities of 5FU and capecitabine are exerted by the irreversible inhibition of thymidylate synthase (TS), a cytosolic enzyme encoded by the TYMS (Thymidylate Synthetase) gene (Fig. 1). TS catalyzes the thymidylate synthesis reaction, which is the sole source of de novo thymidylate and is the rate-limiting step in DNA replication (Longley et al. 2003). In addition, 5FU acts as an antimetabolite to replace thymine and uracil, and thus by fraudulently incorporating fluoropyrimidine metabolites into synthesized nucleic acids, it damages DNA and RNA (Blondy et al. 2020; Longley et al. 2003).
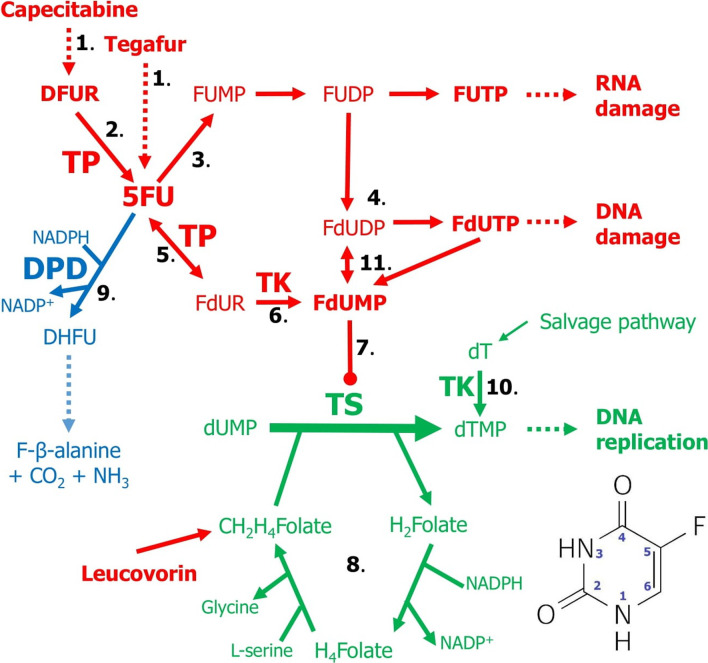
Fig. 1 .
5-fluorouracil (5FU) metabolism pathways. The structure and atom numbering of 5FU is shown in the lower right corner of the figure. Antimetabolite activation pathways are marked in red. The degradation pathway for pyrimidines is marked in blue. The thymidylate synthesis is marked in green. Steps in simplified metabolic pathways: 1. Capecitabine, a 5FU prodrug, is bioactivated in the liver in a two-step process to 5’-deoxy-5-fluorouridine (doxifluridine; DFUR), while tegafur, another 5FU prodrug, is bioactivated by CYP2A6. 2. DFUR is converted to 5FU by thymidine phosphorylase (TP). 3. 5FU is sequentially converted to 5-fluorouridine monophosphate (FUMP), diphosphate (FUDP) and triphosphate (FUTP), which is an active metabolite that is mistakenly incorporated into RNA, causing RNA damage. 4. FUDP is also sequentially converted to 5-fluoro-2’-deoxyuridine diphosphate (FdUDP) and triphosphate (FdUTP), which is an active metabolite that is mistakenly incorporated into DNA, causing DNA damage. 5. TP converts 5FU to 5-fluoro-2’-deoxyuridine (FdUR). 6. Thymidine kinase (TK) converts FdUR to 5-fluoro-2’-deoxyuridine monophosphate (FdUMP). 7. FdUMP is an active metabolite that irreversibly inhibits thymidylate synthase (TS), an enzyme that catalyzes the conversion of 2’-deoxyuridine monophosphate (dUMP) to 2’-deoxythymidine monophosphate (thymidylate; dTMP). 8. Cyclically dihydrofolate (H2Folate) is reduced to tetrahydrofolate (H4Folate) which is then converted to 5,10-methylenetetrahydrofolate (CH2H4Folate). Leucovorin (folinic acid) is converted to CH2H4Folate and then stabilizes the bond formed between TS and FdUMP. 9. Dihydropyrimidine dehydrogenase (DPD) catalyzes the first step of the breakdown of 5FU (mainly in the liver) to 5,6-dihydro-5-fluorouracil (DHFU), which is further broken down into α-fluoro-β-alanine, carbon dioxide and ammonia, excreted from the body. 10. Thymidylate can be salvaged by TK from thymidine (dT) derived from dead cells. 11. FdUTP can be converted to FdUMP by dUTP pyrophosphatase and then converted back to FdUDP by UMP-CMP kinase. Based on information from Longley et al. (2003) and maps in KEGG (Kanehisa et al. 2017)
Metabolic pathways of 5FU and capecitabine
A key enzyme in the catabolic pathway of 5FU is dihydropyrimidine dehydrogenase (DPD), encoded by the DPYD gene, which catalyzes the initial and rate-limiting step in the degradation of pyrimidines, including uracil, thymine, 5FU (Dean and Kane 2016a,b). DPD inactivates 5FU by catalyzing the conversion of 5FU to the non-cytotoxic metabolite 5,6-dihydro-5-fluorouracil (Fig. 1), which occurs mainly in liver cells, but also in other cells, both normal and neoplastic (Longley et al. 2003). Even more than 80% of the administered 5FU is converted in the liver to 5,6-dihydrofluorouracil (Mattison et al. 2002) and excessive DPD activity seems to be one of the mechanisms of resistance to the anti-tumor effects of 5FU (Salonga et al. 2000). On the other hand, however, special care should be taken as patients with DPD enzyme deficiency (Deac et al. 2021; Meulendijks et al. 2016) do not metabolize 5FU at a normal rate and are at risk of severe 5FU toxicity such as mucositis, diarrhea, neutropenia, and neurotoxicity (Dean and Kane 2016a,b).
In the anabolic pathway, 5FU is activated intracellularly by reactions catalyzed by two enzymes: thymidine phosphorylase (TP, encoded by the TYMP gene), a key enzyme that reversibly converts 5FU to 5-fluoro-2’-deoxyuridine (FdUR) and then by thymidine kinase (TK), which converts FdUR to 5-fluoro-2’-deoxyuridine monophosphate, FdUMP (Blondy et al. 2020; Longley et al. 2003), the major active metabolite of 5FU (Fig. 1) that is capable of irreversibly inhibiting TS (Langenbach et al. 1972). In contrast, capecitabine is first converted in the liver to 5’-deoxy-5-fluorouridine (doxifluridine; DFUR), which then forms 5FU upon entry into cells via a TP-catalyzed reaction (Fig. 1), which appears to be more active in cancer cells than in normal cells, possibly leading to selective accumulation of 5FU in tumors (Longley et al. 2003). Also several other enzymes convert 5FU into active antimetabolites such as 5-fluorouridine triphosphate (FUTP) and 5-fluoro-2’-deoxyuridine triphosphate (FdUTP), which due to the ability to compete with endogenous nucleosides can be misincorporated into RNA and DNA, respectively (Blondy et al. 2020; Longley et al. 2003).
TYMS-encoded thymidylate synthase
TS catalyzes reductive methylation of 2’-deoxyuridine monophosphate (dUMP) to 2’-deoxythymidine monophosphate (dTMP; thymidylate) (Fig. 1), with the cofactor 5,10-methylenetetrahydrofolate (C2H4Folate) as a methyl donor (Longley et al. 2003). The TS catalyzed reaction feeds the intracellular pool of thymidylate, which is essential in DNA biosynthesis. Regarding the mechanism of TS inhibition by active fluoropyrimidine, FdUMP enters the active site in TS and forms a stable ternary complex with enzyme and cofactor, thus blocking access of dUMP to the substrate binding site in TS and consequently inhibiting thymidylate synthesis (Longley et al. 2003; Boni et al. 2010). This leads to excessive depletion of the thymidylate pool and disruption of DNA replication and repair (Longley et al. 2003). Ultimately, both thymidylate pool depletion and incorporation of fluoropyrimidine metabolites into nucleic acids lead to cell cycle arrest in S phase (Boni et al. 2010) and apoptosis in response to DNA damage.
In chemotherapy regimens, 5FU is used in combination with folinic acid, also known as leucovorin, which is converted intracellularly to 5,10-methylenetetrahydrofolate, preventing the trace release of free enzyme molecules from the enzyme-cofactor-inhibitor complex, thus enhancing the inhibitory effect of the 5FU metabolite on TS (Fig. 1). Folinic acid and 5FU in combination with oxaliplatin or irinotecan are used as the chemotherapy regimens FOLFOX and FOLFIRI, respectively, in the postoperative treatment of stage II CRC with high risk of recurrence, stage III CRC and palliative chemotherapy of metastatic stage IV CRC (Azwar et al. 2021; Iveson 2020; Taieb and Gallois 2020; Collienne and Arnold 2020; Dienstmann et al. 2015; Labianca et al. 2013; Stec et al. 2011; Grávalos et al. 2009; Van den Eynde and Hendlisz 2009). In turn, capecitabine (Xeloda) is used in combination with oxaliplatin or irinotecan as regimens XELOX (or CAPOX) and XELIRI, respectively. Evaluation of the therapeutic efficacy and toxicity of these regimens is beyond the scope of this article and is partially included in the articles cited above.
Non-coding RNAs regulating pyrimidine metabolism and chemosensitivity to 5FU
The main failure of the fluoropyrimidine-based cancer therapy is the acquisition of drug resistance, which is a multifactorial process (Azwar et al. 2021). Among the mechanisms of resistance to 5FU, there may be changes in drug transport into and out of the cell, in particular overexpression of ATP-binding cassette transporters (Hu et al. 2016), as well as altered drug metabolism, increased level of the molecular target for an active drug, loss of cell cycle checkpoint control, increasing the threshold of apoptosis induction, the process of epithelial-mesenchymal transition (EMT) (Sun et al. 2019b), reprogramming cancer cells into cancer stem-like cells (CSCs) (Shibata et al. 2018), as well as abnormal levels of expression of non-coding RNAs (Wei et al. 2019a).
Cancer patients with low expression of all three genes TYMP, DPYD and TYMS had a longer survival compared to patients with high expression of any of these genes (Salonga et al. 2000). Especially high levels of TYMS-encoded protein in cancerous tissues are predictive of poor response to 5FU-based chemotherapy (Sakatani et al. 2019). On the other hand, higher levels of TYMP-encoded protein in cancer cells appear rather beneficial for capecitabine treatment as thymidine phosphorylase catalyzes the conversion of an inactive prodrug to 5FU inside the cancer cell (Terranova-Barberio et al. 2016). Interestingly, DPYD and TYMS code for enzymes that catalyze rate-limiting steps in enzyme pathways. In particular, expression of TYMS appears to be regulated by various mechanisms, including the involvement of non-coding RNAs. It is postulated that non-coding RNAs form a complex layer of signaling networks, important in both normal and disease processes, including cancer (Chan and Tay 2018; Irminger-Finger et al. 2014). In particular, two classes of non-coding RNAs are distinguished: short non-coding RNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) ranging from 200 to thousands of nucleotides in length (Micallef and Baron 2021; Han et al. 2015).
Mature miRNAs are single-stranded non-coding RNA molecules about 22 nucleotides in length that mediate post-transcriptional gene silencing. Currently, 1917 human hairpin precursor miRNAs and 2654 human mature miRNAs are annotated in the miRBase (22.1 release) (Kozomara et al. 2019). The synthesis of miRNA precursors and their processing in the nuclear and cytoplasmic compartments into mature miRNAs have been thoroughly described (Bartel 2018; Gebert and MacRae 2019; Saliminejad et al. 2019). Briefly, miRNA genes are transcribed into long primary miRNAs (pri-miRNAs) (Gebert and MacRae 2019). The transcripts form stem-loop structures, which are then cleaved in the nucleus by the Drosha RNase III endonuclease (complexed with two DGCR8 protein molecules, forming a so-called Microprocessor complex) into stem-loop molecules of about 70 nucleotides in length, referred to as precursor miRNAs (pre-miRNAs) (Gebert and MacRae 2019; Bartel 2018). Pre-miRNAs are transported into the cytoplasmic compartment by Exportin 5/RAN-GTP. The Dicer RNase III endonuclease then cleaves the pre-miRNA near the loop into a small miRNA:miRNA* duplex, in which the miRNA* is the opposite (passenger) strand of the miRNA (guide strand). After loading into the Argonaute protein (in the ATP-consuming process), the passenger strand of the miRNA:miRNA* duplex is discarded (Bartel 2018). A single mature miRNA is incorporated as a component of an RNA-induced silencing complex (RISC) that binds to a target RNA, primarily messenger RNA (mRNA), through complementary base pairing of a so-called seed region located between nucleotides 2–8 at the 5’-end of the miRNA molecule with a so-called microRNA response element within the target mRNA (Gebert and MacRae 2019; Bartel 2018). Capturing the targeted mRNA by the miRNA-guided Argonaute protein results in inhibition of translation or mRNA degradation mediated by RISC (Gebert and MacRae 2019).
Due to the limited number of molecules of a given type of miRNA in a cell, some lncRNAs can affect protein levels by acting as competing endogenous RNAs (ceRNAs) that bind miRNAs, preventing them from binding to targeted mRNA molecules (Chan and Tay 2018). In other words, lncRNAs can act as sponges to absorb miRNA molecules (Rashid et al. 2016; Salmena et al. 2011). Thus, among the lncRNAs with different roles in the cell (Martino et al. 2021), there may be a subset of the lncRNAs that also act as molecular decoys that sequester specific miRNAs, thereby slowing down the repression of those mRNAs targeted by these miRNAs. It can be assumed that under non-experimental conditions, due to the actual quantitative relationship in the cell between the lncRNA and the sponged miRNA, and between the miRNA and the target mRNA, a low-level miRNA in the cell will be more susceptible to lncRNA acting as a specific ceRNA (Thomson and Dinger 2016).
It was suggested that miRNAs can act as fine tuners of gene expression because targeted miRNAs are typically mildly reduced under physiological conditions (Krützfeldt 2016). A given miRNA can regulate the expression of multiple genes and it is uncertain whether its primary function is to fine-tune the entire set of different mRNAs or to suppress some major mRNAs (Lai 2015). Also, the expression of a given gene can be regulated by several different miRNAs. Accordingly, it can be expected that at least a few different miRNAs regulate the levels of the proteins encoded by TYMS and DPYD, and that altering the levels of these miRNAs may also contribute to reducing the sensitivity of cancer cells to 5FU.
MALAT1 and other lncRNAs affecting the level of TYMS-encoded protein
A precursor transcript of Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1), also known as Nuclear-Enriched Abundant Transcript 2 (NEAT2), is almost 8.8 kb long, from which is derived an over eight kb lncRNA, which is localized in the nucleus (in nuclear specles) and involved in epigenetic modulation of gene expression and alternative splicing (Xu et al. 2022; Uthman et al. 2021). MALAT1 forms a fairly complex secondary structure with exposure of multiple miRNA-binding sites (McCown et al. 2019). Its abnormal overexpression is associated with a higher risk of metastasis in lung cancer (Karimpour et al. 2021). Interestingly, a substantially cytosolic localization of MALAT1 has been demonstrated in three colon cancer lines (HCT116, SW480, SW620) (Sun et al. 2019b), although this result will need to be confirmed in other cell lines and in CRC samples. Especially since MALAT1 is processed in the nucleus and only a short product is exported to the cytosol, while the lncRNA MALAT1 is unlikely to shuttle between the nucleus and the cytoplasmic compartment (Wilusz et al. 2008; Wilusz 2016). Importantly, however, in addition to the formation of RISC complexes in the cytoplasmic compartment, AGO2 (Argonaute protein) is also involved in various processes in the cell nucleus, including interactions with lncRNAs (Meister 2013; Li et al. 2020a) as well as some miRNAs were found in the nucleus (Gebert and MacRae 2019). An exosomal MALAT1 that can be delivered to cancer cells and sponges miRNAs has also been described (Xu et al. 2020d; Poulet et al. 2020). The indication of the cellular compartment in which MALAT1 interacts directly with various miRNAs still needs to be supported by experimental results and is under discussion (Zhou et al. 2021a; Arun et al. 2020; Sun and Ma 2019).
MALAT1 is overexpressed in many types of cancer (Goyal et al. 2021), including CRCs (Younis et al. 2022; Uthman et al. 2021; Hu et al. 2021; Zheng et al. 2020; Xiong et al. 2018; Yang et al. 2015), metastatic lung cancers (Shen et al. 2015), advanced stages of pancreatic cancers (Pang et al. 2015). In general, high levels of MALAT1 expression have been demonstrated in the vast majority of CRC specimens compared to adjacent non-tumor control colorectal tissue specimens in cancer patients (Sun et al. 2019b; Zheng et al. 2020; Luan et al. 2020; Xiong et al. 2018; Li et al. 2017a). Moreover, MALAT1 expression was significantly higher in advanced stages III–IV of tumor-node-metastasis (TNM) (Labianca et al. 2013) than in early stages I–II of CRC (Luan et al. 2020). MALAT1 has also been found to be overexpressed in human colon cancer cell lines including LoVo, SW620, SW1116, HCT116, SW480, HT29, and COLO205 cells compared to the normal human intestinal epithelial cells (Wu et al. 2018a; Tang et al. 2019a; Li et al. 2017a). However, conflicting with these observations are analyzes in another report that suggest decreased levels of MALAT1 in CRC compared to normal colon (Kwok et al. 2018).
Interestingly, the transcriptional regulator YAP1 may be significantly involved in the upregulation of MALAT1 gene expression in cancer cells. YAP1 is an effector of the Hippo pathway and is active while the Hippo pathway is inactive (Dey et al. 2020; Wierzbicki and Rybarczyk 2015). According to the analysis of clinical data, higher levels of YAP1 were statistically significantly associated with higher stages III–IV of CRC, and statistical analysis showed a very strong positive correlation between MALAT1 and YAP1 mRNA levels in CRC tissue specimens from patients (Sun et al. 2019b). The stability and nuclear localization of YAP1 depends on the ANKHD1 protein (ankyrin repeat and KH domain containing 1) which also acts as a YAP1 coactivator, and high levels of ANKHD1 were associated with the invasive properties of CRC cells (Almeida and Machado-Neto 2020). It turned out that nuclear YAP1 in CRC cells binds as a transcription coactivator with the TCF4/β-catenin transcription factor complex in the promoter region of the MALAT1 gene to induce expression of this gene (Sun et al. 2019b). Interestingly, MALAT1 was shown to physically associate with the ANKHD1 protein (Yao et al. 2022), possibly via the KH domain of ANKHD1, which is involved in binding to lncRNAs (Almeida and Machado-Neto 2020). The actual location in the cellular compartment and the importance of this interaction are not fully elucidated in the cited studies of Yao et al. (2022), but it can be speculated that it may stabilize the ANKHD1 and YAP1 complex recruited by TCF4/β-catenin to the promoter region of the MALAT1 gene and enhance gene transcription. Further, upregulated MALAT1 promotes the EMT process of CRC cells and tumor metastasis (Sun et al. 2019b; Chen and Shen 2020). In contrast, silencing MALAT1 reversed the EMT process in HT29 cells (Xiong et al. 2018), strongly arguing for a link between higher MALAT1 expression and CRC metastatic potential. The involvement of YAP1 interacting with TCF4/β-catenin in the regulation of transcription of the MALAT1 gene was previously found in liver cancer cells (Wang et al. 2014a).
The inverse (negative) correlation of MALAT1 with a given miRNA in clinical samples may suggest a direct interaction. For example, a strong negative correlation was observed between MALAT1 and miR-200c-3p in CRCs compared to normal tissues and was associated with the response to cisplatin (Hu et al. 2021). However, clinical data should be supported by the results of reporter assays (Karreth et al. 2011; Tang et al. 2019c) using transfected cultured cells that confirm the importance of a specific site in the MALAT1 sequence for interaction with miRNA. The mentioned interaction of MALAT1 and miR-200c-3p was in fact previously confirmed in 293 T cells (Zhuo et al. 2018). It is worth mentioning here that the upregulation of MALAT1 through miR-200c-3p sponging leads to an increase in the level of the transcriptional repressor ZEB1 and contributes to the promotion of the EMT process (Pretzsch et al. 2019), and thus the migration and invasiveness of cancer cells (Zhuo et al. 2018). MALAT1 in CRCs prevents binding of miRNAs to mRNAs encoding proteins such as β-catenin, c-Myc (in the Wnt/β-catenin signaling pathway), TWIST, SLUG (promote EMT: downregulation of E-cadherin, upregulation of N-cadherin, vimentin) and many others, supported by both extensive analyzes of clinical samples and the results of experiments using established human colon cancer cell lines (Xu et al. 2022; Uthman et al. 2021). At this point, it is also worth suggesting a relationship between the level of TYMS expression and the EMT process in cancer cells. Namely, the results of the analysis of EMT markers in cell lines derived from tumors of various types indicate that the levels of TYMS mRNA are increased in cell lines with a predominance of the mesenchymal-like phenotype (Siddiqui et al. 2017). However, some caution should be exercised when comparing the results of lncRNA interaction in cell lines in vitro with respect to the interaction of the complex network of non-coding RNA in metastatic cancer cells (Witusik-Perkowska et al. 2022).
MALAT1 can also sponge several miRNAs targeting TYMS, as discussed later in this article. Finally, and of particular importance for further consideration, increased levels of MALAT1 in cancer cells may contribute to their resistance to 5FU chemotherapy. Namely, the level of lncRNA MALAT1 was increased in 5FU-resistant colon cancer subline HCT-116/5-FU compared to the parental HCT-116 cell line, while silencing of MALAT1 by siRNA was shown to increase the chemosensitivity of HCT-116/5-FU cells to treatment with 5FU (Tang et al. 2019a). Also in the derived HT-29FUR subline of 5FU-resistant cells, the level of MALAT1 was more than 2 times higher compared to the parent colon cancer HT-29 cell line (Aksoy et al. 2022). However, due to the involvement of MALAT1 in multiple processes, its classification as an oncogene or tumor suppressor is under discussion (Chen et al. 2020a).
Through the use of high-throughput RNA sequencing technology, it was found that several hundred lncRNAs are differentially expressed in 5FU resistant and non-resistant CRC patients. Among them, lncRNA X Inactive Specific Transcript (XIST) was found to promote TS expression through an unknown mechanism, and increased serum XIST levels were associated with lower survival rates in CRC patients receiving 5FU-based therapy (Xiao et al. 2017). The importance of XIST in CRC progression is demonstrated by increased levels of XIST in CRCs and its effect on the Wnt/β-catenin signaling pathway, which promotes elevated c-Myc levels and tumor growth (Sun et al. 2018a). Clinical data indicate that high levels of another lncRNA, namely Taurine Upregulated Gene 1 (TUG1), were also associated with recurrence of CRC in patients receiving 5FU-based chemotherapy (Wang et al. 2019a). In turn, elevated levels of lncRNA HOX Transcript Antisense RNA (HOTAIR) were also found in CRC tissues from patients and in colon cancer cell lines (Lu et al. 2018). High HOTAIR levels were associated with a poor response to 5FU treatment in CRC patients, while silencing HOTAIR in CRC cells improved their sensitivity to 5FU (Li et al. 2017b). A recent review described a number of lncRNAs acting as oncogenes in the CRCs, including HOTAIR and TUG1, which reduce chemosensitivity to 5FU (Yang et al. 2021).
MiRNAs targeting TYMS mRNA
miR-192/215-5p
TYMS was shown as a direct target of miR-192-5p and miR-215-5p (Boni et al. 2010; Song et al. 2010) that share the same seed region (see Table 1). Expression of miR-192/215-5p, contained in two different miRNA clusters (Vychytilova-Faltejskova and Slaby 2019), can be strongly induced in normal colon tissue by activated transcription factor p53 in response to DNA damage (Braun et al. 2008). Subsequently, miR-192/215-5p by targeting MDM2, a key p53 negative regulator (Pichiorri et al. 2010; Sun et al. 2019a), could further potentiate the action of p53, contributing to p53-dependent cell cycle arrest (Braun et al. 2008; Georges et al. 2008). It was also noted that levels of miR-192/215-5p were significantly reduced during colon carcinogenesis (Braun et al. 2008). On the other hand, an almost threefold increase in miR-215-5p levels and a dramatic decrease in the level of TYMS-encoded protein were detected in a small fraction of slowly proliferating colorectal cancer stem-like cells (CRC CSCs) derived from the HCT116 cell line containing wild-type p53 (Song et al. 2010). All of this together may suggest that miR-192/215-5p synchronize thymidylate synthesis with the rate of proliferation. Thus, ectopic miR-192/215-5p expression decreased the amount of TYMS-encoded protein in CRC cells, but this effect did not result in the expected 5FU sensitization, but paradoxically increased their resistance to 5FU treatment, presumably due to cell cycle arrest, thus reducing 5FU-sensitive fraction of cells in the S phase of the cell cycle (Boni et al. 2010). The arrest of the cell cycle was presumably a consequence of miR-215-5p targeting the DTL gene product (Denticleless protein homolog) and p53-dependent p21 upregulation as well (Fesler et al. 2015; Song et al. 2010). Interestingly, however, the increase in endogenous levels of miR-215-5p following treatment of HCT116-5FU and SW480-5FU cells (resistant to 5FU) with melatonin led to reduction in TYMS-encoded protein levels and could also increase the susceptibility of CRC cells to the cytotoxic effects of 5FU (Sakatani et al. 2019). Overall, miR-192/215-5p function as tumor suppressors in human cancers, including preventing the EMT process in CRC (Rokavec et al. 2019; Chen et al. 2017c), which is necessary for the initiation of the processes of migration, invasiveness and metastasis of cancer cells. On the other hand, lowering the levels of miR-192/215-5p blunts their inhibitory effects on the triglyceride synthesis pathway and genes governing extracellular matrix remodeling, which therefore promotes the progression of CRC (Zhao et al. 2019c). It should be noted, however, that miR-195/215-5p belong to several miRNAs that potentially target the largest number of genes active in CRC (Toolabi et al. 2022). Therefore, the impact of changes in these miRNA levels during therapy on tumor progression and chemoresistance may be unpredictable. Moreover, these were the conclusions of experiments carried out in well-controlled in vitro model systems, while in various clinical cases, presumably many different and uncontrolled factors affect the rate of tumor cell proliferation. However, analysis of clinical samples of post-operative CRC tissues showed that low miR-215-5p levels were significantly correlated with a high probability of 3-year recurrence, while high miR-215-5p levels could potentially predict the benefit of 5FU-based chemotherapy after surgery (Li et al. 2013). However, other reports have shown contradictory correlations between the levels of miR-215-5p and the effectiveness in the treatment of patients with CRC, which may be a result of the uncontrolled diversity of research material in many respects, including the genotype of patients and the stage of CRC (Vychytilova-Faltejskova and Slaby 2019).
Table 1.
Confirmed miRNA response elements in the 3’-untranslated region of TYMS mRNA
| miRNA | Sequence (5’- > 3’)† | Targeted sites‡ | References |
|---|---|---|---|
| miR-140-3p | UACCACAGGGUAGAACCACGG | 433–438 | Wan et al. (2021) |
| miR-192-5p | CUGACCUAUGAAUUGACAGCC | 97–103 | Song et al. (2010) |
| miR-197-3p | UUCACCACCUUCUCCACCCAGC | 321–327 | Sun et al (2015); Wang et al (2019a) |
| miR-203a-3p | GUGAAAUGUUUAGGACCACUAG | 291–296, 327–332 | Li et al. (2015a) |
| miR-215-5p | AUGACCUAUGAAUUGACAGAC | 97–103 | Song et al. (2010) |
| miR-330-3p | GCAAAGCACACGGCCUGCAGAGA | 192–197, 506–511 | Xu et al. (2017) |
| miR-375-3p | UUUGUUCGUUCGGCUCGCGUGA | 237–243 | Xu et al. (2020b) |
| miR-433-3p | AUCAUGAUGGGCUCCUCGGUGU | 419–426 | Gotanda et al. (2013) |
| miR-1307-3p | ACUCGGCGUGGCGUCGGUCGUG | 56–61 | Chen et al. (2017a) |
†human miRNA (hsa-miR) sequences were taken from miRBase (www.mirbase.org); seed sequences complementary to the 3’UTR of TYMS mRNA are underlined
‡3’UTR TYMS mRNA sites targeted by miRNA seed sequences; the numbers refer to the position after the STOP codon according to the GenBank sequence accession number NM_001071.4; the sites confirmed by the luciferase reporter assay in the cited publication are underlined
Importantly, it has recently been shown that in highly aggressive hepatocellular carcinoma (HCC), miR-215-5p targets the 3’UTR of mRNA encoding Cell Division Cycle 6 (CDC6), which is involved in the assembly of the pre-replicative complex during the G1 phase of the cell cycle (Xu et al. 2020a). In turn, it was found in breast cancer cells that miR-215-5p targets mRNA encoding RAD54 Homolog B (RAD54B), which is involved in homologous recombination repair of DNA breaks, thus inhibiting proliferation and promoting apoptosis of MCF-7 breast cancer cells (Wang et al. 2021i). It is therefore understood that lowering the levels of both miR-215-5p and miR-192-5p promotes cancer cell proliferation. Interestingly, lncRNA XIST, which is significantly elevated in hepatitis B virus-related HCC compared to adjacent liver tissues, has been found to interact with miR-192-5p and inhibit the activity of this miRNA (Wang et al. 2021h).
Overall, levels of miR-192/215-5p are down-regulated in cancers of various types (Table 2 and Fig. 2), yet up-regulated levels of miR-192/215-5p have been found in esophageal squamous cell carcinoma (ESCC) and gastric cancers (GCs) where these miRNAs also target tumor suppressors, such as mRNAs encoding the pro-apoptotic BIM protein in ESCC (Li et al. 2015c) or the important tumor suppressor RB Transcriptional Corepressor 1 (RB1) and RUNX Family Transcription Factor 1 (RUNX1) in GCs (Chen et al. 2017e; Li et al. 2016) (see also Table 3). )
Table 2.
Expression levels of miRNAs targeting i.a. TYMS mRNA in cancer
| miRNA | Level† | Cancers‡ | References |
|---|---|---|---|
| miR-192-5p | Down | CRC tissues | Zhao et al. (2020b) |
| Down | CRC cell lines (HCT116, SW480, RKO, HT29) | Zheng et al. (2019b) | |
| Down | CRC tissues (TNM stage II) | Braun et al. (2008) | |
| Up | ESCC tissues and four cell lines | Li et al. (2015c) | |
| Down | NSCLC: four cell lines (i.a. A549, H1299) | Zou et al. (2019) | |
| Down | BLCA tissues and five cell lines (i.a. T24) | Ji et al. (2018) | |
| Down | BRCA tissues, MCF-7 and MDA-MB-231 cell lines | Chen et al. (2019d) | |
| Down | HCC tissues | Wang et al. (2021h) | |
| Down | HCC tissues | Lian et al. (2016) | |
| Down | HCC tissues | Ge et al. (2015) | |
| Down | PCa: PC-3 and DU145 cell lines | Sun et al. (2016) | |
| miR-215-5p | Down | CRC tissues stages: III + IV vs I + II | Yan et al. (2020b) |
| Down | CRC tissues stages: II, III, IV vs I | Vychytilova et al. (2017) | |
| Down | CRC tissues stages: IV vs III vs II vs I | Chen et al. (2017c) | |
| Down | CRC tissues: liver metastasis vs without metastasis | Chen et al. (2017c) | |
| Down | CRC tissues and cell lines (SW480, HCT116, LoVo, HT29) | Chen et al. (2016) | |
| Down | CRC tissue stages: III–IV vs I–II | Chen et al. (2016) | |
| Down | CRC tissues (stage II) | Braun et al. (2008) | |
| Down | CRC tissues | Song et al. (2010) | |
| Up | CRC CSCs | Song et al. (2010) | |
| Up | GC tissues, TNM stage III–IV vs I–II | Chen et al. (2017e) | |
| Up | GC tissues, stage III/IV vs I/II | Li et al. (2016) | |
| Up | GC tissues | Deng et al. (2014b) | |
| Down | BRCA tissues | Wang et al. (2021i) | |
| Down | BRCA tissues, stage III–IV vs I–II, three cell lines | Gao et al. (2019) | |
| Down | BRCA tissues, cell lines (i.a. MDA-MB-231) | Yao et al. (2017) | |
| Down | BRCA tissues | Zhou et al. (2014a) | |
| Down | OVCA tissues and three cell lines | Ge et al. (2016) |
†”UP” or “down” in the level of a given miRNA: in cancerous tissues as compared to adjacent non-cancerous tissues (or to tissues of healthy individuals in cases of cervical and prostate cancers); one stage of cancer compared to another stage; cancer cell line compared to a normal epithelial cell line
‡Cancer tissues collected from patients and cancer cell lines of various types: BLCA bladder cancer, BRCA breast cancer (various molecular subtypes including triple-negative breast cancer TNBC), CeCa cervical cancer, CRC colorectal cancer, ESCC esophageal squamous cell carcinoma, GC gastric cancer, HCC hepatocellular carcinoma, LSCC laryngeal squamous cell carcinoma, NSCLC non-small cell lung cancer (including LUAD lung adenocarcinoma SqCLC squamous cell lung carcinoma), OVCA ovarian cancer, OSCC oral squamous cell carcinoma, PCa prostate cancer, PDAC pancreatic ductal adenocarcinoma
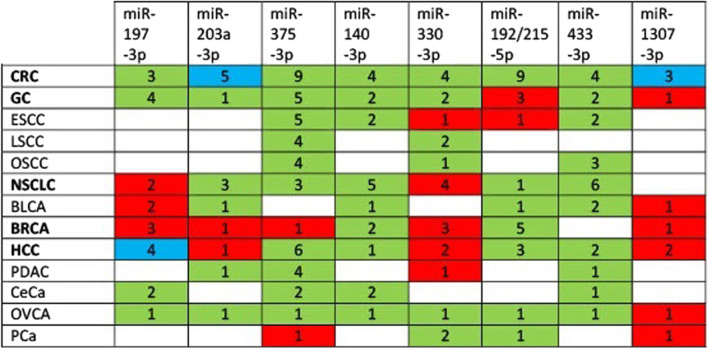
Fig. 2 .
MiRNA levels in various types of cancer. Downregulated miRNAs in a given cancer type are marked green, upregulated marked red, regulated ambiguously marked blue. Cancer abbreviations as in Table 2. The numbers represent the number of the respective reports referred to in Tables 2 to 17
Table 3.
Major genes targeted by miR-192-5p and miR-215-5p in various types of cancer
| Cancers | Targeted gene products† | References |
|---|---|---|
| Cancers with down-regulation of miR-192/215-5p | ||
| Acting as oncogenes or tumor promoters | ||
| CRC | TYMS, EIF5A2, EREG, HOXB9 | Boni et al. (2010); Song et al. (2010); Zhao et al. (2020b); Chen et al. (2016) |
| RAB2A, YY1, ZEB2 | Vychytilova et al. (2017); Zheng et al. (2019b); Chen et al. (2017c) | |
| BRCA | AKT1, RAD54B, SOX9 | Yao et al. (2017); Wang et al. (2021i); Gao et al. (2019) |
| NSCLC | TRIM44 | Zou et al. (2019) |
| BLCA | YY1 | Ji et al. (2018) |
| HCC | CDC6, SLC39A6, TRIM25 | Xu et al. (2020a); Lian et al. (2016); Wang et al. (2021h) |
| PCa | NOB1 | Sun et al. (2016) |
| Cancers with up-regulation of miR-192/215-5p | ||
| Acting as tumor suppressors | ||
| GC | RB1, RUNX1 | Chen et al. (2017e); Deng et al. (2014b); Li et al. (2016 |
| ESCC | BCL2L11 | Li et al. (2015c) |
†mRNAs encoding the following proteins: AKT1 encodes AKT Serine/Threonine Kinase 1; BCL2L11 codes for pro-apoptotic BH3-only protein BIM; CDC6 is an oncogene encoding Cell Division Cycle 6 Homolog; EIF5A2 is an oncogene encoding Eukaryotic Translation Initiation Factor 5A2; EREG codes for Epiregulin, ligand of epidermal growth factor receptor (Cheng et al. (2021); HOXB9 encodes transcription factor Homeobox B9 (Contarelli et al. 2020); NOB1 is an oncogene encoding Nin One Binding Protein; RAB2A is an oncogene encoding a member of the RAS family; RAD54B is an oncogene encoding helicase RAD54 Homolog B involved in homologous recombination and DNA repair (Zhang et al. 2019c); RB1 is a tumor suppressor gene encoding RB Transcriptional Corepressor 1, a key regulator of the G1/S transition in the cell cycle; RUNX1 is tumor suppressor gene coding for RUNX Family Transcription Factor 1; SLC39A6 is an oncogene that encodes a zinc-influx transporter; SOX9 is an oncogene encoding SRY-Box Transcription Factor 9; TRIM25 encodes E3 ubiquitin ligase involved in p53 inactivation (Zhang et al. 2015); TRIM44 is an oncogene encoding a protein involved in the ubiquitination and degradation of target proteins; YY1 is an oncogene encoding Yin Yang 1 transcriptional factor; ZEB2 encodes Zinc Finger E-Box Binding Homeobox 2, a transcriptional corepressor promoting the EMT process in cancer cells
Interestingly, in addition to miR-215-5p targeting TYMS expression, also miR-215-3p derived from the opposite arm of the same primary hairpin microRNA has gene silencing activity that influences 5FU cytotoxic activity since ectopic overexpression of miR-215-3p increased the sensitivity of CRC cells to 5FU, and although the exact mechanism has not been discovered, it has been shown to be at least in part related to targeting the CXCR1 chemokine receptor, which is an interleukin-8 receptor (Li et al. 2018a).
As show in Table 3, miR-192/215-5p down-regulation in CRCs not only promotes an increase in TS, but also an increase in ZEB2, which promotes the EMT process (Pretzsch et al. 2019), and an increase in eIF5A2, which further potentiates the EMT process (Bao et al. 2015). Down-regulation of miR-192/215-5p are generally found in cancers of various types, and in breast cancer it results in an increase in the levels of AKT protein kinase and DNA repair and recombination protein RAD54B.
Finally, it is worth mentioning that p53, which raises the level of miR-192/215-5p, may also act as a transcription repressor of the MALAT1 gene, at least in hematopoietic cells (Ma et al. 2015). As counteraction, MALAT1 may indirectly deacetylate p53 and inhibit p53 transcriptional activity as found in HepG2 cells (Chen et al. 2017h).
Reports also indicate several other miRNAs, which in various types of cancer may regulate the level of protein encoded by TYMS and thus affect the sensitivity of cancer cells to 5FU chemotherapy, including miRNA such as miR-140-3p (Wan et al. 2021), miR-197-3p (Sun et al. 2015), miR-203a-3p (Li et al. 2015a), miR-218-5p (Li et al. 2015b), miR-330-3p (Xu et al. 2017), miR-375-3p (Xu et al. 2020b), miR-433-3p (Gotanda et al. 2013).
MiR-140-3p
MiR-140-3p has been reported to target the 3’UTR of TYMS mRNA in lung adenocarcinoma cells (Wan et al. 2021). Interestingly, miR-140-5p levels were also reported to be decreased in CRC specimens taken from patients, but increased in slowly proliferating and chemoresistant CRC CSCs, and furthermore, CRC cells transfected with miR-140 precursor were resistant to 5FU (Song et al. 2009). However, it is still worth awaiting stronger experimental support for a direct link between levels of miR-140-3p and TYMS-encoded protein, and the chemosensitivity of CRC cells to 5FU. It is worth noting that miR-140-3p also has the potential to target mRNA encoding Phosphatase and TENsin homolog (PTEN) (Yin et al. 2020), an important tumor suppressor that antagonizes the effects of PI3K, thereby activating the PI3K/AKT signaling pathway, although it appears that this putative miR-140-3p function in cancer cells is not likely to be significant. As shown in Table 4, miR-140-3p is generally down-regulated in various types of cancer, causing, in addition to the expected increase in TS, also an increase in the anti-apoptotic protein BCL-2 in CRC and GC cells and activation of the WNT/β-catenin pathway in CRC (see Table 5).
Table 4.
Expression levels of miRNAs targeting i.a. TYMS mRNA in cancer
| miRNA | Level† | Cancers‡ | References |
|---|---|---|---|
| miR-140-3p | Down | CRC tissues | Chen et al. (2020c) |
| Down | CRC tissues, SW480 and HCT116 cell lines | Jiang et al. (2019a) | |
| Down | Primary tumor tissues of CRC patients | Piepoli et al. (2012) | |
| Down | Liver metastatic vs none-metastatic CRC | Liu et al. (2021) | |
| Down | GC tissues and cell lines | Wang et al. (2021a) | |
| Down | GC tissues and five cell lines | Chen et al. (2021) | |
| Down | ESCC tissues and five cell lines | Wang et al. (2021d) | |
| Down | ESCC tissues and five cell lines | Chen et al. (2020d) | |
| Down | NSCLC (LUAD) tissues and five cell lines (i.a. A549) | Wang et al. (2021f) | |
| Down | NSCLC (LUAD): four cell lines (i.a. A549, H1299) | Wu et al. (2020) | |
| Down | NSCLC tissues and three cell lines (i.a. A549, H1299) | Hu et al. (2020) | |
| Down | NSCLC (SqCLC) tissues, stage III vs II vs I, four cell lines | Huang et al. (2019a) | |
| Down | NSCLC tissues | Dong et al. (2016) | |
| Down | Lung cancer tissues and six cell lines (i.a. A549) | Kong et al. (2015) | |
| Down | BLCA tissues and four cell lines (i.a. T24) | Yuan et al. (2021) | |
| Down | BRCA tissues stage I-IV | Dou et al. (2021) | |
| Down | BRCA tissues, MCF-7 (ER+) and MDA-MB-453 (HER2+) | Zhou et al. (2019) | |
| Down | HCC tissues | Gao et al. (2021b) | |
| Down | CeCa tissues | Wang et al. (2022a) | |
| Down | CeCa tissues | Wang et al. (2021g) | |
| Down | OVCA tissues | Qiao et al. (2021) |
†,‡For description see footnote to Table 2
Table 5.
Major genes targeted by miR-140-3p in various types of cancer
| Cancers | Targeted gene products† | References |
|---|---|---|
| Cancers with down-regulation of miR-140-3p | ||
| Acting as oncogenes or tumor promoters | ||
| CRC | BCL2, BCL9 | Liu et al. (2021) |
| GC | BCL2 | Chen et al. (2021) |
| ESCC | E2F3, NRIP1 | Wang et al. (2021d); Chen et al. (2020d) |
| LUAD | TYMS | Wan et al. (2021) |
| Lung cancer | ATP6AP2, BRD9, JAK1 | Kong et al. (2015); Huang et al. (2019a); Hu et al. (2020) |
| BLCA | ANXA8 | Yuan et al. (2021) |
| HCC | GRN, VEGFA | Gao et al. (2021b); Hou et al. (2020) |
| OVCA | AGTR1 | Qiao et al. (2021) |
| Acting both as oncogenes and tumor suppressors, or unclassified | ||
| LUAD | E2F7 | Wang et al. (2021f) |
| BRCA | TRIM28 | Zhou et al. (2019) |
| CeCa | ELOA (TCEB3), PDZK1 | Wang et al. (2021g); Wang et al. (2022a) |
†mRNAs encoding the following proteins: AGTR1 encodes the Angiotensin II Receptor Type I and is recognized as an oncogene in various cancers; ANXA8 encodes Annexin A8 that is highly expressed in some cancers; ATP6AP2 is a proto-oncogene coding for ATPase H + Transporting Accessory Protein 2 (Renin/Prorenin Receptor); BCL2 codes for anti-apoptotic protein; BCL9 codes for the coactivator involved in β-catenin mediated transcription; BRD9 encodes Bromodomain-Containing Protein 9 involved in chromatin remodeling complexes; E2F3 encodes E2F Transcription Factor 3; E2F7 is a tumor suppressor gene that encodes atypical E2F Transcription Factor 7; ELOA encodes Elongin A, also known as RNA polymerase II Transcription Elongation Factor B Subunit 3; GRN encodes Granulin Precursor that is presumably involved in tumorigenesis; JAK1 encodes Janus Kinase 1 phosphorylating a tyrosine residue; NRIP1 encodes Nuclear Receptor Interacting Protein 1 which is elevated in tumors; PDZK1 is a tumor suppressor gene encoding PDZ Domain Containing 1, scaffolding protein; TRIM28 encodes a protein that may have both oncogenic and tumor suppressor effects; VEGFA encodes Vascular Endothelial Growth Factor A that promotes angiogenesis
Potentially both miR-140-5p and miR-140-3p could be sponged by lncRNA MALAT1, although this has not been shown in CRC, while the direct interaction of miR-140-5p with MALAT1 has been shown experimentally in cancers of various types such, as prostate cancer (Hao et al. 2020), hepatocellular carcinoma (Fan et al. 2020; Hou et al. 2020), osteosarcoma (Sun and Qin 2018), tongue squamous cell carcinoma (Zhu et al. 2019). MiR-140-3p levels can also be regulated by lncRNA TUG1 and this has already been demonstrated in bladder cancer (Yuan et al. 2021). It was also confirmed that TUG1 sponges miR-140-5p in osteosarcoma cells (Zhao et al. 2019a).
MiR-197-3p
One report showed that miR-197-3p directly targets the 3’UTR of TYMS mRNA, which led to a reduction in TS protein level, while increasing the sensitivity of CRC cells to the cytotoxic effects of 5FU (Sun et al. 2015). On the other hand, lncRNA TUG1 can act as a ceRNA that sponges miR-197-3p and thereby increases the level of TYMS-encoded protein, mediating the acquisition of 5FU resistance by CRC cells (Wang et al. 2019a). HOTAIR, which is elevated in CRC tissues, has also been shown to sponge miR-197-3p (Lu et al. 2018). Importantly, miR-197-3p has been found to be sponged also by MALAT1 in NSCLC, contributing to the process of EMT and cancer cell resistance to treatment with cisplatin, adriamycin, gefitinib and paclitaxel (Yang et al. 2019). The direct interaction of miR-197-3p with the miRNA response elements in MALAT1 was confirmed by the results of reporter assays (Yang et al. 2019). In this context, it would be particularly valuable to determine whether the increase in the level of MALAT1 in CRCs contributes to the reduction of their chemosensitivity to 5FU, e.g. by sponging miR-197-3p. According to old measurements, the level of miR-197-3p was decreased in the HCT116 colorectal cancer cell line after treatment with 5FU (Zhou et al. 2010). It is worth noting at this point that SGC7901/5-FU gastric cancer cells partially resistant to 5FU also had decreased level of miR-197-3p compared to parental SGC7901 line, while transfection of the miR-197 mimic into SGC7901/5-FU cells restored sensitivity to the growth inhibitory effects of 5FU (Xiong et al. 2015). Importantly, miR-197-3p, whose levels are lower in CRCs (Lu et al. 2018), in turn in NSCLC, acts as an oncomiR targeting mRNAs encoding pro-apoptotic BH3-only proteins NOXA and BMF (Fiori et al. 2014).
As shown in Tables 6 and 7, miR-197-3p is down-regulated in CRC and GC, where it targets TYMS and MTDH, which encodes a protein also involved in TYMS induction and PTEN repression, while in NSCLC and BRCA miR-197-3p is up-regulated because it also targets suppressor genes that are important in these cancers. Reports on the level of miR-197-3p in HCC seem contradictory, but it is worth paying attention to the up-regulation of the level of miR-197-3p in metastatic HCC, which results in a decrease in the level of negative regulators of the WNT/β-catenin pathway (AXIN2, NKD1, DKK2) in HCC clinical tissues (Hu et al. 2018a).
Table 6.
Expression levels of miRNAs targeting i.a. TYMS mRNA in cancer
| miRNA | Level† | Cancers‡ | References |
|---|---|---|---|
| miR-197-3p | Down | HCT8Fu (5FU resistant) vs HCT8 (parental sensitive) | Wang et al. (2019a) |
| Down | CRC tissues, HCT116, LoVo, HT29, SW480 cell lines | Lu et al. (2018) | |
| Down | after treatment with 5FU in HCT116 cell line | Zhou et al. (2010) | |
| Down | GC tissues and two cell lines | Han and Liu (2021) | |
| Down | GC tissues | Niu et al. (2020) | |
| Down | GC tissues | Chen et al. (2019c) | |
| Down | GC tissues and four cell lines | Liao et al. (2018) | |
| Up | NSCLC tissues and four cell lines (i.a. A549, H460, H1299) | Yang et al. (2019) | |
| Up | NSCLC (LUAD) tissues | Chen and Yang (2018) | |
| Up | BLCA tissues and four cell lines (i.a. T24) | Jiang et al. (2019b) | |
| Up | BLCA tissues and three cell lines (i.a. T24) | Wang et al. (2016a) | |
| Up | BRCA tissues and MCF-7, T47D cell lines (ER+ luminal A) | Li et al. (2021d) | |
| Up | BRCA 11 cell lines, TNBC vs luminal | Ye et al. (2019) | |
| Up | BRCA (TNBC) tissues | Tang et al. (2018) | |
| Down | HCC tissues | Bi et al. (2021) | |
| Down | HCC tissues and three cell lines | Ni et al. (2019) | |
| Up | HCC tissues with metastasis vs non-metastasis | Hu et al. (2018a) | |
| Up | HCC cell lines | Dai et al. (2014) | |
| Down | CeCa tissues | Gu et al. (2021) | |
| Down | CeCa tissues and four cell lines | Hu et al. (2018b) | |
| Down | OVCA tissues and four cell lines | Xie et al. (2020) |
†,‡For description see footnote to Table 2
Table 7.
Major genes targeted by miR-197-3p in various types of cancer
| Cancers | Targeted gene products† | References |
|---|---|---|
| Cancers with down-regulation of miR-197-3p | ||
| Acting as oncogenes or tumor promoters | ||
| CRC | TYMS | Sun et al. (2015); Wang et al. (2019a) |
| GC | PRKCB | Chen et al. (2019c) |
| GC | CXCR6, MAPK1, MTDH | Han and Liu (2021); Xiong et al. (2015); Liao et al. (2018) |
| HCC | AGR2 | Bi et al. (2021) |
| CeCa | E2F6, FOXM1 | Gu et al. (2021); Hu et al. (2018b) |
| Acting both as oncogenes and tumor suppressors, or unclassified | ||
| HCC | ZIK1 | Ni et al. (2019) |
| OVCA | ABCA7 | Xie et al. (2020) |
| Cancers with up-regulation of miR-197-3p | ||
| Acting as tumor suppressors | ||
| NSCLC | BMF, PMAIP1 (NOXA) | Fiori et al. (2014) |
| LUAD | CYLD | Chen and Yang (2018) |
| BRCA | FBXW7, HIPK3, NLK | Ye et al. (2019); Li et al. (2021d); Tang et al. (2018) |
| HCC | AXIN2, NKD1, DKK2, CD82 | Hu et al. (2018a); Dai et al. (2014) |
| Acting both as oncogenes and tumor suppressors, or unclassified | ||
| NSCLC | CTNND1 | Yang et al. (2019) |
†mRNAs encoding the following proteins: ABCA7 encodes ATP Binding Cassette Subfamily A Member 7 (Muriithi et al. 2020); AGR2 is an oncogene that encodes a protein disulphide isomerase; AXIN2, NKD1 and DKK2 function as tumor suppressors and regulators of the WNT/β-catenin pathway; BMF encodes pro-apoptotic BH3-only protein; CD82 codes for metastasis suppressor; CTNND1 encodes Catenin Delta 1 which interacts with cadherins and acts in adhesion between cells; CXCR6 encodes C-X-C Motif Chemokine Receptor 6; CYLD is a tumor suppressor gene that codes for CYLD Lysine 63 Deubiquitinase; E2F6 encodes E2F Transcription Factor 6; FBXW7 is a tumor suppressor gene encoding F-Box and WD Repeat Domain Containing 7 which targets cyclin E and c-Myc for ubiquitin-mediated degradation; FOXM1 functions as an oncogene encoding Forkhead Box M1 protein; HIPK3 codes for Homeodomain Interacting Protein Kinase 3, serine/threonine protein kinase; MAPK1 encodes Mitogen-Activated Protein Kinase 1 (p42-MAPK/ERK2); MTDH (Metadherin) acts as an oncogene and is involved in induction of TYMS (Yoo et al. 2009); NLK codes for Nemo-Like Kinase, serine/threonine protein kinase and acts as tumor suppressor in breast cancer; PMAIP1 codes for NOXA, which is a p53-induced pro-apoptotic BH3-only protein; PRKCB codes for Protein Kinase Cβ; ZIK1 encodes the Zinc Finger Protein Interacting With K Protein 1, which probably acts as a transcriptional repressor
MiR-203a-3p
Levels of miR-203a-3p were also found to be decreased in 5FU-resistant CRC cells (see Table 8). Mir-203a-3p can target several sites in the 3’UTR of TYMS mRNA (Table 1) and it was found that silencing of miR-203a-3p by an antisense oligonucleotide increased the level of the TYMS-encoded protein and decreased the sensitivity of CRC cells to the cytotoxic effects of 5FU, and conversely, as a result of miR-203a mimic transfection, a decreased level of TS protein and an increased sensitivity to 5FU were observed (Li et al. 2015a). More importantly, miR-203a-3p increased the anti-tumor activity of 5FU also after injection of the miR-203 precursor along with 5FU into NOD/SCID mice with CRC (Li et al. 2015a). Importantly, however, miR-203a-3p can also target mRNA encoding PTEN, thereby activating the PI3K/AKT signaling pathway, and its involvement in stimulating cell proliferation and inhibiting apoptosis has been demonstrated in hepatocytes (Zhang et al. 2020).
Table 8.
Expression levels of miRNAs targeting i.a. TYMS mRNA in cancer
| miRNA | Level† | Cancers‡ | References |
|---|---|---|---|
| miR-203a-3p | Down | CRC tissues, SW480, HT29, SW620, HCT15 cell lines | Qian et al. (2019) |
| Up | CRC tissues, HCT116, HT29, LoVo, SW1116 cell lines | Chen et al. (2018) | |
| Down | CRC tissues | Xiao et al. (2018) | |
| Down | LoVo/5-FU (5FU resistant) vs LoVo (parental sensitive) | Li et al. (2015a) | |
| Up | in serum of CRC patients | Huang et al. (2020a) | |
| Down | GC tissues and two cell lines | Wang et al. (2018a) | |
| Down | NSCLC tissues | Liang et al. (2020) | |
| Down | NSCLC tissues | Yang et al. (2020) | |
| Down | NSCLC (LUAD) tissues, TNM stage III–IV vs I–II | Wang et al. (2020a) | |
| Down | BLCA tissues and four cell lines (i.a. T24) | Na et al. (2019) | |
| Up | BRCA tissues (mainly luminal A/B) | Gomes et al. (2016) | |
| Up | HCC tissues | Huo et al. (2017) | |
| Down | PDAC four cell lines (i.a. PANC-1, SW 1990) | An and Zheng (2020) | |
| Down | OVCA tissues | Liu et al. (2019a) |
†,‡For description see footnote to Table 2
Moreover, it has been shown in ovarian cancer and earlier in glioblastomas that miR-203a-3p can target mRNA encoding ataxia-telangiectasia (ATM) (Liu et al. 2019a; Yang et al. 2017), a serine-threonine kinase activated by a double-strand DNA break and involved in the checkpoint of DNA damage response processes. This has potential implications for therapy as inhibition of ATM has been shown to sensitize gliomas to chemotherapy (Yang et al. 2017).
In addition to the brief summary in Table 9, it is worth noting that miR-203a-3p targets TYMS mRNA and a number of oncogenes in various types of cancer, such as BIRC5 encoding the anti-apoptotic Survivin and SNAI2/ZEB2 encoding transcriptional repressors that promote EMT (Pretzsch et al. 2019). On the other hand, miR-203a-3p can also target tumor suppressors and is up-regulated in HCC (Tables 8, 9).
Table 9.
Major genes targeted by miR-203a-3p in various types of cancer
| Cancers | Targeted gene products† | References |
|---|---|---|
| Cancers with down-regulation of miR-203a-3p | ||
| Acting as oncogenes or tumor promoters | ||
| CRC | TYMS | Li et al. (2015a) |
| Glioblastoma | TYMS | Chen et al. (2017b) |
| NSCLC | AVL9, BIRC5, DLX5, E2F1, ZEB2 | Liang et al. (2020); Yang et al. (2020); Wang et al. (2020a) |
| BLCA | SIX4 | Na et al. (2019) |
| PDAC | SNAI2 | An and Zheng (2020 |
| Acting both as oncogenes and tumor suppressors, or unclassified | ||
| CRC | THBS2 | Qian et al. (2019) |
| GC | IGF1R | Wang et al. (2018a) |
| OVCA | ATM | Liu et al. (2019a) |
| Cancers with up-regulation of miR-203a-3p | ||
| Acting as tumor suppressors | ||
| HCC | IL24, PTEN | Huo et al. (2017); Zhang et al. (2020) |
| Acting both as oncogenes and tumor suppressors, or unclassified | ||
| CRC | PDE4D | Chen et al. (2018) |
†mRNAs encoding the following proteins: ATM codes for ATM Serine/Threonine Kinase (Ataxia Telangiectasia Mutated), although it is considered as a tumor suppressor, ATM signaling can be involved in chemoresistance of cancer cells (Cremona and Behrens 2014) as well as promote the EMT process (Liu et al. 2019a); AVL9 codes for cell migration associated protein and is an oncogene in NSCLC; BIRC5 can be regarded as an oncogene that codes for apoptosis inhibitor Survivin; DLX5 codes for transcriptional activator and acts as oncogene (Tan and Testa 2021); E2F1 encodes E2F Transcription Factor 1; IGF1R encodes the Insulin-like Growth Factor 1 Receptor (Werner et al. 2016); IL24 codes for interleukin 24, which can induce apoptosis in a variety of cancer cells, thereby acting as a tumor suppressor; PDE4D encodes cAMP-specific Phosphodiesterase 4D; PTEN is a tumor suppressor gene encoding Phosphatase and Tensin Homolog; SIX4 codes for transcriptional regulator and acts as oncogene; THBS2 codes for Thrombospondin 2; SNAI2 codes for SLUG which represses the gene encoding E-cadherin and thus promotes the EMT process of cancer cells
Interestingly, lncRNA MALAT1 was shown to downregulate miR-203a-3p levels in glioblastoma multiforme cells, thereby promoting TYMS expression (Chen et al. 2017b). The direct interaction of MALAT1 and miRNA was indicated by the significant upregulation of miR-203a-3p in si-MALAT1 transfected human glioblastoma cells resistant to temozolomide (Chen et al. 2017b). Other studies confirmed that MALAT1 sponges miR-203a-3p in renal cell carcinoma (Zhang et al. 2019a). Luciferase reporter assays confirmed a targeted relationship between MALAT1 and miR-203a-3p, and expression level analyzes showed significant upregulation of miR-203a-3p in si-MALAT1 transfected renal cell carcinoma lines (Zhang et al. 2019a). The direct interaction of miR-203a-3p with two sites in MALAT1 was also demonstrated in luciferase reporter assays in human retinal microvascular endothelial cells (Yu et al. 2020). RT-PCR analysis of CRC samples from 85 patients also showed an inverse correlation between MALAT1 and miR-203a-3p (Wu et al. 2018b). However, the question of localization in the cellular compartment where the demonstrated direct interaction of MALAT1-miRNA takes place still needs to be answered.
MiR-218-5p
MiR-218-5p was found to be downregulated in primary CRC tissues and its expression in CRC cell lines was significantly decreased after treatment with 5FU, while ectopic overexpression of miR-218-5p directly suppressed BIRC5-encoded Survivin as well as indirectly decreased TS levels through an unknown molecular mechanism (Li et al. 2015b). MiR-218-5p can be derived from two stem-loop sequences, mir-218-1 and mir-218-2, which are transcribed from two loci located on chromosomes 4p15.31 and 5q35.1, respectively (Guan et al. 2013), and its level may be partially regulated by lncRNA HOTAIR which can recruit EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit) to the promoter of the SLIT3 gene and repress its transcription along with the mir-218-2 contained therein (Li et al. 2017b). Importantly, increased levels of MALAT1 in CRCs have been shown to directly inhibit the activity of miR-218-5p as well as cause EZH2-dependent repression of the CDH1 gene encoding E-cadherin (Li et al. 2017a). Thus, by repressing the CDH1 gene and sponging the miR-218-5p, MALAT1 can promote the EMT process and cancer cell metastasis, lead to an increase in Survivin levels and cancer cell resistance to 5FU and oxaliplatin treatment.
MiR-330-3p
Other studies found that miR-330-3p levels were reduced in CRC tissues (see Table 10) compared to adjacent normal tissues in patients, suggesting that miR-330-3p may act as a tumor suppressor (Xu et al. 2017). Importantly, miR-330-3p levels were also decreased in 5FU-resistant CRC tissues compared to 5FU-sensitive CRC tissues after surgery (Gao et al. 2021a). Moreover, it was shown that ectopic expression of miR-330 mimics directly decreased the level of TYMS-encoded protein and increased the sensitivity of CRC cells to the cytotoxic effect of 5FU (Xu et al. 2017). Also, the second strand of the mature miRNA, namely miR-330-5p, has the potential to regulate TYMS expression because the miRNA response element is located in the mRNA coding region, but the direct interaction of miR-330-5p with TYMS mRNA still needs to be supported by experimental results. MiR-330-5p levels have been found to be reduced in CRC tissues and two colon cancer cell lines (Huang et al. 2022). MiR-330-5p/3p levels can be regulated by MALAT1 and it has already been shown that MALAT1 can sponge miR-330-5p (Shi et al. 2021). The functions of miR-330-5p and miR-330-3p in various types of cancer, not yet taking into account the interaction with MALAT1, have been discussed in a recent review article (Jafarzadeh et al. 2022).
Table 10.
Expression levels of miRNAs targeting i.a. TYMS mRNA in cancer
| miRNA | Level† | Cancers‡ | References |
|---|---|---|---|
| miR-330-3p | Down | CRC tissues and HCT116, SW480 cell lines | Wang et al. (2022b) |
| Down | CRC tissues: 5FU resistant vs sensitive (after surgery) | Gao et al. (2021a) | |
| Down | CRC tissues and SW480, SW620 cell lines | Huang et al. (2020b) | |
| Down | CRC tissues and HCT116, HT29, SW620, SW480 cell lines | Xu et al. (2017) | |
| Down | GC tissues and cell lines | Ma et al. (2020) | |
| Down | GC tissues and cell lines | Guan et al. (2016) | |
| Up | ESCC: two cell lines | Meng et al. (2015) | |
| Down | LSCC tissues and cell lines | Cheng et al. (2020a) | |
| Down | LSCC tissues and two cell lines | Fan and Zhu (2022) | |
| Down | OSCC tissues | Qian et al. (2021) | |
| Up | NSCLC tissues | Wang et al. (2021e) | |
| Up | NSCLC primary tissues with vs without brain metastasis | Wei et al. (2019b) | |
| Up | NSCLC tissues and four cell lines (i.a. A549) | Chen et al. (2019b) | |
| Up | NSCLC tissues | Liu et al. (2015) | |
| Up | BRCA tissues, four cell lines (i.a. MCF-7, MDA-MB-231) | Ji et al. (2021) | |
| Up | BRCA tissues (ER/HER2 ±), metastatic vs non-metastatic | Zhang et al. (2019b) | |
| Up | BRCA tissues (ER/PR ±) | Wang et al. (2018b) | |
| Down | BRCA (TNBC) tissues (stage I–III) | He et al. (2020) | |
| Up | HCC tissues and four cell lines | Zhao et al. (2019b) | |
| Up | HCC tissues, TNM stage II + III vs I, six cell lines | Hu et al. (2017a) | |
| Up | PDAC cell lines (i.a. PANC-1, SW 1990) | Xiong et al. (2019) | |
| Down | OVCA tissues, stage III–IV vs I–II, three cell lines | Cai et al. (2021) | |
| Down | PCa tissues and four cell lines (i.a. PC-3, DU145) | Li et al. (2020b) | |
| Down | PCa: four cell lines (i.a. PC-3, DU145) | Lee et al. (2009) |
†,‡For description see footnote to Table 2
Using A549 cells, it was shown that miR-330-3p can also target PTEN mRNA and consequently promote AKT phosphorylation, which partly explains the selective pressure to increase miR-330-3p levels in NSCLC (Wang et al. 2021e). In turn, in highly invasive triple negative breast cancers, miR-330-3p targets the proto-oncogene c-Myc (He et al. 2020). Moreover, in prostate cancer cells, miR-330-3p was found to act as a tumor suppressor that targets the 3’UTR of mRNA encoding the transcription factor E2F1 (Lee et al. 2009). Thus, it can be seen that the selection pressure in cancer cells to decrease or increase the level of this miRNA is strongly dependent on the type of tumor.
Although miR-330-3p can target a variety of tumor suppressor genes, including PTEN and PDCD4, and is therefore up-regulated in various cancer types (see Table 11). In CRCs, however, miR-330-3p is down-regulated and targets TYMS mRNA and HK2 mRNA (Gao et al. 2021a). HK2 encodes Hexokinase 2 catalyzing the rate-limiting step of glucose metabolism and is therefore an enzyme that is highly expressed in rapidly growing cancer cells (Pedersen 2007; Sun et al. 2018b).
Table 11.
Major genes targeted by miR-330-3p in various types of cancer
| Cancers | Targeted gene products† | References |
|---|---|---|
| Cancers with down-regulation of miR-330-3p | ||
| Acting as oncogenes or tumor promoters | ||
| CRC | TYMS, HK2 | Xu et al. (2017); Gao et al. (2021a) |
| GC | MSI1, PRRX1 | Guan et al. (2016); Ma et al. (2020) |
| LSCC | SLC7A11, TRA2B | Fan and Zhu (2022); Cheng et al. (2020a) |
| OSCC | GLS | Qian et al. (2021) |
| BRCA | MYC | He et al. (2020) |
| PCa | BMI1, E2F1 | Li et al. (2020b); Lee et al. (2009) |
| Acting both as oncogenes and tumor suppressors, or unclassified) | ||
| CRC | MYO6, PFN1 | Wang et al. (2022b); Huang et al. (2020b) |
| NSCLC | GRIA3 | Wei et al. (2019b) |
| OVCA | RIPK4 | Cai et al. (2021) |
| Cancers with up-regulation of miR-330-3p | ||
| Acting as tumor suppressors | ||
| ESCC | PDCD4 | Meng et al. (2015) |
| NSCLC | EGR2, PTEN, RASSF1A | Liu et al. (2015); Wang et al. (2021e); Chen et al. (2019b) |
| BRCA | PDCD4 | Ji et al. (2021) |
| HCC | BTG1, ING4 | Zhao et al. (2019b); Hu et al. (2017a) |
†mRNAs encoding the following proteins: BMI1 encodes BMI1 Proto-Oncogene, Polycomb Ring Finger; BTG1 is a tumor suppressor gene that codes for BTG Anti-Proliferation Factor 1; EGR2 is a tumor suppressor gene that codes for transcription factor Early Growth Response 2; GLS encodes GLS1 (Glutaminase) which promotes proliferation as opposed to proliferation-inhibiting GLS2 (Kim and Kim 2013); GRIA3 encodes Glutamate Ionotropic Receptor AMPA Type Subunit 3; HK2 encodes Hexokinase 2 catalyzing the rate-limiting step of glucose metabolism and is highly expressed in rapidly growing cancer cells; ING4 encodes tumor suppressor protein that can bind p53; MSI1 encodes Musashi RNA-binding protein that is involved in tumorigenesis; MYC encodes transcription factor MYC Proto-Oncogene (c-Myc); MYO6 encodes motor protein Myosin VI; PFN1 encodes Profilin 1 that binds actin monomers and mediates actin polymerization but also inhibit formation of IP3 from PIP2; PDCD4 encodes programmed cell death protein 4 acting as a tumor suppressor; PRRX1 codes for Paired Related Homeobox 1 protein that can promote the EMT process in cancer cells (Du et al. 2021); RASSF1A is a tumor suppressor gene encoding Ras Association Domain Family Member 1; RIPK4 encodes Receptor Interacting Serine/Threonine Kinase 4 and acts as a tumor suppressor or promoter depending on the type of cancer (Xu et al. 2020c); SLC7A11 encodes Cystine/Glutamate Transporter (Koppula et al. 2021); TRA2B is an oncogene encoding Transformer 2 β Homolog
MiR-375-3p
Also for miR-375-3p there is a miRNA response element in the 3’UTR of TYMS mRNA (see Table 1) and overexpression of miR-375 mimics in CRC cell lines increased their sensitivity to cytotoxic activity of 5FU in vitro and in tumor-bearing mice (Xu et al. 2020b). As the transcription factor FOXM1 has been shown to up-regulate TYMS expression in CRCs (Varghese et al. 2019), it is wort noting that miR-375-3p also targets FOXM1 mRNA (Chen et al. 2020b).
MiR-375-3p is down-regulated in CRC and analysis of clinical data showed a statistically significant reduction in miR-375-3p levels in stages III–IV compared to stages I–II (Mao et al. 2016), which is inverse to higher YAP1 levels in stages III–IV than in stages I–II (Sun et al. 2019b). Notes that miR-375-3p targets YAP1, a nuclear effector of the Hippo pathway, and the down-regulation of miR-375-3p in CRCs also leads to increased expression of Cyclin D1 and Survivin, which promotes proliferation and chemoresistance of cancer cells (Xu et al. 2019a). The works cited in Table 12 also contain data indicating a reduction in miR-375-3p levels in 5FU-resistant CRCs compared to 5FU-sensitive CRCs (Chen et al. 2020b; Xu et al. 2019a).
Table 12.
Expression levels of miRNAs targeting i.a. TYMS mRNA in cancer
| miRNA | Level† | Cancers‡ | References |
|---|---|---|---|
| miR-375-3p | Down | CRC tissues, Caco2, HCT116, SW480, HT29 cell lines | Xu et al. (2020b) |
| Down | CRC tissues | Liu et al. (2020a) | |
| Down | CRC tissues: 5FU-resistant vs sensitive | Chen et al. (2020b) | |
| Down | CRC tissues, Caco2, HCT116, SW480, HT29, SW620 lines | Xu et al. (2019a) | |
| Down | HCT8/FU (5FU resistant) vs HCT8 (parental sensitive) | Xu et al. (2019a) | |
| Down | HCT116/FU (5FU resistant) vs HCT116 (sensitive) | Xu et al. (2019a) | |
| Down | CRC tissues: 5FU-resistant vs sensitive | Xu et al. (2019a) | |
| Down | in serum of CRC patients | Huang et al. (2020a) | |
| Down | CRC tissues, HCT116, SW480, HT29, SW620 cell lines | Xu et al. (2016a) | |
| Down | CRC tissues, SW480, HT29, SW620, HCT116, HCT8 | Mao et al. (2016) | |
| Down | CRC stage III–IV vs stage I–II | Mao et al. (2016) | |
| Down | CRC tissues | Wang et al. (2014b) | |
| Down | CRC tissues, HT29, SW620, HCT116 cell lines | Dai et al. (2012) | |
| Down | GC tissues and three cell lines | Liu et al. (2019b) | |
| Down | GC tissues and two cell lines | Huang et al. (2019b) | |
| Down | GC tissues and ten cell lines | Kang et al. (2018) | |
| Down | GC tissues | Chen et al. (2017d) | |
| Down | GC tissues | Yuan et al. (2018) | |
| Down | ESCC tissues and four cell lines | Li et al. (2021a) | |
| Down | ESCC tissues | Cheng et al. (2020b) | |
| Down | ESCC tissues and two cell lines | Xu et al. (2019b) | |
| Down | ESCC tissues and one cell line | Hu et al. (2017b) | |
| Down | ESCC tissues and cell lines | Kong et al. (2012) | |
| Down | LSCC and three cell lines | Chang et al. (2020) | |
| Up | LSCC: III/IV vs I/II TNM stage | Wu et al. (2016) | |
| Down | LSCC: III–IV vs I–II clinical stage | Guo et al. (2016) | |
| Down | LSCC tissues and two cell lines | Wang et al. (2016b) | |
| Down | LSCC tissues, UICC advanced III–IV vs early I–II stage | Luo et al. (2014) | |
| Down | OSCC tissues and four cell lines | Tong et al. (2021) | |
| Down | OSCC tissues and four cell lines | Wu et al. (2017) | |
| Down | OSCC tissues, with vs without lymph node metastasis | Zhang et al. (2017) | |
| Down | OSCC tissues | Shi et al. (2015) | |
| Down | NSCLC tissues and A549, H1299 cell lines | Jin et al. (2021) | |
| Down | NSCLC tissues, stages IV vs III vs II vs I | Chen et al. (2017f) | |
| Down | NSCLC (SqCLC) tissues, stage III vs II | Chen et al. (2017g) | |
| Up | BRCA tissues (luminal A/B, HER2+), three cell lines | Guan et al. (2021) | |
| Down | HCC tissues and four cell lines (i.a. Huh7) | Xu et al. (2021) | |
| Down | HCC tissues and five cell lines | Li et al. (2021b) | |
| Down | HCC tissues and four cell lines (i.a. Huh7) | Li et al. (2021c) | |
| Down | HCC tissues and five cell lines (i.a. Huh7) | Li et al. (2018b) | |
| Down | HCC tissues | He et al. (2012) | |
| Down | HCC tissues | Liu et al. (2010) | |
| Down | PDAC tissues and two cell lines (PANC-1, SW 1990) | Yonemori et al. (2017) | |
| Up | PDAC tissues and ten cell lines (i.a. PANC-1, SW 1990) | Yang et al. (2016) | |
| Down | PDAC tissues | Zhou et al. (2014b) | |
| Down | PDAC tissues | Song et al. (2013a) | |
| Down | PDAC tissues and four cell lines (i.a. PANC-1, SW 1990) | Song et al. (2013b) | |
| Down | CeCa tissues (stage I-IV) and four cell lines | Cao et al. (2021) | |
| Up | CeCa: PTX-resistant vs pre-chemotherapy tissues | Shen et al. (2013) | |
| Down | CeCa tissues, FIGO stage IIA vs IB1/IB2 | Wang et al. (2011) | |
| Down | OVCA tissues and four cell lines | Shu et al. (2021) | |
| Up | PCa tissues | Porzycki et al. (2018) |
†,‡For description see footnote to Table 2
In contrast, in breast cancers (both luminal A/B and HER2-positive) the up-regulated mir-375-3p targets the mRNA encoding the tumor suppressor FOXO1 (forkhead box protein O1), which activates the p53 signaling pathway and indeed p53 tumor suppressor was found to be decreased along with FOXO1 in breast cancer cell lines (Guan et al. 2021). Also in paclitaxel-resistant cervical cancer, miR-375-3p targeting CDH1 mRNA encoding E-cadherin was found to be up-regulated, therefore miR-375-3p may facilitate the EMT process of cervical cancer cells (Shen et al. 2013, 2014).
The direct interaction of miR-375-3p with the miRNA response element in MALAT1 was supported by the results of luciferase reporter assays performed with 293 T fibroblast cells, while the results of the RNA pull-down assays using hepatocellular carcinoma cells showed a direct interaction of MALAT1-miRNA dependent on the miR-375-3p seed sequence (Zhao et al. 2020a).
As a brief summary, Tables 12 and 13 show that down-regulation of miR-375-3p targeting FOXM1, TYMS, YAP1, PIK3CA, FZD8 can promote 5FU resistance of CRC cells, tumor growth and CRC metastasis by activating the PI3K/AKT and WNT/β-catenin signaling pathways. In CRC cells, YAP1 promotes proliferation and inhibits apoptosis by upregulating Survivin (Xu et al. 2019a). Besides CRCs, miR-375-3p targets the Hippo pathway effector YAP1 also in other cancers such as GC, HCC and OVCA (see Table 13 and Fig. 3).
Table 13.
Major genes targeted by miR-375-3p in various types of cancer
| Cancers | Targeted gene products† | References |
|---|---|---|
| Cancers with down-regulation of miR-375-3p | ||
| Acting as oncogenes or tumor promoters | ||
| CRC | TYMS, CBX3, FOXM1, | Xu et al. (2020b); Liu et al. (2020a); Chen et al. (2020b); |
| FZD8, PIK3CA, YAP1 | Xu et al. (2016a); Wang et al. (2014b Xu et al. (2019a | |
| ESCC | MTDH | Hu et al. (2017b) |
| GC | PDPK1, TEAD4, YAP1, | Chen et al. (2017d); Kang et al. (2018) |
| YBX1, YWHAZ | Huang et al. (2019b); Liu et al. (2019b) | |
| OSCC | SLC7A11 | Wu et al. (2017) |
| NSCLC | SPIN1 | Jin et al. (2021) |
| HCC | ERBB2, HMGA2, JAK2 | Li et al. (2018b); Xu et al. (2021); Li et al. (2021b) |
| HCC | MTDH, PDGFC, YAP1 | He et al. (2012); Li et al. (2021c); Liu et al. (2010) |
| HCC CSCs | YAP1 | Zhao et al. (2020a) |
| PDAC | PDPK1, ZFP36L2 | Zhou et al. (2014b); Song et al. (2013a); Yonemori et al. (2017) |
| OVCA | YAP1 | Shu et al. (2021) |
| Acting both as oncogenes and tumor suppressors, or unclassified | ||
| CRC | KLF4, SP1 | Mao et al. (2016); Xu et al. (2019a) |
| GC | CCN2 | Kang et al. (2018) |
| ESCC | HMGB1, IGF1R, SESN3, | Cheng et al. (2020b); Kong et al. (2012); Li et al. (2021a) |
| SP1 | Xu et al. (2019b) | |
| LSCC | HNF1B, IGF1R | Chang et al. (2020); Luo et al. (2014) |
| OSCC | KLF5, IGF1R, PAX6 | Shi et al. (2015); Zhang et al. (2017); Tong et al. (2021) |
| CeCa | SP1 | Wang et al. (2011) |
| Cancers with up-regulation of miR-375-3p | ||
| Acting as tumor suppressors | ||
| BRCA | FOXO1 | Guan et al. (2021) |
| CeCa | CDH1 | Shen et al. (2014) |
| Acting both as oncogenes and tumor suppressors, or unclassified | ||
| PDAC | HOXB3 | Yang et al. (2016) |
†mRNAs encoding the following proteins: CBX3 codes for protein involved in transcriptional silencing (upregulated in cancers); CCN2 codes for connective tissue growth factor; CDH1 is a tumor suppressor gene encoding E-cadherin; ERBB2 codes for HER-2 Receptor Tyrosine Kinase; FOXO1 is a tumor suppressor gene; FZD8 encodes Frizzled Class Receptor 8 which is a receptor for Wnt proteins (Sun et al. (2021); HMGA2 encodes HMG AT-Hook Protein 2, which functions in the regulation of the cell cycle; HMGB1 encodes HMG Box 1, which has both oncogenic and tumor suppression functions; HNF1B encodes Hepatocyte Nuclear Factor 1-β and functions as a tumor suppressor gene or oncogene (Chandra et al. (2021); HOXB3 encodes transcription factor Homeobox B3 (Li et al. (2019b); JAK2 encodes Janus Kinase 2 phosphorylating a tyrosine residue; KLF4 is a tumor suppressor gene encoding Kruppel Like Factor 4, but may also act as an oncogene depending on the cellular context; KLF5 encodes Kruppel Like Factor 5 and acts as an oncogene or tumor suppressor gene depending on the cellular context; PAX6 is a tumor suppressor gene encoding transcription factor Paired Box 6; PDGFC codes for Platelet Derived Growth Factor C (promotes angiogenesis); PDPK1 encodes 3-Phosphoinositide-Dependent Protein Kinase-1 (Domrachev et al. (2021); PIK3CA is an oncogene encoding PI3K, subunit p110α; SESN3 encodes sestrin3, stress-induced protein; SP1 encodes Sp1 Transcription Factor that regulates oncogenes and tumor suppressor genes; SPIN1 encodes Spindlin 1 and is considered an oncogene (Janecki et al. (2018); TEAD4 is an oncogene encoding TEA Domain Transcription Factor 4 (Hippo pathway); YBX1 is an oncogene encoding Y-Box Binding Protein 1; YAP1 is an oncogene encoding Yes1 Associated Transcriptional Regulator (Hippo pathway); ZFP36L2 is an oncogene encoding ZFP36 Ring Finger Protein Like 2; YWHAZ is functioning as an oncogene and codes for an adapter protein belonging to the 14-3-3 family
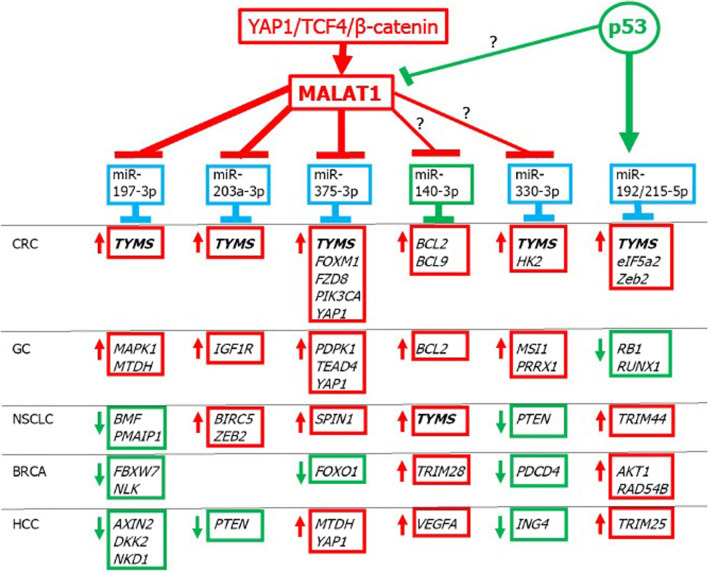
Fig. 3 .
MALAT1-miRNAs network regulating the expression level of TYMS along with other cancer-related genes in various types of cancer. Cancer type designation: CRC, colorectal cancer; GC, gastric cancer; NSCLC, non-small cell lung cancer; BRCA, breast cancer; HCC, hepatocellular carcinoma. Red indicates oncogenic effects, green indicates tumor suppressor effects, blue indicates tumor suppressor or oncogenic effects depending on the type of cancer. YAP1 in complex with TCF4/β-catenin upregulates the expression of the MALAT1 gene in CRC and HCC (Sun et al. 2019b; Wang et al. 2014a)
MiR-433-3p
In turn, overexpression of miR-433-3p in human cervical cancer HeLa cells resulted in a reduction of TYMS mRNA and protein levels, and sensitized cells to treatment with 5FU (Gotanda et al. 2013). The set of oncogenes targeted by miR-433-3p that is down-regulated in various types of cancer (Table 14) is listed in Table 15. From this overview, it can be seen that miR-433-3p targeting mRNA encoding the cAMP Response Element-Binding protein 1 (CREB1) has been found in cancer cells of various types including CRC, bladder cancer and HCC (Yan et al. 2018; Xu et al. 2016b; Yang et al. 2013).
Table 14.
Expression levels of miRNAs targeting i.a. TYMS mRNA in cancer
| miRNA | Level† | Cancers‡ | References |
|---|---|---|---|
| miR-433-3p | Down | CRC tissues, LoVo, SW620, SW480, HCT116 cell lines | Zhang et al. (2018) |
| Down | CRC: stage II vs stage I | Zhang et al. (2018) | |
| Down | CRC tissues, SW480, LoVo, HT29, Caco-2, SW620 lines | Li et al. (2018c) | |
| Down | CRC tissues and cell lines | Yan et al. (2018) | |
| Down | CRC tissues, SW620, HCT116, SW480, LoVo, HT29 lines | Li et al. (2017c) | |
| Down | GC: recurrent vs non-recurrent | Wang et al. (2021c) | |
| Down | GC tissues: stage IV vs stage II | Guo et al. (2013) | |
| Down | ESCC tissues and two cell lines | Li and Li (2021) | |
| Down | ESCC tissues | Cheng et al. (2020b) | |
| Down | OSCC tissues | Liu et al. (2020b) | |
| Down | OSCC tissues, TNM stage III/IV vs I/II | Wang et al. (2017) | |
| Down | OSCC tissues and five cell lines | Wang et al. (2015) | |
| Down | NSCLC tissues and A549, H1975 cell lines | Yu et al. (2022) | |
| Down | NSCLC tissues and A549, H522 cell lines | Guo et al. (2022) | |
| Down | NSCLC tissues and A549, H1299, H23, H1581 cell lines | Zhang et al. (2021) | |
| Down | NSCLC tissues | Liu et al. (2020c) | |
| Down | NSCLC tissues | Li et al. (2019a) | |
| Down | NSCLC tissues and four cell lines (A549, H460, H522) | Liu et al. (2018) | |
| Down | BLCA tissues and five cell lines (i.a. T24) | Wang et al. (2020b) | |
| Down | BLCA tissues and three cell lines (i.a. T24) | Xu et al. (2016b) | |
| Down | HCC tissues, stage III/IV vs I/II, four cell lines | Song et al. (2021) | |
| Down | HCC tissues | Ma et al. (2021a) | |
| Down | PDAC tissues and two cell lines (PANC-1, SW 1990) | Zhou et al. (2021b) | |
| Down | CeCa tissues and four cell lines | Liang et al. (2017) | |
| Down | OVCA tissue | Liang et al. (2016) |
†,‡For description see footnote to Table 2
Table 15.
Major genes targeted by miR-433-3p in various types of cancer
| Cancers | Targeted gene products† | References |
|---|---|---|
| Cancers with down-regulation of miR-433-3p | ||
| Acting as oncogenes or tumor promoters | ||
| CRC | CREB1, HOXA1, MACC1, MAPK8 | Yan et al. (2018); Li et al. (2018c); Li et al. (2017c) |
| GC | KRAS | Guo et al. (2013) |
| ESCC | REV3L | Li and Li (2021) |
| OSCC | HDAC6, PAK4, PTK2 | Wang et al. (2015); Liu et al. (2020b); Wang et al. (2017) |
| NSCLC | E2F3, LPCAT1, SMAD2, NUCKS1 | Liu et al. (2018); Guo et al. (2022); Li et al. (2019a); Yu et al. (2022) |
| BLCA | CREB1 | Xu et al. (2016b) |
| HCC | CREB1 | Yang et al. (2013) |
| PDAC | GOT1 | Zhou et al. (2021b) |
| CeCa | TYMS, MTDH | Gotanda et al. (2013); Liang et al. (2017) |
| Acting both as oncogenes and tumor suppressors, or unclassified | ||
| CRC | CCAR1 | Yan et al. (2018) |
| GC | ATG5, ATG12 | Wang et al. (2021c) |
| ESCC | HMGB1 | Cheng et al. (2020b) |
| NSCLC | TIPRL | Zhang et al. (2021) |
| BLCA | CCAR1 | Wang et al. (2020b) |
| HCC | CBX3, FXYD3 | Song et al. (2021); Ma et al. (2021a) |
| OVCA | NOTCH1 | Liang et al. (2016) |
†mRNAs encoding the following proteins: ATG5 and ATG12 code for Autophagy-Related Proteins 5 and 12, respectively; CBX3 encodes Chromobox Protein Homolog 3 and functions as both an oncogene and a tumor suppressor; CCAR1 codes for Cell Division Cycle And Apoptosis Regulator 1 that functions as a transcriptional coactivator; CREB1 codes for transcription factor cAMP Response Element Binding Protein 1; E2F3 is an oncogene coding for E2F Transcription Factor 3; FXYD3 codes for Sodium/Potassium-Transporting ATPase Subunit FXYD3; GOT1 codes for Glutamic-Oxaloacetic Transaminase 1 that functions as a tumor promoter in pancreatic cancer growth; HDAC6 encodes Histone Deacetylase 6; HOXA1 is an oncogene coding for transcription factor Homeobox A1 (Li et al. 2019b); KRAS encodes KRAS Proto-Oncogene (GTPase K-Ras); LPCAT1 is an oncogene encoding Lysophosphatidylcholine Acetyltransferase 1 (Wei et al. 2019c); MACC1 codes for Metastasis-Associated in Colon Cancer protein 1; MAPK8 codes for Mitogen-Activated Protein Kinase 8 and is involved in colon cancer chemoresistance (Wu et al. 2019); NOTCH1 encoding Notch Receptor 1 acts as an oncogene or tumor suppressor gene, depending on the cellular context; NUCKS1 is recognized as oncogene encoding Nuclear Casein Kinase and cyclin dependent Kinase Substrate 1 that is involved in DNA repair and promotes proliferation, invasion and migration of NSCLC (Zhao et al. 2020c); PAK4 gene codes for P21 (RAC1) Activated Kinase 4, serine/threonine protein kinase; PTK2 encodes non-receptor Protein Tyrosine Kinase 2, also known as Focal Adhesion Kinase; REV3L codes for catalytic subunit of the DNA polymerase zeta complex, involved in the chemoresistance of ESCC (Zhu et al. 2016); SMAD2 encodes the SMAD Family Member 2 protein, which can act as an oncogenic protein in the TGF-β pathway (Pefani et al. 2016); TIPRL codes for TOR Signaling Pathway Regulator that was found as metastasis suppressor in gastric cancer
MiR-1307-3p
Mir-1307-3p targets a variety of tumor suppressor genes and is rather up-regulated in various types of cancer (see Tables 16, 17). However, miR-1307-3p also directly targets 3’UTR of TYMS mRNA and is significantly involved in the regulation of TS protein levels, as in CRC patients with low miR-1307-3p levels (due to T > C SNP in the terminal-loop of pre-miRNA) (Tang et al. 2015) elevated TS levels were reported along with insensitivity to capecitabine chemotherapy (Chen et al. 2017a). Moreover, relatively high levels of miR-1307-3p and low levels of TYMS-encoded protein were found in Caco-2 and HCT116 cell lines, while low levels of miR-1307-3p along with high levels of TYMS-encoded protein were found in SW480, LoVo and DLD1 cell lines (Chen et al. 2017a). This suggests a significant involvement of miR-1307-3p in the regulation of TYMS expression in at least some colon cancers. However, this effect does not appear to be of any importance in other cancers. For example, in the early stages of breast cancer of a various molecular subtypes (luminal, triple negative), miR-1307-3p significantly targets the mRNA encoding the SMYD4 protein (Han et al. 2019) identified as a tumor suppressor in breast cancer (Hu et al. 2009).
Table 16.
Expression levels of miRNAs targeting i.a. TYMS mRNA in cancer
| miRNA | Level† | Cancers‡ | References |
|---|---|---|---|
| miR-1307-3p | Down | CRC tissues, HT29, SW480, SW620, HCT116 cell lines | Zheng et al. (2019a) |
| Down | CRC cell lines: LoVo, DLD1, SW620, SW480 | Chen et al. (2017a) | |
| Up | SW480 and SW620 colon cancer cell lines | Yue et al. (2020) | |
| Up | GC tissues and cell lines | Ma et al. (2021b) | |
| Up | BLCA tissues | Wang et al. (2021b) | |
| Up | BRCA tissues (stage I), cell lines (MCF-7, MDA-MB-231) | Han et al. (2019) | |
| Up | HCC tissues | Wang et al. (2019b) | |
| Up | HCC tissues and four cell lines | Chen et al. (2019a) | |
| Up | OVCA: PTX-resistant vs sensitive cancer tissue | Zhou et al. (2015) | |
| Up | PCa tissue and five cell lines (i.a. PC-3, DU145) | Qiu and Dou (2017) |
†,‡For description see footnote to Table 2
Table 17.
Major genes targeted by miR-1307-3p in various types of cancer
| Cancers | Targeted gene products† | References |
|---|---|---|
| Cancers with down-regulation of miR-1307-3p | ||
| Acting as oncogenes or tumor promoters | ||
| CRC | TYMS | Chen et al. (2017a) |
| Acting as tumor suppressors | ||
| CRC | ISM1 | Zheng et al. (2019a); Yue et al. (2020) |
| Cancers with up-regulation of miR-1307-3p | ||
| Acting as tumor suppressors | ||
| CRC | TRARG1 | Zheng et al. (2019a) |
| GC | DAB2IP | Ma et al. (2021b) |
| BLCA | SMG1 | Wang et al. (2021b) |
| BRCA | SMYD4 | Han et al. (2019) |
| HCC | DAB2IP, ISM1 | Chen et al. (2019a); Wang et al. (2019b) |
| OVCA | DAPK3 | Zhou et al. (2015) |
| PCa | FOXO3 | Qiu and Dou (2017) |
†mRNAs encoding the following proteins: DAB2IP is a tumor suppressor gene encoding DAB2 Interacting Protein, a Ras GTPase-activating protein; DAPK3 is a tumor suppressor gene that codes for serine/threonine Death Associated Protein Kinase 3; FOXO3 is a tumor suppressor gene encoding a transcription factor that activates genes encoding i.a. pro-apoptotic BIM and PUMA; ISM1 codes for Isthmin 1 that acts as an angiogenesis inhibitor; SMG1 is a tumor suppressor gene encoding a protein that can enhance p53 activity (Gewandter et al. 2011); SMYD4 is a tumor suppressor gene involved in the development of breast cancer; TRARG1 codes for Trafficking Regulator of GLUT4
In summary, a series of at least a few miRNAs (see Table 1 and Fig. 2) acting directly on TYMS mRNA fine-tune TS enzyme protein levels according to cell needs and may contribute to cancer cell resistance to the cytotoxic effects of 5FU. In addition, at least two lncRNAs, MALAT1 and TUG1, affect TS levels by sponging the respective miRNAs (see Table 18). Other lncRNAs, such as HOTAIR and XIST, also appear to affect TS levels and the sensitivity of cancer cells to 5FU, although the mechanisms of action are still poorly understood.
Table 18.
lncRNA MALAT1 sponging miRNAs targeting TYMS
| lncRNA | Confirmed or putative binding sites for miRNA† | References |
|---|---|---|
| MALAT1 | Confirmed as ceRNA for miRNAs: | |
| miR-197-3p at 2775–2781, 2814–2820, 2854–2864 | Yang et al. (2019) | |
| miR-203a-3p at 5414–5419, 6400–6405 | Yu et al. (2020); Zhang et al. (2019a); Chen et al. (2017b) | |
| miR-375-3p at 772–778 | Zhao et al. (2020a) | |
| MALAT1 | Presumably can sponge miRNAs: | |
| miR-140-3p at 683–689, 4525–4531, 8737–8743 | – | |
| miR-330-3p at 8439–8445, and other | – | |
| TUG1 | Confirmed as ceRNA for miRNAs: | |
| miR-140-3p at 2096–2101, 6438–6443, 6728–6733 | Yuan et al. (2021) | |
| miR-197-3p at 1438–1444, 2098–2103, 3549–3554 | Wang et al. (2019a); Tang et al. (2018) | |
†Sites in MALAT1 and TUG1 (transcript variant 8) according to the GenBank sequence accession numbers NR_002819.4, NM_001398480.1, respectively; sites confirmed by the luciferase reporter assay are underlined
MiRNAs targeting DPYD mRNA
Two highly homologous miRNAs, miR-27a-3p and miR-27b-3p, have been shown to directly reduce the levels of DPYD mRNA and DPD protein in CRC cells, and ectopic expression of miR-27a-3p and miR-27b-3p significantly increased the sensitivity of cancer cells to the cytotoxic effect of 5FU (Offer et al. 2014). Another report showed that ectopic overexpression of miR-302b-3p led to an increase in the sensitivity of HCC cell lines to 5FU by negatively regulating DPD protein levels, as well as inhibiting entry into the S phase of the cell cycle and promoting apoptosis by targeting mRNA encoding the anti-apoptotic protein MCL-1 (Cai et al. 2015). MiR-494-3p was also found to directly target DPYD mRNA and miR-494-3p was shown to be reduced in CRC cells selected for 5FU resistance, compared to 5FU sensitive parental cells, while miR-494 mimic caused restoration of cell chemosensitivity to 5FU (Chai et al. 2015).
It should also be noted that the levels of enzymes encoded by the TYMS, TYMP and DPYD genes are indirectly regulated by miR-21-5p targeting the human mutS homolog2 (hMSH2) in CRC cells (Deng et al. 2014a). Finally, it is worth mentioning that both miR-215-5p and miR-21-5p, regulating the TYMS-encoded protein directly or indirectly, are included in a set of six miRNAs whose levels were tested in CRC surgical specimens for verification as a diagnostic tool to predict which stage II CRC patients may benefit from chemotherapy following radical surgery for CRC. That clinical trial project was registered at ClinicalTrial.gov as NCT02635087. According to published data, miR-215-5p along with miR-103a-3p and miR-143-5p, both alone and in combination, have proven to be reliable stage II CRC biomarkers that can help stratify patients into risk groups and identify patients who could potentially benefit from postoperative chemotherapy (Caritg et al. 2016).
Conclusions and future perspectives
Putting it all together, it can be seen that, at least in colon cancer, several miRNAs targeting TYMS mRNA have been identified, the levels of which can be regulated to varying degrees by long non-coding RNAs, creating a complex regulatory network. Due to their negative regulation by MALAT1, which can consequently increase the level of the TS enzyme and decrease the susceptibility to 5FU treatment, these miRNAs can be divided into three groups (Fig. 3).
The first group of miRNAs targeting TYMS mRNA consists of miR-197-3p, miR-203a-3p and miR-375-3p which are negatively regulated by MALAT1 as confirmed experimentally. The levels of these miRNAs are actually reduced in colon and gastric cancers. On the other hand, the selection pressure to increase miR-197-3p levels may be suggested in non-small cell lung cancer, bladder cancer, breast cancer and hepatocellular carcinoma (Table 6 and Fig. 2), presumably because miR-197-3p also targets mRNAs encoding proteins that function as tumor suppressors in these types of cancer, such as apoptosis initiators in lung cancers (Fiori et al. 2014) or negative regulators of the Wnt/β-catenin pathway in hepatocellular carcinoma (Hu et al. 2018a). On the other hand, decreased levels of miR-375-3p were found in most of the cancer types shown in Fig. 2, and apart from the targeted TYMS mRNA, the targeting of YAP1 mRNA by miR-375-3p has also been found in various cancers (Table 13).
The second group of miRNAs targeting TYMS mRNAs consists of miR-140-3p and miR-330-3p, which could potentially be sponged by MALAT1, but the inhibition of these miRNAs by MALAT1 has not yet been supported by experimental results. While miR-140-3p levels have been found to be down-regulated in all the cancer types shown in Fig. 2, miR-330-3p levels have also been found to be reduced in colon and gastric cancers, but the influence of selection pressure on the elevation of miR-330-3p levels may be suggested in non-small cell lung cancer, breast cancer and hepatocellular carcinoma, in which miR-330-3p seems to be significantly involved also in targeting tumor suppressor genes such as PTEN, PDCD4, and ING4 (Table 11 and Fig. 3).
The third group of miRNAs targeting TYMS mRNAs consists of miR-192-5p, miR-215-5p, miR-433-3p and miR-1307-3p whose seed sequences do not recognize complementary miRNA response elements within MALAT1. MiR-192-5p and miR-215-5p are up-regulated by p53 in response to DNA damage and target mRNAs encoding i.a. proliferation promoting proteins including TYMS mRNA (Fig. 3). MiR-192/215-5p also target mRNA encoding the transcriptional corepressor ZEB2 (Chen et al. 2017c), which is involved in the transformation of epithelial cancer cells into cells capable of migration, invasion and metastasis. MiR-192/215-5p are down-regulated in most types of cancers (Fig. 2), but in the invasive and metastatic stages of gastric cancers there appears to be a selection pressure on the up-regulation of these miRNA, presumably because they also target mRNA encoding a significant tumor suppressor and Retinoblastoma-associated protein (Chen et al. 2017e). In turn, the potential involvement of miR-433-3p and miR-1307-3p in the regulation of TS levels in colon cancers does not seem to be well documented. Although miR-433 is down-regulated in all the cancer types analyzed (Fig. 2), miR-433-3p targeting TYMS mRNA and its contribution to increasing 5FU susceptibility was demonstrated primarily in cervical cancer HeLa cells (Gotanda et al. 2013). To conclude this part of the discussion, although one report showed that miR-1307-3p targets TYMS mRNA in colon cancer cells (Chen et al. 2017a), the biological significance of this interaction is uncertain as miR-1307-3p is elevated in most of the cancer types analyzed here (Fig. 2).
Considering the putative MALAT1-miRNAs interaction network presented in Fig. 3, a potential positive feedback loop is seen to the upregulate MALAT1 expression in colon cancer as well as in hepatocellular carcinoma (Fig. 4). In both colon cancer and hepatocellular carcinoma, it was found that YAP1 can complex with TCF4/β-catenin at the promoter region of the MALAT1 gene and enhance the expression of this gene (Sun et al. 2019b; Wang et al. 2014a). Further, the lncRNA MALAT1 can directly interact with miR-375-3p as revealed by the results of the RNA pull-down assay in Huh7 hepatocellular carcinoma cells, thus counteracting the down-regulation of YAP1 by miR-375-3p (Zhao et al. 2020a). Thus, YAP1, by activating the BIRC5 gene encoding the anti-apoptotic Survivin, contributes, along with the increase in the TS enzyme level, to reducing the sensitivity of colorectal cancer cells to the 5FU treatment (Xu et al. 2019a). YAP1 also promotes the EMT process which enables the cancer metastasis process (Ling et al. 2017). Interestingly, the direct interaction of YAP1 with the TCF4/β-catenin complex was also found in the nuclei of placenta-derived mesenchymal stem cells (Feng et al. 2020). It would be valuable to try to determine whether the involvement of YAP1 in the upregulation of MALAT1 gene expression is unique in colon cancer and hepatocellular carcinoma, or whether a similar upregulation mechanism of MALAT1 gene expression is involved in various types of cancer. The exact mechanism leading to aberrant expression of MALAT1 is still unknown, but various transcription factors, coactivators and RNA-binding proteins may be involved (Xu et al. 2022). Therefore, it may also be the starting point for the search for small molecule inhibitors of the YAP1/TCF4/β-catenin transcription complex (Yong et al. 2021; Yan et al. 2020a; Tang et al. 2019b). This could be a fruitful area for further research in the context of the lack of approved therapy targeting the abnormal overexpression of MALAT1 (Uthman et al. 2021). Presumably, at least partially suppressing MALAT1 overexpression by influencing the miRNA interaction network could both reduce tumor metastatic potential and increase the sensitivity of cancer cells to 5FU.
Fig. 4 .
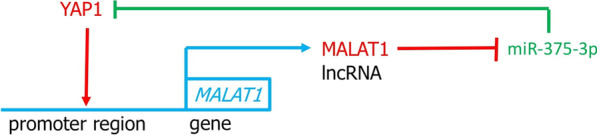
Hypothetical feedback between lncRNA MALAT1 and YAP1 via miR-375-3p enhancing MALAT1 gene expression
Finally, it would also be important to estimate the weight of the individual elements of the MALAT1-miRNAs network, taking into account their actual concentrations in a 5FU-sensitive cancer cell compared to a 5FU resistant cell, as this could contribute to the construction of a diagnostic tools to monitor the therapeutic process.
This article focuses on those non-coding RNAs that, by regulating the level of thymidylate synthase, can influence the sensitivity of cancer cells to 5FU treatment. It is worth noting, however, that the MALAT1 and miRNAs network outlined here regulating the level of the protein encoded by TYMS is rather a subset of a much larger set of non-coding RNA networks involving MALAT1 (Su et al. 2021; Poursheikhani et al. 2020). The effectiveness of 5FU therapy may also depend on other non-coding RNAs, not mentioned in this article, that may affect the concentration of fluoropyrymidines inside the cell, as well as affect glucose metabolism in the cancer cell (Marjaneh et al. 2019), DNA damage repair, cell cycle regulation, inducing and executing apoptosis. While the main goal of looking for interactions between non-coding RNAs in cancer cells is to open up new possibilities for diagnosis and therapy, identifying potential MALAT1 and miRNAs interaction networks is also interesting in itself, as like the Rosetta Stone mentioned earlier (Salmena et al. 2011), it points to complex mechanisms of fine-tuning the expression level of key genes involved in the proliferation of cancer cells.
Acknowledgements
Not applicable.
Abbreviations
- 5FU
5-Fluorouracil
- BLCA
Bladder cancer
- BRCA
Breast cancer
- CeCa
Cervical cancer
- ceRNA
Competing endogenous RNA
- CRC
Colorectal cancer
- CSC
Cancer stem-like cell
- DPD
Dihydropyrimidine dehydrogenase
- EMT
Epithelial-mesenchymal transition
- ESCC
Esophageal squamous cell carcinoma
- GC
Gastric cancer
- HCC
Hepatocellular carcinoma
- lncRNA
Long non-coding RNA
- LSCC
Laryngeal squamous cell carcinoma
- LUAD
Lung adenocarcinoma
- miRNA
MicroRNA
- mRNA
Messenger RNA
- NSCLC
Non-small-cell lung cancer
- OSCC
Oral squamous cell carcinoma
- OVCA
Ovarian cancer
- PCa
Prostate cancer
- PDAC
Pancreatic ductal adenocarcinoma
- RISC
RNA-induced silencing complex
- SqCLC
Squamous cell lung carcinoma
- TNBC
Triple-negative breast cancer
- TNM
Tumor-node-metastasis
- TS
Thymidylate synthase
Author contributions
JM: designed and wrote the manuscript; the author read and approved the final manuscript.
Funding
None.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The author declares no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ak Aksoy S, Tunca B, Erçelik M, Tezcan G, Ozturk E, Cecener G, Ugras N, Yilmazlar T, Yerci O. Early-stage colon cancer with high MALAT1 expression is associated with the 5-Fluorouracil resistance and future metastasis. Mol Biol Rep. 2022 doi: 10.1007/s11033-022-07680-y. [DOI] [PubMed] [Google Scholar]
- Almeida BO, Machado-Neto JA. Emerging functions for ANKHD1 in cancer-related signaling pathways and cellular processes. BMB Rep. 2020;53:413–418. doi: 10.5483/BMBRep.2020.53.8.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An N, Zheng B. MiR-203a-3p inhibits pancreatic cancer cell proliferation, EMT, and apoptosis by regulating SLUG. Technol Cancer Res Treat. 2020;19:1533033819898729. doi: 10.1177/1533033819898729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun G, Aggarwal D, Spector DL. MALAT1 long non-coding RNA: functional implications. Noncoding RNA. 2020;6:22. doi: 10.3390/ncrna6020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azwar S, Seow HF, Abdullah M, Faisal Jabar M, Mohtarrudin N. Recent updates on mechanisms of resistance to 5-fluorouracil and reversal strategies in colon cancer treatment. Biology (basel) 2021;10:854. doi: 10.3390/biology10090854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Lu Y, Wang X, Feng W, Sun X, Guo H, Tang C, Zhang X, Shi Q, Yu H. Eukaryotic translation initiation factor 5A2 (eIF5A2) regulates chemoresistance in colorectal cancer through epithelial mesenchymal transition. Cancer Cell Int. 2015;15:109. doi: 10.1186/s12935-015-0250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. Metazoan microRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Guo Y, Li Q, Liu L, Bao S, Xu P. Role of long intergenic non-protein coding RNA 01857 in hepatocellular carcinoma malignancy via the regulation of the microRNA-197-3p/anterior GRadient 2 axis. PLoS ONE. 2021;16:e0258312. doi: 10.1371/journal.pone.0258312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondy S, David V, Verdier M, Mathonnet M, Perraud A, Christou N. 5-Fluorouracil resistance mechanisms in colorectal cancer: from classical pathways to promising processes. Cancer Sci. 2020;111:3142–3154. doi: 10.1111/cas.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, Garcia-Foncillas J, Bandres E. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther. 2010;9:2265–2275. doi: 10.1158/1535-7163.MCT-10-0061. [DOI] [PubMed] [Google Scholar]
- Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Ørntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, He K, Chang S, Tong D, Huang C. MicroRNA-302b enhances the sensitivity of hepatocellular carcinoma cell lines to 5-FU via targeting Mcl-1 and DPYD. Int J Mol Sci. 2015;16:23668–23682. doi: 10.3390/ijms161023668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Ye L, Hu X, He W, Zhuang D, Guo Q, Shu K, Jie Y. MicroRNA miR-330-3p suppresses the progression of ovarian cancer by targeting RIPK4. Bioengineered. 2021;12:440–449. doi: 10.1080/21655979.2021.1871817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Li H, Li L. LncRNA SNHG17 contributes to the progression of cervical cancer by targeting microRNA-375-3p. Cancer Manag Res. 2021;13:4969–4978. doi: 10.2147/CMAR.S312469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caritg O, Navarro A, Moreno I, Martínez-Rodenas F, Cordeiro A, Muñoz C, Ruiz-Martinez M, Santasusagna S, Castellano JJ, Monzó M. Identifying high-risk stage II colon cancer patients: a three-microRNA-based score as a prognostic biomarker. Clin Colorectal Cancer. 2016;15:e175–e182. doi: 10.1016/j.clcc.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Chai J, Dong W, Xie C, Wang L, Han DL, Wang S, Guo HL, Zhang ZL. MicroRNA-494 sensitizes colon cancer cells to fluorouracil through regulation of DPYD. IUBMB Life. 2015;67:191–201. doi: 10.1002/iub.1361. [DOI] [PubMed] [Google Scholar]
- Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19:1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Srinivasan S, Batra J. Hepatocyte nuclear factor 1 beta: a perspective in cancer. Cancer Med. 2021;10:1791–1804. doi: 10.1002/cam4.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Wei Z, Cao H. miR-375-3p inhibits the progression of laryngeal squamous cell carcinoma by targeting hepatocyte nuclear factor-1β. Oncol Lett. 2020;20:80. doi: 10.3892/ol.2020.11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol Cancer. 2020;19:167. doi: 10.1186/s12943-020-01287-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang C. miR-197-3p-induced downregulation of lysine 63 deubiquitinase promotes cell proliferation and inhibits cell apoptosis in lung adenocarcinoma cell lines. Mol Med Rep. 2018;17:3921–3927. doi: 10.3892/mmr.2017.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Han S, Huang W, Wu J, Liu Y, Cai S, He Y, Wu S, Song W. MicroRNA-215 suppresses cell proliferation, migration and invasion of colon cancer by repressing Yin-Yang 1. Biochem Biophys Res Commun. 2016;479:482–488. doi: 10.1016/j.bbrc.2016.09.089. [DOI] [PubMed] [Google Scholar]
- Chen Q, Mao Y, Meng F, Wang L, Zhang H, Wang W, Hua D. Rs7911488 modified the efficacy of capecitabine-based therapy in colon cancer through altering miR-1307-3p and TYMS expression. Oncotarget. 2017;8:74312–74319. doi: 10.18632/oncotarget.19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Xu XK, Li JL, Kong KK, Li H, Chen C, He J, Wang F, Li P, Ge XS, Li FC. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017;8:22783–22799. doi: 10.18632/oncotarget.15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DL, Lu YX, Zhang JX, Wei XL, Wang F, Zeng ZL, Pan ZZ, Yuan YF, Wang FH, Pelicano H, Chiao PJ, Huang P, Xie D, Li YH, Ju HQ, Xu RH. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836–4849. doi: 10.7150/thno.20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Liu H, Yang H, Gao Y, Zhang G, Hu J. The long noncoding RNA, TINCR, functions as a competing endogenous RNA to regulate PDK1 expression by sponging miR-375 in gastric cancer. Onco Targets Ther. 2017;10:3353–3362. doi: 10.2147/OTT.S137726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Liu K, Li L, Chen Y, Du S. miR-215 promotes cell migration and invasion of gastric cancer by targeting Retinoblastoma tumor suppressor gene 1. Pathol Res Pract. 2017;213:889–894. doi: 10.1016/j.prp.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Chen LJ, Li XY, Zhao YQ, Liu WJ, Wu HJ, Liu J, Mu XQ, Wu HB. Down-regulated microRNA-375 expression as a predictive biomarker in non-small cell lung cancer brain metastasis and its prognostic significance. Pathol Res Pract. 2017;213:882–888. doi: 10.1016/j.prp.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Gan TQ, Qin H, Huang SN, Yang LH, Fang YY, Li ZY, Pan LJ, Chen G. Implication of downregulation and prospective pathway signaling of microRNA-375 in lung squamous cell carcinoma. Pathol Res Pract. 2017;213:364–372. doi: 10.1016/j.prp.2017.01.007. [DOI] [PubMed] [Google Scholar]
- Chen R, Liu Y, Zhuang H, Yang B, Hei K, Xiao M, Hou C, Gao H, Zhang X, Jia C, Li L, Li Y, Zhang N. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017;45:9947–9959. doi: 10.1093/nar/gkx600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gao H, Liang J, Qiao J, Duan J, Shi H, Zhen T, Li H, Zhang F, Zhu Z, Han A. miR-203a-3p promotes colorectal cancer proliferation and migration by targeting PDE4D. Am J Cancer Res. 2018;8:2387–2401. [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang L, Yao B, Liu Q, Guo C. miR-1307-3p promotes tumor growth and metastasis of hepatocellular carcinoma by repressing DAB2 interacting protein. Biomed Pharmacother. 2019;117:109055. doi: 10.1016/j.biopha.2019.109055. [DOI] [PubMed] [Google Scholar]
- Chen T, Yang Z, Liu C, Wang L, Yang J, Chen L, Li W. Circ_0078767 suppresses non-small-cell lung cancer by protecting RASSF1A expression via sponging miR-330-3p. Cell Prolif. 2019;52:e12548. doi: 10.1111/cpr.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ju H, Zhao T, Yu S, Li P, Jia J, Li N, Jing X, Tan B, Li Y. hsa_circ_0092306 targeting miR-197-3p promotes gastric cancer development by regulating PRKCB in MKN-45 Cells. Mol Ther Nucleic Acids. 2019;18:617–626. doi: 10.1016/j.omtn.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Feng Y, Zhang H, Shi X, Li B, Ju W, Yu X, Zhang N, Luo X. MicroRNA-192 inhibits cell proliferation and induces apoptosis in human breast cancer by targeting caveolin 1. Oncol Rep. 2019;42:1667–1676. doi: 10.3892/or.2019.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhu C, Jin Y. The oncogenic and tumor suppressive functions of the long noncoding RNA MALAT1: an emerging controversy. Front Genet. 2020;11:93. doi: 10.3389/fgene.2020.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Pei L, Xie P, Guo G. Circ-PRKDC contributes to 5-fluorouracil resistance of colorectal cancer cells by regulating miR-375/FOXM1 axis and Wnt/β-Catenin pathway. Onco Targets Ther. 2020;13:5939–5953. doi: 10.2147/OTT.S253468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Lin CH, Huang LY, Qiu XM. CircRNA SMARCC1 sponges MiR-140-3p to regulate cell progression in colorectal cancer. Cancer Manag Res. 2020;12:4899–4910. doi: 10.2147/CMAR.S254185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Jiang J, Zhao Y, Wang X, Zhang C, Zhuan L, Zhang D, Zheng Y. Circular RNA circNTRK2 facilitates the progression of esophageal squamous cell carcinoma through up-regulating NRIP1 expression via miR-140-3p. J Exp Clin Cancer Res. 2020;39:133. doi: 10.1186/s13046-020-01640-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Cai S, Gu T, Song F, Xue Y, Sun D. MiR-140-3p impedes gastric cancer progression and metastasis by regulating BCL2/BECN1-mediated autophagy. Onco Targets Ther. 2021;14:2879–2892. doi: 10.2147/OTT.S299234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhu H, Gao W. MicroRNA-330-3p represses the proliferation and invasion of laryngeal squamous cell carcinoma through downregulation of Tra2β-mediated Akt signaling. Mol Cell Probes. 2020;52:101574. doi: 10.1016/j.mcp.2020.101574. [DOI] [PubMed] [Google Scholar]
- Cheng H, Jiang W, Song Z, Li T, Li Y, Zhang L, Wang G. Circular RNA circLPAR3 facilitates esophageal squamous cell carcinoma progression through upregulating HMGB1 via sponging miR-375/miR-433. Onco Targets Ther. 2020;13:7759–7771. doi: 10.2147/OTT.S244699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WL, Feng PH, Lee KY, Chen KY, Sun WL, Van Hiep N, Luo CS, Wu SM. The role of EREG/EGFR pathway in tumor progression. Int J Mol Sci. 2021;22:12828. doi: 10.3390/ijms222312828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collienne M, Arnold D. The optimal duration of adjuvant chemotherapy in colon cancer. Cancers (basel) 2020;12:2509. doi: 10.3390/cancers12092509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contarelli S, Fedele V, Melisi D. HOX genes family and cancer: a novel role for homeobox B9 in the resistance to anti-angiogenic therapies. Cancers (BASEL) 2020;12:3299. doi: 10.3390/cancers12113299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona CA, Behrens A. ATM signalling and cancer. Oncogene. 2014;33:3351–3360. doi: 10.1038/onc.2013.275. [DOI] [PubMed] [Google Scholar]
- Dai X, Chiang Y, Wang Z, Song Y, Lu C, Gao P, Xu H. Expression levels of microRNA-375 in colorectal carcinoma. Mol Med Rep. 2012;5:1299–1304. doi: 10.3892/mmr.2012.815. [DOI] [PubMed] [Google Scholar]
- Dai W, Wang C, Wang F, Wang Y, Shen M, Chen K, Cheng P, Zhang Y, Yang J, Zhu R, Zhang H, Li J, Zheng Y, Lu J, Zhou Y, Xu L, Guo C. Anti-miR-197 inhibits migration in HCC cells by targeting KAI1/CD82. Biochem Biophys Res Commun. 2014;446:541–548. doi: 10.1016/j.bbrc.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Deac AL, Burz CC, Militaru C, Bocșan IC, Pop RM, Achimaș-Cadariu P, Buzoianu AD. Role of microRNAs in fluoropyrimidine-related toxicity: what we know. Eur Rev Med Pharmacol Sci. 2021;25:3306–3315. doi: 10.26355/eurrev_202104_25742. [DOI] [PubMed] [Google Scholar]
- Dean L, Kane M. (2016a) Fluorouracil therapy and DPYD genotype. In: Pratt VM, Scott SA, Pirmohamed M, Esquivel B, Kane MS, Kattman BL, Malheiro AJ, editors. Medical genetics summaries [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2012–2016a Nov 3. Accessed 11 Jan 2021.
- Dean L, Kane M. (2016b) Capecitabine therapy and DPYD genotype. In: Pratt VM, Scott SA, Pirmohamed M, Esquivel B, Kane MS, Kattman BL, Malheiro AJ, editors. Medical genetics summaries [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2012–2016b Sep 15. Accessed 02 Nov 2020.
- Deng J, Lei W, Fu JC, Zhang L, Li JH, Xiong JP. Targeting miR-21 enhances the sensitivity of human colon cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys Res Commun. 2014;443:789–795. doi: 10.1016/j.bbrc.2013.11.064. [DOI] [PubMed] [Google Scholar]
- Deng Y, Huang Z, Xu Y, Jin J, Zhuo W, Zhang C, Zhang X, Shen M, Yan X, Wang L, Wang X, Kang Y, Si J, Zhou T. MiR-215 modulates gastric cancer cell proliferation by targeting RB1. Cancer Lett. 2014;342:27–35. doi: 10.1016/j.canlet.2013.08.033. [DOI] [PubMed] [Google Scholar]
- Dey A, Varelas X, Guan KL. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov. 2020;19:480–494. doi: 10.1038/s41573-020-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol. 2015;33:1787–1796. doi: 10.1200/JCO.2014.60.0213. [DOI] [PubMed] [Google Scholar]
- Domrachev B, Singh S, Li D, Rudloff U. Mini-review: PDPK1 (3-phosphoinositide dependent protein kinase-1), an emerging cancer stem cell target. J Cancer Treat Diagn. 2021;5:30–35. doi: 10.29245/2578-2967/2021/1.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Yao C, Teng X, Chai J, Yang X, Li B. MiR-140-3p suppressed cell growth and invasion by downregulating the expression of ATP8A1 in non-small cell lung cancer. Tumour Biol. 2016;37:2973–2985. doi: 10.1007/s13277-015-3452-9. [DOI] [PubMed] [Google Scholar]
- Dou D, Ren X, Han M, Xu X, Ge X, Gu Y, Wang X, Zhao S. Circ_0008039 supports breast cancer cell proliferation, migration, invasion, and glycolysis by regulating the miR-140-3p/SKA2 axis. Mol Oncol. 2021;15:697–709. doi: 10.1002/1878-0261.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Liu X, Yang M, Wang W, Sun J. The regulatory role of PRRX1 in cancer epithelial-mesenchymal transition. Onco Targets Ther. 2021;14:4223–4229. doi: 10.2147/OTT.S316102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D, Zhu Y. Circ_0120175 promotes laryngeal squamous cell carcinoma development through up-regulating SLC7A11 by sponging miR-330-3p. J Mol Histol. 2022 doi: 10.1007/s10735-022-10061-1. [DOI] [PubMed] [Google Scholar]
- Fan L, Huang X, Chen J, Zhang K, Gu YH, Sun J, Cui SY. Long noncoding RNA MALAT1 contributes to sorafenib resistance by targeting miR-140-5p/Aurora-A signaling in hepatocellular carcinoma. Mol Cancer Ther. 2020;19:1197–1209. doi: 10.1158/1535-7163.MCT-19-0203. [DOI] [PubMed] [Google Scholar]
- Feng X, Liu J, Xu Y, Zhu J, Chen W, Feng B, Pan Q, Yu J, Shi X, Yang J, Li Y, Li L, Cao H. Molecular mechanism underlying the difference in proliferation between placenta-derived and umbilical cord-derived mesenchymal stem cells. J Cell Physiol. 2020;235:6779–6793. doi: 10.1002/jcp.29572. [DOI] [PubMed] [Google Scholar]
- Fesler A, Xu X, Zheng X, Li X, Jiang J, Russo JJ, Ju J. Identification of miR-215 mediated targets/pathways via translational immunoprecipitation expression analysis (TrIP-chip) Oncotarget. 2015;6:24463–24473. doi: 10.18632/oncotarget.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori ME, Barbini C, Haas TL, Marroncelli N, Patrizii M, Biffoni M, De Maria R. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ. 2014;21:774–782. doi: 10.1038/cdd.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchino F, Rudà R, Soffietti R. Mechanisms and therapy for cancer metastasis to the brain. Front Oncol. 2018;8:161. doi: 10.3389/fonc.2018.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao JB, Zhu MN, Zhu XL. miRNA-215-5p suppresses the aggressiveness of breast cancer cells by targeting Sox9. FEBS Open Bio. 2019;9:1957–1967. doi: 10.1002/2211-5463.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SJ, Ren SN, Liu YT, Yan HW, Chen XB. Targeting EGFR sensitizes 5-Fu-resistant colon cancer cells through modification of the lncRNA-FGD5-AS1-miR-330-3p-Hexokinase 2 axis. Mol Ther Oncolytics. 2021;23:14–25. doi: 10.1016/j.omto.2021.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Wen Y, Jiang F, Gu X, Zhu X. Circular RNA circ_0008274 upregulates granulin to promote the progression of hepatocellular carcinoma via sponging microRNA -140-3p. Bioengineered. 2021;12:1890–1901. doi: 10.1080/21655979.2021.1926195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L, Han S, Yuan Q, Yang M. MiRNA-192 and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet. 2015;11:e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge G, Zhang W, Niu L, Yan Y, Ren Y, Zou Y. miR-215 functions as a tumor suppressor in epithelial ovarian cancer through regulation of the X-chromosome-linked inhibitor of apoptosis. Oncol Rep. 2016;35:1816–1822. doi: 10.3892/or.2015.4482. [DOI] [PubMed] [Google Scholar]
- Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, Chau BN. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- Gewandter JS, Bambara RA, O'Reilly MA. The RNA surveillance protein SMG1 activates p53 in response to DNA double-strand breaks but not exogenously oxidized mRNA. Cell Cycle. 2011;10:2561–2567. doi: 10.4161/cc.10.15.16347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes BC, Martins M, Lopes P, Morujão I, Oliveira M, Araújo A, Rueff J, Rodrigues AS. Prognostic value of microRNA-203a expression in breast cancer. Oncol Rep. 2016;36:1748–1756. doi: 10.3892/or.2016.4913. [DOI] [PubMed] [Google Scholar]
- Gotanda K, Hirota T, Matsumoto N, Ieiri I. MicroRNA-433 negatively regulates the expression of thymidylate synthase (TYMS) responsible for 5-fluorouracil sensitivity in HeLa cells. BMC Cancer. 2013;13:369. doi: 10.1186/1471-2407-13-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal B, Yadav SRM, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188502. doi: 10.1016/j.bbcan.2021.188502. [DOI] [PubMed] [Google Scholar]
- Grávalos C, García-Escobar I, García-Alfonso P, Cassinello J, Malón D, Carrato A. Adjuvant chemotherapy for stages II, III and IV of colon cancer. Clin Transl Oncol. 2009;11:526–533. doi: 10.1007/s12094-009-0397-8. [DOI] [PubMed] [Google Scholar]
- Gu Q, Hou W, Shi L, Liu H, Zhu Z, Ye W. Circular RNA ZNF609 functions as a competing endogenous RNA in regulating E2F transcription factor 6 through competitively binding to microRNA-197-3p to promote the progression of cervical cancer progression. Bioengineered. 2021;12:927–936. doi: 10.1080/21655979.2021.1896116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Wei G, Wu J, Fang D, Liao Z, Xiao H, Li M, Li Y. Down-regulation of miR-218-2 and its host gene SLIT3 cooperate to promote invasion and progression of thyroid cancer. J Clin Endocrinol Metab. 2013;98(8):E1334–E1344. doi: 10.1210/jc.2013-1053. [DOI] [PubMed] [Google Scholar]
- Guan A, Wang H, Li X, Xie H, Wang R, Zhu Y, Li R. MiR-330–3p inhibits gastric cancer progression through targeting MSI1. Am J Transl Res. 2016;8:4802–4811. [PMC free article] [PubMed] [Google Scholar]
- Guan X, Shi A, Zou Y, Sun M, Zhan Y, Dong Y, Fan Z. EZH2-mediated microRNA-375 upregulation promotes progression of breast cancer via the inhibition of FOXO1 and the p53 signaling pathway. Front Genet. 2021;12:633756. doi: 10.3389/fgene.2021.633756. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guo LH, Li H, Wang F, Yu J, He JS. The tumor suppressor roles of miR-433 and miR-127 in gastric cancer. Int J Mol Sci. 2013;14:14171–14184. doi: 10.3390/ijms140714171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, An R, Zhao R, Sun Y, Liu M, Tian L. miR-375 exhibits a more effective tumor-suppressor function in laryngeal squamous carcinoma cells by regulating KLF4 expression compared with simple co-transfection of miR-375 and miR-206. Oncol Rep. 2016;36:952–960. doi: 10.3892/or.2016.4852. [DOI] [PubMed] [Google Scholar]
- Guo Y, Xue W, Sun S, Chen X, Li H, Yan C. Circular RNA circZCCHC6 contributes to tumorigenesis by regulating LPCAT1 via miR-433-3p in non-small cell lung cancer. Clin Exp Med. 2022 doi: 10.1007/s10238-021-00780-2. [DOI] [PubMed] [Google Scholar]
- Han X, Liu Z. Long non-coding RNA JPX promotes gastric cancer progression by regulating CXCR6 and autophagy via inhibiting miR-197. Mol Med Rep. 2021;23:60. doi: 10.3892/mmr.2020.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Wang M, Ma N, Xu Y, Jiang Y, Gao X. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361:13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Han S, Zou H, Lee JW, Han J, Kim HC, Cheol JJ, Kim LS, Kim H. miR-1307-3p stimulates breast cancer development and progression by targeting SMYD4. J Cancer. 2019;10:441–448. doi: 10.7150/jca.30041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao T, Wang Z, Yang J, Zhang Y, Shang Y, Sun J. MALAT1 knockdown inhibits prostate cancer progression by regulating miR-140/BIRC6 axis. Biomed Pharmacother. 2020;123:109666. doi: 10.1016/j.biopha.2019.109666. [DOI] [PubMed] [Google Scholar]
- He XX, Chang Y, Meng FY, Wang MY, Xie QH, Tang F, Li PY, Song YH, Lin JS. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- He D, Yang X, Kuang W, Huang G, Liu X, Zhang Y. The novel circular RNA Circ-PGAP3 promotes the proliferation and invasion of triple negative breast cancer by regulating the miR-330-3p/Myc Axis. Onco Targets Ther. 2020;13:10149–10159. doi: 10.2147/OTT.S274574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff PM, Cassidy J, Schmoll HJ. The evolution of fluoropyrimidine therapy: from intravenous to oral. Oncologist. 2001;6(Suppl 4):3–11. doi: 10.1634/theoncologist.6-suppl_4-3. [DOI] [PubMed] [Google Scholar]
- Hou ZH, Xu XW, Fu XY, Zhou LD, Liu SP, Tan DM. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am J Physiol Cell Physiol. 2020;318:C649–C663. doi: 10.1152/ajpcell.00510.2018. [DOI] [PubMed] [Google Scholar]
- Hu L, Zhu YT, Qi C, Zhu YJ. Identification of Smyd4 as a potential tumor suppressor gene involved in breast cancer development. Cancer Res. 2009;69:4067–4072. doi: 10.1158/0008-5472.CAN-08-4097. [DOI] [PubMed] [Google Scholar]
- Hu T, Li Z, Gao CY, Cho CH. Mechanisms of drug resistance in colon cancer and its therapeutic strategies. World J Gastroenterol. 2016;22:6876–6889. doi: 10.3748/wjg.v22.i30.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Feng Y, Sun L, Qu L, Sun C. Roles of microRNA-330 and its target gene ING4 in the development of aggressive phenotype in hepatocellular carcinoma cells. Dig Dis Sci. 2017;62:715–722. doi: 10.1007/s10620-016-4429-2. [DOI] [PubMed] [Google Scholar]
- Hu C, Lv L, Peng J, Liu D, Wang X, Zhou Y, Huo J. MicroRNA-375 suppresses esophageal cancer cell growth and invasion by repressing metadherin expression. Oncol Lett. 2017;13:4769–4775. doi: 10.3892/ol.2017.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Wang P, Lin J, Zheng X, Yang F, Zhang G, Chen D, Xie J, Gao Z, Peng L, Xie C. MicroRNA-197 promotes metastasis of hepatocellular carcinoma by activating Wnt/β-catenin signaling. Cell Physiol Biochem. 2018;51:470–486. doi: 10.1159/000495242. [DOI] [PubMed] [Google Scholar]
- Hu Q, Du K, Mao X, Ning S. miR-197 is downregulated in cervical carcinogenesis and suppresses cell proliferation and invasion through targeting forkhead box M1. Oncol Lett. 2018;15:10063–10069. doi: 10.3892/ol.2018.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Zou Y, Jing LL. miR-140-3p inhibits progression of non-small cell lung cancer by targeting Janus kinase 1. J Biosci. 2020;45:48. doi: 10.1007/s12038-020-0003-3. [DOI] [PubMed] [Google Scholar]
- Hu H, Liu S, Chu A, Chen J, Xing C, Jing J. Comprehensive analysis of ceRNA network of ERCC4 in colorectal cancer. PeerJ. 2021;9:e12647. doi: 10.7717/peerj.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Wang Y, Li Q, Fei X, Ma H, Hu R. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Lett. 2019;446:81–89. doi: 10.1016/j.canlet.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Huang S, Zhang X, Guan B, Sun P, Hong CT, Peng J, Tang S, Yang J. A novel circular RNA hsa_circ_0008035 contributes to gastric cancer tumorigenesis through targeting the miR-375/YBX1 axis. Am J Transl Res. 2019;11:2455–2462. [PMC free article] [PubMed] [Google Scholar]
- Huang G, Wei B, Chen Z, Wang J, Zhao L, Peng X, Liu K, Lai Y, Ni L. Identification of a four-microRNA panel in serum as promising biomarker for colorectal carcinoma detection. Biomark Med. 2020;14:749–760. doi: 10.2217/bmm-2019-0605. [DOI] [PubMed] [Google Scholar]
- Huang Y, Sun H, Ma X, Zeng Y, Pan Y, Yu D, Liu Z, Xiang Y. HLA-F-AS1/miR-330-3p/PFN1 axis promotes colorectal cancer progression. Life Sci. 2020;254:117180. doi: 10.1016/j.lfs.2019.117180. [DOI] [PubMed] [Google Scholar]
- Huang Y, Chen Z, Zhou X, Huang H. Circ_0000467 exerts an oncogenic role in colorectal cancer via miR-330-5p-dependent regulation of TYRO3. Biochem Genet. 2022 doi: 10.1007/s10528-021-10171-7. [DOI] [PubMed] [Google Scholar]
- Huo W, Du M, Pan X, Zhu X, Gao Y, Li Z. miR-203a-3p.1 targets IL-24 to modulate hepatocellular carcinoma cell growth and metastasis. FEBS Open Bio. 2017;7:1085–1091. doi: 10.1002/2211-5463.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irminger-Finger I, Kargul J, Laurent GJ. Non-coding RNAs: a novel level of genome complexity. Int J Biochem Cell Biol. 2014;54:286. doi: 10.1016/j.biocel.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Iveson T. Adjuvant chemotherapy in colon cancer: state of the art and future perspectives. Curr Opin Oncol. 2020;32:370–376. doi: 10.1097/CCO.0000000000000640. [DOI] [PubMed] [Google Scholar]
- Jafarzadeh A, Paknahad MH, Nemati M, Jafarzadeh S, Mahjoubin-Tehran M, Rajabi A, Shojaie L, Mirzaei H. Dysregulated expression and functions of microRNA-330 in cancers: a potential therapeutic target. Biomed Pharmacother. 2022;146:112600. doi: 10.1016/j.biopha.2021.112600. [DOI] [PubMed] [Google Scholar]
- Janecki DM, Sajek M, Smialek MJ, Kotecki M, Ginter-Matuszewska B, Kuczynska B, Spik A, Kolanowski T, Kitazawa R, Kurpisz M, Jaruzelska J. SPIN1 is a proto-oncogene and SPIN3 is a tumor suppressor in human seminoma. Oncotarget. 2018;9:32466–32477. doi: 10.18632/oncotarget.25977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji D, Jiang L, Li Y. MiR-192-5p suppresses the growth of bladder cancer cells via targeting Yin Yang 1. Hum Cell. 2018;31:210–219. doi: 10.1007/s13577-018-0201-6. [DOI] [PubMed] [Google Scholar]
- Ji C, Hu J, Wang X, Zheng W, Deng X, Song H, Yu Y, Luo Q, Hua K, Zhou X, Fang L. Hsa_circ_0053063 inhibits breast cancer cell proliferation via hsa_circ_0053063/hsa-miR-330-3p/PDCD4 axis. Aging (albany NY) 2021;13:9627–9645. doi: 10.18632/aging.202707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Li T, Wang J, Jiao R, Shi X, Huang X, Ji G. miR-140-3p suppresses cell growth and induces apoptosis in colorectal cancer by targeting PD-L1. Onco Targets Ther. 2019;12:10275–10285. doi: 10.2147/OTT.S226465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wei T, Li W, Zhang R, Chen M. Circular RNA hsa_circ_0002024 suppresses cell proliferation, migration, and invasion in bladder cancer by sponging miR-197–3p. Am J Transl Res. 2019;11:1644–1652. [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Su Z, Sheng H, Li K, Yang B, Li S. Circ_0086720 knockdown strengthens the radiosensitivity of non-small cell lung cancer via mediating the miR-375/SPIN1 axis. Neoplasma. 2021;68:96–107. doi: 10.4149/neo_2020_200331N333. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Huang T, Zhou Y, Zhang J, Lung RWM, Tong JHM, Chan AWH, Zhang B, Wong CC, Wu F, Dong Y, Wang S, Yang W, Pan Y, Chak WP, Cheung AHK, Pang JCS, Yu J, Cheng ASL, To KF. miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis. 2018;9:92. doi: 10.1038/s41419-017-0134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimpour M, Ravanbakhsh R, Maydanchi M, Rajabi A, Azizi F, Saber A. Cancer driver gene and non-coding RNA alterations as biomarkers of brain metastasis in lung cancer: a review of the literature. Biomed Pharmacother. 2021;143:112190. doi: 10.1016/j.biopha.2021.112190. [DOI] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, Krauthammer M, Halaban R, Provero P, Adams DJ, Tuveson DA, Pandolfi pp. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Kim H. Oncogenes and tumor suppressors regulate glutamine metabolism in cancer cells. J Cancer Prev. 2013;18:221–226. doi: 10.15430/jcp.2013.18.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, Qin YR, Guan XY. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- Kong XM, Zhang GH, Huo YK, Zhao XH, Cao DW, Guo SF, Li AM, Zhang XR. MicroRNA-140-3p inhibits proliferation, migration and invasion of lung cancer cells by targeting ATP6AP2. Int J Clin Exp Pathol. 2015;8:12845–12852. [PMC free article] [PubMed] [Google Scholar]
- Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12:599–620. doi: 10.1007/s13238-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J. Strategies to use microRNAs as therapeutic targets. Best Pract Res Clin Endocrinol Metab. 2016;30:551–561. doi: 10.1016/j.beem.2016.07.004. [DOI] [PubMed] [Google Scholar]
- Kwok ZH, Roche V, Chew XH, Fadieieva A, Tay Y. A non-canonical tumor suppressive role for the long non-coding RNA MALAT1 in colon and breast cancers. Int J Cancer. 2018;143:668–678. doi: 10.1002/ijc.31386. [DOI] [PubMed] [Google Scholar]
- Labianca R, Nordlinger B, Beretta GD, Mosconi S, Mandalà M, Cervantes A, Arnold D, ESMO Guidelines Working Group Early colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):vi64–72. doi: 10.1093/annonc/mdt354. [DOI] [PubMed] [Google Scholar]
- Lai EC. Two decades of miRNA biology: lessons and challenges. RNA. 2015;21:675–677. doi: 10.1261/rna.051193.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenbach RJ, Danenberg PV, Heidelberger C. Thymidylate synthetase: mechanism of inhibition by 5-fluoro-2'-deoxyuridylate. Biochem Biophys Res Commun. 1972;48:1565–1571. doi: 10.1016/0006-291X(72)90892-3. [DOI] [PubMed] [Google Scholar]
- Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT, Goan YG, Lu PJ. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene. 2009;28:3360–3370. doi: 10.1038/onc.2009.192. [DOI] [PubMed] [Google Scholar]
- Li T, Li S. Circ_0023984 facilitates esophageal squamous cell carcinoma progression by regulating miR-433-3p/REV3L axis. Dig Dis Sci. 2021 doi: 10.1007/s10620-021-06916-4. [DOI] [PubMed] [Google Scholar]
- Li S, Gao J, Gu J, Yuan J, Hua D, Shen L. MicroRNA-215 inhibits relapse of colorectal cancer patients following radical surgery. Med Oncol. 2013;30:549. doi: 10.1007/s12032-013-0549-0. [DOI] [PubMed] [Google Scholar]
- Li T, Gao F, Zhang XP. miR-203 enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Oncol Rep. 2015;33:607–614. doi: 10.3892/or.2014.3646. [DOI] [PubMed] [Google Scholar]
- Li PL, Zhang X, Wang LL, Du LT, Yang YM, Li J, Wang CX. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis. 2015;36:1484–1493. doi: 10.1093/carcin/bgv145. [DOI] [PubMed] [Google Scholar]
- Li S, Li F, Niu R, Zhang H, Cui A, An W, Wang X. Mir-192 suppresses apoptosis and promotes proliferation in esophageal aquamous cell caicinoma by targeting Bim. Int J Clin Exp Pathol. 2015;8:8048–8056. [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang QY, Zou JL, Li ZW, Tian TT, Dong B, Liu XJ, Ge S, Zhu Y, Gao J, Shen L. miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget. 2016;7:4817–4828. doi: 10.18632/oncotarget.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Zhang X, Wang H, Wang L, Liu T, Du L, Yang Y, Wang C. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol Cancer Ther. 2017;16:739–751. doi: 10.1158/1535-7163.MCT-16-0591. [DOI] [PubMed] [Google Scholar]
- Li P, Zhang X, Wang L, Du L, Yang Y, Liu T, Li C, Wang C. lncRNA HOTAIR contributes to 5FU resistance through suppressing miR-218 and activating NF-κB/TS signaling in colorectal cancer. Mol Ther Nucleic Acids. 2017;15(8):356–369. doi: 10.1016/j.omtn.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mao X, Wang X, Miao G, Li J. miR-433 reduces cell viability and promotes cell apoptosis by regulating MACC1 in colorectal cancer. Oncol Lett. 2017;13:81–88. doi: 10.3892/ol.2016.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XW, Qiu SJ, Zhang X. Overexpression of miR-215-3p sensitizes colorectal cancer to 5-fluorouracil induced apoptosis through regulating CXCR1. Eur Rev Med Pharmacol Sci. 2018;22:7240–7250. doi: 10.26355/eurrev_201811_16258. [DOI] [PubMed] [Google Scholar]
- Li L, Jia L, Ding Y. Upregulation of miR-375 inhibits human liver cancer cell growth by modulating cell proliferation and apoptosis via targeting ErbB2. Oncol Lett. 2018;16:3319–3326. doi: 10.3892/ol.2018.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li J, Yang T, Lin S, Li H. MicroRNA-433 represses proliferation and invasion of colon cancer cells by targeting homeobox A1. Oncol Res. 2018;26:315–322. doi: 10.3727/096504017X15067856789781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen M, Yu B. miR-433 suppresses tumor progression via Smad2 in non-small cell lung cancer. Pathol Res Pract. 2019;215:152591. doi: 10.1016/j.prp.2019.152591. [DOI] [PubMed] [Google Scholar]
- Li B, Huang Q, Wei GH. The role of HOX transcription factors in cancer predisposition and progression. Cancers (basel) 2019;11:528. doi: 10.3390/cancers11040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, Cheng Z, Zhu Q. AGO2 and its partners: a silencing complex, a chromatin modulator, and new features. Crit Rev Biochem Mol Biol. 2020;55:33–53. doi: 10.1080/10409238.2020.1738331. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang W, Zhang M, Sun W, Shi W, Li F. Circular RNA circ-0016068 promotes the growth, migration, and invasion of prostate cancer cells by regulating the miR-330-3p/BMI-1 axis as a competing endogenous RNA. Front Cell Dev Biol. 2020;8:827. doi: 10.3389/fcell.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Li P, Lv H, Wu Y, Xu K, Xu M, Ma Y. E2F transcription factor 1 is involved in the phenotypic modulation of esophageal squamous cell carcinoma cells via microRNA-375. Bioengineered. 2021;12:10047–10062. doi: 10.1080/21655979.2021.1996510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xiao C, He K, Xiang G. Circ_0072088 promotes progression of hepatocellular carcinoma by activating JAK2/STAT3 signaling pathway via miR-375. IUBMB Life. 2021;73(9):1153–1165. doi: 10.1002/iub.2520. [DOI] [PubMed] [Google Scholar]
- Li D, Wang T, Sun FF, Feng JQ, Peng JJ, Li H, Wang C, Wang D, Liu Y, Bai YD, Shi ML, Zhang T. MicroRNA-375 represses tumor angiogenesis and reverses resistance to sorafenib in hepatocarcinoma. Cancer Gene Ther. 2021;28:126–140. doi: 10.1038/s41417-020-0191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li Q, He S. Hsa_circ_0025202 suppresses cell tumorigenesis and tamoxifen resistance via miR-197-3p/HIPK3 axis in breast cancer. World J Surg Oncol. 2021;19:39. doi: 10.1186/s12957-021-02149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J, Jing Y, Dong Q, Huan L, Chen D, Bao C, Wang Q, Zhao F, Li J, Yao M, Qin L, Liang L, He X. miR-192, a prognostic indicator, targets the SLC39A6/SNAIL pathway to reduce tumor metastasis in human hepatocellular carcinoma. Oncotarget. 2016;7:2672–2683. doi: 10.18632/oncotarget.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Guo Q, Li L, Cheng Y, Ren C, Zhang G. MicroRNA-433 inhibits migration and invasion of ovarian cancer cells via targeting Notch1. Neoplasma. 2016;63:696–704. doi: 10.4149/neo_2016_506. [DOI] [PubMed] [Google Scholar]
- Liang C, Ding J, Yang Y, Deng L, Li X. MicroRNA-433 inhibits cervical cancer progression by directly targeting metadherin to regulate the AKT and β-catenin signalling pathways. Oncol Rep. 2017;38:3639–3649. doi: 10.3892/or.2017.6049. [DOI] [PubMed] [Google Scholar]
- Liang J, Sun T, Wang G, Zhang H. Clinical significance and functions of miR-203a-3p/AVL9 axis in human non-small-cell lung cancer. Per Med. 2020;17:271–282. doi: 10.2217/pme-2019-0108. [DOI] [PubMed] [Google Scholar]
- Liao Z, Li Y, Zhou Y, Huang Q, Dong J. MicroRNA-197 inhibits gastric cancer progression by directly targeting metadherin. Mol Med Rep. 2018;17:602–611. doi: 10.3892/mmr.2017.7908. [DOI] [PubMed] [Google Scholar]
- Ling HH, Kuo CC, Lin BX, Huang YH, Lin CW. Elevation of YAP promotes the epithelial-mesenchymal transition and tumor aggressiveness in colorectal cancer. Exp Cell Res. 2017;350:218–225. doi: 10.1016/j.yexcr.2016.11.024. [DOI] [PubMed] [Google Scholar]
- Liu AM, Poon RT, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun. 2010;394:623–627. doi: 10.1016/j.bbrc.2010.03.036. [DOI] [PubMed] [Google Scholar]
- Liu X, Shi H, Liu B, Li J, Liu Y, Yu B. miR-330-3p controls cell proliferation by targeting early growth response 2 in non-small-cell lung cancer. Acta Biochim Biophys Sin (shanghai) 2015;47:431–440. doi: 10.1093/abbs/gmv032. [DOI] [PubMed] [Google Scholar]
- Liu N, Liu Z, Zhang W, Li Y, Cao J, Yang H, Li X. MicroRNA-433 reduces cell proliferation and invasion in non-small cell lung cancer via directly targeting E2F transcription factor 3. Mol Med Rep. 2018;18:1155–1164. doi: 10.3892/mmr.2018.9020. [DOI] [PubMed] [Google Scholar]
- Liu HY, Zhang YY, Zhu BL, Feng FZ, Zhang HT, Yan H, Zhou B. MiR-203a-3p regulates the biological behaviors of ovarian cancer cells through mediating the Akt/GSK-3β/Snail signaling pathway by targeting ATM. J Ovarian Res. 2019;12:60. doi: 10.1186/s13048-019-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Song S, Lin S, Zhang M, Du Y, Zhang D, Xu W, Wang H. Circ-SERPINE2 promotes the development of gastric carcinoma by sponging miR-375 and modulating YWHAZ. Cell Prolif. 2019;52:e12648. doi: 10.1111/cpr.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhan Y, Wang J, Wang J, Guo J, Kong D. lncRNA-SNHG17 promotes colon adenocarcinoma progression and serves as a sponge for miR-375 to regulate CBX3 expression. Am J Transl Res. 2020;12:5283–5295. [PMC free article] [PubMed] [Google Scholar]
- Liu D, Jian X, Xu P, Zhu R, Wang Y. Linc01234 promotes cell proliferation and metastasis in oral squamous cell carcinoma via miR-433/PAK4 axis. BMC Cancer. 2020;20:107. doi: 10.1186/s12885-020-6541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang S, Guan Y, Zhang Q. Long noncoding RNA OSER1-AS1 promotes the malignant properties of non-small cell lung cancer by sponging microRNA-433-3p and thereby increasing Smad2 expression. Oncol Rep. 2020;44:599–610. doi: 10.3892/or.2020.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Chen C, Cui M, Zhang H. miR-140-3p inhibits colorectal cancer progression and its liver metastasis by targeting BCL9 and BCL2. Cancer Med. 2021;10:3358–3372. doi: 10.1002/cam4.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- Lu X, Liu Z, Ning X, Huang L, Jiang B. The long noncoding RNA HOTAIR promotes colorectal cancer progression by sponging miR-197. Oncol Res. 2018;26:473–481. doi: 10.3727/096504017X15105708598531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan C, Li Y, Liu Z, Zhao C. Long noncoding RNA MALAT1 promotes the development of colon cancer by regulating miR-101-3p/STC1 Axis. Onco Targets Ther. 2020;13:3653–3665. doi: 10.2147/OTT.S242300. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Luo J, Wu J, Li Z, Qin H, Wang B, Wong TS, Yang W, Fu QL, Lei W. miR-375 suppresses IGF1R expression and contributes to inhibition of cell progression in laryngeal squamous cell carcinoma. Biomed Res Int. 2014;2014:374598. doi: 10.1155/2014/374598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XY, Wang JH, Wang JL, Ma CX, Wang XC, Liu FS. Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genomics. 2015;16:676. doi: 10.1186/s12864-015-1881-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Ma J, Yang Y, He X, Pan X, Wang Z, Qian Y. Effects of miR-330-3p on invasion, migration and EMT of gastric cancer cells by targeting PRRX1-mediated Wnt/β-catenin signaling pathway. Onco Targets Ther. 2020;13:3411–3423. doi: 10.2147/OTT.S238665. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ma YS, Wu TM, Qian B, Liu YS, Ding H, Fan MM, Liu JB, Yu F, Wang HM, Shi Y, Gu LP, Li L, Tian LL, Wang PY, Wang GR, Wu ZJ, Zou QF, Ling CC, Fu D. KDM5A silencing transcriptionally suppresses the FXYD3-PI3K/AKT axis to inhibit angiogenesis in hepatocellular cancer via miR-433 up-regulation. J Cell Mol Med. 2021;25:4040–4052. doi: 10.1111/jcmm.16371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhou A, Song J. Upregulation of miR-1307-3p and its function in the clinical prognosis and progression of gastric cancer. Oncol Lett. 2021;21:91. doi: 10.3892/ol.2020.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q, Quan T, Luo B, Guo X, Liu L, Zheng Q. MiR-375 targets KLF4 and impacts the proliferation of colorectal carcinoma. Tumour Biol. 2016;37:463–471. doi: 10.1007/s13277-015-3809-0. [DOI] [PubMed] [Google Scholar]
- Marjaneh RM, Khazaei M, Ferns GA, Avan A, Aghaee-Bakhtiari SH. The role of microRNAs in 5-FU resistance of colorectal cancer: possible mechanisms. J Cell Physiol. 2019;234:2306–2316. doi: 10.1002/jcp.27221. [DOI] [PubMed] [Google Scholar]
- Martino M, Esposito F, Pallante P. Long non-coding RNAs regulating multiple proliferative pathways in cancer cell. Transl Cancer Res. 2021;10:3140–3157. doi: 10.21037/tcr-21-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison LK, Soong R, Diasio RB. Implications of dihydropyrimidine dehydrogenase on 5-fluorouracil pharmacogenetics and pharmacogenomics. Pharmacogenomics. 2002;3:485–492. doi: 10.1517/14622416.3.4.485. [DOI] [PubMed] [Google Scholar]
- McCown PJ, Wang MC, Jaeger L, Brown JA. Secondary structural model of human MALAT1 reveals multiple structure–function relationships. Int J Mol Sci. 2019;20:5610. doi: 10.3390/ijms20225610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- Meng H, Wang K, Chen X, Guan X, Hu L, Xiong G, Li J, Bai Y. MicroRNA-330–3p functions as an oncogene in human esophageal cancer by targeting programmed cell death 4. Am J Cancer Res. 2015;5:1062–1075. [PMC free article] [PubMed] [Google Scholar]
- Meulendijks D, Cats A, Beijnen JH, Schellens JH. Improving safety of fluoropyrimidine chemotherapy by individualizing treatment based on dihydropyrimidine dehydrogenase activity—ready for clinical practice? Cancer Treat Rev. 2016;50:23–34. doi: 10.1016/j.ctrv.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Micallef I, Baron B. The mechanistic roles of ncRNAs in promoting and supporting chemoresistance of colorectal cancer. Noncoding RNA. 2021;7:24. doi: 10.3390/ncrna7020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriithi W, Macharia LW, Heming CP, Echevarria JL, Nyachieo A, Filho PN, Neto VM. ABC transporters and the hallmarks of cancer: roles in cancer aggressiveness beyond multidrug resistance. Cancer Biol Med. 2020;17:253–269. doi: 10.20892/j.issn.2095-3941.2019.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na XY, Shang XS, Zhao Y, Ren PP, Hu XQ. MiR-203a functions as a tumor suppressor in bladder cancer by targeting SIX4. Neoplasma. 2019;66:211–221. doi: 10.4149/neo_2018_180512N312. [DOI] [PubMed] [Google Scholar]
- Ni JS, Zheng H, Huang ZP, Hong YG, Ou YL, Tao YP, Wang MC, Wang ZG, Yang Y, Zhou WP. MicroRNA-197-3p acts as a prognostic marker and inhibits cell invasion in hepatocellular carcinoma. Oncol Lett. 2019;17:2317–2327. doi: 10.3892/ol.2018.9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Guo LY, Zhang JY, Ji T, Mao D, Li XF, Du XX. E2F1-induced upregulation of lncRNA HCG18 stimulates proliferation and migration in gastric cancer by binding to miR-197-3p. Eur Rev Med Pharmacol Sci. 2020;24:9949–9956. doi: 10.26355/eurrev_202010_23207. [DOI] [PubMed] [Google Scholar]
- Offer SM, Butterfield GL, Jerde CR, Fossum CC, Wegner NJ, Diasio RB. microRNAs miR-27a and miR-27b directly regulate liver dihydropyrimidine dehydrogenase expression through two conserved binding sites. Mol Cancer Ther. 2014;13:742–751. doi: 10.1158/1535-7163.MCT-13-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Fukuoka M. S-1: a new oral fluoropyrimidine in the treatment of patients with advanced non-small-cell lung cancer. Clin Lung Cancer. 2009;10:290–294. doi: 10.3816/CLC.2009.n.040. [DOI] [PubMed] [Google Scholar]
- Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36:2403–2407. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- Pedersen PL. Warburg, me and Hexokinase 2: multiple discoveries of key molecular events underlying one of cancers' most common phenotypes, the "Warburg Effect", i.e., elevated glycolysis in the presence of oxygen. J Bioenerg Biomembr. 2007;39:211–222. doi: 10.1007/s10863-007-9094-x. [DOI] [PubMed] [Google Scholar]
- Pefani DE, Pankova D, Abraham AG, Grawenda AM, Vlahov N, Scrace S, O’Neill E. TGF-β targets the hippo pathway scaffold RASSF1A to facilitate YAP/SMAD2 nuclear translocation. Mol Cell. 2016;63:156–166. doi: 10.1016/j.molcel.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, Zhou W, Benson DM, Jr, Hofmainster C, Alder H, Garofalo M, Di Leva G, Volinia S, Lin HJ, Perrotti D, Kuehl M, Aqeilan RI, Palumbo A, Croce CM. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18:367–381. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Piepoli A, Tavano F, Copetti M, Mazza T, Palumbo O, Panza A, di Mola FF, Pazienza V, Mazzoccoli G, Biscaglia G, Gentile A, Mastrodonato N, Carella M, Pellegrini F, di Sebastiano P, Andriulli A. Mirna expression profiles identify drivers in colorectal and pancreatic cancers. PLoS ONE. 2012;7:e33663. doi: 10.1371/journal.pone.0033663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porzycki P, Ciszkowicz E, Semik M, Tyrka M. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int Urol Nephrol. 2018;50:1619–1626. doi: 10.1007/s11255-018-1938-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet C, Njock MS, Moermans C, Louis E, Louis R, Malaise M, Guiot J. Exosomal long non-coding RNAs in lung diseases. Int J Mol Sci. 2020;21:3580. doi: 10.3390/ijms21103580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poursheikhani A, Abbaszadegan MR, Nokhandani N, Kerachian MA. Integration analysis of long non-coding RNA (lncRNA) role in tumorigenesis of colon adenocarcinoma. BMC Med Genomics. 2020;13:108. doi: 10.1186/s12920-020-00757-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretzsch E, Bösch F, Neumann J, Ganschow P, Bazhin A, Guba M, Werner J, Angele M. Mechanisms of metastasis in colorectal cancer and metastatic organotropism: hematogenous versus peritoneal spread. J Oncol. 2019;2019:7407190. doi: 10.1155/2019/7407190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Gong L, Mou Y, Han Y, Zheng S. MicroRNA-203a-3p is a candidate tumor suppressor that targets thrombospondin 2 in colorectal carcinoma. Oncol Rep. 2019;42:1825–1832. doi: 10.3892/or.2019.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Chen S, Li S, Wang Y, Yao J. Circ_0000003 regulates glutamine metabolism and tumor progression of tongue squamous cell carcinoma via the miR-330-3p/GLS axis. Oncol Rep. 2021;45:45. doi: 10.3892/or.2021.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao ZW, Jiang Y, Wang L, Wang L, Jiang J, Zhang JR, Mu P. LINC00852 promotes the proliferation and invasion of ovarian cancer cells by competitively binding with miR-140-3p to regulate AGTR1 expression. BMC Cancer. 2021;21:1004. doi: 10.1186/s12885-021-08730-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Dou Y. miR-1307 promotes the proliferation of prostate cancer by targeting FOXO3A. Biomed Pharmacother. 2017;88:430–435. doi: 10.1016/j.biopha.2016.11.120. [DOI] [PubMed] [Google Scholar]
- Rashid F, Shah A, Shan G. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinform. 2016;14:73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokavec M, Bouznad N, Hermeking H. Paracrine induction of epithelial-mesenchymal transition between colorectal cancer cells and its suppression by a p53/miR-192/215/NID1 Axis. Cell Mol Gastroenterol Hepatol. 2019;7:783–802. doi: 10.1016/j.jcmgh.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakatani A, Sonohara F, Goel A. Melatonin-mediated downregulation of thymidylate synthase as a novel mechanism for overcoming 5-fluorouracil associated chemoresistance in colorectal cancer cells. Carcinogenesis. 2019;40:422–431. doi: 10.1093/carcin/bgy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451–5465. doi: 10.1002/jcp.27486. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonga D, Danenberg KD, Johnson M, Metzger R, Groshen S, Tsao-Wei DD, Lenz HJ, Leichman CG, Leichman L, Diasio RB, Danenberg PV. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- Shen Y, Wang P, Li Y, Ye F, Wang F, Wan X, Cheng X, Lu W, Xie X. miR-375 is upregulated in acquired paclitaxel resistance in cervical cancer. Br J Cancer. 2013;109:92–99. doi: 10.1038/bjc.2013.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Zhou J, Li Y, Ye F, Wan X, Lu W, Xie X, Cheng X. miR-375 mediated acquired chemo-resistance in cervical cancer by facilitating EMT. PLoS ONE. 2014;9:e109299. doi: 10.1371/journal.pone.0109299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- Shi W, Yang J, Li S, Shan X, Liu X, Hua H, Zhao C, Feng Z, Cai Z, Zhang L, Zhou D. Potential involvement of miR-375 in the premalignant progression of oral squamous cell carcinoma mediated via transcription factor KLF5. Oncotarget. 2015;6:40172–40185. doi: 10.18632/oncotarget.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Zheng Z, Lin X, Ma H. Long noncoding RNA MALAT1 regulates the progression of atherosclerosis by miR-330-5p/NF-κB signal pathway. J Cardiovasc Pharmacol. 2021;78:235–246. doi: 10.1097/FJC.0000000000001061. [DOI] [PubMed] [Google Scholar]
- Shibata M, Ham K, Hoque MO. A time for YAP1: tumorigenesis, immunosuppression and targeted therapy. Int J Cancer. 2018;143:2133–2144. doi: 10.1002/ijc.31561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J, Xiao L, Yan S, Fan B, Zou X, Yang J. Mechanism of microRNA-375 promoter methylation in promoting ovarian cancer cell malignancy. Technol Cancer Res Treat. 2021;20:1533033820980115. doi: 10.1177/1533033820980115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A, Vazakidou ME, Schwab A, Napoli F, Fernandez-Molina C, Rapa I, Stemmler MP, Volante M, Brabletz T, Ceppi P. Thymidylate synthase is functionally associated with ZEB1 and contributes to the epithelial-to-mesenchymal transition of cancer cells. J Pathol. 2017;242:221–233. doi: 10.1002/path.4897. [DOI] [PubMed] [Google Scholar]
- Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, Fodstad O, Ju J. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28:4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Wang Y, Titmus MA, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9:96. doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SD, Zhou J, Zhou J, Zhao H, Cen JN, Li DC. MicroRNA-375 targets the 3-phosphoinositide-dependent protein kinase-1 gene in pancreatic carcinoma. Oncol Lett. 2013;6:953–959. doi: 10.3892/ol.2013.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Zhou J, He S, Zhu D, Zhang Z, Zhao H, Wang Y, Li D. Expression levels of microRNA-375 in pancreatic cancer. Biomed Rep. 2013;1:393–398. doi: 10.3892/br.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wang S, Cheng X. LINC01006 regulates the proliferation, migration and invasion of hepatocellular carcinoma cells through regulating miR-433-3p/CBX3 axis. Ann Hepatol. 2021;25:100343. doi: 10.1016/j.aohep.2021.100343. [DOI] [PubMed] [Google Scholar]
- Stec R, Bodnar L, Smoter M, Mączewski M, Szczylik C. Metastatic colorectal cancer in the elderly: an overview of the systemic treatment modalities (review) Oncol Lett. 2011;2:3–11. doi: 10.3892/ol.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su K, Wang N, Shao Q, Liu H, Zhao B, Ma S. The role of a ceRNA regulatory network based on lncRNA MALAT1 site in cancer progression. Biomed Pharmacother. 2021;137:111389. doi: 10.1016/j.biopha.2021.111389. [DOI] [PubMed] [Google Scholar]
- Sun Y, Ma L. New insights into long non-coding RNA MALAT1 in cancer and metastasis. Cancers (BASEL) 2019;11:216. doi: 10.3390/cancers11020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Qin B. Long noncoding RNA MALAT1 regulates HDAC4-mediated proliferation and apoptosis via decoying of miR-140-5p in osteosarcoma cells. Cancer Med. 2018;7:4584–4597. doi: 10.1002/cam4.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Zhou N, Han Q, Zhao L, Bai C, Chen Y, Zhou J, Zhao RC. MicroRNA-197 influences 5-fluorouracil resistance via thymidylate synthase in colorectal cancer. Clin Transl Oncol. 2015;17:876–883. doi: 10.1007/s12094-015-1318-7. [DOI] [PubMed] [Google Scholar]
- Sun J, Fan Z, Lu S, Yang J, Hao T, Huo Q. MiR-192 suppresses the tumorigenicity of prostate cancer cells by targeting and inhibiting nin one binding protein. Int J Mol Med. 2016;37:485–492. doi: 10.3892/ijmm.2016.2449. [DOI] [PubMed] [Google Scholar]
- Sun N, Zhang G, Liu Y. Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/β-catenin signaling pathway. Gene. 2018;665:141–148. doi: 10.1016/j.gene.2018.04.014. [DOI] [PubMed] [Google Scholar]
- Sun L, Suo C, Li ST, Zhang H, Gao P. Metabolic reprogramming for cancer cells and their microenvironment: beyond the Warburg Effect. Biochim Biophys Acta Rev Cancer. 2018;1870:51–66. doi: 10.1016/j.bbcan.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Sun CY, Zhang XP, Wang W. Coordination of miR-192 and miR-22 in p53-mediated cell fate decision. Int J Mol Sci. 2019;20:E4768. doi: 10.3390/ijms2019a4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang S, Li G, Wang G, Song J, Li Z, Zhang Z, Yuan W, Li X. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38:2627–2644. doi: 10.1038/s41388-018-0628-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sun Y, Wang W, Zhao C. Frizzled receptors in tumors, focusing on signaling, roles, modulation mechanisms, and targeted therapies. Oncol Res. 2021;28:661–674. doi: 10.3727/096504020X16014648664459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taieb J, Gallois C. Adjuvant chemotherapy for stage III colon cancer. Cancers (basel) 2020;12:2679. doi: 10.3390/cancers12092679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Testa JR. DLX genes: roles in development and cancer. Cancers (basel) 2021;13:3005. doi: 10.3390/cancers13123005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R, Qi Q, Wu R, Zhou X, Wu D, Zhou H, Mao Y, Li R, Liu C, Wang L, Chen W, Hua D, Zhang H, Wang W. The polymorphic terminal-loop of pre-miR-1307 binding with MBNL1 contributes to colorectal carcinogenesis via interference with Dicer1 recruitment. Carcinogenesis. 2015;36:867–875. doi: 10.1093/carcin/bgv066. [DOI] [PubMed] [Google Scholar]
- Tang T, Cheng Y, She Q, Jiang Y, Chen Y, Yang W, Li Y. Long non-coding RNA TUG1 sponges miR-197 to enhance cisplatin sensitivity in triple negative breast cancer. Biomed Pharmacother. 2018;107:338–346. doi: 10.1016/j.biopha.2018.07.076. [DOI] [PubMed] [Google Scholar]
- Tang D, Yang Z, Long F, Luo L, Yang B, Zhu R, Sang X, Cao G. Inhibition of MALAT1 reduces tumor growth and metastasis and promotes drug sensitivity in colorectal cancer. Cell Signal. 2019;57:21–28. doi: 10.1016/j.cellsig.2019.01.013. [DOI] [PubMed] [Google Scholar]
- Tang Z, Ma Q, Wang L, Liu C, Gao H, Yang Z, Liu Z, Zhang H, Ji L, Jiang G. A brief review: some compounds targeting YAP against malignancies. Future Oncol. 2019;15:1535–1543. doi: 10.2217/fon-2019-0035. [DOI] [PubMed] [Google Scholar]
- Tang D, Yang Z, Long F, Luo L, Yang B, Zhu R, Sang X, Cao G, Wang K. Long noncoding RNA MALAT1 mediates stem cell-like properties in human colorectal cancer cells by regulating miR-20b-5p/Oct4 axis. J Cell Physiol. 2019;234:20816–20828. doi: 10.1002/jcp.28687. [DOI] [PubMed] [Google Scholar]
- Tanner JA, Tyndale RF. Variation in CYP2A6 activity and personalized medicine. J Pers Med. 2017;7:18. doi: 10.3390/jpm7040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova-Barberio M, Roca MS, Zotti AI, Leone A, Bruzzese F, Vitagliano C, Scogliamiglio G, Russo D, D'Angelo G, Franco R, Budillon A, Di Gennaro E. Valproic acid potentiates the anticancer activity of capecitabine in vitro and in vivo in breast cancer models via induction of thymidine phosphorylase expression. Oncotarget. 2016;7:7715–7731. doi: 10.18632/oncotarget.6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- Tong F, Guo J, Miao Z, Li Z. LncRNA SNHG17 promotes the progression of oral squamous cell carcinoma by modulating miR-375/PAX6 axis. Cancer Biomark. 2021;30:1–12. doi: 10.3233/CBM-191070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toolabi N, Daliri FS, Mokhlesi A, Talkhabi M. Identification of key regulators associated with colon cancer prognosis and pathogenesis. J Cell Commun Signal. 2022;16:115–127. doi: 10.1007/s12079-021-00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uthman YA, Ibrahim KG, Abubakar B, Bello MB, Malami I, Imam MU, Qusty N, Cruz-Martins N, Batiha GE, Abubakar MB. MALAT1: a promising therapeutic target for the treatment of metastatic colorectal cancer. Biochem Pharmacol. 2021;190:114657. doi: 10.1016/j.bcp.2021.114657. [DOI] [PubMed] [Google Scholar]
- Van den Eynde M, Hendlisz A. Treatment of colorectal liver metastases: a review. Rev Recent Clin Trials. 2009;4:56–62. doi: 10.2174/157488709787047558. [DOI] [PubMed] [Google Scholar]
- Varghese V, Magnani L, Harada-Shoji N, Mauri F, Szydlo RM, Yao S, Lam EW, Kenny LM. FOXM1 modulates 5-FU resistance in colorectal cancer through regulating TYMS expression. Sci Rep. 2019;9:1505. doi: 10.1038/s41598-018-38017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vychytilova-Faltejskova P, Slaby O. MicroRNA-215: From biology to theranostic applications. Mol Aspects Med. 2019;70:72–89. doi: 10.1016/j.mam.2019.03.002. [DOI] [PubMed] [Google Scholar]
- Vychytilova-Faltejskova P, Merhautova J, Machackova T, Gutierrez-Garcia I, Garcia-Solano J, Radova L, Brchnelova D, Slaba K, Svoboda M, Halamkova J, Demlova R, Kiss I, Vyzula R, Conesa-Zamora P, Slaby O. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis. 2017;6:399. doi: 10.1038/s41389-017-0006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S, Liu Z, Chen Y, Mai Z, Jiang M, Di Q, Sun B. MicroRNA-140-3p represses the proliferation, migration, invasion and angiogenesis of lung adenocarcinoma cells via targeting TYMS (thymidylate synthetase) Bioengineered. 2021;12:11959–11977. doi: 10.1080/21655979.2021.2009422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Li Y, Zhou J, Xu J, Peng C, Ye F, Shen Y, Lu W, Wan X, Xie X. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;179:2580–2588. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang H, Zhang Y, Zhen N, Zhang L, Qiao Y, Weng W, Liu X, Ma L, Xiao W, Yu W, Chu Q, Pan Q, Sun F. Mutual inhibition between YAP and SRSF1 maintains long non-coding RNA, Malat1-induced tumourigenesis in liver cancer. Cell Signal. 2014;26:1048–1059. doi: 10.1016/j.cellsig.2014.01.022. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang Q, Li M, Jiang S, Wang X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem Biophys Res Commun. 2014;444:199–204. doi: 10.1016/j.bbrc.2014.01.028. [DOI] [PubMed] [Google Scholar]
- Wang XC, Ma Y, Meng PS, Han JL, Yu HY, Bi LJ. miR-433 inhibits oral squamous cell carcinoma (OSCC) cell growth and metastasis by targeting HDAC6. Oral Oncol. 2015;51:674–682. doi: 10.1016/j.oraloncology.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Wang YY, Wu ZY, Wang GC, Liu K, Niu XB, Gu S, Meng JS. LINC00312 inhibits the migration and invasion of bladder cancer cells by targeting miR-197-3p. Tumour Biol. 2016;37:14553–14563. doi: 10.1007/s13277-016-5303-8. [DOI] [PubMed] [Google Scholar]
- Wang B, Lv K, Chen W, Zhao J, Luo J, Wu J, Li Z, Qin H, Wong TS, Yang W, Fu QL, Lei W. miR-375 and miR-205 regulate the invasion and migration of laryngeal squamous cell carcinoma synergistically via AKT-mediated EMT. Biomed Res Int. 2016;2016:9652789. doi: 10.1155/2016/9652789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Zhang ZF, Fan SH, Zhuang J, Shan Q, Han XR, Wen X, Li MQ, Hu B, Sun CH, Qiao B, Tao Q, Wu DM, Lu J, Zheng YL. MicroRNA-433 inhibits oral squamous cell carcinoma cells by targeting FAK. Oncotarget. 2017;8:100227–100241. doi: 10.18632/oncotarget.22151. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Z, Zhao Z, Yang Y, Luo M, Zhang M, Wang X, Liu L, Hou N, Guo Q, Song T, Guo B, Huang C. MiR-99b-5p and miR-203a-3p function as tumor suppressors by targeting IGF-1R in gastric cancer. Sci Rep. 2018;8:10119. doi: 10.1038/s41598-018-27583-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chen SH, Kong P, Zhang LY, Zhang LL, Zhang NQ, Gu H. Increased expression of miR-330-3p: a novel independent indicator of poor prognosis in human breast cancer. Eur Rev Med Pharmacol Sci. 2018;22:1726–1730. doi: 10.26355/eurrev_201803_14587. [DOI] [PubMed] [Google Scholar]
- Wang M, Hu H, Wang Y, Huang Q, Huang R, Chen Y, Ma T, Qiao T, Zhang Q, Wu H, Chen Q, Han D, Wang G, Wang X. Long non-coding RNA TUG1 mediates 5-fluorouracil resistance by acting as a ceRNA of miR-197-3p in colorectal cancer. J Cancer. 2019;10:4603–4613. doi: 10.7150/jca.32065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YG, Wang T, Ding M, Xiang SH, Shi M, Zhai B. hsa_circ_0091570 acts as a ceRNA to suppress hepatocellular cancer progression by sponging hsa-miR-1307. Cancer Lett. 2019;460:128–138. doi: 10.1016/j.canlet.2019.06.007. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang X, Liu Y, Xu S. Long noncoding RNA FBXL19-AS1 induces tumor growth and metastasis by sponging miR-203a-3p in lung adenocarcinoma. J Cell Physiol. 2020;235:3612–3625. doi: 10.1002/jcp.29251. [DOI] [PubMed] [Google Scholar]
- Wang F, Fan M, Cai Y, Zhou X, Tai S, Yu Y, Wu H, Zhang Y, Liu J, Huang S, He N, Hu Z, Jin X. Circular RNA circRIMS1 acts as a sponge of miR-433-3p to promote bladder cancer progression by regulating CCAR1 expression. Mol Ther Nucleic Acids. 2020;22:815–831. doi: 10.1016/j.omtn.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chen K, Li D, Chen M, Li A, Wang J. miR-140-3p is involved in the occurrence and metastasis of gastric cancer by regulating the stability of FAM83B. Cancer Cell Int. 2021;21:537. doi: 10.1186/s12935-021-02245-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhang Y, Zhou X, Chen X, Xiang J, Fan M, Yu Y, Cai Y, Wu H, Huang S, He N, Hu Z, Ding G, Jin X. Circular RNA CircPPP1CB suppresses tumorigenesis by interacting with the MiR-1307-3p/SMG1 axis in human bladder cancer. Front Cell Dev Biol. 2021;9:704683. doi: 10.3389/fcell.2021.704683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun Y, Zhang X, Cai H, Zhang C, Qu H, Liu L, Zhang M, Fu J, Zhang J, Wang J, Zhang G. Oxidative stress activates NORAD expression by H3K27ac and promotes oxaliplatin resistance in gastric cancer by enhancing autophagy flux via targeting the miR-433-3p. Cell Death Dis. 2021;12:90. doi: 10.1038/s41419-020-03368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang Q, Gong Y, Hu Q, Zhang H, Ke S, Chen Y. Knockdown of circRNA circ_0087378 represses the tumorigenesis and progression of esophageal squamous cell carcinoma through modulating the miR-140-3p/E2F3 Axis. Front Oncol. 2021;10:607231. doi: 10.3389/fonc.2020.607231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Z, Lu J, Zhang H. Circular RNA circ-PTEN elevates PTEN inhibiting the proliferation of non-small cell lung cancer cells. Hum Cell. 2021;34:1174–1184. doi: 10.1007/s13577-021-00526-y. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wo Y, Lu T, Sun X, Liu A, Dong Y, Du W, Su W, Huang Z, Jiao W. Circ-AASDH functions as the progression of early stage lung adenocarcinoma by targeting miR-140-3p to activate E2F7 expression. Transl Lung Cancer Res. 2021;10:57–70. doi: 10.21037/tlcr-20-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Miao C, Gao X. TCEB3 is regulated by Circ-0000212/miR-140-3p axis to promote the progression of cervical cancer. Onco Targets Ther. 2021;14:2853–2865. doi: 10.2147/OTT.S278710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yin G, Bian H, Yang J, Zhou P, Yan K, Liu C, Chen P, Zhu J, Li Z, Xue T. LncRNA XIST upregulates TRIM25 via negatively regulating miR-192 in hepatitis B virus-related hepatocellular carcinoma. Mol Med. 2021;27:41. doi: 10.1186/s10020-021-00278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Liao J, Tan C, Zhou H, Wang J, Wang K, Li Y, Wu W. Integrated study of miR-215 promoting breast cancer cell apoptosis by targeting RAD54B. J Cell Mol Med. 2021;25:3327–3338. doi: 10.1111/jcmm.16402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zheng Y, Zhang X, Xu SW, Li XH, Meng YL, Liu JQ. Roles of circ_0000135/miR-140-3p/PDZK1 network in cervical cancer. Clin Transl Oncol. 2022 doi: 10.1007/s12094-021-02751-1. [DOI] [PubMed] [Google Scholar]
- Wang J, Ke S, Gong Y, Cai Y, Xia L, Shi Z, Qiu H, Shi W, Wang Q, Chen Y. Circ_0011385 knockdown inhibits cell proliferation, migration and invasion, whereas promotes cell apoptosis by regulating miR-330-3p/MYO6 axis in colorectal cancer. Biomed J. 2022 doi: 10.1016/j.bj.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Wang X, Lv L, Zheng Y, Zhang N, Yang M. The emerging role of noncoding RNAs in colorectal cancer chemoresistance. Cell Oncol (dordr) 2019;42:757–768. doi: 10.1007/s13402-019-00466-8. [DOI] [PubMed] [Google Scholar]
- Wei C, Zhang R, Cai Q, Gao X, Tong F, Dong J, Hu Y, Wu G, Dong X. MicroRNA-330-3p promotes brain metastasis and epithelial-mesenchymal transition via GRIA3 in non-small cell lung cancer. Aging (albany NY) 2019;11:6734–6761. doi: 10.18632/aging.102201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Dong X, Lu H, Tong F, Chen L, Zhang R, Dong J, Hu Y, Wu G, Dong X. LPCAT1 promotes brain metastasis of lung adenocarcinoma by up-regulating PI3K/AKT/MYC pathway. J Exp Clin Cancer Res. 2019;38:95. doi: 10.1186/s13046-019-1092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner H, Sarfstein R, LeRoith D, Bruchim I. Insulin-like growth factor 1 signaling axis meets p53 genome protection pathways. Front Oncol. 2016;6:159. doi: 10.3389/fonc.2016.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki PM, Rybarczyk A. The Hippo pathway in colorectal cancer. Folia Histochem Cytobiol. 2015;53:105–119. doi: 10.5603/FHC.a2015.0015. [DOI] [PubMed] [Google Scholar]
- Wilusz JE. Long noncoding RNAs: re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859:128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. (2008) 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135(5):919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witusik-Perkowska M, Jaskólski DJ, Liberski PP, Szemraj J. If artificial in vitro microenvironment can influence tumor drug resistance network via modulation of lncRNA expression?-Comparative analysis of glioblastoma-derived cell culture models and initial tumors in vivo. Cell Mol Neurobiol. 2022;42:1005–1020. doi: 10.1007/s10571-020-00991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yu J, Ma Y, Wang F, Liu H. miR-148a and miR-375 may serve as predictive biomarkers for early diagnosis of laryngeal carcinoma. Oncol Lett. 2016;12:871–878. doi: 10.3892/ol.2016.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sun X, Song B, Qiu X, Zhao J. MiR-375/SLC7A11 axis regulates oral squamous cell carcinoma proliferation and invasion. Cancer Med. 2017;6:1686–1697. doi: 10.1002/cam4.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Meng WY, Jie Y, Zhao H. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol. 2018;233:6750–6757. doi: 10.1002/jcp.26383. [DOI] [PubMed] [Google Scholar]
- Wu C, Zhu X, Tao K, Liu W, Ruan T, Wan W, Zhang C, Zhang W. MALAT1 promotes the colorectal cancer malignancy by increasing DCP1A expression and miR203 downregulation. Mol Carcinog. 2018;57:1421–1431. doi: 10.1002/mc.22868. [DOI] [PubMed] [Google Scholar]
- Wu Q, Wu W, Fu B, Shi L, Wang X, Kuca K. JNK signaling in cancer cell survival. Med Res Rev. 2019;39:2082–2104. doi: 10.1002/med.21574. [DOI] [PubMed] [Google Scholar]
- Wu S, Wang H, Pan Y, Yang X, Wu D. miR-140-3p enhances cisplatin sensitivity and attenuates stem cell-like properties through repressing Wnt/β-catenin signaling in lung adenocarcinoma cells. Exp Ther Med. 2020;20:1664–1674. doi: 10.3892/etm.2020.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Yurievich UA, Yosypovych SV. Long noncoding RNA XIST is a prognostic factor in colorectal cancer and inhibits 5-fluorouracil-induced cell cytotoxicity through promoting thymidylate synthase expression. Oncotarget. 2017;8:83171–83182. doi: 10.18632/oncotarget.20487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, Guo X, Zhang Y. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via MiR-203a-3p-mediated Wnt/ß-catenin signaling pathway. Cell Physiol Biochem. 2018;46:1275–1285. doi: 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- Xie W, Shui C, Fang X, Peng Y, Qin L. miR-197-3p reduces epithelial-mesenchymal transition by targeting ABCA7 in ovarian cancer cells. 3 Biotech. 2020;10:375. doi: 10.1007/s13205-020-02362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong HL, Zhou SW, Sun AH, He Y, Li J, Yuan X. MicroRNA-197 reverses the drug resistance of fluorouracil-induced SGC7901 cells by targeting mitogen-activated protein kinase 1. Mol Med Rep. 2015;12:5019–5025. doi: 10.3892/mmr.2015.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Wang J, Zhu H, Liu L, Jiang Y. Chronic oxymatrine treatment induces resistance and epithelial-mesenchymal transition through targeting the long non-coding RNA MALAT1 in colorectal cancer cells. Oncol Rep. 2018;39:967–976. doi: 10.3892/or.2018.6204. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xiong X, Shi Q, Yang X, Wang W, Tao J. LINC00052 functions as a tumor suppressor through negatively modulating miR-330-3p in pancreatic cancer. J Cell Physiol. 2019;234:15619–15626. doi: 10.1002/jcp.28209. [DOI] [PubMed] [Google Scholar]
- Xu L, Wen T, Liu Z, Xu F, Yang L, Liu J, Feng G, An G. MicroRNA-375 suppresses human colorectal cancer metastasis by targeting Frizzled 8. Oncotarget. 2016;7:40644–40656. doi: 10.18632/oncotarget.9811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhu Y, Liang Z, Li S, Xu X, Wang X, Wu J, Hu Z, Meng S, Liu B, Qin J, Xie L, Zheng X. c-Met and CREB1 are involved in miR-433-mediated inhibition of the epithelial-mesenchymal transition in bladder cancer by regulating Akt/GSK-3β/Snail signaling. Cell Death Dis. 2016;7:e2088. doi: 10.1038/cddis.2015.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jiang H, Zhang F, Gao J, Hou J. MicroRNA-330 inhibited cell proliferation and enhanced chemosensitivity to 5-fluorouracil in colorectal cancer by directly targeting thymidylate synthase. Oncol Lett. 2017;13:3387–3394. doi: 10.3892/ol.2017.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen X, Xu M, Liu X, Pan B, Qin J, Xu T, Zeng K, Pan Y, He B, Sun H, Sun L, Wang S. miR-375-3p suppresses tumorigenesis and partially reverses chemoresistance by targeting YAP1 and SP1 in colorectal cancer cells. Aging (albany NY) 2019;11:7357–7385. doi: 10.18632/aging.102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jiang J, Zhang J, Cheng L, Pan S, Li Y. MicroRNA-375 inhibits esophageal squamous cell carcinoma proliferation through direct targeting of SP1. Exp Ther Med. 2019;17:1509–1516. doi: 10.3892/etm.2018.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Huang J, Hua S, Liang L, He X, Zhan M, Lu L, Chu J. Interactome analysis of gene expression profiles identifies CDC6 as a potential therapeutic target modified by miR-215-5p in hepatocellular carcinoma. Int J Med Sci. 2020;17:2926–2940. doi: 10.7150/ijms.51145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Ye ML, Zhang YP, Li WJ, Li MT, Wang HZ, Qiu X, Xu Y, Yin JW, Hu Q, Wei WH, Chang Y, Liu L, Zhao Q. MicroRNA-375-3p enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Cancer Sci. 2020;111:1528–1541. doi: 10.1111/cas.14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wei Q, He Z. Insight into the function of RIPK4 in keratinocyte differentiation and carcinogenesis. Front Oncol. 2020;10:1562. doi: 10.3389/fonc.2020.01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xiao Y, Liu B, Pan S, Liu Q, Shan Y, Li S, Qi Y, Huang Y, Jia L. Exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of colorectal cancer via FUT4 enhanced fucosylation and PI3K/Akt pathway. J Exp Clin Cancer Res. 2020;39:54. doi: 10.1186/s13046-020-01562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Yin S, Zheng M, Pei X, Ji X. Circular RNA circZFR promotes hepatocellular carcinoma progression by regulating miR-375/HMGA2 axis. Dig Dis Sci. 2021;66:4361–4373. doi: 10.1007/s10620-020-06805-2. [DOI] [PubMed] [Google Scholar]
- Xu WW, Jin J, Wu XY, Ren QL, Farzaneh M. MALAT1-related signaling pathways in colorectal cancer. Cancer Cell Int. 2022;22:126. doi: 10.1186/s12935-022-02540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, You WQ, Sheng NQ, Gong JF, Hu LD, Tan GW, Chen HQ, Wang ZG. A CREB1/miR-433 reciprocal feedback loop modulates proliferation and metastasis in colorectal cancer. Aging (albany NY) 2018;10:3774–3793. doi: 10.18632/aging.101671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Qian M, He Q, Zhu H, Yang B. The posttranslational modifications of Hippo-YAP pathway in cancer. Biochim Biophys Acta Gen Subj. 2020;1864:129397. doi: 10.1016/j.bbagen.2019.07.006. [DOI] [PubMed] [Google Scholar]
- Yan J, Wei R, Li H, Dou Y, Wang J. miR-452-5p and miR-215-5p expression levels in colorectal cancer tissues and their relationship with clinicopathological features. Oncol Lett. 2020;20:2955–2961. doi: 10.3892/ol.2020.11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem. 2013;288:28893–28899. doi: 10.1074/jbc.M113.502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Yan R, Zhang X, Zhu Z, Wang C, Liang C, Zhang X. Deregulation of MicroRNA-375 inhibits cancer proliferation migration and chemosensitivity in pancreatic cancer through the association of HOXB3. Am J Transl Res. 2016;8:1551–1559. [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Wang Y, Sims M, Cai C, He P, Häcker H, Yue J, Cheng J, Boop FA, Pfeffer LM. MicroRNA203a suppresses glioma tumorigenesis through an ATM-dependent interferon response pathway. Oncotarget. 2017;8:112980–112991. doi: 10.18632/oncotarget.22945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Li H, Chen T, Ren H, Shi P, Chen M. LncRNA MALAT1 depressed chemo-sensitivity of NSCLC cells through directly functioning on miR-197-3p/p120 catenin axis. Mol Cells. 2019;42:270–283. doi: 10.14348/molcells.2019.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li D, Li Y, Jiang Q, Sun R, Liu J, Wu F, Miao J, Ni L, Shi X, Huang C. Low-temperature plasma suppresses proliferation and induces apoptosis in lung cancer cells by regulating the miR-203a/BIRC5 axis. Onco Targets Ther. 2020;13:5145–5153. doi: 10.2147/OTT.S244853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yan X, Li X, Ma Y, Goel A. Long non-coding RNAs in colorectal cancer: novel oncogenic mechanisms and promising clinical applications. Cancer Lett. 2021;504:67–80. doi: 10.1016/j.canlet.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Zhang P, Li J, Xu W. MicroRNA-215 acts as a tumor suppressor in breast cancer by targeting AKT serine/threonine kinase 1. Oncol Lett. 2017;14:1097–1104. doi: 10.3892/ol.2017.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao PA, Wu Y, Zhao K, Li Y, Cao J, Xing C. The feedback loop of ANKHD1/lncRNA MALAT1/YAP1 strengthens the radioresistance of CRC by activating YAP1/AKT signaling. Cell Death Dis. 2022;13:103. doi: 10.1038/s41419-022-04554-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X, Xie X, Tang H. circFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol Ther Nucleic Acids. 2019;18:88–98. doi: 10.1016/j.omtn.2019.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Jiang J, Deng H, Wang Z, Gu R, Wang F. miR-140-3p aggregates osteoporosis by targeting PTEN and activating PTEN/PI3K/AKT signaling pathway. Hum Cell. 2020;33:569–581. doi: 10.1007/s13577-020-00352-8. [DOI] [PubMed] [Google Scholar]
- Yonemori K, Seki N, Kurahara H, Osako Y, Idichi T, Arai T, Koshizuka K, Kita Y, Maemura K, Natsugoe S. ZFP36L2 promotes cancer cell aggressiveness and is regulated by antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer Sci. 2017;108:124–135. doi: 10.1111/cas.13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Li Y, Lin S, Wang Z, Xu Y. Inhibitors targeting YAP in gastric cancer: current status and future perspectives. Drug Des Devel Ther. 2021;15:2445–2456. doi: 10.2147/DDDT.S308377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BK, Gredler R, Vozhilla N, Su ZZ, Chen D, Forcier T, Shah K, Saxena U, Hansen U, Fisher PB, Sarkar D. Identification of genes conferring resistance to 5-fluorouracil. Proc Natl Acad Sci U S A. 2009;106:12938–12943. doi: 10.1073/pnas.0901451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis NS, AlMasoud ES, Al Khawajah F, Alghazal FJ, AlMofarfesh HM, Al-Khalaf LH, Al Otaibi MS, Alkhamis SM, Al Naser ZA, Al Mousa ZH, Alabdulaziz ZI, Mohamed ME. Potential genetic biomarker of Saudi Arabian patients with colorectal cancer. Eur Rev Med Pharmacol Sci. 2022;26:3109–3126. doi: 10.26355/eurrev_202205_28728. [DOI] [PubMed] [Google Scholar]
- Yu L, Fu J, Yu N, Wu Y, Han N. Long noncoding RNA MALAT1 participates in the pathological angiogenesis of diabetic retinopathy in an oxygen-induced retinopathy mouse model by sponging miR-203a-3p. Can J Physiol Pharmacol. 2020;98:219–227. doi: 10.1139/cjpp-2019-0489. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang H, Zhao C, Li G, Zhang Y, Sun Y. CircRNA circ_0008037 facilitates tumor growth and the Warburg effect via upregulating NUCKS1 by binding to miR-433-3p in non-small cell lung cancer. Thorac Cancer. 2022;13:162–172. doi: 10.1111/1759-7714.14235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan KT, Li BX, Yuan YJ, Tan M, Tan JF, Dai WG, Feng WD, Zuo JD. Deregulation of MicroRNA-375 inhibits proliferation and migration in gastric cancer in association with autophagy-mediated AKT/mTOR signaling pathways. Technol Cancer Res Treat. 2018;17:1533033818806499. doi: 10.1177/1533033818806499. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yuan JB, Gu L, Chen L, Yin Y, Fan BY. Annexin A8 regulated by lncRNA-TUG1/miR-140-3p axis promotes bladder cancer progression and metastasis. Mol Ther Oncolytics. 2021;22:36–51. doi: 10.1016/j.omto.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue N, Ye M, Zhang R, Wang M. MicroRNA-1307-3p accelerates the progression of colorectal cancer via regulation of TUSC5. Exp Ther Med. 2020;20:1746–1751. doi: 10.3892/etm.2020.8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Elabd S, Hammer S, Solozobova V, Yan H, Bartel F, Inoue S, Henrich T, Wittbrodt J, Loosli F, Davidson G, Blattner C. TRIM25 has a dual function in the p53/Mdm2 circuit. Oncogene. 2015;34(46):5729–5738. doi: 10.1038/onc.2015.21. [DOI] [PubMed] [Google Scholar]
- Zhang B, Li Y, Hou D, Shi Q, Yang S, Li Q. MicroRNA-375 inhibits growth and enhances radiosensitivity in oral squamous cell carcinoma by targeting insulin like growth factor 1 receptor. Cell Physiol Biochem. 2017;42:2105–2117. doi: 10.1159/000479913. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang L, Zhang T, Dong XM, Zhu Y, Chen LH. Reduced miR-433 expression is associated with advanced stages and early relapse of colorectal cancer and restored miR-433 expression suppresses the migration, invasion and proliferation of tumor cells in vitro and in nude mice. Oncol Lett. 2018;15:7579–7588. doi: 10.3892/ol.2018.8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li W, Gu W, Yan Y, Yao X, Zheng J. MALAT1 accelerates the development and progression of renal cell carcinoma by decreasing the expression of miR-203 and promoting the expression of BIRC5. Cell Prolif. 2019;52:e12640. doi: 10.1111/cpr.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Wang XX, Zhang F. Correlations of the MiR-330 expression with the pathogenesis and prognosis of breast cancer. Eur Rev Med Pharmacol Sci. 2019;23:1584–1590. doi: 10.26355/eurrev_201902_17117. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li X, Han Y, Ji T, Huang X, Gao Q, Ma D. RAD54B potentiates tumor growth and predicts poor prognosis of patients with luminal A breast cancer. Biomed Pharmacother. 2019;118:109341. doi: 10.1016/j.biopha.2019.109341. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zheng Y, Zhu G. MiR-203a-3p targets PTEN to promote hepatocyte proliferation by regulating PI3K/Akt pathway in BRL-3A cells. Biosci Biotechnol Biochem. 2020;84:725–733. doi: 10.1080/09168451.2019.1694860. [DOI] [PubMed] [Google Scholar]
- Zhang F, Cheng R, Li P, Lu C, Zhang G. Hsa_circ_0010235 functions as an oncogenic drive in non-small cell lung cancer by modulating miR-433-3p/TIPRL axis. Cancer Cell Int. 2021;21:73. doi: 10.1186/s12935-021-01764-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZY, Zhao YC, Liu W. Long non-coding RNA TUG1 regulates the progression and metastasis of osteosarcoma cells via miR-140-5p/PFN2 axis. Eur Rev Med Pharmacol Sci. 2019;23:9781–9792. doi: 10.26355/eurrev_201911_19541. [DOI] [PubMed] [Google Scholar]
- Zhao X, Chen GQ, Cao GM. Abnormal expression and mechanism of miR-330-3p/BTG1 axis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(16):6888–6898. doi: 10.26355/eurrev_201908_18728. [DOI] [PubMed] [Google Scholar]
- Zhao H, Chen J, Chen J, Kong X, Zhu H, Zhang Y, Dong H, Wang J, Ren Q, Wang Q, Chen S, Deng Z, Chen Z, Cui Q, Zheng J, Lu J, Wang S, Tan J. miR-192/215-5p act as tumor suppressors and link Crohn's disease and colorectal cancer by targeting common metabolic pathways: an integrated informatics analysis and experimental study. J Cell Physiol. 2019;234:21060–21075. doi: 10.1002/jcp.28709. [DOI] [PubMed] [Google Scholar]
- Zhao L, Lou G, Li A, Liu Y. lncRNA MALAT1 modulates cancer stem cell properties of liver cancer cells by regulating YAP1 expression via miR-375 sponging. Mol Med Rep. 2020;22:1449–1457. doi: 10.3892/mmr.2020.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Ye Z, Li Y, Li C, Yang X, Chen Q, Xing C. LncRNA FTX contributes to the progression of colorectal cancer through regulating miR-192-5p/EIF5A2 Axis. Onco Targets Ther. 2020;13:2677–2688. doi: 10.2147/OTT.S241011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Wang B, Ma Y, Kuang J, Liang J, Yuan Y. NUCKS1 promotes proliferation, invasion and migration of non-small cell lung cancer by upregulating CDK1 expression. Cancer Manag Res. 2020;12:13311–13323. doi: 10.2147/CMAR.S282181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zheng Y, Lei W, Xiang L, Chen M. miR-1307-3p overexpression inhibits cell proliferation and promotes cell apoptosis by targeting ISM1 in colon cancer. Mol Cell Probes. 2019;48:101445. doi: 10.1016/j.mcp.2019.101445. [DOI] [PubMed] [Google Scholar]
- Zheng XF, Liu KX, Wang XM, Zhang R, Li X. MicroRNA-192 acts as a tumor suppressor in colon cancer and simvastatin activates miR-192 to inhibit cancer cell growth. Mol Med Rep. 2019;19:1753–1760. doi: 10.3892/mmr.2019.9808. [DOI] [PubMed] [Google Scholar]
- Zheng X, Ren J, Peng B, Ye J, Wu X, Zhao W, Li Y, Chen R, Gong X, Bai C, Wang Y, Zhao H, Zhang Y. MALAT1 overexpression promotes the growth of colon cancer by repressing β-catenin degradation. Cell Signal. 2020;73:109676. doi: 10.1016/j.cellsig.2020.109676. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhou Y, Yin B, Hao W, Zhao L, Ju W, Bai C. 5-Fluorouracil and oxaliplatin modify the expression profiles of microRNAs in human colon cancer cells in vitro. Oncol Rep. 2010;23:121–128. doi: 10.3892/or_00000613. [DOI] [PubMed] [Google Scholar]
- Zhou SW, Su BB, Zhou Y, Feng YQ, Guo Y, Wang YX, Qi P, Xu S. Aberrant miR-215 expression is associated with clinical outcome in breast cancer patients. Med Oncol. 2014;31:259. doi: 10.1007/s12032-014-0259-2. [DOI] [PubMed] [Google Scholar]
- Zhou J, Song S, He S, Zhu X, Zhang Y, Yi B, Zhang B, Qin G, Li D. MicroRNA-375 targets PDK1 in pancreatic carcinoma and suppresses cell growth through the Akt signaling pathway. Int J Mol Med. 2014;33:950–956. doi: 10.3892/ijmm.2014.1638. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wang M, Wu J, Jie Z, Chang S, Shuang T. The clinicopathological significance of miR-1307 in chemotherapy resistant epithelial ovarian cancer. J Ovarian Res. 2015;8:23. doi: 10.1186/s13048-015-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang B, Wang Y, Chen G, Lian Q, Wang H. miR-140-3p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28. Oncol Lett. 2019;17:3835–3841. doi: 10.3892/ol.2019.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Liu L, Zhou J, Chen Y, Xie D, Yao Y, Cui D. Novel insights into MALAT1 function as a MicroRNA sponge in NSCLC. Front Oncol. 2021;11:758653. doi: 10.3389/fonc.2021.758653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liu K, Cui J, Xiong J, Wu H, Peng T, Guo Y. Circ-MBOAT2 knockdown represses tumor progression and glutamine catabolism by miR-433-3p/GOT1 axis in pancreatic cancer. J Exp Clin Cancer Res. 2021;40:124. doi: 10.1186/s13046-021-01894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zou S, Zhou J, Zhu H, Zhang S, Shang Z, Ding WQ, Wu J, Chen Y. REV3L, the catalytic subunit of DNA polymerase ζ, is involved in the progression and chemoresistance of esophageal squamous cell carcinoma. Oncol Rep. 2016;35:1664–1670. doi: 10.3892/or.2016.4549. [DOI] [PubMed] [Google Scholar]
- Zhu M, Zhang C, Chen D, Chen S, Zheng H. lncRNA MALAT1 potentiates the progression of tongue squamous cell carcinoma through regulating miR-140-5p-PAK1 pathway. Onco Targets Ther. 2019;19(12):1365–1377. doi: 10.2147/OTT.S192069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo M, Yuan C, Han T, Cui J, Jiao F, Wang L. A novel feedback loop between high MALAT-1 and low miR-200c-3p promotes cell migration and invasion in pancreatic ductal adenocarcinoma and is predictive of poor prognosis. BMC Cancer. 2018;18:1032. doi: 10.1186/s12885-018-4954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P, Zhu M, Lian C, Wang J, Chen Z, Zhang X, Yang Y, Chen X, Cui X, Liu J, Wang H, Wen Q, Yi J. miR-192-5p suppresses the progression of lung cancer bone metastasis by targeting TRIM44. Sci Rep. 2019;9:19619. doi: 10.1038/s41598-019-56018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.