Abstract
Background
Prognostic models are typically developed to estimate the risk that an individual in a particular health state will develop a particular health outcome, to support (shared) decision making. Systematic reviews of prognostic model studies can help identify prognostic models that need to further be validated or are ready to be implemented in healthcare.
Objectives
To provide a step-by-step guidance on how to conduct and read a systematic review of prognostic model studies and to provide an overview of methodology and guidance available for every step of the review progress.
Sources
Published, peer-reviewed guidance articles.
Content
We describe the following steps for conducting a systematic review of prognosis studies: 1) Developing the review question using the Population, Index model, Comparator model, Outcome(s), Timing, Setting format, 2) Searching and selection of articles, 3) Data extraction using the Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies (CHARMS) checklist, 4) Quality and risk of bias assessment using the Prediction model Risk Of Bias ASsessment (PROBAST) tool, 5) Analysing data and undertaking quantitative meta-analysis, and 6) Presenting summary of findings, interpreting results, and drawing conclusions. Guidance for each step is described and illustrated using a case study on prognostic models for patients with COVID-19.
Implications
Guidance for conducting a systematic review of prognosis studies is available, but the implications of these reviews for clinical practice and further research highly depend on complete reporting of primary studies.
Keywords: Meta-analysis, Prediction model, Prognosis, Prognostic model, Systematic review
Introduction
There has been a growing demand for personalized, risk-based, or stratified medicine. This implies that medical decisions on treatment and further diagnostic tests are ideally tailored to the patient rather than based on a ‘one size fits all’ approach. Information on the prognosis of the individual patient is therefore crucial [[1], [2], [3], [4]]. The number of studies investigating biomarkers, prognostic factors, and prognostic models has been increasing rapidly. Systematic reviews are needed to summarize the information from their primary publications [5,6].
We distinguish between three types of prognosis studies [7]:
-
1.
Overall prognosis studies give insight in the occurrence of certain outcome(s) in a particular time frame for a group of individuals with a particular health condition (not necessarily a disease) [8].
-
2.
Prognostic factor studies aim to identify characteristics that are associated with the occurrence of certain outcome(s) in a particular time frame for individuals with a particular health condition [9].
-
3.
Prognostic model studies combine multiple prognostic factors in one multivariable prognostic model aimed at making predictions for occurrence of a certain outcome in a particular time frame in individuals with a particular health condition [10]. Studies on prognostic models can be further categorized as model development, model validation, or a combination of these [2,[11], [12], [13], [14]].
It may be clear that these different types of prognosis studies are designed to address different prognosis questions, and, as such, different types of systematic reviews can be conducted in the field of prognosis research [4,15]. Herein we focus on type 3, systematic reviews of prognostic model studies, but most of the principles and guidance can easily be adapted to the other types of prognosis study reviews. An example of such a prognostic model review was published in this journal in 2021 [16]. All guidance about reviews and meta-analysis of prognostic prediction models (estimating the probability of future occurrence of outcomes) also directly applies to reviews of diagnostic prediction models (estimating the probability of current presence of outcomes) [17].
Prognostic models are developed and validated to estimate the risk (i.e. probability) that an individual in a particular health state will develop a particular health outcome. These risk estimates are based on patient information, such as from demographics, medical history, comorbidities, imaging, lab and omics data, and previous treatments. The estimated risk by prognostic models can be used to make healthcare decisions, such as starting, stopping or refraining from treatment, or selecting patients that need more extensive care, to inform patients and family members about likely outcomes and/or to create risk stratifications for randomized intervention trials [1,2,4].
For many diseases, target populations, and outcomes, multiple prognostic models have already been developed. For example, there are >400 prediction models for prognosis of chronic obstructive pulmonary disease [18], 363 models for predicting cardiovascular disease occurrence in the general population [19], 232 models for diagnosis and prognosis of COVID-19 [20], 37 models for predicting pulmonary tuberculosis treatment outcomes [21], and 27 models for the clinical management of malaria [22]. Systematic reviews of prognostic models provide an overview of the existing models, their quality (risk of bias), and their predictive performance. These reviews can serve as a valuable tool to decide which prognostic model(s) should be further evaluated or implemented in medical practice or public health. Possible aims of a systematic review of prognostic model studies include the following [4,23,24]:
-
1.
To identify all existing prognostic models—developed or validated—for a particular target population, condition, or prognostic outcome.
-
2.
To summarize the predictive performance of a specific prognostic model and to identify sources of heterogeneity in its performance across multiple external validation studies of that model (Table 1 ).
-
3.
To summarize and compare the predictive performance of several prognostic models across multiple external validation studies of those models, for a certain target population, condition or outcome.
-
4.
To identify whether particular predictors, when added to a specific existing prognostic model, improve the predictive performance of that specific model.
Table 1.
Glossary
| Definition | |
|---|---|
| Calibration | Agreement between observed outcome risks and the risks predicted by the model. |
| Calibration slope | Slope of the linear predictor in case you would fit a regression line. The calibration slope ideally equals 1. A calibration slope <1 indicates that predictions are too extreme (e.g. low-risk individuals have a predicted risk that is too low, and high-risk individuals are given a predicted risk that is too high). Conversely, a slope >1 indicates that predictions are not extreme enough [26]. |
| Concordance c-statistic | Statistic that quantifies the chance that for any two individuals of which one developed the outcome and the other did not, the former has a higher predicted risk according to the model than the latter. A c-statistic of 1 means perfect discriminative ability, whereas a model with a c-statistic of 0.5 is not better than flipping a coin [27]. C-statistic is highly dependent on case-mix in the population (i.e. in homogeneous populations c-statistics are in general lower compared to heterogeneous populations) [28,29]. |
| Discrimination | Ability of the model to distinguish between people who did and did not develop the outcome of interest, often quantified by the concordance c-statistic. |
| External validation | Evaluating the predictive performance of a prediction model in a study population other than the population from which the model was developed. |
| OE ratio | The ratio of the total number of actual observed participants with the outcome in a specific time frame (e.g. in 1 y) and the total number of participants with the outcome as predicted by the model. |
| Prediction horizon | Time frame over which the model predicts the outcome (e.g. predicting 10-y risk of developing cardiovascular disease). |
| Predictive performance | Accuracy of the predictions made by a prediction model, often expressed in terms of calibration and discrimination. |
OE, observed expected.
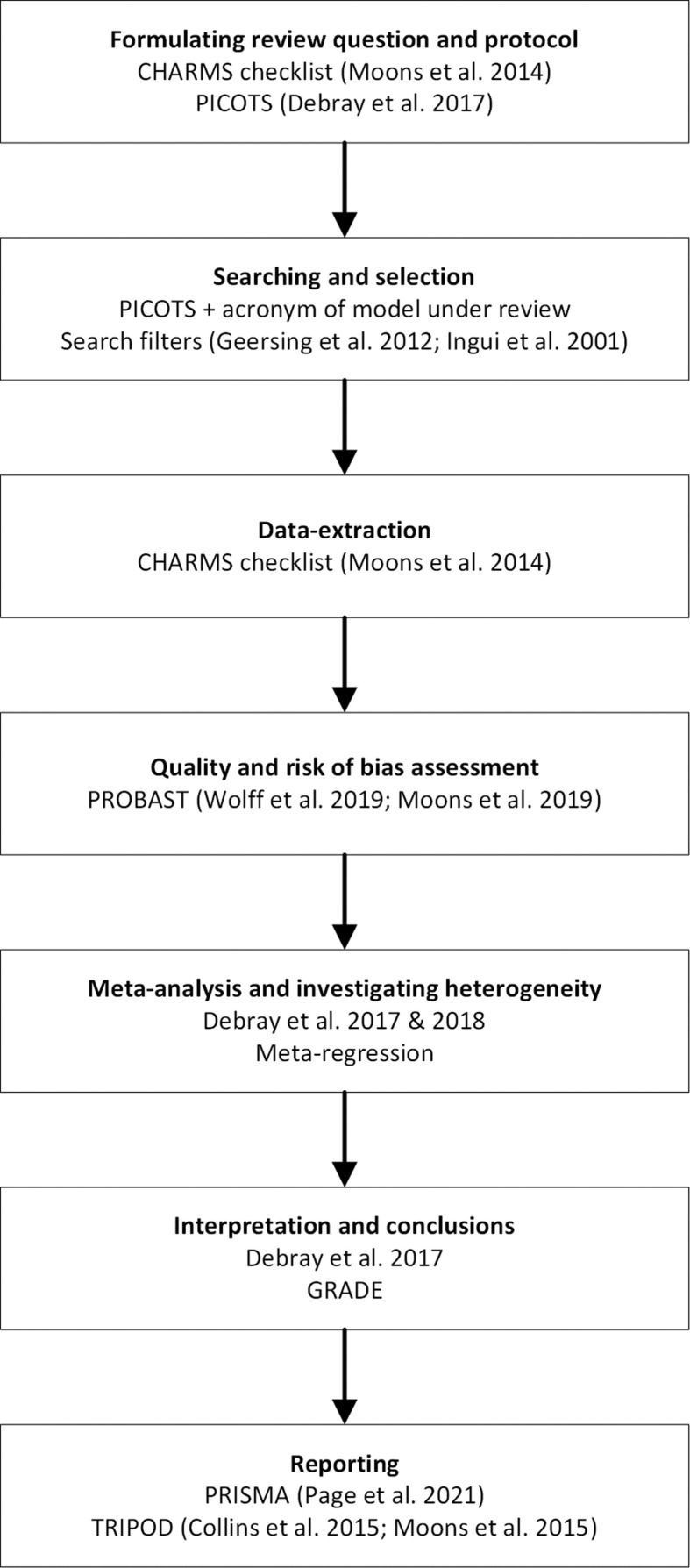
The aim of this paper was to provide a step-by-step guidance on how to conduct and read a systematic review of prognostic model studies (regardless the specific aim) and to provide an overview of methodology available for every step of the review progress (Fig. 1 ). We hereby did not differentiate between prognostic models developed by prevailing regression modelling techniques (e.g. time to event models or logistic regression models) or by using modern techniques based on artificial intelligence or machine learning. The method of model development does not change the necessary steps of the systematic review. We illustrated every step using a case study: the third update of the currently ongoing COVID-PRECISE living review on models for predicting the prognosis of individuals with COVID-19 (see https://www.covprecise.org/) [20]. We chose this example as many of the published systematic reviews of prognostic models have similar aims to this COVID-PRECISE living review (i.e. identifying all prediction models available for a specific population or a specific outcome). We also referred to the Cochrane Prognosis Methods Group website (https://methods.cochrane.org/prognosis/tools) for detailed guidance on every step of a prognostic model review that is discussed below [25].
Fig. 1.
Review steps. References: the Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies [23], Population, Index model, Comparartor model, Outcome(s) model, Timing, Setting [30], search filters [31,32], Prediction model Risk Of Bias ASsessment tool [24,33], meta-analysis [30,34], the Preferred Items for Systematic Reviews and Meta-analyses guidelines [35], Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis statement [36,37].
Step 1: developing the review question
The first step when conducting a systematic review is to formulate a review question. This is an important step, as all subsequent steps of the review process are dictated by the question, including the search strategy, the eligibility criteria, the items for which to extract data from included studies, the choice of meta-analysis methods, and the interpretation of results. Guidance for formulating a review question for reviews of prognosis studies is provided in the Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies (CHARMS) checklist [23] and was subsequently further developed [30]. These papers, as well as the Cochrane Prognosis Methods Group guidance, advise to use the PICOTS system for formulating the review question (Table 2 ) [25]. This is an adaption and extension of the PICO (Population, Intervention, Comparator, Outcome) system, which is commonly used for systematic reviews of intervention and diagnostic test accuracy studies [38,39]. Systematic reviews of prognosis studies are advised to also explicitly consider the Timing (moment at which a prognostication is to be made and the time period over which the prognostication is done, i.e. the prediction horizon) and the Setting (the context in which the model is intended to be used).
Table 2.
PICOTS system
| PICOTS | Explanation |
|---|---|
| Population | Target population in which the prognostic model will be used. |
| Index model | Prognostic model(s) under review. |
| Comparator model | If applicable (depending on the review question), competing prognostic models for the index model. |
| Outcome(s) | Outcome(s) of interest that is (are) predicted by the prognostic model(s). |
| Timing | 1. Moment in time the prognostic model is to be used 2. Time period over which the outcome occurrence is predicted by the prognostic model (prediction horizon). |
| Setting | Intended setting or context of the prognostic model(s) under review. |
PICOTS, Population, Index model, Comparator model, Outcome(s), Timing, Setting.
Case study
In the case study we aimed to present a broad overview of all prognostic models available for patients diagnosed with COVID-19. The review question therefore was “Which models—developed and/or validated—are currently available to predict the prognosis or course of infection in patients with COVID-19, and how valid and useful are these models?” Using the PICOTS format:
-
•
Population: Patients with confirmed or suspected COVID-19
-
•
Index model: All available prognostic models
-
•
Comparator model: Not applicable
-
•
Outcome: All outcomes (e.g. mortality, ICU admission, and progression to severe disease)
-
•
Timing: (1) Moment of prediction is at the moment of COVID-19 diagnosis or shortly thereafter; (2) all prediction horizons
-
•
Setting: Inpatients and outpatients
As the aim of this scoping review was to present an overview of all available models for a specific group of patients (i.e. patients diagnosed with COVID-19), we did not limit to specific index models, outcomes, prediction horizons, and settings.
Step 2: Searching and selection of articles
Searching for prognostic model studies often includes databases such as MEDLINE and Embase. It can be challenging as publications are often not indexed as prognosis study and are not restricted to a unique study design [31,32,40]. For example, researchers may adopt terms like “prognosis,” “prediction,” “predictive,” “risk factors,” “models,” or “algorithms” to describe their objectives, methods, and results. Furthermore, prognostic model studies can be based on data from prospective or retrospective cohort studies, from randomized trials, from routine care data registries, and many other research designs [1,11,13,14,41]. For this reason, it can be difficult to determine from the title and abstract whether a study is about a prognostic model or not. Search strategies for identifying these studies are therefore very broad and usually combine elements of the PICOTS. As a result, the number of papers that need to be screened on title, abstract, and full text may sometimes be relatively high. Search filters to narrow the search have been developed and validated [31,32,40,42]. For example, the Geersing filter combined with the Ingui filter showed a sensitivity of 0.95 in identifying prognosis papers [31].
In specific situations, it may be possible to substantially reduce the search space of a systematic review. For instance, systematic reviews that focus on one specific prediction model (e.g. EuroSCORE [43]) may add the name of the model as requirement in the search query [44]. Alternatively, it is possible to perform a citation search for studies citing the original development paper of the model [45]. As for other types of systematic reviews, snowballing is always an important step to identify all relevant studies [46]. This means that reference lists of related systematic reviews and of included primary studies should be screened to identify studies potentially missed by the search strategy [46]. In general, we advise authors to seek help from an information specialist when developing a search strategy for a review involving prognostic model studies.
After running the search strategy, the identified references must be separated into relevant studies matching the review question versus irrelevant studies. Ideally, each reference is reviewed by two or more reviewers independently, first on title and abstract and later based on full text. Discrepancies should be solved by discussion or by involving a third reviewer. Eligibility criteria for study selection have to be formulated in advance, based on elements of the PICOTS and generic elements such as language, and pilot tested on a part of the identified studies.
Case study
In the case study, the publicly available living evidence collection on COVID-19 was searched up to 1 July 2020, using a semi-automated search string consisting of search terms related to SARS and COVID-19. Details of the search strategy are available on the website of this initiative [47]. As this review was conducted in the beginning of the COVID-19 pandemic, we also searched for preprints published on bioRxiv, medRxiv, and arXiv; abstracts and full texts were screened in duplicate by independent reviewers for eligibility. Discrepancies were resolved through discussion. Studies in which prognostic models were developed and/or validated that were written in English and met the PICOTS, were included. The search identified 37 421 records, of which 444 were screened on full text for eligibility and 107 prognostic model studies were included.
Step 3: Data extraction
After the relevant studies have been identified and selected, the next step is to extract the necessary data from the reports of the included studies. This is ideally done independently by two or more reviewers to avoid mistakes and missing relevant information. Data extraction provides the necessary information for presenting a descriptive table of the included studies and allows qualitative and, if desired and if possible, quantitative (i.e. meta-analysis) summary of the findings of the included studies. The CHARMS checklist has been developed to guide data extraction for reviews of prognostic model studies [23].
Critical information to extract is defined by the PICOTS, i.e. included participants, outcome and predictor definition and measurement, details on timing of the prediction and outcome assessment, and setting. Furthermore, information needs to be collected about source of data, sample size and number of participants with the outcome(s), details on statistical analyses such as handling of missing data and selection of predictors, and predictive performance of the model(s), including discrimination and calibration performance and their corresponding standard errors or confidence intervals (Table 1). Furthermore, if presented in the primary studies, measures related to the clinical utility of a prognostic model, such as results from decision curve analysis and net benefit [48,49], should be extracted and presented in the review.
In many situations, reviewers will face the problem that information they are interested in, is not reported in sufficient detail [50]. It may therefore be necessary to contact the study authors to avoid bias. Alternatively, it is possible to restore the missing information upon data extraction. Methods for this purpose have been described in detail [4,30,34].
Case study
Data were extracted using a standardized form based on the CHARMS checklist. Data were extracted with regards to population (e.g. confirmed or suspected COVID-19), setting (e.g. hospitalized patients or outpatients), predictors included in the models (e.g. patient characteristics, imaging, or blood biomarkers), outcome (e.g. mortality, ICU admission, or progression to severe disease), timing (e.g. in hospital or within 30 days), number of participants and outcomes, analyses (e.g. type of model, handling of missing data) and predictive performance measures.
Step 4: Quality and risk of bias assessment
Risk of bias occurs when the study has shortcomings or flaws in the design or analyses that are likely to result in invalid or distorted results. Study quality and risk of bias assessment is ideally done by two reviewers independently, with discrepancies discussed between the two reviewers and/or solved by a third reviewer. The applicability of a study to the review question needs to be addressed as it is possible that a study does meet the eligibility criteria but does not completely fit the PICOTS of the review. For example, a prediction model might be developed for the prediction of the combined outcome severe anaemia and development of sepsis in children with malaria, while the systematic review is focusing on the prediction of sepsis only.
For studies of prognostic models, the Prediction model Risk Of Bias ASsessment (PROBAST) Tool should be used to assess the risk of bias and the applicability of the included studies (www.probast.org) [24,33]. This quality assessment tool can be used for studies on prognostic (and diagnostic) model development, validation, and updating, as well as for studies that aim to quantify whether particular predictor(s) have added value to an existing prediction model. The studies are assessed for four domains: participants, predictors, outcomes, and analysis. Each domain contains signaling questions, that can be scored with “Yes,” “Probably yes,” “Probably no,” “No,” or “No information.” All signaling questions are formulated so that “Yes” indicates absence of bias. Applicability is judged for the first three domains. Risk of bias and concern for applicability can be graded as “Low,” “High,” or “Unclear.” An adaptation of PROBAST for prediction models developed using artificial intelligence or machine learning (PROBAST-AI) is currently being developed [51]. For prognostic factor studies, the QUIPS tool is available for risk of bias assessment [52]. As this tool is focussing on prognostic factor studies, its use is not recommended for prognostic model studies.
Case study
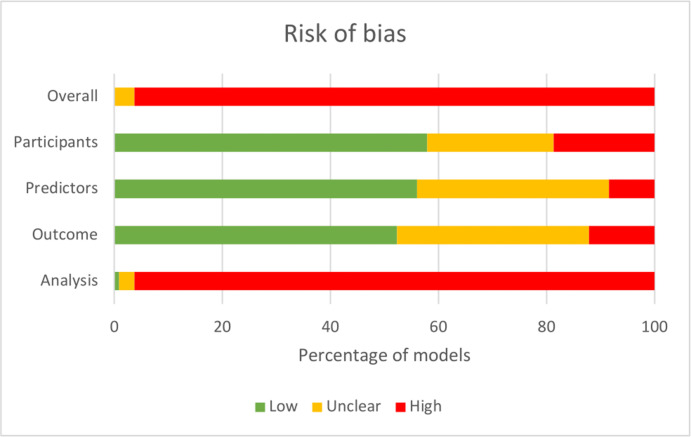
In our case study the PROBAST tool was used for assessing risk of bias of the included prognostic COVID-19 models. Instructions on how to operationalize items were provided to all reviewers. Overall risk of bias was high for most studies (Fig. 2 ). This was mainly driven by a high risk of bias for the analysis domain due to amongst others a low sample size and lack of internal or external validation.
Fig. 2.
Risk of bias as assessed using the Prediction model Risk Of Bias Assessment tool. The figure represents the percentage of studies scoring a low (green), high (red), or unclear (orange) risk of bias for each of the four Prediction model Risk Of Bias ASsessment domains and the overall risk of bias.
Step 5: Analysing data and undertaking quantitative meta-analysis
After identifying all studies that fit the PICOTS of the review and collecting the relevant data from the included studies, authors can consider the feasibility of performing a meta-analysis. Meta-analysis of a prediction model's performance is only advisable if there are more than five external validation studies available for the same index prognostic model [53]. This is similar to a meta-analysis of intervention or diagnostic test accuracy studies, where also multiple studies of the same intervention or index test are required to allow for a meta-analysis. Meta-analysis involves calculating a weighted average of a prediction model's performance, where study weights are (to some extent) defined by the standard error of a study and thus the sample size [30,34].
For prognostic model reviews focusing on identifying all developed prognostic models for a particular target population, condition, or outcome, a meta-analysis is not applicable because, as said, one requires multiple validation studies of the same model. In case meta-analysis is considered not to be of added value or if it is not feasible to conduct a meta-analysis (e.g. due to a too limited number of validation studies of the same prognostic model) results can be summarized in the form of descriptive statistics, tables, and figures.
Returning to the situation with a prognostic model being evaluated on its predictive performance across multiple different studies, these so-called external validation studies will likely differ in many aspects, such as population characteristics, definition and measurement of predictors and outcomes, and applied study designs or data sources. This is called between-study heterogeneity. Because of this between-study heterogeneity, a random effects meta-analysis is often recommended over a fixed effects meta-analysis [30,34]. Meta-analysis of the discrimination performance (e.g. c-statistic or area under the receiver operating characteristic curve) and the calibration (e.g. observed expected [OE] ratio, calibration slope) can be performed if studies are sufficiently similar (as preferably judged by clinical expert) or in case there is heterogeneity but researchers have reasons to conduct a meta-analysis (e.g. studies are heterogeneous but model performance is not). The R packages (R Foundation for Statisitical Computing, Vienna, Austria) such as metamisc [54] and metafor [55] are available for this. Main interest is in the prediction interval surrounding the pooled discrimination and calibration estimate. The prediction interval indicates the likely performance that will be found in a new study. A prediction interval does not only include uncertainty around the pooled estimate, but also between-study heterogeneity [56]. Often this prediction interval is broader than a confidence interval, indicating existing heterogeneity between studies. Sources of this heterogeneity should be further explored using subgroup analyses and meta-regression [30,34].
Case study
In the case study, which aimed to identify all existing developed and validated prognostic models for COVID-19 patients, a meta-analysis was not possible because there was not one model that was validated in multiple studies. A descriptive summary of the identified models could thus only be given, including characteristics on eligibility criteria, predictors included in the models, predicted outcomes, analysis methods, and performance measures.
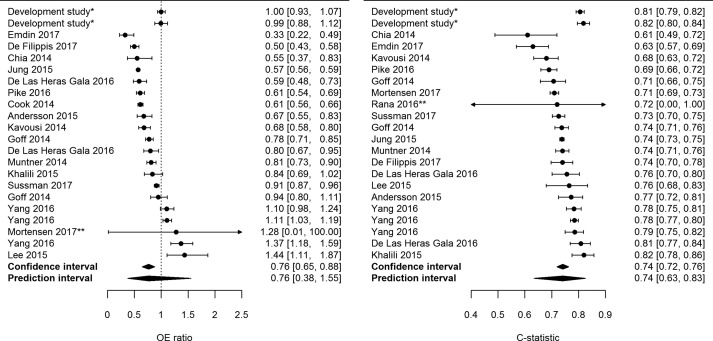
However, for illustration purposes, in another systematic review on the performance of the Pooled Cohort Equations for predicting the future occurrence of cardiovascular disease in the adult general population, meta-analysis of the c-statistic and the OE ratio was performed (Fig. 3 ) [6]. Meta-analysis of the OE ratio included 20 external validations and resulted in a pooled estimate of 0.76, indicating that on average the model overestimates the number of observed outcomes. The prediction interval is broad, ranging from 0.38 to 1.55. This indicates that future studies might also find overestimation of observed outcomes but that it is also possible that there will be studies that find underestimation of observed outcomes. For the c-statistic, also 20 external validations were included, and this resulted in a pooled estimate of 0.74 with a prediction interval ranging from 0.63 to 0.83.
Fig. 3.
Forest plots of the Observed Expected ratio and c-statistic of the Pooled Cohort Equations for predicting 10-year risk of cardiovascular disease in women in the general population. ∗Performance of the model in the development study after internal validation. The first row contains the performance of the White model, the second the African American model (not included in the pooled estimate of performance).
Step 6: presenting summary of findings, interpreting results, and drawing conclusions
The last step of a systematic review is a clear presentation of the findings (e.g. in a summary of findings table), the interpretation of the results, and the authors' conclusions [30]. The following items can give the review author guidance to communicate the results and conclusions of the review effectively, thereby increasing the usability of the reviews' evidence: 1) was all necessary information given on the PICOTS and the performance of the prognostic models; 2) was the summarized performance of the prognostic model(s) sufficient in terms of calibration and discrimination; 3) what was the certainty of the summarized evidence for each of these models with regards to specific populations and specific outcomes. To be able to draw valid conclusions about the certainty of the evidence regarding the generalizability of a prediction model, ideally multiple external validation studies of the same prediction model and of sufficient quality are available for the same population. A method developed to assess the certainty of the overall evidence from systematic reviews is the Grading of Recommendations Assessment, Development and Evaluation (GRADE) approach. For systematic reviews of prognostic models, GRADE is not available yet, but it is currently being developed. Until GRADE for prognostic models becomes available, it is advised to adapt the GRADE guidance for overall prognosis studies and for prognostic factor studies [[57], [58], [59]] (by changing measures for association into performance measures of models and changing the exploratory and confirmatory phases of a prognostic factor into development and validation of a model).
Case study
The GRADE approach was not used in the case study. Results of the systematic review were therefore discussed in the light of the continuously evolving COVID-19 pandemic. For example, the authors concluded that most prediction models are poorly reported and at high risk of bias. Furthermore, they identified one promising prognostic model, for which further external validation by independent researchers is advised.
Concluding remarks
Systematic reviews of prognostic models are an important tool to decide on further validation or evaluation and, if applicable, implementation of the most relevant or accurate models. Notably in the past decade much guidance for conducting systematic review and meta-analysis of prognostic model studies has been developed by investigators that are also associated to the Cochrane Prognosis Methods Group [25]. To make such reviews possible and to draw valuable conclusions, first and foremost complete and transparent reporting of the primary prognostic model studies is essential. Therefore, the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) statement has been published [36,37]. Adhering to the TRIPOD statement is required for informative reviews and should be promoted. An update of the TRIPOD statement for prediction models developed using artificial intelligence (TRIPOD-AI) is currently under development [51,60], as well as TRIPOD-SRMA for the reporting of systematic reviews and meta-analysis of prediction model studies. For now, we advise to follow the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement [35] and add relevant items from the TRIPOD statement.
Transparency declaration
JAAD, KGMM, MvS, and LH have nothing to disclose. No external funding was received for any part of this manuscript.
Author contributions
All authors conceptualized the manuscript. JAAD wrote the first draft of the manuscript that was subsequently revised by KGMM, MvS, and LH.
Editor: M. Leeflang
References
- 1.Moons K.G., Royston P., Vergouwe Y., Grobbee D.E., Altman D.G. Prognosis and prognostic research: what, why, and how? BMJ. 2009;338:b375. doi: 10.1136/bmj.b375. [DOI] [PubMed] [Google Scholar]
- 2.Moons K.G., Kengne A.P., Woodward M., Royston P., Vergouwe Y., Altman D.G., et al. Risk prediction models: I. Development, internal validation, and assessing the incremental value of a new (bio)marker. Heart. 2012;98:683–690. doi: 10.1136/heartjnl-2011-301246. [DOI] [PubMed] [Google Scholar]
- 3.Steyerberg E. Springer Science & Business Media; Berlin, Germany: 2008. Clinical prediction models: a practical approach to development, validation, and updating. [Google Scholar]
- 4.Riley R.D., van der Windt D., Croft P., Moons K.G. Oxford University Press; Oxford, UK: 2019. Prognosis research in healthcare: concepts, methods, and impact. [Google Scholar]
- 5.Moons K.G., Hooft L., Williams K., Hayden J.A., Damen J.A., Riley R.D. Implementing systematic reviews of prognosis studies in Cochrane. Cochrane Database Syst Rev. 2018;10:Ed000129. doi: 10.1002/14651858.ED000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damen J.A., Pajouheshnia R., Heus P., Moons K.G.M., Reitsma J.B., Scholten R., et al. Performance of the Framingham risk models and pooled cohort equations for predicting 10-year risk of cardiovascular disease: a systematic review and meta-analysis. BMC Med. 2019;17:109. doi: 10.1186/s12916-019-1340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riley R.D., Ridley G., Williams K., Altman D.G., Hayden J., de Vet H.C. Prognosis research: toward evidence-based results and a Cochrane methods group. J Clin Epidemiol. 2007;60:863. doi: 10.1016/j.jclinepi.2007.02.004. –5; author reply 5–6. [DOI] [PubMed] [Google Scholar]
- 8.Hemingway H., Croft P., Perel P., Hayden J.A., Abrams K., Timmis A., et al. Prognosis research strategy (PROGRESS) 1: a framework for researching clinical outcomes. BMJ. 2013;346:e5595. doi: 10.1136/bmj.e5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riley R.D., Hayden J.A., Steyerberg E.W., Moons K.G., Abrams K., Kyzas P.A., et al. Prognosis Research Strategy (PROGRESS) 2: prognostic factor research. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steyerberg E.W., Moons K.G., van der Windt D.A., Hayden J.A., Perel P., Schroter S., et al. Prognosis Research Strategy (PROGRESS) 3: prognostic model research. PLoS Med. 2013;10 doi: 10.1371/journal.pmed.1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moons K.G., Kengne A.P., Grobbee D.E., Royston P., Vergouwe Y., Altman D.G., et al. Risk prediction models: II. External validation, model updating, and impact assessment. Heart. 2012;98:691–698. doi: 10.1136/heartjnl-2011-301247. [DOI] [PubMed] [Google Scholar]
- 12.Moons K.G., Altman D.G., Vergouwe Y., Royston P. Prognosis and prognostic research: application and impact of prognostic models in clinical practice. BMJ. 2009;338:b606. doi: 10.1136/bmj.b606. [DOI] [PubMed] [Google Scholar]
- 13.Royston P., Moons K.G., Altman D.G., Vergouwe Y. Prognosis and prognostic research: developing a prognostic model. BMJ. 2009;338:b604. doi: 10.1136/bmj.b604. [DOI] [PubMed] [Google Scholar]
- 14.Altman D.G., Vergouwe Y., Royston P., Moons K.G. Prognosis and prognostic research: validating a prognostic model. BMJ. 2009;338:b605. doi: 10.1136/bmj.b605. [DOI] [PubMed] [Google Scholar]
- 15.Riley R.D., Moons K.G.M., Snell K.I.E., Ensor J., Hooft L., Altman D.G., et al. A guide to systematic review and meta-analysis of prognostic factor studies. BMJ. 2019;364:k4597. doi: 10.1136/bmj.k4597. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Felix B.M., Barca L.V., Garcia-Esquinas E., Correa-Pérez A., Fernández-Hidalgo N., Muriel A., et al. Prognostic models for mortality after cardiac surgery in patients with infective endocarditis: a systematic review and aggregation of prediction models. Clin Microbiol Infect. 2021;27:1422–1430. doi: 10.1016/j.cmi.2021.05.051. [DOI] [PubMed] [Google Scholar]
- 17.van Smeden M., Reitsma J.B., Riley R.D., Collins G.S., Moons K.G. Clinical prediction models: diagnosis versus prognosis. J Clin Epidemiol. 2021;132:142–145. doi: 10.1016/j.jclinepi.2021.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Bellou V., Belbasis L., Konstantinidis A.K., Tzoulaki I., Evangelou E. Prognostic models for outcome prediction in patients with chronic obstructive pulmonary disease: systematic review and critical appraisal. BMJ. 2019;367:l5358. doi: 10.1136/bmj.l5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damen J.A., Hooft L., Schuit E., Debray T.P., Collins G.S., Tzoulaki I., et al. Prediction models for cardiovascular disease risk in the general population: systematic review. BMJ. 2016;353:i2416. doi: 10.1136/bmj.i2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wynants L., Van Calster B., Collins G.S., Riley R.D., Heinze G., Schuit E., et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peetluk L.S., Ridolfi F.M., Rebeiro P.F., Liu D., Rolla V.C., Sterling T.R. Systematic review of prediction models for pulmonary tuberculosis treatment outcomes in adults. BMJ Open. 2021;11 doi: 10.1136/bmjopen-2020-044687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Njim T., Tanyitiku B.S. Prognostic models for the clinical management of malaria and its complications: a systematic review. BMJ Open. 2019;9 doi: 10.1136/bmjopen-2019-030793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moons K.G., de Groot J.A., Bouwmeester W., Vergouwe Y., Mallett S., Altman D.G., et al. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med. 2014;11 doi: 10.1371/journal.pmed.1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moons K.G.M., Wolff R.F., Riley R.D., Whiting P.F., Westwood M., Collins G.S., et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. 2019;170 doi: 10.7326/M18-1377. W1–33. [DOI] [PubMed] [Google Scholar]
- 25.Cochrane Methods Prognosis. Prognosis; [cited] https://methods.cochrane.org/prognosis/
- 26.Van Calster B., McLernon D.J., van Smeden M., Wynants L., Steyerberg E.W. Calibration: the Achilles heel of predictive analytics. BMC Med. 2019;17:230. doi: 10.1186/s12916-019-1466-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrell F. Springer; Switzerland: 2015. Regression modeling strategies: with Applications to linear models, Logistic and ordinal regression, and survival analysis. City, country/state. [Google Scholar]
- 28.Vergouwe Y., Moons K.G., Steyerberg E.W. External validity of risk models: use of benchmark values to disentangle a case-mix effect from incorrect coefficients. Am J Epidemiol. 2010;172:971–980. doi: 10.1093/aje/kwq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steyerberg E.W., Vickers A.J., Cook N.R., Gerds T., Gonen M., Obuchowski N., et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Debray T.P., Damen J.A., Snell K.I., Ensor J., Hooft L., Reitsma J.B., et al. A guide to systematic review and meta-analysis of prediction model performance. BMJ. 2017;356:i6460. doi: 10.1136/bmj.i6460. [DOI] [PubMed] [Google Scholar]
- 31.Geersing G.J., Bouwmeester W., Zuithoff P., Spijker R., Leeflang M., Moons K.G. Search filters for finding prognostic and diagnostic prediction studies in Medline to enhance systematic reviews. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0032844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingui B.J., Rogers M.A. Searching for clinical prediction rules in MEDLINE. J Am Med Inform Assoc. 2001;8:391–397. doi: 10.1136/jamia.2001.0080391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolff R.F., Moons K.G.M., Riley R.D., Whiting P.F., Westwood M., Collins G.S., et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med. 2019;170:51–58. doi: 10.7326/M18-1376. [DOI] [PubMed] [Google Scholar]
- 34.Debray T.P., Damen J.A., Riley R.D., Snell K., Reitsma J.B., Hooft L., et al. A framework for meta-analysis of prediction model studies with binary and time-to-event outcomes. Stat Methods Med Res. 2018;28:2768–2786. doi: 10.1177/0962280218785504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins G.S., Reitsma J.B., Altman D.G., Moons K.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med. 2015;162:55–63. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 37.Moons K.G., Altman D.G., Reitsma J.B., Ioannidis J.P., Macaskill P., Steyerberg E.W., et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162 doi: 10.7326/M14-0698. W1–73. [DOI] [PubMed] [Google Scholar]
- 38.Thomas J., Kneale D., McKenzie J.E., Brennan S.E., Bhaumik S. In: Cochrane handbook for systematic reviews of interventions. 2nd ed. Higgins J.P.T., Thomas J., Chandler J., Cumpston M., Li T., Page M.J., et al., editors. John Wiley & Sons; Chichester (UK): 2019. Determining the scope of the review and the questions it will address. [Google Scholar]
- 39.Leeflang M.M., Davenport C., Bossuyt P.M. In: Cochrane handbook for systematic reviews of diagnostic test accuracy. 2nd ed. Deeks J.J., Bossuyt P.M., Leeflang M.M., Takwoingi Y., editors. John Wiley & Sons; Chichester (UK): 2019. Defining the review question. [Google Scholar]
- 40.Kavanagh P.L., Frater F., Navarro T., LaVita P., Parrish R., Iorio A. Optimizing a literature surveillance strategy to retrieve sound overall prognosis and risk assessment model papers. J Am Med Inform Assoc. 2021;28:766–771. doi: 10.1093/jamia/ocaa232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pajouheshnia R., Peelen L.M., Moons K.G.M., Reitsma J.B., Groenwold R.H.H. Accounting for treatment use when validating a prognostic model: a simulation study. BMC Med Res Methodol. 2017;17:103. doi: 10.1186/s12874-017-0375-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilczynski N.L., Haynes R.B. Developing optimal search strategies for detecting clinically sound prognostic studies in MEDLINE: an analytic survey. BMC Med. 2004;2:23. doi: 10.1186/1741-7015-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nashef S.A., Roques F., Michel P., Gauducheau E., Lemeshow S., Salamon R. European system for cardiac operative risk evaluation (EuroSCORE) Eur J Cardiothorac Surg. 1999;16:9–13. doi: 10.1016/s1010-7940(99)00134-7. [DOI] [PubMed] [Google Scholar]
- 44.Siregar S., Groenwold R.H., de Heer F., Bots M.L., van der Graaf Y., van Herwerden L.A. Performance of the original EuroSCORE. Eur J Cardiothorac Surg. 2012;41:746–754. doi: 10.1093/ejcts/ezr285. [DOI] [PubMed] [Google Scholar]
- 45.Tzoulaki I., Liberopoulos G., Ioannidis J.P. Assessment of claims of improved prediction beyond the Framingham risk score. JAMA. 2009;302:2345–2352. doi: 10.1001/jama.2009.1757. [DOI] [PubMed] [Google Scholar]
- 46.Lefebvre C., Glanville J., Briscoe S., Littlewood A., Marshall C., Metzendorf M.I., et al. In: Cochrane handbook for systematic reviews of interventions. 2nd ed. Higgins J.P.T., Thomas J., Chandler J., Cumpston M., Li T., Page M.J., et al., editors. John Wiley & Sons; Chichester (UK): 2019. Searching for and selecting studies. [Google Scholar]
- 47.Institute of Social and Preventive Medicine . 2020. Living evidence on COVID-19.https://ispmbern.github.io/covid-19/living-review/index.html [cited ]. Available from: [Google Scholar]
- 48.Vickers A.J., Van Calster B., Steyerberg E.W. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ. 2016;352:i6. doi: 10.1136/bmj.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Calster B., Wynants L., Verbeek J.F.M., Verbakel J.Y., Christodoulou E., Vickers A.J., et al. Reporting and interpreting decision curve analysis: a guide for investigators. Eur Urol. 2018;74:796–804. doi: 10.1016/j.eururo.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heus P., Damen J.A.A.G., Pajouheshnia R., Scholten R.J.P.M., Reitsma J.B., Collins G.S., et al. Poor reporting of multivariable prediction model studies: towards a targeted implementation strategy of the TRIPOD statement. BMC Med. 2018;16:120. doi: 10.1186/s12916-018-1099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins G.S., Dhiman P., Andaur Navarro C.L., Ma J., Hooft L., Reitsma J.B., et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open. 2021;11 doi: 10.1136/bmjopen-2020-048008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayden J.A., van der Windt D.A., Cartwright J.L., Cote P., Bombardier C. Assessing bias in studies of prognostic factors. Ann Intern Med. 2013;158:280–286. doi: 10.7326/0003-4819-158-4-201302190-00009. [DOI] [PubMed] [Google Scholar]
- 53.IntHout J., Ioannidis J.P., Borm G.F. The Hartung-Knapp-Sidik-Jonkman method for random effects meta-analysis is straightforward and considerably outperforms the standard DerSimonian-Laird method. BMC Med Res Methodol. 2014;14:25. doi: 10.1186/1471-2288-14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Debray T.P. 2017. Metamisc: diagnostic and prognostic meta-analysis.https://cran.r-project.org/package=metamisc [Google Scholar]
- 55.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1–48. [Google Scholar]
- 56.Riley R.D., Higgins J.P., Deeks J.J. Interpretation of random effects meta-analyses. BMJ. 2011;342:d549. doi: 10.1136/bmj.d549. [DOI] [PubMed] [Google Scholar]
- 57.Huguet A., Hayden J.A., Stinson J., McGrath P.J., Chambers C.T., Tougas M.E., et al. Judging the quality of evidence in reviews of prognostic factor research: adapting the GRADE framework. Syst Rev. 2013;2:71. doi: 10.1186/2046-4053-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foroutan F., Guyatt G., Zuk V., Vandvik P.O., Alba A.C., Mustafa R., et al. Use of GRADE for assessment of evidence about prognostic factors: rating certainty in identification of groups of patients with different absolute risks. J Clin Epidemiol. 2020;121:62–70. doi: 10.1016/j.jclinepi.2019.12.023. [DOI] [PubMed] [Google Scholar]
- 59.Iorio A., Spencer F.A., Falavigna M., Alba C., Lang E., Burnand B., et al. Use of GRADE for assessment of evidence about prognosis: rating confidence in estimates of event rates in broad categories of patients. BMJ. 2015;350:h870. doi: 10.1136/bmj.h870. [DOI] [PubMed] [Google Scholar]
- 60.Collins G.S., Moons K.G.M. Reporting of artificial intelligence prediction models. Lancet. 2019;393:1577–1579. doi: 10.1016/S0140-6736(19)30037-6. [DOI] [PubMed] [Google Scholar]