Significance
In type 1 diabetes (T1D), immune cells destroy pancreatic β-cells, and the trigger of this response is unknown. Some amino acid sequences (epitopes) in the insulin protein are a major target for this autoimmune response. We identified a sequence in the human gut bacterium Parabacteroides distasonis that mimics an important insulin epitope (insB:9-23). Human and mouse immune cells specific to insB:9-23 cross-react with this bacterial mimic. Further, P. distasonis can accelerate diabetes onset in a mouse model of T1D, inducing destructive immune cells and decreasing protective immune cells. We found a significant association between the presence of this bacterial mimic and development of T1D in children. Taken together, our results suggest this mimic has the potential to trigger/modify T1D onset.
Keywords: type 1 diabetes, Parabacteroides distasonis, insB:9-23 epitope, molecular mimicry, gut microbiome
Abstract
Type 1 diabetes (T1D) is an autoimmune disease characterized by the destruction of pancreatic β-cells. One of the earliest aspects of this process is the development of autoantibodies and T cells directed at an epitope in the B-chain of insulin (insB:9–23). Analysis of microbial protein sequences with homology to the insB:9–23 sequence revealed 17 peptides showing >50% identity to insB:9–23. Of these 17 peptides, the hprt4–18 peptide, found in the normal human gut commensal Parabacteroides distasonis, activated both human T cell clones from T1D patients and T cell hybridomas from nonobese diabetic (NOD) mice specific to insB:9–23. Immunization of NOD mice with P. distasonis insB:9–23 peptide mimic or insB:9–23 peptide verified immune cross-reactivity. Colonization of female NOD mice with P. distasonis accelerated the development of T1D, increasing macrophages, dendritic cells, and destructive CD8+ T cells, while decreasing FoxP3+ regulatory T cells. Western blot analysis identified P. distasonis–reacting antibodies in sera of NOD mice colonized with P. distasonis and human T1D patients. Furthermore, adoptive transfer of splenocytes from P. distasonis–treated mice to NOD/SCID mice enhanced disease phenotype in the recipients. Finally, analysis of human children gut microbiome data from a longitudinal DIABIMMUNE study revealed that seroconversion rates (i.e., the proportion of individuals developing two or more autoantibodies) were consistently higher in children whose microbiome harbored sequences capable of producing the hprt4–18 peptide compared to individuals who did not harbor it. Taken together, these data demonstrate the potential role of a gut microbiota-derived insB:9–23-mimic peptide as a molecular trigger of T1D pathogenesis.
1. Introduction
Type 1 diabetes (T1D) is an autoimmune disease characterized by selective destruction of pancreatic β-cells by autoreactive T cells (1). Genome-wide association studies have identified 152 genetic regions that influence the risk of developing T1D (2); however, multiple studies have shown that the incidence rate of T1D in children is rising at rates exceeding what can be explained on a genetic basis alone (3). Indeed, even among identical twins, the concordance of T1D is only 65% (4). Likewise, there is a sixfold difference in incidence of T1D in neighboring regions of Karelia in Russia and Finland, despite the very similar genetic background of the inhabitants (5).
Various environmental factors have been studied as potential modifiers or triggers of the autoimmune response in T1D, including diet, birth mode, infection, and antibiotics. Viral infections have been suggested to play a role in T1D pathogenesis, but most of these viruses have been proposed to act by direct infection of the β-cell (6). Recently, attention has been focused on the gut microbiome as a potential disease modifier through its effects on metabolite composition, intestinal permeability, and regulation of the immune response in subjects with T1D (7, 8). However, the exact environmental modifiers and how they might affect T1D pathogenesis remain largely unknown (9).
One of the earliest markers of T1D is the development of islet autoantibodies (AABs) (10). These AABs target several autoantigens including insulin, glutamic acid decarboxylase (GAD), insulinoma-associated autoantigen-2 (IA-2), zinc transporter-8 (ZnT8), and an islet-specific glucose‐6‐phosphatase catalytic subunit related protein (IGRP or G6PC2) (11). Among these, insulin autoantibodies (IAAs) are usually the first to be detected, and insulin is the only autoantigen exclusively expressed by β-cells (12). In humans, IAAs may develop years before the onset of overt diabetes (13) and are especially prominent in early-onset T1D (14). They also show a significant correlation with the rate of progression from prediabetes to overt disease (15). More importantly, insulin or insulin-derived peptides are a target of disease pathogenetic T cells in both humans with T1D and in the most established murine model, the nonobese diabetic (NOD) mouse. In NOD mice, over 90% of anti-insulin T cell clones target a single 15–amino acid peptide corresponding to the insulin B-chain 9–23 sequence (insB:9–23) (15). InsB:9–23–specific T cells have also been identified in islets (16) and peripheral blood lymphocytes of T1D patients (17, 18).
In the present study, we hypothesized that exposure to a microbial peptide that resembles the insulin epitope, insB:9–23, could stimulate or modify the autoimmune response initiating T1D (19). To address this hypothesis, we analyzed bacterial, viral, and fungal genome databases to identify microbial proteins that have >50% sequence homology to human insB:9–23. Of these, 17 were synthesized and tested for the ability to activate insB:9–23–specific T cells. Herein, we demonstrate that one of these peptides, a peptide from the gut commensal organism Parabacteroides distasonis can activate both human and NOD mouse insB:9–23–specific T cells ex vivo to the same extent as the human insulin B:9–23 peptide. This bacterial peptide also cross-reacts with immune cells obtained from mice immunized with the human insB:9–23 peptide. Furthermore, administration of P. distasonis bacteria by oral gavage accelerated T1D progression in NOD mice in vivo by stimulating innate immune cells and CD8+ T cells and decreasing regulatory T cells. Analysis of human gut microbiota datasets from the longitudinal DIABIMMUNE study revealed that being exposed to this bacterial peptide is a predictor of the development of seroconversion. Seroconversion rates were significantly higher in subjects who harbored the microbiota that can encode this bacterial peptide in their gut. Furthermore, this bacterial peptide was enriched in children who later developed AABs and were diagnosed with T1D in all three countries analyzed. Taken together, we demonstrate that an insB:9–23–like microbial peptide in normal gut microbiota may mimic the native insulin peptide and has the potential to play a role in the onset or progression of T1D.
2. Results
2.1 A. P. distasonis insB:9–23–like Peptide Stimulates NOD Mouse Hybridomas and Human T Cell Clones Specific to insB:9–23.
To determine if any microbe might have DNA-encoding proteins with sequences resembling the dominant T cell epitope involved in the autoimmune response to the insulin B:9–23 peptide linked to T1D (SHLVEALYLVCGERG) (15–18), we used BlastP to search National Center for Biotechnology Information (NCBI) databases for predicted proteomes of all sequenced viruses (taxid:10239), bacteria (taxid:2), and fungi (taxid:4751). We identified 47 microbial peptides with over eight residues identical to the insulin peptide (supplementary information (SI) Appendix, Table S1). Among these 47 microbial peptides, we selected 17 bacterial and viral peptides that contained the largest number of previously identified residues in the insB:9–23 peptide critical for the synthesis of this interaction (18) (Fig. 1A).
Fig. 1.
P. distasonis hprt4-18 peptide mimic can stimulate both human and NOD mice T cells specific to insB:9–23. (A) Sequence alignment of 17 microbial peptides tested in this study. The amino acids highlighted in red are identical to insB:9–23. (B) Treatment of insB:11–23–specific human T cell clones with each of selected 17 microbial peptides, where, negative control and irrelevant peptides were used as the negative controls and insB:9-23 and insB:9–23 R22E were used as positive controls. ELISpot was used to determine IFN-γ secretion (P < 0.05) (n = 3). (C) Dose–response of IIT-3 hybridomas to insB:9–23 (black) or to hprt4-18 peptides (red), where peptides were presented to hybridomas as covalently linked to I-Ag7 expressed on macrophages (C3g7 cells). Data are presented as IL-2–induced proliferation in CTLL-2 cells. (D and E) Mice (n = 3/group) were immunized with either hprt4-18 peptide (D) or insB:9–23 peptide (E). After 7 d of immunization, popliteal lymph nodes were stimulated either by hprt4-18 peptide, insB:9–23, ConA (positive control), or scrambled peptide (negative control). Secretion of IL-2 (D and E) was determined by ELISpot. All samples in each panel are biologically independent. Data are expressed as means ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001. Statistical analysis was performed by one-way ANOVA using Dunnett’s multiple comparisons test.
These 17 microbial insB:9–23–like peptides, as well as a negative control (irrelevant/scramble) and positive control (insB:9–23), were then tested for their ability to stimulate an insB:11–23–specific human T cell clone isolated from a T1D patient (18) (Fig. 1B). In addition, we included a second positive control, a variant of the insB:9–23 peptide in which R at position 22 is substituted by E (insB:9–23R22E) and which has been shown to be even more potent than native insB:9–23 in stimulating insB:9–23 hybridomas from NOD mice (20) and human T cell clones (18). This insB:9–23–specific human T cell clone (T1D#3 C8) was previously generated from the peripheral blood samples of a T1D patient with HLA-DQB1*03:02 (DQ8) haplotypes (18). We stimulated this T cell clone with either insB:9–23R22E or insB:9–23 or each of 17 microbial peptides in the presence of HLA-DQ8-cis– or DQ8-trans–expressing HEK293 antigen-presenting cells (APCs). As expected, in the human T cell assay, both positive controls stimulated interferon-gamma (IFN-γ) secretion, with insB:9–23R22E being more active than the wild-type sequence. Among the 17 microbial peptides tested, only one could stimulate human T cells to produce IFN-γ (Fig. 1B). This peptide (hprt4-18) represents amino acids 4 to 18 in the N terminus of the hypoxanthine phosphoribosyltransferase (hprt) protein of P. distasonis 33B and D13 strains (formerly known as Bacteroides sp. 2_1_33B/Parabacteroides sp. D13) (21). Interestingly, P. distasonis 33B and D13 are the only organisms in the NCBI dataset (as of October 2021) that possess this insB:9–23 mimic sequence in their genomes.
Next, we tested the same set of microbial insB:9–23–like peptides and controls on NOD IIT-3 T cell hybridomas, previously shown to recognize the insB:9–23 epitope (22). In this assay, C3g7 cells that have high expression of MHC-II I-Ag7 were used as APCs and treated with each peptide at concentrations from 0.01 to 10 μM. Coculturing of these cells with IIT-3 T cell hybridomas stimulated interleukin (IL)-2 secretion only in the presence of insB:9–23 and the hprt4-18 peptide and produced similar dose–response curves, with the bacterial peptide being only slightly less potent (Fig. 1C). Thus, both human T cell clones and NOD mouse T cell hybridomas specific to insB:9–23 were reactive to hprt4-18, raising the possibility that hprt4-18 may have the potential to modulate the development of T1D.
2.2. P. distasonis hprt4-18 Peptide Stimulates a T Cell Response to insB:9–23 In Vivo.
To further explore the cross-reactivity of the microbial and insulin-derived peptide sequences, NOD mice were immunized with either insB:9–23 peptide or the hprt4-18 peptide in Complete Freund’s Adjuvant. After 7 d, lymphocytes were isolated from the popliteal lymph nodes and stimulated with either insB:9–23, hprt4-18, or control peptides. T cell activation was assessed using an enzyme-linked immunospot (ELISpot) assay. Consistent with the in vitro results above, lymphocytes from mice immunized with the hprt4-18 peptide exhibited a strong immune response to both hprt4-18 peptide and insB:9–23 as measured by IL-2 and IFN-γ secretion (Fig. 1D and SI Appendix, Fig. S1A). Conversely, lymphocytes from mice immunized with insB:9–23 peptide showed a strong response to both insB:9–23 peptide and the microbial hprt4-18 peptide (Fig. 1E and SI Appendix, Fig. S1B). Taken together with the assays using the hybridoma T cell lines, these data indicate that the microbial peptide hprt4-18 strongly cross-reacts with the recognition and signaling machinery for human insB:9–23 in stimulating an immune response.
2.3. P. distasonis Colonization Accelerates Diabetes Onset in NOD Mice.
Based on this cross-reactivity, we hypothesized that P. distasonis colonization and potential exposure to the microbial hprt4-18 peptide could trigger and/or modify the immune response and stimulate autoimmunity in NOD mice. To test this hypothesis, both male and female NOD mice were orally gavaged either with a saline suspension of P. distasonis (108 colony-forming units [CFUs]/mouse/day) or saline daily for 4 wk, starting at 3 wk of age (i.e., after weaning) and followed for 30 wk (Fig. 2A; n = 40/group). Two weeks after the last oral gavage, qPCR using fecal DNA samples revealed significant levels of P. distasonis in the feces of the treated mice, whereas neither male nor female control NOD mice housed under specific pathogen-free conditions had P. distasonis in their gut microbiome (Fig. 2B). At 12 wk of age, when mice are in the prediabetic stage, three to five mice from each group were randomly selected and examined histologically for insulitis. In the P. distasonis–colonized female NOD mice, there was more than a twofold increase in severe insulitis scores compared to controls (representative images in Fig. 2 C and D). Next, to determine whether this effect was specific to P. distasonis and not a nonspecific effect stimulated by a new prokaryotic product, we colonized NOD mice with another common gram-negative human gut commensal, Bacteroides fragilis (23). To this end, we orally gavaged a new cohort of female NOD mice either with B. fragilis (108 CFU/mouse/daily) or saline for 4 wk (n = 3 or 4). Two weeks after the final oral gavage, colonization was confirmed using qPCR. Since NOD mice in our facility do not have B. fragilis in their normal microbiome and since B. fragilis colonizes the mice well, it is an ideal control (SI Appendix, Fig. S1C). At 12 wk of age (before diabetes onset), we killed the mice and analyzed the islets for insulitis (Fig. 2D). Notably, we did not find any significant differences in the development of insulitis in mice colonized with B. fragilis compared to the control group, indicating that the increased infiltration of the immune cells into the islets of the NOD mice was specifically stimulated by P. distasonis.
Fig. 2.
P. distasonis colonization enhances disease onset in female NOD mice, and T1D patients have a stronger immune response to P. distasonis than healthy individuals. (A) Schematic overview of the P. distasonis (P.dis) oral gavage experiments (n = 40/group/sex). Blue arrows show the time points (week 10) of fecal sample collection for qPCR experiments for P. distasonis or B. fragilis colonization, and the red arrow shows the time point (week 12) for pancreata collection for insulitis analysis (n = 5 mice/group/sex). (B) Relative abundance of P. distasonis in fecal samples determined by qPCR (week 12, n = 10 to 13/male, n = 12 to 17/female). (C) Representative images of islets insulitis score data where islets were scored as no insulitis, peri-insulitis, moderate insulitis, or having severe infiltration as shown in images. (D) Quantification of insulitis scores obtained from B. fragilis colonized, P. distasonis colonized, or saline-gavaged female NOD mice at week 12 (n = 3 to 5/group, n = 100 to 150 islets/group). (E) Diabetes incidence in female NOD mice (n = 35/group/sex) after daily oral gavage with either saline or P. distasonis for 4 wk after weaning (P < 0.05). (F) Diabetes incidence in the recipient NOD/SCID mice after adoptive transfer of 5 × 107 splenocytes/mouse from individual female NOD mice to female NOD/SCID mice at 6 wk of age (1:1 ratio, same sex, n = 15). All samples in each panel are biologically independent. Log-rank (Mantel-Cox) test was used for survival curves and adoptive transfer experiment. Data are expressed as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.01. Statistical analysis was performed by two-way ANOVA using Sídak multiple comparisons test or two-tailed unpaired Student t test. ns, not significant.
Consistent with the increase in insulitis, female NOD mice subjected to P. distasonis colonization showed significantly accelerated onset of T1D, with only 19.5% disease-free in the P. distasonis–colonized group versus 42% disease-free in the control group (Fig. 2E). This effect was specific to the female mice, with no differences in insulitis scores or T1D incidence in male NOD mice with P. distasonis colonization (SI Appendix, Fig. S1 D and E).
To determine whether P. distasonis colonization stimulates an immune response against the bacterium, we performed Western blot on P. distasonis and B. fragilis cellular protein lysates probed with serum samples from either 12-wk-old P. distasonis, B. fragilis–colonized or control NOD mice. P. distasonis and B. fragilis colonization of the NOD mice gut stimulated an antibody response to multiple bacterial proteins in both female and male NOD mice (SI Appendix, Fig. S1 F–H). This response was specific; Western blots using protein lysates from B. fragilis produced only a weak humoral immune response in both the saline- and P. distasonis–treated groups (SI Appendix, Fig. S1 I and J). Importantly, serum lipopolysaccharide (LPS) levels were not significantly different between the groups, indicating that the P. distasonis humoral response was not the result of general gut barrier dysfunction stimulated by bacterial colonization (SI Appendix, Fig. S1K).
To determine whether a similar enrichment in the humoral immune responses against P. distasonis occurs in humans with T1D, Western blot analysis was performed using serum samples from 12 T1D patients (median age, 17.5 y; median disease duration, 8 y; males) with age-, sex-, and ethnicity-matched controls. As shown in SI Appendix, Fig. S2 A and C, there was a very weak humoral response against P. distasonis in healthy subjects, and strong reactivity was observed in 7 out of 12 T1D patients. Thus, consistent with our findings in NOD mice, when assessed in a random subset of T1D patients there appears to be a more general humoral immune response to P. distasonis proteins. Because systemic antibody responses to intestinal commensal bacteria in T1D patients might be related to some level of gut dysbiosis (24), we tested whether this enriched humoral immune response was specific to P. distasonis or was a result of a more generalized leaky gut. To this end, we performed an additional Western blot analysis against B. fragilis lysate using the same patients’ sera. Unlike with P. distasonis, we did not observe an enrichment in reactivity against B. fragilis in T1D patients compared to serum from the healthy controls (SI Appendix, Fig. S2 B and D). These results suggest that the humoral response against P. distasonis is not part of a nonspecific response related to gut dysbiosis.
2.4. Adoptive Transfer of Splenocytes Enhances T1D Onset in NOD/SCID Mice.
To determine if T1D accerlation in the P. distasonis–colonized NOD mice was T cell mediated, we performed an adoptive transfer experiment using immunodeficient NOD/SCID mice, which lack functional B and T cells, as recipients (25). To this end, a new cohort of NOD mice was orally gavaged with P. distasonis or saline (n = 15 to 20/group) as described above. At week 15, these mice were killed, and 5 × 106 splenocytes were transferred from individual NOD diabetes-free donors to 9-wk-old NOD/SCID recipients (1:1 ratio, sex matched), after which the recipients were followed for 10 wk (SI Appendix, Fig. S2 E and F). Consistent with the effect of P. distasonis colonization on spontaneous T1D in NOD mice (Fig. 2), female NOD/SCID mice that received splenocytes from the P. distasonis–treated group developed T1D at a higher rate than those receiving splenocytes from control NOD mice. Thus, at 10 wk following transfer, 62.5% of recipients recieving control splenocytes remained disease-free, and this was decreased to 31% in mice receiving splenocytes from P. distasonis colonized NOD donors (Fig. 2). Hence, the immune response stimulated by P. distasonis in female NOD mice was sufficient to accelerate T1D in NOD/SCID mice.
2.5. P. distasonis Colonization Increases CD8+ T Cells and Decreases Foxp3+ Regulatory T Cells in the Splenocytes of Female NOD Mice.
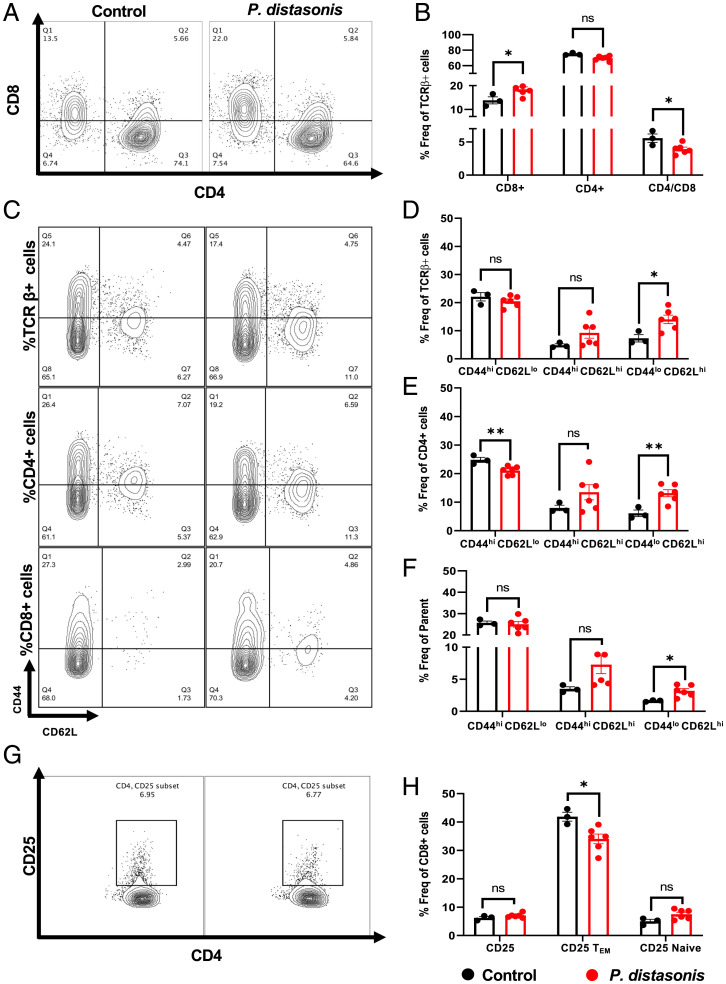
To determine the effects of P. distasonis treatment on NOD mice immune cell composition, a new cohort of 12-wk-old NOD mice were orally gavaged with either P. distasonis or saline, and the T cell populations in splenocytes and pancreatic lymph nodes (PLNs) were assessed by flow cytometry. The gating strategy for different T cell subsets and innate immune cells is described in SI Appendix, Fig. S3 A and B. We found that in the spleens of P. distasonis–colonized NOD mice, there was a significant 31% increase in CD8+ T cells, leading to a 30% decrease in the CD4/CD8 ratio (Fig. 3 A and B). PLN cells isolated from the P. distasonis–colonized mice showed similar trends in CD4+ T cells, CD8+ T cells, and CD4/CD8 ratio, but these did not quite reach statistical significance (SI Appendix, Fig. S4 A and B). Using fluorescence-activated cell sorter (FACS) analysis, we also determined the various subsets of cells including naive cells (CD44lo CD62Lhi), effector memory cells (TEM; CD44hi CD62Llo), and central memory cells (TCM; CD62Lhi CD44hi) in the TCRβ+/CD8+/CD4+ cell population in both the spleen and PLNs (Fig. 3 C–F). This revealed a significant increase in the TCRβ+/CD8+/CD4+ naive T cell population (Fig. 3 D and E) and a decrease in TEM CD4+ T cells in splenocytes (Fig. 3 C–F) but no alterations in TCRβ+/CD4+ naive cells in PLNs of P. distasonis–colonized mice (SI Appendix, Fig. S4 C–F). There was also a twofold increase in the naive cell population in both CD4+ and CD8+ T cells in the spleen (Fig. 3 E and F) and in the CD8+ naive cell population in PLNs (SI Appendix, Fig. S4F). There were no differences in the TCM and TEM populations in TCRβ+/CD8+ cells in splenocytes (Fig. 3 D and F) or in TCRβ+/CD8+/CD4+ cells in PLNs (SI Appendix, Fig. S4 D–F).
Fig. 3.
P. distasonis colonization increased CD8+ T cell and naive cell phenotype. Mice were orally gavaged with either P. distasonis or saline for 4 wk after weaning and were analyzed at 12 wk of age. (A) Representative image from a single experimental cohort of FACS analyses for splenic CD4+ and CD8+ T cells from saline- and P. distasonis–gavaged mice. (B) CD4+ and CD8+ cells as percentage of TCR-β+ immune cell subsets and ratio of splenic CD4+ to CD8+ T cells. (C) CD44lo/hi and CD62Llo/hi T cells in TCR-β+, CD4+, and CD8+ T cells. (D) Percentage of CD44hi CD62Llo (TEM), CD44hi CD62Lhi (TCM), CD44lo CD62Lhi (Naive) in TCR-β+ immune cell subsets. (E) Percentage of CD44hi CD62Llo (TEM), CD44hi CD62Lhi (TCM), CD44lo CD62Lhi (Naive) in CD4+ immune T cell subsets. (F) Percentage of CD44hi CD62Llo (TEM), CD44hi CD62Lhi (TCM), CD44lo CD62Lhi (Naive) in CD8+ immune T cell subsets. (G) Representative image of FACS analyses of CD4+, CD25+ Treg cell population in saline- and P. distasonis–gavaged mice. (H) Percentage of CD4+ CD25+ cells in CD4+ single-cell subsets, CD44hi CD62Llo (TEM), and CD44lo CD62Lhi (Naive) population in percentage of CD4+ CD25+ single-cell subsets. All samples in each panel are biologically independent. Spleens were obtained from female NOD mice oral gavaged with either P. distasonis (n = 6) or saline (n = 3). Data are expressed as mean ± SEM. *P < 0.05, **P < 0.01. Statistical analysis was performed by two-tailed, unpaired Student t test. Freq, frequency; ns, not significant.
T-regulatory cells (Treg cells) play a key role in modulating T1D autoimmunity by suppressing self-reactive T cell proliferation (26). This is modulated by Forkhead box protein P3 (Foxp3) expression and IL-10 secretion. It was previously shown that CD4+ CD25+ CD44+ TEM cells positively correlate with Foxp3 expression and production of IL-10 (27). We found a 15% decrease of CD4+ CD25+ CD44+ TEM cells in both splenocytes (Fig. 3 G and H) and PLN cells (SI Appendix, Fig. S4 G and H) and a 1.6-fold increase in the total CD4+ CD25+ PLN cell population (SI Appendix, Fig. S4H) of P. distasonis–colonized mice, while there were no differences in other cell subsets (Fig. 3H). In the CD4+ T cell population, the percentages of CD25+Foxp3+ cells were significantly decreased in the splenocytes and PLNs of P. distasonis–colonized NOD mice by 21% and 17.8%, respectively (Figs. 4 A and B and SI Appendix, Fig. S5A; representative images). There was no difference in the pancreas and spleen weight or in the total number of spleen and PLN cells between P. distasonis–colonized mice and control mice (SI Appendix, Fig. 5B). Thus, P. distasonis colonization decreased anti-inflammatory CD4+ CD25+ CD44+ TEM cells and CD25+Foxp3+ Treg cells and increased inflammatory CD8+ T cells in splenocytes and PLNs, which could contribute to the enhancement of T1D in female NOD mice.
Fig. 4.
P. distasonis colonization decreases Foxp3+ cells and increases dendritic and macrophage populations. (A) Representative image of FACS analyses of splenic Foxp3+ cells in saline- and P. distasonis–gavaged mice. (B) Percentage of Foxp3+ cells in CD4+ T cell subsets. (C) Representative image of FACS analyses of dendritic cells (CD11b−CD11c+ and CD11c+ CD11b+) in saline- and P. distasonis–gavaged mice. (D) Percentage of CD11b+/−CD11c+/− population in splenic single-cell subsets. (E) Representative image of FACS analyses of macrophages (F4/80+cells) in spleen of saline- and P. distasonis–gavaged mice. (F) Percentage of F4/80+ cells in CD11b+ cell subsets. (G) Percentage of F4/80+ cell subsets, where SSCloLy6Clo represents residential macrophage, and SSCloLy6Chi represents circulatory macrophages. All samples in each panel are biologically independent. Spleens were obtained from female NOD mice oral gavaged with either P. distasonis (n = 6) or saline (n = 3). Data are expressed as means ± SEM. *P < 0.05, ****P < 0.0001. Statistical analysis was performed by two-tailed, unpaired Student t test. Freq, frequency; FSC-A, forward scatter-A; ns, not significant.
2.6. P. distasonis Colonization in Female NOD Mice Increases Dendritic Cells and Macrophages in the Splenocytes.
To further explore the mechanisms that led to an increase in CD8+ T cells in P. distasonis–colonized NOD female mice, we assessed the number of APCs in spleens and PLNs. CD11c+ CD11b+ dendritic cells and F4/80+ macrophages play an essential role in accelerating diabetes in NOD mice (28, 29). FACS analysis revealed a 1.5-fold increase in CD11b+ CD11c+ dendritic cells and a 1.6-fold increase in F4/80+ macrophages in the splenocytes of P. distasonis–colonized mice (Fig. 4 C–F), with no significant differences in dendritic cells in PLNs (SI Appendix, Fig. S5 D–G). Moreover, CD11c+ CD11b− dendritic cells (30) increased 1.8-fold in the spleen of P. distasonis–colonized mice (Fig. 4D). There was also a 1.3-fold increase in circulatory macrophages (SSCloLy6Chi cells), while there was no change in the number of residential macrophages (SSCloLy6Clo) in this population (Fig. 4G). However, there was a sevenfold increase in F4/80+ macrophage population in PLNs in P. distasonis–colonized mice (SI Appendix, Fig. S5G). These findings are consistent with previous studies demonstrating a role for CD11c+ CD11b+ dendritic cells and F4/80+ macrophages in accelerating diabetes in NOD mice (28, 29) and demonstrated that P. distasonis colonization can stimulate both innate and adaptive immune responses.
2.7. P. distasonis hprt4-18 Is a Predictor of Seroconversion and Is Enriched in the Gut Microbiome of Children Developing AABs.
To investigate the potential role of the P. distasonis insB:9–23 mimetic peptide in human T1D, we reanalyzed human gut microbiome data from the DIABIMMUNE study (31) using shotgun metagenomic sequencing performed on stool samples collected monthly from children 0 to 3 y of age who were genetically predisposed to T1D living in Finland, Russia, and Estonia (n = 269 children) and correlated this with the development of two or more islet AABs (i.e., seroconversion, which occurred in 15 of these children) or development of T1D (which occurred in 6 of the 15 seroconverted children during the follow-up period). In both cases, we used the metagenomic data to search the microbiome for the specific DNA sequence encoding the P. distasonis hprt4-18 peptide RILVELLYLVCSEYL. Importantly, the hprt4-18 sequence of P. distasonis is unique, making this analysis possible. We could therefore use mixed-effects logistic regression to test for associations between the presence of this sequence and AAB seropositivity or T1D while correcting for stool collection age and country as fixed effects and subject IDs as random effects.
Notably, seroconversion rates (i.e., the proportion of individuals developing two or more AABs during the follow-up of the study) were consistently higher in children whose microbiome harbored a genetic sequence capable of producing the hprt4-18 peptide compared to individuals who did not harbor the sequence (Fig. 5A). Thus, the seroconversion rate was significantly higher in children between age 1 and 2 y who were hprt4-18 (+) than in those who were hprt4-18 (−) (8.24% versus only 1.03%, P = 0.029). We observed similar results for age 2 to 3 y, where the seroconversion rate was 8.64% for hprt4-18 (+) individuals and only 1.28% for hprt4-18 (−) individuals (P = 0.055). A similar trend was observed in the 0- to 1-y age group, although it did not reach statistical significance. When all age groups were combined, the seroconversion rate was significant [i.e., 9.93% for an hprt4-18 (+) children during the first 2 y of life (years 0 to 2) compared to 1.20% for an hprt4-18 (−) individual (P = 0.025). However, the seroconversion rate was not significant when all 3 y of observation were combined (12.20% for hprt4-18 (+) individuals and 2.44% for hprt4-18 (−) individuals; P = 0.146). These results indicate that being exposed to hprt4-18 peptide, especially between ages 1 and 2 y, is a strong predictor of seroconversion for these children.
Fig. 5.
Seroconversion rates are significantly higher in children who harbor microbiota encoding the hprt4-18 peptide. (A and B) Seroconversion rate (A; i.e., the proportion of individuals developing two or more AABs) and T1D development rate (B) in subjects with or without hprt4-18. The analysis was completed on a cohort of 270 children (126 from Finland, 74 from Estonia, and 70 from Russia) and 1,168 stool samples/metagenomes (624 from Finland, 221 from Estonia, and 323 from Russia). Of these children, 15 were seroconverted and 6 were diagnosed with T1D. *P < 0.05.
Although the number of total cases of T1D development in the DIABIMMUNE study was small, we also calculated the T1D development rate for this cohort. Again, the T1D development rate ranged from 3.24 to 4.88% for the various hprt4-18 (+) subgroups, while for the hprt4-18 (−) children, it was 0% in all subgroups except the age 0 to 1 y subgroup, where it was 1.55% (Fig. 5B).
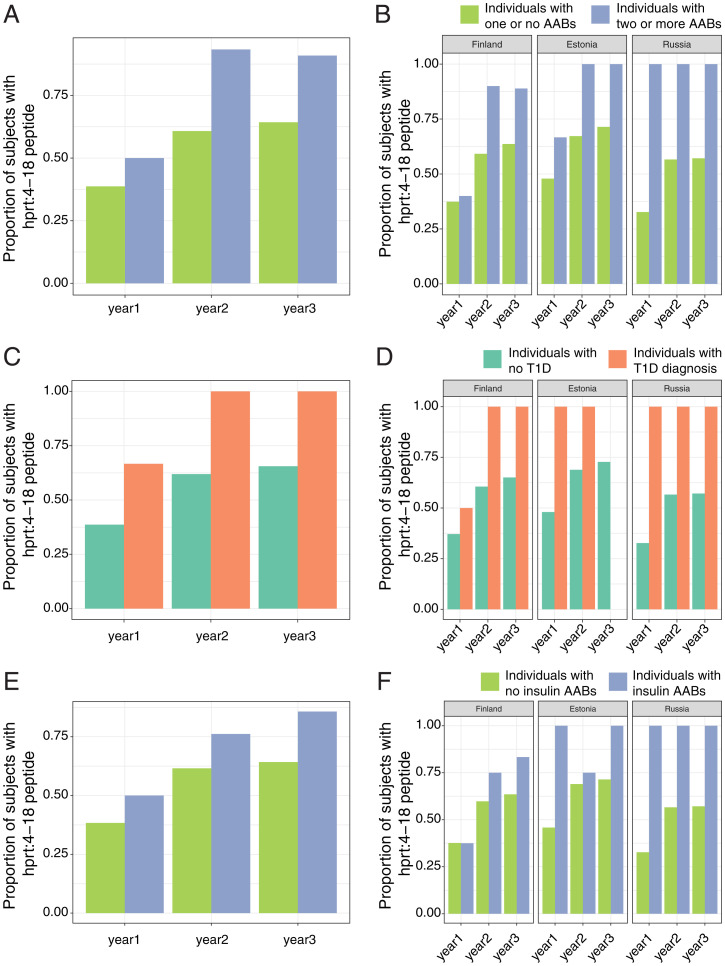
We then used mixed-effects logistic regression to test for associations between the presence of hprt4-18 and AAB seropositivity, T1D, or IAA presence, while correcting for age at stool collection and country as fixed effects and subject IDs as random effects. In addition to notable differences in the seroconversion rate, having a microbiota capable of producing the hprt4-18 peptide was significantly associated with the development of seropositivity (i.e., having two or more T1D-associated AABs; mixed-effects logistic regression, P = 0.0017; Fig. 6 A and B). In addition, hprt4-18 presence was also significantly associated with T1D onset (P = 0.020; Fig. 6 C and D) and IAA positivity (P = 0.047; Fig. 6 E and F). No individuals in the Estonia population of our cohort developed diabetes in year 3(Fig. 6D). Thus, the occurrence of this peptide was consistently more frequent in individuals with two or more AABs and individuals who developed T1D during the first, second, and third year of life in all three countries compared to healthy individuals in the same cohort. These results indicate that exposure to the P. distasonis hprt4-18 peptide in the first 3 y of life is a predictor of seroconversion and T1D onset and suggest that the presence of this peptide in the microbiome may trigger or potentiate the autoimmune response that leads to the development of T1D in genetically susceptible individuals.
Fig. 6.
Reanalysis of DIABIMMUNE gut microbiome data. (A and B) The percentage of subjects having sequence encoding hprt4-18 in the gut in all three countries (A; Finland, Estonia, and Russia) and in individual countries (B) at different phases of gut microbiota development (age 0 to 3 y). Blue represents subjects who ultimately developed two or more AABs (seroconverted), and green represents subjects who remained AAB negative throughout the study (nonseroconverted). (C and D) The percentage of subjects having sequence encoding hprt4-18 in the gut in all three countries (C; Finland, Estonia, and Russia) and in individual countries (D) at different phases of gut microbiota development (age 0 to 3 y). Green represents subjects who did not develop T1D, and orange represents the subjects who were diagnosed with T1D during the course of the study. (E and F) The percentage of subjects having sequence encoding hprt4-18 in the gut in all three countries (E; Finland, Estonia, and Russia) and in individual countries (F) at different phases of gut microbiota development (age 0 to 3 y). Green represents subjects without IAAs, and blue represents subjects with IAAs. The analysis was completed on a cohort of 270 infants (126 from Finland, 74 from Estonia, and 70 from Russia) and 1,168 stool samples/metagenomes (624 from Finland, 221 from Estonia, and 323 from Russia). Of these children, 15 were seroconverted and 6 were diagnosed with T1D.
3. Discussion
Despite major increases in our understanding of the role of autoimmunity in the pathogenesis of T1D, the triggering events that lead to disease development remain poorly understood. It is well known that an important component of the early autoimmune response in individuals who ultimately develop T1D is the development of AABs and T cell reactivity to islet proteins, especially insulin. While there are multiple potential epitopes, the dominant sequence within the insulin molecule to which reactivity occurs is a sequence in the B-chain involving amino acids 9 to 23 (14). This B:9–23 peptide (SHLVEALYLVCGERG) can bind to DQ8 molecules utilizing three different registers, with the amino acid V at position 12, E at position 13, or A at position 14 (32) as the first anchor for binding register 1, 2, and 3, respectively. It has been shown that an R→E substitution at position 22 of the B:9–23 peptide creates an even more potent agonist for activating B:9–23 T cell clones, implicating the A at position 14 as the p1 anchor (18).
In this study, we investigated the potential role of molecular mimicry as a link between microbial flora and this component of T1D development. Molecular mimicry mechanisms are based on the degeneracy of T cell recognition (33, 34) and can be either pathogenic (35) or protective (36). While molecular mimicry has long been postulated as a potential factor in autoimmune diseases, including T1D (37, 38), progress in this area has been limited due to a lack of identification of microbial sequences that might trigger this response (39). Taking advantage of the growing genome databases for microbes in the environment, we identified 47 microbial peptides with high sequence homology to insB:9–23. We demonstrated that of these, a peptide with the sequence of hprt4-18 from P. distasonis can be recognized by and activate murine IIT-3 T cell hybridomas known to respond to insB:9–23. In addition, presentation of hprt4-18 by DQ8cis or DQ8trans molecules activated a human DQ8-restricted insB:9–23–specific T cell clone generated from peripheral blood mononuclear cells (PBMCs) of a T1D patient. The DQ8-restricted insulin clone recognized the insB:9–23 bound to DQ8 in register 3. Amino acid sequence alignment of hprt4-18 and inB:9–23 implicated that the L at position 9 of the hprt4-18 peptide acts as the first anchor for binding to DQ8.
Colonization of the NOD mice gut with P. distasonis results in increased insulitis and accelerates diabetes onset. Our data show that P. distasonis stimulated an increase in CD8+ T cells, naive CD4+ and CD8+ T cells, macrophages, and dendritic cells and decreased Treg (FoxP3+) cells, while further studies are required to determine whether disease incidence after adoptive transfer of splenocytes from P. distasonis–colonized mice is specifically due to transfer of hprt 4–18 peptide–specific CD8+ T cells. However, our cross-reactivity data suggest that hprt4-18 has the potential to play a key role in this process. Previous studies have shown that CD8+ T cells, Treg cells, and naive T cells can all play a role in the adoptive transfer of disease phenotype to female NOD/SCID mice (40–42).
Microbiota and gut microbiota in particular have been shown to play a role in modulating T1D onset in NOD mice (7, 43–45), but previous reports identified primarily protective effects, most through somewhat nonspecific mechanisms. For example, NOD mice reared in germ-free environments have higher rates of development of diabetes than those raised in conventional facilities (44). In terms of protective effects of some specific bacterial species, the presence of segmented filamentous bacteria in the gut correlates with diabetes protection in NOD female mice, which normally have a high incidence of disease (46). Likewise, female NOD mice colonized at 3 to 10 wk of age with Akkermensia muciniphila showed delayed diabetes development (47), and oral administration of heat-killed B. fragilis has been shown to suppress autoimmunity in NOD mice when administered under conditions that induce increased gut permeability (23). Hundreds of treatments have been shown to decrease the development of diabetes in NOD mice (48, 49); however, the specificity of these effects has been questioned (50). By contrast, in the present study, we showed that exposure to P. distasonis can accelerate disease development in NOD mice and that a specific peptide encoded in their genome, hprt4-18, can serve as a mimic of the major insulin auto-epitope at position B9-23. This peptide and the insulin peptide can also stimulate a bidirectional cross-reactive immune response, proving its nature as a molecular mimic.
In addition to immunologic cross-reactivity, our study also provides insight into the mechanisms of immune cell regulation by this bacterium. Colonization of NOD mice with P. distasonis increases the CD8+ T cell population and decreases the CD4/CD8 T cell ratio. A reduction in this ratio was previously shown to be associated with increased T1D in humans (51). G9C8 CD8+ T cell clones originally isolated from the islets of young NOD mice cells are activated by an insB:15–23 peptide (i.e., the C-terminal fragment of the insB:9–23 peptide), and this accelerates diabetes when adoptively transferred in NOD/SCID mice, even in the absence of CD4+ T-cells (52, 53). Wong et al. (52) showed that cross-presentation of insulin by dendritic cells can stimulate G9C8 T cell clones, and both insulin and the insB:15–23 peptide can stimulate proliferation of the naive cell phenotype in NOD mice. The P. distasonis sequence is identical to this peptide in six of its nine residues. We also observed that P. distasonis colonization in NOD mice decreased Foxp3+ Treg cells in the spleen and PLNs. This effect on Foxp3+ Treg cells is particularly interesting because Foxp3+ Treg cells are dysregulated in newly diagnosed T1D patients (54), and increasing Foxp3+ Treg cells can delay the onset of diabetes in NOD mice (26, 55).
A plethora of human gut microbiome studies have demonstrated that the composition of gut microbiota in patients with autoimmune diseases, including multiple sclerosis (56), systemic lupus erythematosus, anti-phospholipid syndrome (57), Crohn disease (58–60), ulcerative colitis (61), inflammatory bowel diseases (62, 63), and celiac disease (64), is significantly different from that in healthy controls. Although the methodologies and conclusions differ in studies of microbiota from subjects with or at risk for T1D (8, 65, 66), most show that the diversity of gut microbiota is decreased in T1D patients, with increased prevalence of Bacteroidetes species. This is also associated with an altered serum metabolomic profile compared to healthy controls (7). While most of these studies were not able to define any mechanism by which this might relate to disease development, data in the DIABIMMUNE study has suggested that the higher T1D rates in Finnish Karelia and Estonia compared to Russian Karelia may be related, in part, to the action of different LPSs in the gut microbiota on the immune response.
The Environmental Determinants of Diabetes in the Young (TEDDY) study, which has collected over 12,000 fecal samples from 903 children at risk for T1D in four countries, has identified Parabacteroides as the genus most significantly associated with T1D (67). This is consistent with our findings that a P. distasonis peptide can induce T cells and antibodies that cross-react with the major epitope in insulin involved in the autoimmune response. Reanalysis of the DIABIMMUNE metagenomic data showed that seroconversion rates were consistently higher in children whose microbiome harbored a genetic sequence capable of producing the hrpt4–18 peptide compared to individuals who did not harbor the sequence. Further, a significant number of the children becoming seropositive for AABs related to T1D have the microbiota encoding the P. distasonis hprt4-18 peptide in their gut microbiota in the first 3 y of life compared to fewer than 20% of children remaining seronegative. Moreover, although our sample size is small for this analysis (n = 6), 100% of the children developing T1D harbored hprt4-18 peptide in their gut. These results show that harboring hprt4-18 is a predictor of seroconversion, and our data suggest that it might also be the trigger of the autoimmunity.
The TEDDY study showed that human gut microbiota development can be divided into three phases: a developmental phase (months 3 to 14), a transitional phase (months 15 to 30), and a stable phase (months 31 to 46) (67). Our data suggest that exposure to P. distasonis in the developmental and transitional phases may be critical in T1D pathogenesis. Further studies will be needed to directly investigate the link between human T1D and P. distasonis, but the cross-reactivity of the P. distasonis hprt4–18 peptide with insB:9–23–activated human T cell clones represents a potential mechanism.
In this study, we also showed that there is an enriched humoral immune response in T1D patients against P. distasonis compared to B. fragilis. While daily insulin injections can result in low titers of antibodies to insulin in T1D patients (68), we do not believe our findings are related to insulin antibody (IA) development. In a previous study using phage display to distinguish the binding epitope of IAA from IA idiotypes, Devendra et al. (69) showed that the sequence LGRGGSK was a major epitope for IAA but not IA in patients with newly diagnosed T1D, whereas KRSRLDV showed strong binding to IA but not with IAA. Likewise, Jaeger et al. (70) showed different binding characteristics between IAA and IA, consistent with different epitope specificities.
While we have focused only on insulin and the insB:9–23 peptide as an antigenic determinant of disease, there are clearly other epitopes recognized by both circulating and islet-infiltrating T cells in the insulin B-chain (71), A-chain (72, 73), or C-peptides (16, 74). The recent discovery that hybrid insulin peptides (HIPs) may be novel epitopes formed by posttranslational modifications formed in the β-cell (75, 76) also opens the possibility of microbial sequences that may more closely mimic these hybrid peptides. Defective ribosomal products of insulin have also been shown to lead to aberrant insulin polypeptides rendering β-cells immunogenic (77). We must also keep in mind that insulin is not the only antigen to which antibodies or reactive T cell lines are formed in T1D. Other islet antigens include GAD, IA-2, and HIPs (78). Given the enormous number of microbial peptides produced by the gut microbiota, we expect that other microbial peptides may exist with the potential to mimic these epitopes and/or trigger related autoantigen reactive T cells. Indeed, Tai et al. (79) identified a microbial peptide mimic produced by Fusobacteria that can stimulate IGRP-specific mouse T cells and promote diabetes development in a new Toll-like receptor (TLR)–deficient (TLR−/−) and MyD88−/− NY8.3 NOD mouse model.
In summary, our data define a molecular mimicry mechanism in which a specific sequence in a normal commensal gut microbe can mimic a sequence in the insulin B-chain and trigger or modify the immune response involved in the development of T1D. This finding may provide a target for treatment and a window of opportunity to prevent or delay T1D development. These data also have implications for other diseases with an autoimmune component. Today, we have databases with enormous amounts of microbial and microbiome sequence data, which can be leveraged to address the role of molecular mimicry in the autoimmunity not only of T1D but also of lupus erythematosus (80), inflammatory cardiomyopathy (81), and multiple sclerosis (82). Our findings demonstrate a molecular mimicry link to gut antigens and autoimmune diseases with the potential to ultimately provide tools, including vaccines, antibiotics, or probiotics, for the prevention and treatment of autoimmune diseases.
4. Material and Methods
4.1. Human T Cell Clone Stimulation.
Twenty 15-mer peptides (Fig. 1A) used in human and mouse T cell stimulation experiments were chemically synthesized by Genscript (trifluoroacetic acid [TFA] removal, >85% purity). The peptides were dissolved in dimethyl sulfoxide (DMSO) at 20 mg/mL (∼12 mM). Human T cell clone stimulation assays were performed as previously described (83). Briefly, insB:11–23–specific T cell clones were stimulated in 96-well round-bottom plates with irrelevant, specific, or microbial mimotope peptides in the presence of irradiated DQ8cis- or DQ8trans-expressing HEK293 cells as APCs. Fifty microliters of supernatants from cultures of T cell clones were collected after 48 h of stimulation and added to each well of 96-well round-bottom plates precoated with IFN-γ (clone MD-1) capturing antibodies (BioLegend). After overnight incubation, bound cytokines were detected by biotinylated anti–IFN-γ (clone 4s.B3) and quantified using a Victor2 D time-resolved fluorometer (PerkinElmer). The % activity was calculated by dividing the SI of mutated peptides by the SI of the wild-type peptide. Experiments were performed in the presence of 1 μg/mL of anti-CD28 antibodies. Unless otherwise stated, peptide concentrations are 2.5 μM.
4.2. NOD Mice T cell Stimulation and Antigen Presentation Assay
This experiment was performed as described previously (22). Briefly, peptides were ordered from Genscript (TFA removal, >85% purity) and dissolved in a base buffer consisting of 50 mM NaCl and 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 6.8, with 200 µM (tris(2-carboxyethyl)phosphine) (TCEP). P. distasonis hprt4-18 peptide (RILVELLYLVCSEYL) was dissolved in 5% DMSO (the final DMSO concentration was less than 1%). The C3g7 cell line, which expresses an abundance of MHC-II I-Ag7, was used as APCs and was treated with 1/2log dilutions of peptide (starting from 10 µM). These cells were then cultured with the IIT-3 T cell hybridomas for 18 h. Culture supernatants were assayed for IL-2 by incubation with the CTLL-2 cell line. CTLL-2 cells are responsive to IL-2 and only actively divide in the presence of IL-2. Next, CTLL cell proliferation was determined by 3H uptake count per minute (CPM) using a scintillation counter.
4.3. Immunization of NOD Mice
The experiments were performed as described previously (84). Briefly, 13-wk-old male NOD mice (n = 3 mice per group) were immunized in the footpad with either hprt4-18 peptide or insB:9–23 peptide (10 nmol/mouse). After 7 d, the draining (popliteal) lymph nodes were removed and pooled for examination by ELISpot. In the ELISpot assay, node cells were recalled with the various peptides to elicit either an IL-2 or IFN-γ response. Spots were analyzed by Immunospot 5.0 (C.T.L.)
4.4. Analysis of hprt:4–18 Presence in the DIABIMMUNE Cohort.
From 269 individuals in the DIABIMMUNE cohort, 1,164 metagenomes were assembled as described previously (31). Briefly, assembly was conducted with megaHIT (85), and open reading frames were predicted using Prodigal (86). Translated protein sequences of the cluster centroids were screened for protein sequence RILVELLYLVCSEYL. Assembled genes were clustered into gene families at 95% identity using CD-HIT (87). Genes were further binned into metagenomic species using canopy clustering. Seroconversion rate is the proportion of individuals developing two or more AABs during a given time period. Seroconversion rates were calculated for individuals who did or did not harbor hprt4-18 in three different time windows: during the first year of life (year 0 to 1), the first 2 y of life (years 1 to 2), and the first 3 y of life (years 2 to 3). Seroconversion rates between groups of individuals with or without hprt4-18 were compared using the test of proportions in R. All statistical analyses regarding the DIABIMMUNE cohort data were conducted in R v.3.6.3. The code used for this analysis is available at https://github.com/tvatanen/p_distansonis_peptide.
4.5. FACS and Flow Cytometry.
All experiments complied with regulations and ethics guidelines of the National Institute of Health and were approved by the IACUC of Boston College (Protocol No.#B2019-003 and 2019-004). We collected spleen and PLN cells from saline-treated and P. distasonis–colonized female NOD mice at 12 wk of age. Single-cell suspension was obtained by mechanical disruption of spleen and PLNs using an Ammonium-Chloride-Potassium lysis buffer for 5 min followed by filtration with a 70-µm filter. For staining of surface markers, cells were incubated in fluorescently labeled antibodies (SI Appendix, Table S4) for 15 min in phosphate-buffered saline containing 2% fetal bovine serum at room temperature. Cells were then washed with 2 mL staining buffer and fixed with 1% paraformaldehyde. For Foxp3 staining, fix and perm permeabilization kit (Thermo Fisher Scientific) was used per manufacturer’s instructions. All the flow cytometry antibodies were obtained from BioLegend, and flow cytometry was performed using BD FACSAria III sorter (BD Biosciences) at the Boston College Core facility. The data were analyzed using FlowJo 10.0 software. Gating strategy for the determination of different cells subsets are described in SI Appendix, Fig. S3.
4.6. Human Plasma Samples.
Peripheral blood was collected from living subjects following the provision of written informed consent (and assent in the case of minors) in accordance with Institutional Review Board–approved protocols (University of Florida IRB# 201400703) and the Declaration of Helsinki. The samples were deidentified prior to use in this study. The patient demographics are outlined in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We first want to thank Prof. Emil Unanue and Dr. Anthony Vomund (University of Washington, St. Louis) for testing microbial insulins on T cell hybridomas, immunization experiments, and their help in interpreting the data. Thanks to Jonathan Dreyfuss and Hui Pan (Joslin Bioinformatics Core) and Dogus Yusuf Dogru for bioinformatics analysis. Thanks to Babak Momeni (Boston College) for sharing his laboratory’s anaerobic chamber. We thank Bret Judson and the Boston College Imaging Core for infrastructure and support. We acknowledge our undergraduate students Tu Tran, Ruixu Huang, David Kim, Scott Hsu, Kaan Sevgi, and Maximilian Figura, Yena Sung, Jaewon Oh and Typhania Zanou for their help with the microscopy work and the animal experiments. Thanks to Nancy McGilloway and Todd Gaines for their support in the Boston College Animal Care Facility. We thank Alex Kostic, Tao Xu, Thomas Serwold, and Shio Kobayashi (Joslin Diabetes Center) for valuable discussion. This work was supported by (i) NIH Grant Nos. 1K01DK117967-01 (to E.A.), R01DK031036 and R01121967 (to C.R.K.), and NIH P01 AI042288 (to M.A.A) and (ii) The G. Harold and Leila Y. Mathers Charitable Foundation Research Grant No. MF-1905-00311 and a Juvenile Diabetes Research Foundation Grant No. 1-INO-2022-1108-A-N (to E.A).
Footnotes
Reviewers: D.H., University of Massachusetts Medical School; and L.H., University of Melbourne.
Competing interest statement: While the authors do not have any conflict of interest related to this study, M.A.A. was a coauthor related to a large clinical study (Type 1 Diabetes TrialNet Study Group) with several coauthors, including the reviewers, in the last four years. M.A.A. has neither communicated with the reviewers related to this project nor discussed the findings or conclusions of this study with the reviewers before or after the review process.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120028119/-/DCSupplemental.
Data Availability
Because the DIABIMMUNE study metagenomics sequencing reads were not ideal for a peptide search, we first assembled the reads using SPAdes (11), and they are available at https://github.com/ablab/spades. All other study data are included inthe article and/or supporting information.
References
- 1.Pugliese A., Autoreactive T cells in type 1 diabetes. J. Clin. Invest. 127, 2881–2891 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robertson C. C., et al. ; Type 1 Diabetes Genetics Consortium, Fine-mapping, trans-ancestral and genomic analyses identify causal variants, cells, genes and drug targets for type 1 diabetes. Nat. Genet. 53, 962–971 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayer-Davis E. J., et al. ; SEARCH for Diabetes in Youth Study, Incidence trends of type 1 and type 2 diabetes among youths, 2002-2012. N. Engl. J. Med. 376, 1419–1429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redondo M. J., Jeffrey J., Fain P. R., Eisenbarth G. S., Orban T., Concordance for islet autoimmunity among monozygotic twins. N. Engl. J. Med. 359, 2849–2850 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Kondrashova A., et al. , A six-fold gradient in the incidence of type 1 diabetes at the eastern border of Finland. Ann. Med. 37, 67–72 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Coppieters K. T., Boettler T., von Herrath M., Virus infections in type 1 diabetes. Cold Spring Harb. Perspect. Med. 2, a007682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dedrick S., et al. , The role of gut microbiota and environmental factors in type 1 diabetes pathogenesis. Front. Endocrinol. (Lausanne) 11, 78 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostic A. D., et al. ; DIABIMMUNE Study Group, The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boljat A., et al. , Environmental risk factors for type 1 diabetes mellitus development. Exp. Clin. Endocrinol. Diabetes 125, 563–570 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Ziegler A. G., et al. , Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 309, 2473–2479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atkinson M. A., Eisenbarth G. S., Michels A. W., Type 1 diabetes. Lancet 383, 69–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L., Nakayama M., Eisenbarth G. S., Insulin as an autoantigen in NOD/human diabetes. Curr. Opin. Immunol. 20, 111–118 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer J. P., et al. , Insulin antibodies in insulin-dependent diabetics before insulin treatment. Science 222, 1337–1339 (1983). [DOI] [PubMed] [Google Scholar]
- 14.Harrison L. C., The dark side of insulin: A primary autoantigen and instrument of self-destruction in type 1 diabetes. Mol. Metab. 52, 101288 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steck A. K., et al. , Age of islet autoantibody appearance and mean levels of insulin, but not GAD or IA-2 autoantibodies, predict age of diagnosis of type 1 diabetes: Diabetes autoimmunity study in the young. Diabetes Care 34, 1397–1399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michels A. W., et al. , Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes 66, 722–734 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayama M., et al. , Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects. Proc. Natl. Acad. Sci. U.S.A. 112, 4429–4434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., et al. , Autoreactive T cells specific for insulin B:11-23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc. Natl. Acad. Sci. U.S.A. 111, 14840–14845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cusick M. F., Libbey J. E., Fujinami R. S., Molecular mimicry as a mechanism of autoimmune disease. Clin. Rev. Allergy Immunol. 42, 102–111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stadinski B. D., et al. , Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc. Natl. Acad. Sci. U.S.A. 107, 10978–10983 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakamoto M., Benno Y., Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int. J. Syst. Evol. Microbiol. 56, 1599–1605 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Wan X., et al. , Pancreatic islets communicate with lymphoid tissues via exocytosis of insulin peptides. Nature 560, 107–111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sofi M. H., et al. , Polysaccharide a-dependent opposing effects of mucosal and systemic exposures to human gut commensal Bacteroides fragilis in type 1 diabetes. Diabetes 68, 1975–1989 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paun A., et al. , Association of HLA-dependent islet autoimmunity with systemic antibody responses to intestinal commensal bacteria in children. Sci. Immunol. 4, eaau8125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Füchtenbusch M., Larger E., Thebault K., Boitard C., Transfer of diabetes from prediabetic NOD mice to NOD-SCID/SCID mice: Association with pancreatic insulin content. Horm. Metab. Res. 37, 63–67 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Tang Q., et al. , In vitro-expanded antigen-specific regulatory T cells suppress autoimmune diabetes. J. Exp. Med. 199, 1455–1465 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu T., Soong L., Liu G., König R., Chopra A. K., CD44 expression positively correlates with Foxp3 expression and suppressive function of CD4 Treg cells. Biol. Direct 4, 40 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxena V., Ondr J. K., Magnusen A. F., Munn D. H., Katz J. D., The countervailing actions of myeloid and plasmacytoid dendritic cells control autoimmune diabetes in the nonobese diabetic mouse. J. Immunol. 179, 5041–5053 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Carrero J. A., et al. , Resident macrophages of pancreatic islets have a seminal role in the initiation of autoimmune diabetes of NOD mice. Proc. Natl. Acad. Sci. U.S.A. 114, E10418–E10427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klementowicz J. E., et al. , Cutting edge: Origins, recruitment, and regulation of CD11c+ cells in inflamed islets of autoimmune diabetes mice. J. Immunol. 199, 27–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vatanen T., et al. , Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat. Microbiol. 4, 470–479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., et al. , C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes. Proc. Natl. Acad. Sci. U.S.A. 115, 162–167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mason D., A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today 19, 395–404 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Calis J. J., de Boer R. J., Keşmir C., Degenerate T-cell recognition of peptides on MHC molecules creates large holes in the T-cell repertoire. PLOS Comput. Biol. 8, e1002412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wucherpfennig K. W., Strominger J. L., Molecular mimicry in T cell-mediated autoimmunity: Viral peptides activate human T cell clones specific for myelin basic protein. Cell 80, 695–705 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mañá P., et al. , Tolerance induction by molecular mimicry: Prevention and suppression of experimental autoimmune encephalomyelitis with the milk protein butyrophilin. Int. Immunol. 16, 489–499 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Tian J., Lehmann P. V., Kaufman D. L., T cell cross-reactivity between coxsackievirus and glutamate decarboxylase is associated with a murine diabetes susceptibility allele. J. Exp. Med. 180, 1979–1984 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atkinson M. A., et al. , Cellular immunity to a determinant common to glutamate decarboxylase and coxsackie virus in insulin-dependent diabetes. J. Clin. Invest. 94, 2125–2129 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honeyman M. C., Stone N. L., Falk B. A., Nepom G., Harrison L. C., Evidence for molecular mimicry between human T cell epitopes in rotavirus and pancreatic islet autoantigens. J. Immunol. 184, 2204–2210 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Gearty S. V., et al. , An autoimmune stem-like CD8 T cell population drives type 1 diabetes. Nature 602, 156–161 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong F. S., Visintin I., Wen L., Flavell R. A., Janeway C. A. Jr, CD8 T cell clones from young nonobese diabetic (NOD) islets can transfer rapid onset of diabetes in NOD mice in the absence of CD4 cells. J. Exp. Med. 183, 67–76 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tonkin D. R., He J., Barbour G., Haskins K., Regulatory T cells prevent transfer of type 1 diabetes in NOD mice only when their antigen is present in vivo. J. Immunol. 181, 4516–4522 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Wen L., et al. , Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455, 1109–1113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markle J. G., et al. , Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Sun J., et al. , Pancreatic β-cells limit autoimmune diabetes via an immunoregulatory antimicrobial peptide expressed under the influence of the gut microbiota. Immunity 43, 304–317 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Kriegel M. A., et al. , Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. U.S.A. 108, 11548–11553 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hänninen A., et al. , Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut 67, 1445–1453 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Atkinson M. A., Roep B. O., Posgai A., Wheeler D. C. S., Peakman M., The challenge of modulating β-cell autoimmunity in type 1 diabetes. Lancet Diabetes Endocrinol. 7, 52–64 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roep B. O., Atkinson M., von Herrath M., Satisfaction (not) guaranteed: Re-evaluating the use of animal models of type 1 diabetes. Nat. Rev. Immunol. 4, 989–997 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Skyler J. S., Hope vs hype: Where are we in type 1 diabetes? Diabetologia 61, 509–516 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Al-Sakkaf L., et al. , Persistent reduction of CD4/CD8 lymphocyte ratio and cell activation before the onset of type 1 (insulin-dependent) diabetes. Diabetologia 32, 322–325 (1989). [DOI] [PubMed] [Google Scholar]
- 52.Wong F. S., et al. , Activation of insulin-reactive CD8 T-cells for development of autoimmune diabetes. Diabetes 58, 1156–1164 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong F. S., et al. , Identification of an MHC class I-restricted autoantigen in type 1 diabetes by screening an organ-specific cDNA library. Nat. Med. 5, 1026–1031 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Viisanen T., et al. , FOXP3+ regulatory T cell compartment is altered in children with newly diagnosed type 1 diabetes but not in autoantibody-positive at-risk children. Front. Immunol. 10, 19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grinberg-Bleyer Y., et al. , IL-2 reverses established type 1 diabetes in NOD mice by a local effect on pancreatic regulatory T cells. J. Exp. Med. 207, 1871–1878 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maruvada P., Leone V., Kaplan L. M., Chang E. B., The human microbiome and obesity: Moving beyond associations. Cell Host Microbe 22, 589–599 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Rinaldi M., Perricone R., Blank M., Perricone C., Shoenfeld Y., Anti-saccharomyces cerevisiae autoantibodies in autoimmune diseases: From bread baking to autoimmunity. Clin. Rev. Allergy Immunol. 45, 152–161 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Gevers D., et al. , The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamps L. W., et al. , Pathogenic Yersinia DNA is detected in bowel and mesenteric lymph nodes from patients with Crohn’s disease. Am. J. Surg. Pathol. 27, 220–227 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Chassaing B., et al. , Crohn disease–associated adherent-invasive E. coli bacteria target mouse and human Peyer’s patches via long polar fimbriae. J. Clin. Invest. 121, 966–975 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carding S., Verbeke K., Vipond D. T., Corfe B. M., Owen L. J., Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 26, 26191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frank D. N., et al. , Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Navaneethan U., Venkatesh P. G., Shen B., Clostridium difficile infection and inflammatory bowel disease: Understanding the evolving relationship. World J. Gastroenterol. 16, 4892–4904 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quagliariello A., et al. , Effect of Bifidobacterium breve on the intestinal microbiota of coeliac children on a gluten free diet: A pilot study. Nutrients 8, 660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vatanen T., et al. , The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vatanen T., et al. ; DIABIMMUNE Study Group, Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 1551 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Stewart C. J., et al. , Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu X., Chen F., Exogenous insulin antibody syndrome (EIAS): A clinical syndrome associated with insulin antibodies induced by exogenous insulin in diabetic patients. Endocr. Connect. 7, R47–R55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Devendra D., et al. , The use of phage display to distinguish insulin autoantibody (IAA) from insulin antibody (IA) idiotypes. Diabetologia 46, 802–809 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Jaeger C., et al. , Binding characteristics and crossreactivity of insulin autoantibodies and insulin antibodies directed to three different insulin molecules. Acta Diabetol. 45, 191–194 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Higashide T., et al. , T cell epitope mapping study with insulin overlapping peptides using ELISPOT assay in Japanese children and adolescents with type 1 diabetes. Pediatr. Res. 59, 445–450 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Kent S. C., et al. , Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature 435, 224–228 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Mannering S. I., et al. , The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J. Exp. Med. 202, 1191–1197 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pathiraja V., et al. , Proinsulin-specific, HLA-DQ8, and HLA-DQ8-transdimer-restricted CD4+ T cells infiltrate islets in type 1 diabetes. Diabetes 64, 172–182 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Mannering S. I., Di Carluccio A. R., Elso C. M., Neoepitopes: A new take on beta cell autoimmunity in type 1 diabetes. Diabetologia 62, 351–356 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Delong T., et al. , Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 351, 711–714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kracht M. J., et al. , Autoimmunity against a defective ribosomal insulin gene product in type 1 diabetes. Nat. Med. 23, 501–507 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Babon J. A., et al. , Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat. Med. 22, 1482–1487 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tai N., et al. , Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J. Exp. Med. 213, 2129–2146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Greiling T. M., et al. , Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci. Transl. Med. 10, eaan2306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gil-Cruz C., et al. , Microbiota-derived peptide mimics drive lethal inflammatory cardiomyopathy. Science 366, 881–886 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Harkiolaki M., et al. , T cell-mediated autoimmune disease due to low-affinity crossreactivity to common microbial peptides. Immunity 30, 348–357 (2009). [DOI] [PubMed] [Google Scholar]
- 83.Chow I. T., et al. , Discriminative T cell recognition of cross-reactive islet-antigens is associated with HLA-DQ8 transdimer-mediated autoimmune diabetes. Sci. Adv. 5, eaaw9336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mohan J. F., et al. , Unique autoreactive T cells recognize insulin peptides generated within the islets of Langerhans in autoimmune diabetes. Nat. Immunol. 11, 350–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li D., Liu C. M., Luo R., Sadakane K., Lam T. W., MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 86.Hyatt D., et al. , Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Because the DIABIMMUNE study metagenomics sequencing reads were not ideal for a peptide search, we first assembled the reads using SPAdes (11), and they are available at https://github.com/ablab/spades. All other study data are included inthe article and/or supporting information.