Significance
The human zinc finger antiviral protein ZAP potently restricts many important viruses such as SARS-CoV-2 and HIV. ZAP comes in two flavors, with a long and a short variant exhibiting slightly different functions that demand characterization. We carefully dissected how these ZAP variants are made, and revealed that the ZAP variants regulate the RNAs of many genes involved in new cellular pathways, including a cellular stress response and a pathway that induces cell migration. Hence, ZAP emerges as a regulator of important cellular processes with relevance to cancer. Our elucidation of the synthesis, cellular targets and pathways of its variants is paramount to pharmacologically target ZAP for immunomodulation or cancer treatment, of which we also provide proof of concept.
Keywords: zinc finger antiviral protein, alternative splicing, splice-switching antisense oligonucleotides, unfolded protein response, epithelial-mesenchymal transition
Abstract
Human ZAP inhibits many viruses, including HIV and coronaviruses, by binding to viral RNAs to promote their degradation and/or translation suppression. However, the regulatory role of ZAP in host mRNAs is largely unknown. Two major alternatively spliced ZAP isoforms, the constitutively expressed ZAPL and the infection-inducible ZAPS, play overlapping yet different antiviral and other roles that need further characterization. We found that the splicing factors hnRNPA1/A2, PTBP1/2, and U1-snRNP inhibit ZAPS production and demonstrated the feasibility to modulate the ZAPL/S balance by splice-switching antisense oligonucleotides in human cells. Transcriptomic analysis of ZAP-isoform–specific knockout cells revealed uncharacterized host mRNAs targeted by ZAPL/S with broad cellular functions such as unfolded protein response (UPR), epithelial-mesenchymal transition (EMT), and innate immunity. We established that endogenous ZAPL and ZAPS localize to membrane compartments and cytosol, respectively, and that the differential localization correlates with their target-RNA specificity. We showed that the ZAP isoforms regulated different UPR branches under resting and stress conditions and affected cell viability during ER stress. We also provided evidence for a different function of the ZAP isoforms in EMT-related cell migration, with effects that are cell-type dependent. Overall, this study demonstrates that the competition between splicing and IPA is a potential target for the modulation of the ZAPL/S balance, and reports new cellular transcripts and processes regulated by the ZAP isoforms.
The RNA-binding protein (RBP) zinc finger antiviral protein (ZAP, alias PARP13/ZC3HAV1) binds to many viral RNAs, including alphaviruses, filoviruses, and retroviruses, and targets them for RNA degradation or translational suppression (1–5). The two major isoforms ZAP-Long (ZAPL) and ZAP-Short (ZAPS) are, respectively, generated by alternative splicing and intronic polyadenylation (IPA) (SI Appendix, Fig. S1A). ZAPL and ZAPS share the same RNA-binding domain, but ZAPS lacks the enzymatically inactive poly(ADP ribose) polymerase (PARP)-like domain and the C-terminal prenylation motif (6–8). Although the cis-acting elements responsible for the infection-mediated transcription induction reside in the ZAP promoter, only ZAPS is up-regulated upon infection (9). Knowledge of the regulation of ZAPL/S synthesis is limited to the transcription termination factors SCAF4/8, the cleavage factor CPSF2, and the ZAP-binding partner TRIM25 (10–12).
The distinct functions of ZAPL and ZAPS are poorly understood. All ZAP isoforms have antiviral activity, and among these, ZAPL appears to be the most potent (13), yet ZAPS exerts a stronger severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) suppression (14, 15). ZAPS was reported to both augment and mediate the resolution of interferon (IFN) responses (11, 16). Most studies focused on the function of ZAP in immunity, and little is known about its roles in other processes. Only few cellular RNA targets of ZAP have been identified: TRAILR4 (the decoy receptor of proapoptotic cytokine TRAIL [tumor necrosis factor-related apoptosis-inducing ligand]), the host IFNs (IFNB and IFNL1/2/3), and ZMAT3 (11, 17, 18). Identifying more cellular target RNAs of ZAPL/S and their isoform specificity are essential to understanding their distinct functions.
Cells activate different adaptive responses to cope with various stressors, such as the unfolded protein response (UPR) upon protein misfolding in the endoplasmic reticulum (ER) and the epithelial-mesenchymal transition (EMT) during wound healing. UPR signals via the IRE1, PERK, and/or ATF6 arms to induce the transcription of UPR target genes such as ER chaperones, ER-associated degradation, and quality control factors, to attenuate global translation and enhance IRE1-dependent decay of ER-bound mRNAs (19). UPR ensures cell survival in the case of manageable stress, but it activates apoptosis in prolonged ER stress. EMT is the process by which epithelial/endothelial cells transdifferentiate into mesenchymal/fibroblast cells, and involves the loss of epithelial/endothelial markers and cellular contacts and the simultaneous acquisition of mesenchymal/fibroblast markers and migration capability (20). The roles of ZAP in the regulation of cellular responses such as UPR and EMT are completely unexplored.
Here, we demonstrated that splicing-versus-IPA competition determines the ZAPL/S balance, and that the targeting of cis-acting elements of splicing or IPA by splice-switching antisense oligonucleotides (ssASOs) enables a shift in ZAPL/S production. We also identified several splicing factors—heterogeneous nuclear ribonucleoprotein A1/A2 (hnRNPA1/A2), polypyrimidine tract-binding protein 1/2 (PTBP1/2), and U1 small nuclear RNP (snRNP)—as suppressors of ZAPS production. Using ZAPS and ZAPL knockout cells, we identified new host mRNAs regulated by these isoforms, with roles in UPR, EMT, and innate immunity. We established that endogenous ZAPL and ZAPS respectively localize to membranes and cytosol, and the disparities in subcellular localization correlate well with their target RNA selectivity. Lastly, we provided experimental evidence for the role of ZAP isoforms in UPR by regulating various arms and in EMT through affecting cell migration.
Results
Competition between Splicing and IPA Regulates ZAPL and ZAPS Balance.
We used the exon/intron annotations and polyadenylation signals of the canonical ZAPL isoform as reference. ZAPL is generated by inclusion of all exons 1–13, while ZAPS is formed by IPA using an IPA signal (iPAS) within intron 9; hence, ZAPS lacks exons 10–13. We hypothesize that the ZAPL/S ratio is dictated by the competition between intron 9 splicing and IPA. The sequences of the 5′ splice site (5′ss, AGA/gtaagt) and the iPAS (agtaaa) in ZAP intron 9, henceforth called ZAP I9-5′ss and iPAS, differ from the consensus 5′ss (CAG/gtaagt) and PAS (aataaa) (SI Appendix, Fig. S1A). To test whether these sequences regulate the ZAPL/S ratio, we mutated the ZAP I9-5′ss and iPAS in a ZAP-splicing minigene (ZAPmg) (Fig. 1A). As expected, upon strengthening of the 5′ss (CAG) or weakening of the iPAS (AGG), the percentage of ZAPS strongly decreased in HEK293T cells (Fig. 1A). Conversely, weakening of the 5′ss (5C) or strengthening of the iPAS (AAT) led to predominant ZAPS (Fig. 1A).
Fig. 1.
The competition between splicing and IPA determines the balance of ZAP isoforms. (A) Left, schematic of the ZAP minigene to test the role of cis-acting elements in the generation of ZAPL or ZAPS. Boxes, exons; black lines, introns; black box, sequence before IPA; arrows, the strengths of mutated sequences. Right, qRT-PCR results of ZAPL/S in HEK293T cells transfected with the indicated ZAPmg versions. We normalized the %ZAPS mRNA to HPRT1 and SDHA as reference genes. (B) Schematic of ssASOs targeting ZAP intron 9 5′ss (I9-5), intron 9 3′ss (I9-3), intron 10 5′ss (I10-5), the cryptic 5′ss in exon 9 (I9-Cry5), and iPAS (iPAS1 and iPAS2). (C, D) Representative blots of ZAP proteins in MCF7 (C) or MDA-MB-231 (D) cell lines transfected with ctrl-ssASO or ssASOs blocking ZAP-iPAS (iPAS2) or ZAP splice sites (I9-5 and I9-3). We treated cells with IFN-β for 24 h as indicated by +. GAPDH, protein loading control; We quantified the density of protein bands using a densitometer, normalized to that of the loading control. %ZAPS as ZAPS over sum of ZAPL and ZAPS (mRNA or protein). One-way ANOVA with Dunnett’s test; n ≥ 3; means ± SDs. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; ns P > 0.05.
Moreover, we tested whether the binding of U1 to the ZAP I9-5′ss suppressed IPA in addition to promoting splicing. For loss-of-function studies, we overexpressed a U1-decoy, which bears a consensus 5′ss, to bind and block endogenous U1 (21). While transient HEK293T transfection with the plasmid encoding U1-decoy increased ZAPS, the control decoy with a mutation that reduces binding to U1 had no effect (SI Appendix, Fig. S1B and C). For gain-of-function experiments, we transiently overexpressed a suppressor-U1 with perfect base pairing to ZAP I9-5′ss, and observed a reduction in the percentage of ZAPS, while an unrelated suppressor-U1 did not have any effect (SI Appendix, Fig. S1D and E). Similar results were observed with the ZAPmg upon U1-decoy or suppressor-U1 overexpression (SI Appendix, Fig. S1F). Together, our data show that the competition between splicing and IPA is fine-tuned by the sequences of ZAP I9-5′ss and iPAS.
Next, we designed various ssASOs to block the ZAP iPAS or the nearby splice sites to shift toward either splicing or IPA, respectively (Fig. 1B). On one hand, ssASOs blocking ZAP-iPAS (iPAS1 or iPAS2 or in combination) suppressed the percentage of ZAPS in both untreated and IFN-β-treated HEK293T cells (SI Appendix, Fig. S2A and B). On the other hand, blocking ZAP exon 9–10 splicing by various ssASOs against the I9-5′ss, I9-3′ss, or I10-5′ss alone, or against the I9-3′ss in combination with either the ssASOs for I9-5′ss or I10-5′ss, showed that the I10-5 and the combination of the I9-5 plus I9-3 significantly increased the percentage of ZAPS (SI Appendix, Fig. S2C). This enhancement of the I9-5 plus I9-3 combination over single ssASOs was not due to a higher overall dose, because the half-dose of each ssASO showed similar effects (SI Appendix, Fig. S2D). We inadvertently discovered a cryptic 5′ss located 30 nucleotides upstream of the authentic 5′ss, being activated in the I9-5 ssASO treatment, and this cryptic 5′ss could be blocked by an ssASO as well (SI Appendix, Fig. S2E and F). In the human breast cancer cells MCF7 and MDA-MB-231, the ssASO blocking ZAP iPAS (iPAS2) also efficiently suppressed the ZAPS percentage (Fig. 1C and D, except in MCF7 without IFN-β), while the I9-5 and I9-3 ssASOs increased the ZAPS percentage (Fig. 1C and D, except in MDA-MB-231 with IFN-β). Experiments in the human muscle cells SJCRH30 showed the same trend (SI Appendix, Fig. S2G).
Overall, these results suggest the importance of the competition between splicing and IPA in the ZAPL/S balance. ZAPL was reduced by some of the splicing ssASOs in some cells, and ZAPS was either reduced or increased by some of the iPAS or splicing ssASOs, respectively. However, the iPAS ssASOs never raised ZAPL. Despite the slight differences in the activity of the ssASOs in different cell lines, we conclude that our ssASOs targeting the ZAP cis-acting sequences effectively modulate the ZAPS percentage by blocking the iPAS or the splice sites.
Splicing Factors PTBP1/2 and hnRNPA1/A2 Repress ZAP IPA.
To identify trans-acting RBPs regulating ZAP splicing and IPA, we mined the Cross-Linking and Immuno-Precipitation databases (22) for RBPs bound to the ZAP intron 9 and with roles in alternative splicing and polyadenylation (SI Appendix, Fig. S3A). Then, we used loss- and gain-of-function experiments of a few candidate splicing factors in HEK293T. Transient knockdown of PTBP1/2 or hnRNPA1/A2 by RNA interference consistently up-regulated the percentage of ZAPS mRNA and protein (Fig. 2A and B and SI Appendix, Fig. S3B). Furthermore, transient overexpression of PTBP1 and/or hnRNPA1 was sufficient to suppress the percentage of ZAPS on both mRNA and protein levels (Fig. 2C). These results strongly suggest that PTBP1/2 and hnRNPA1/A2 suppress ZAP IPA, thus downregulating the ZAPS mRNA and protein. PTBP1 and hnRNPA1 localize mainly in the nucleus but translocate to the cytoplasm upon certain stimulations such as viral infection (23, 24). We observed a predominant nuclear staining of both RBPs in untreated cells, but IFN-β treatment neither altered the protein levels of PTBP1 or hnRNPA1 significantly, nor induced their cytoplasmic translocation (SI Appendix, Fig. S3C and D). In summary, we identified PTBP1/2 and hnRNPA1/A2 as suppressors of ZAP IPA.
Fig. 2.
PTBP1/2 and hnRNPA1/A2 suppress ZAP IPA. (A, B) Representative blots (left) and quantification (right) of %ZAPS mRNA and protein upon knockdown of PTBP1/2 by two shRNA mixtures (s1 or s2) (A) or hnRNPA1/A2 by DsiRNAs (B) in HEK293T cells. Scrb, scrambled sh/DsiRNA. DsiRNA, Dicer-Substrate Short Interfering RNA. Cells were also treated with IFN-β for 8 h. (C) Representative blots (left) and quantification (right) of %ZAPS mRNA and protein upon overexpression of PTBP1 or hnRNPA1 or both in HEK293T cells treated with IFN-β for 24 h. We used β-act as the western blot loading control of ZAPL/S proteins (A, B) and vinculin (Vin) as loading control for ZAPL/S (C), PTBP1, and hnRNA1/A2 proteins (A–C). We quantified the density of protein bands using a densitometer, normalized to that of the loading control. We used HPRT1 and SDHA as reference genes ZAPL/S mRNAs for qRT-PCR. %ZAPS as ZAPS over sum of ZAPL and ZAPS (mRNA or protein). One-way ANOVA with Dunnett’s test (A, C) and two-tailed t test (B); n ≥ 3; means ± SDs. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ns P > 0.05.
ZAPL and ZAPS Target Different Cellular Transcripts.
We generated ZAP-isoform-specific knockout (KO) HEK293T cells that produce only ZAPL or ZAPS by removing essential sequences for IPA or generating unstable transcripts upon splicing (SI Appendix, Fig. S4A). We obtained two ZAPS-KO and three ZAPL-KO clones that respectively expressed undetectable ZAPS and ZAPL, whose deletion sites were verified by sequencing (SI Appendix, Fig. S4B–M). In the ZAPL/S-KO clones, the level of the remaining ZAP isoform increased, consistent with splicing-versus-IPA competition (SI Appendix, Fig. S4B–G), and these cells retained the selective up-regulation of ZAPS by IFN-β (SI Appendix, Fig. S4N). RNA sequencing (RNA-seq) analyses confirmed the strong up-regulation of many IFN-stimulated genes (ISGs) among the total list of 389 ISGs (25) in all genotypes upon IFN-β stimulation, indicating a robust IFN response (SI Appendix, Fig. S5, ISGs labeled in blue and green). While the IFN mRNAs (IFNB, IFNL1/2/3) previously found as ZAPS targets (11) were expressed too low for statistical analysis, the known ZAPL-targeted transcript TRAILR4 (alias TNFRSF10D) (17) was among the differentially expressed genes (DEGs) upon ZAPL loss (Fig. 3A), supporting the reliability of our RNA-seq dataset.
Fig. 3.
Different transcripts targeted by ZAPL and ZAPS. (A) Heatmap displaying the expression of DEGs (P ≤ 0.05 and |log2FC| ≥ 0.5) between the ZAPL- or ZAPS-KOs and WT in both unstimulated and IFN-β-treated conditions. (B) Correlation of gene expression between RNA-seq versus RT-qPCR using Pearson’s R2 value. (C) Venn diagram showing the overlaps of DEGs identified from the isoform-specific KO versus WT in both untreated and IFN-β-treated cells, compared with the DEGs for the full ZAP-KO in GSE159853 (RNA-seq, ZAP-KO g1/g3 versus WT, uninfected, or HCMV-infected human umbilical vein endothelial cells for 24 h). (D) nRIP using whole-cell lysates from IFN-β-treated WT cells followed by RT-qPCR for the indicated genes, classified as DEGs for ZAPL or ZAPS. Multiple t test with FDR of 1%. *, confirmed interaction. n ≥ 3, means ± SDs.
Next, we derived the DEGs of either ZAPL- or ZAPS-KO compared to wild-type (WT) cells, and either with or without IFN-β. The loss of ZAPL or ZAPS modestly altered the RNA abundance on a small fold change (FC) and/or on a small number of host genes, likely due to the partially compensative effects of the remaining ZAP isoform, so we used moderate cutoff values (P ≤ 0.05 and |log2FC| ≥ 0.5). The loss of ZAPL or ZAPS significantly changed the expression of 11 (excluding ZAP/ZC3HAV1) or 7 cellular transcripts, respectively (Fig. 3A and SI Appendix, Fig. S6A–D). We validated most DEGs of the ZAPL/S-KO (SI Appendix, Fig. S6E–H), and the relative gene expression changes of our RNA-seq highly aligned with the RT-qPCR data (Fig. 3B). In addition, 7 of 12 DEGs from our ZAPL-KO were also found in the total ZAP-KO dataset on uninfected or human cytomegalovirus (HCMV)-infected human umbilical vein endothelial cells (18) (Fig. 3C; full gene list in SI Appendix, Dataset S1). Since ZAP destabilizes the target RNAs through direct binding (26), we performed native RNA-protein immunoprecipitation (nRIP) to examine the physical interaction between ZAP and up-regulated DEG transcripts (SI Appendix, Fig. S6I). Using a stringent cutoff (false discovery rate [FDR] ≤ 1%), we detected enrichment for several ZAPL-KO DEGs (MYEF2, GET1, NT5DC2) and ZAPS-KO DEGs (HSPA8, ZNF121) (Fig. 3D). The interaction between most DEGs and ZAP remained upon the loss of either ZAPL or ZAPS, suggesting that both ZAPL/S are capable of binding transcripts under nRIP conditions (SI Appendix, Fig. S6J–O). Together, we identified and validated new host transcripts regulated by ZAPL/S, several of which are directly bound by ZAP.
ZAPL and ZAPS Localize to Different Subcellular Compartments.
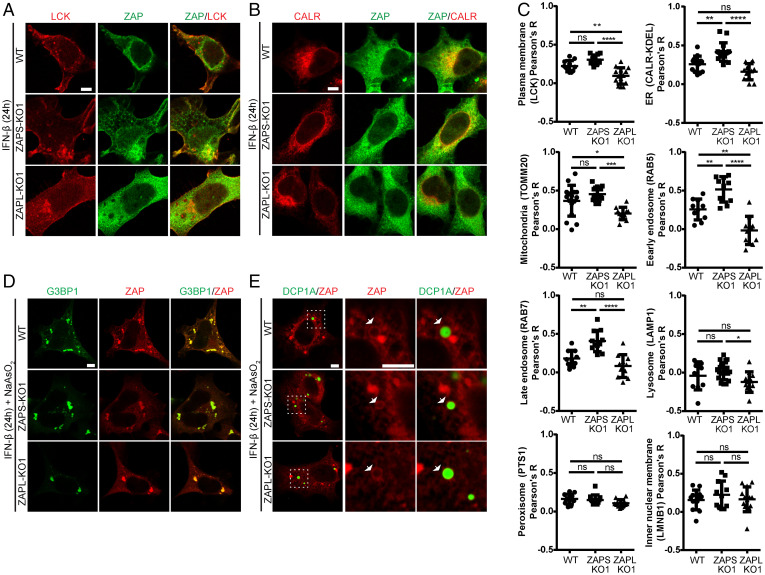
We sought to examine the localization of endogenous ZAPL and ZAPS using the ZAPS-KO and ZAPL-KO cells, respectively, upon IFN-β treatment to induce the expression of ZAPS. We performed immunofluorescence with anti-ZAP antibody in cells transfected with fluorescent protein–tagged markers for various cellular organelles. We observed that ZAPL localized to plasma membranes (labeled by LCK), ER (CALR), and mitochondria (TOMM20), more than ZAPS (Fig. 4A–C and SI Appendix, Fig. S7A). ZAPL, but not ZAPS, localized to early and late endosomes respectively labeled by RAB5 and RAB7A, but neither localized to lysosomes (LAMP1) to peroxisomes (PTS1), or to nuclear inner membranes (LMNB1) (Fig. 4C and SI Appendix, Fig. S7B–F). Upon oxidative stress by sodium arsenite, both ZAP isoforms were recruited to stress granules (SGs) marked by G3BP1 (Fig. 4D and SI Appendix, Fig. S7G). Unexpectedly, both ZAPL/S formed a ring surrounding the P-bodies (PBs) marked by DCP1A (Fig. 4E and SI Appendix, Fig. S7H). Overall, ZAPL localizes to some but not all membranous compartments, while ZAPS localizes throughout the cytoplasm, and both isoforms were recruited to RNA granules upon cellular stress.
Fig. 4.
Different subcellular localization of endogenous ZAPL and ZAPS. (A, B) Images of cells with the indicated genotypes stained with anti-ZAP antibody (green) and mScarlet- or mCherry-tagged markers of plasma membrane LCK (A) or ER CALR (B). (C) Quantification of the colocalization of ZAP with the indicated organelle’s marker-positive pixels using Pearson’s R values. (D, E) Images of cells with indicated genotypes stained with ZAP (red) and GFP-tagged markers of SGs G3BP1 (D) or PBs DCP1A (E) in sodium arsenite–stressed cells. In (E), the center and left panels showed the enlarged views of white dotted boxes in the right panels. Arrows, ring-like structures surrounding PBs. n ≥ 10 images/genotype. Scale bars, 5 μm. One-way ANOVA with Tukey’s test; means ± SDs. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; ns P > 0.05.
Should the ZAPS cytosolic and the ZAPL membrane localization confer their target-mRNA selectivity, we may observe an increase in mRNAs encoding proteins residing in cytosol or membrane in the ZAPS-KO or ZAPL-KO cells, respectively. Gene Set Enrichment Analysis (GSEA) for Gene Ontology Cellular Component (GOCC) using the expression of all of the genes showed that compared to the ZAPL-KO cells, the ZAPS-KO cells up-regulated genes in ribosome-related terms (cytoplasmic ribosome and polysome) and nuclear component terms (chromosomal telomeric region and DNA package complex), while down-regulated genes in secretory and membrane component terms (SI Appendix, Fig. S8A and B and Dataset S2). Given the reported cytosolic localization of the ribosomal protein encoded mRNAs (27) and the ER association of most membrane/secretory protein mRNAs (28), these enrichments support that the differential subcellular localization of ZAPL/S contribute to their preference on host mRNA targets.
ZAP Regulates UPR.
The total of 35 (excluding ZAP/ZC3HAV1) genes that were differentially regulated between the ZAPL-KO and ZAPS-KO cells were enriched in the hallmarks of UPR, UV response, and EMT, using low stringent cutoff values (P ≤ 0.1 and |log2FC| ≥ 0.5) to obtain significant gene numbers for reliable enrichment analysis (Fig. 5A and SI Appendix, Dataset S3 and S4). Since ZAP was already implicated in the DNA damage response upon UV (29), we tested the ZAP functions in UPR and EMT. We confirmed that the UPR DEGs, including the transcription factor downstream of the PERK arm ATF4, the cap-dependent translation suppressor EIF4EBP1 (alias 4E-BP1), the SKI RNA exosome component TTC37 (alias SKI3), the ER-to-Golgi transporter COP-II complex component SEC31A necessary for the activation of the ATF6 arm of UPR, were up-regulated in ZAPS-KO cells, while slightly reduced in ZAPL-KO cells (Fig. 5B). Moreover, GSEA for Hallmark gene sets using the expression of all genes without any cutoff revealed an overall down-regulation of genes in the UPR hallmark of the ZAPL-KO relative to WT cells as indicated by a negative Normalized Enrichment Score (NES) of −1.7 (SI Appendix, Fig. S9 A and B and Dataset S5). We observed an elevated basal UPR in ZAPS-KO but not ZAPL-KO cells, as indicated by a size expansion of ER but not of mitochondria or lysosomes (Fig. 5C and SI Appendix, Fig. S9C). Together, these results suggest different functions of the ZAP isoforms in the regulation of basal UPR.
Fig. 5.
ZAP isoforms regulate UPR. (A) A volcano plot showing the transcriptome-wide log2FC of expression in untreated ZAPS-KO relative to ZAPL-KO cells. Red gene symbols, UPR factors; purple, EMT factors. Red dots, low cutoff DEGs (P ≤ 0.1 and |log2FC| ≥ 0.5). The dotted vertical lines mark the |log2FC| of 1 (outer lines) and 0.5 (inner lines), while the dotted horizontal lines mark the −log10 (adjusted P value) of 1 (P = 0.1, lower lines) and 1.30 (P = 0.05, upper line). (B) RT-qPCR validation of UPR DEGs in the ZAPL/S-KO cells from (A). (C) Mean fluorescence intensity of live HEK293T cells stained with ER tracker (PE-CF594). (D, E) qRT-PCR for factors in the different arms of UPR upon Tg-induced acute (6 h Tg, D) or prolonged (24 h Tg, E) ER stress for different genotypes. We normalized the cycle threshold values to the reference genes (HPRT1 and SDHA), and then further normalized to the respective WT. The dotted horizontal lines in (B, D, and E) mark the |log2FC| value of 1. (F) MTS assay to measure cell viability during ER stress upon ZAPL or ZAPS loss. We normalized the absorbance to that of the WT cells of the corresponding treatments. We treated cells with Tg for 24 h in the presence of 4μ8C or GSK2656157 to inhibit IRE1 or PERK, respectively. n ≥ 3; means ± SDs; One-way ANOVA with Dunnett’s test (B–F). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; ns P > 0.05.
We next examined which UPR branches (IRE1/XBP1s, PERK/CHOP, and ATF6) were affected by the loss of ZAP isoforms. We triggered ER stress with Thapsigargin (Tg), an ER Ca2+ channel inhibitor, to induce the expression of UPR reporters, whose expression is normally kept low. We observed a general trend toward increased expression of reporters from the IRE1 (XBP1s) and the PERK (CHOP) arms (except one clone with 24 h Tg) in the ZAPS-KO cells during either acute or prolonged ER stress (Fig. 5D and E). The ZAPS-KO in the ATF6 (EDEM1) arm and two ZAPL-KO clones in all arms did not show consistent changes (Fig. 5D and E). These results suggest a role of ZAPS in dampening the UPR response during stress.
Since prolonged ER stress triggers cell death, mainly through the PERK/ATF4/CHOP and IRE1 pathways (19), we examined the cell viability upon the loss of ZAP isoforms during prolonged ER stress. Without stress, ZAPL-KO cells displayed a mild reduction in cell viability which could be due to other pathways beyond UPR (Fig. 5F). Upon ER stress, the cell viability in the ZAPS-KO cells remained reduced, while the viability of ZAPL-KO cells showed a slight recovery compared to WT (Fig. 5F). The recovery of cell viability in the ZAPL-KO cells was lost with the PERK inhibitor but not the IRE1 inhibitor (Fig. 5F), suggesting that ZAPL regulates cell viability during stress via PERK. The impaired cell viability of ZAPS-KO cells remained with IRE1 or PERK inhibitor, likely due to the up-regulation of proapoptotic CHOP independent of PERK (SI Appendix, Fig. S9D). Together, these results showed opposite roles of ZAPL and ZAPS in cell viability during ER stress.
Overall, these results indicate that the ZAP isoforms regulate different UPR branches under resting and stress conditions, as well as affect cell viability during ER stress.
ZAP Regulates EMT.
Last, we examined the EMT phenotypes in the ZAPL/S-KO cells. We confirmed the expression changes of important EMT factors (the transcription factors SNAI2/SLUG, the adaptor DAB2, and the matrix metalloprotease inhibitor TIMP3) in the ZAPL/S-KO clones (SI Appendix, Fig. S10A). Moreover, GSEA revealed an overall down-regulation trend in the EMT hallmark for the ZAPL-KO cells relative to WT (NES of −1.6), although the ZAPL loss reduced the expression of both EMT promoters (SNAI2, DAB2, and Vimentin/VIM) and suppressors (TIMP3) (SI Appendix, Fig. S10B and C and Dataset S5). Importantly, collective cell migration toward the cell-free area was impaired in the ZAPL-KO cells, while it increased in ZAPS-KO compared to WT cells (Fig. 6A and B and SI Appendix, Movies S1–S3). We also observed a reduction in or an enhancement of chemotaxis-driven single-cell migration in the ZAPL-KO and ZAPS-KO cells, respectively (Fig. 6C and D). Together, these results indicate the differential roles of ZAP isoforms in regulating cell migration in HEK293T, in which ZAPL promotes EMT while ZAPS suppresses it.
Fig. 6.
ZAP isoforms regulate EMT. (A, E) Images of collective cell migration toward the cell-free area (wound) at the indicated time points in HEK293T (A) or MCF7 (E) cells upon the loss of ZAPS or ZAPL. 0 h refers to the time of removal of the molds. (B, F) Left graphs show the percentage of the closing wound at indicated time points for the indicated genotypes from A (B) and E (F). Right graphs plot the wound closure rates (slopes) obtained by linear fitting of the data from the left graphs. (C, G) Representative images of migrated cells in the Transwell assay of respective genotypes in HEK293T (C) and MCF7 (G) upon the loss of either ZAPS or ZAPL. (D, H) Quantification of cell migration in C (D) and G (H). n ≥ 3; means ± SDs; One-way ANOVA with Dunnett’s test. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. Scale bars, 100 μm.
Since HEK293T cells are semiepithelial and express both epithelial and mesenchymal markers (30), we tested the role of ZAPL/S on cell migration in MCF7, a low-migrative and noninvasive epithelial breast cancer cell line and a well-established EMT model. We efficiently knocked down either ZAPL or ZAPS by the stable integration of isoform-specific small hairpin RNAs (shRNAs) (SI Appendix, Fig. S10D). Compared with HEK293T, the MCF7 cells mainly migrate collectively as a sheet at a lower migration rate and display a low level of single-cell migration (Fig. 6E–H). Unexpectedly, we observed a clear reduction of collective cell migration upon ZAPS loss (Fig. 6E and F and SI Appendix, Movies S4–S6), and a markedly diminished single-cell migration upon the loss of either isoform (Fig. 6G and H). Since collective and single-cell migration are characteristic of partial and full EMT (31), respectively, our results suggest different roles of ZAPL and ZAPS in different EMT stages in MCF7 cells, and that ZAPS promotes migration.
Overall, we provided the first evidence that there was a different function of the ZAP isoforms in EMT-related cell migration and that the effects are cell-type dependent.
Discussion
We demonstrated the importance of the competition between splicing and IPA to fine-tune the ZAPL/S ratio, and that blocking the ZAP-iPAS or splice sites with ssASOs shifts the production of ZAP isoforms in human cells. These ssASOs may be relevant for disorders in which one ZAP isoform is more beneficial than the other. While increasing ZAPL may help fight against alphaviruses (7, 32) and sensitize cancer cells to TRAIL-mediated apoptosis (17), elevating ZAPS may suppress SARS-CoV-2 via interfering with viral RNA-dependent RNA-polymerase production (14, 15). Future studies are warranted to test the effectiveness of the ZAP ssASOs on ZAP-sensitive viruses or diseases. We also identified the splicing factors hnRNPA1/A2 and PTBP1/2 that suppress ZAPS, and added ZAP to the list of U1-repressed IPA events (33, 34).
We generated the ZAP-isoform specific knockout cells, in which only ZAPL or ZAPS are expressed under endogenous regulation, to avoid potential complications associated with overexpressed ZAP isoforms. Not only did we confirm the findings from previous studies using overexpressed ZAP isoforms upon total ZAP-KO such as localization, responsiveness to IFNs, and their reported target RNAs and functions (8, 11, 13, 17, 35) but we also uncovered unexpected transcripts and processes regulated by ZAPL/S. Individual RT-qPCR validations in the isoform-specific knockout cells expanded the number of targets from those DEGs identified by RNA-seq (TNFRSF10D, CERK, and SEC31A for ZAPS and HSPA8 for ZAPL), likely because of the higher sensitivity of this assay.
Previous work and our study showed that ZAPS was predominantly free in the cytosol, while ZAPL preferentially localized to membrane organelles, but the identity of these organelles varied among studies (8, 11, 17). Here, we showed that endogenous ZAPL localized to some of the reported organelles (endosome and ER) as well as mitochondria and plasma membrane, yet we did not see any ZAPL localization to lysosomes, as opposed to two studies (8, 11). The differences across studies may reflect cell-type heterogeneity or arise from overexpressed ZAPL overloading the folding and transport system, thus being misfolded or missorted into different compartments. Although ZAP was known to be recruited to SGs upon cellular stress, its localization to PBs remained unclear (17, 35–38), so our finding that ZAPL/S formed rings around PBs may explain the previous discrepancies.
Schwerk et al. showed that different subcellular location and expression kinetics of ZAPL/S contributed to their selectivity against viral or IFN RNAs (11). Our transcriptomic analysis further expanded the preferential ZAPL and ZAPS targets to many host transcripts encoding secretory/transmembrane proteins or cytosolic/nuclear components, respectively. Moreover, our transcriptomics of isoform-specific KOs revealed that ZAPL/S target uncharacterized cellular mRNAs, which function in cellular responses to various stressors such as ER stress (UPR) and tissue damage (EMT), in addition to viral infections. We showed that ZAP has mild effects on many transcripts, with the exception of TRAILR4 mRNA, such that the net effects of the ZAP isoforms on UPR and EMT depended on the overall expression profile of the cells. Our result is in line with previous work showing that ZAP binds weakly to many host transcripts in addition to a few ones with strong binding (18). Since UPR and EMT have strong implications in cancer, our study suggests that ZAP may affect the properties of cancer cells. The role of ZAP in cancer remains controversial: while ZAP low expression is associated with poorer prognosis for liver cancer patients (39), ZAP was only shown to promote invasiveness and metastasis of pancreatic cancer (40). Our data suggest that the distinct functions of the ZAP isoforms and cell-type heterogeneity may account for these apparently contradicting observations. Lastly, the recent finding of the RNA-decay Ski2-Ski3-Ski8 (SKI) complex possessing antiviral activity against several ZAP-sensitive viruses (41), and our findings of ZAPL/S regulating the SKI component TTC37 in the context of UPR may locate ZAP as a missing link between RNA decay, UPR, and immunity. Further studies should address the mechanisms of how ZAP isoforms regulate UPR and EMT, perhaps via decay or translation modulation of mRNA targets in these pathways.
Overall, this study demonstrates that the competition between splicing and IPA is crucial and thus a potential target for the modulation of the ZAPL/S balance and discovers new cellular transcripts and processes regulated by ZAPL/S.
Materials and Methods
We generated the ZAP(L/S)-KO cells using CRISPR-Cas9-mediated deletion of genomic DNA on HEK293T using a pair of guide RNA-containing PX458 plasmids followed by the selection of GFP+ cells. To remove ZAPS, we deleted the ZAP-iPAS. To knock out ZAPL, we deleted exon 10 to generate an unstable ZAPL containing a premature stop codon. We generated the stable ZAP(L/S) knockdowns by transducing MCF7 cells with lentiviruses carrying PLKO.1 plasmids expressing shRNAs against ZAPL or ZAPS (11, 17). Details can be found in the supplementary information, including cell culture, plasmids, ssASOs, protein analyses, immunofluorescence, RNA-seq, UPR experiments, and migration assays. We performed statistical tests on GraphPad Prism. We showed all of the data as means ± SDs, unless indicated otherwise. Oligonucleotide sequences are in SI Appendix, Table S1-4. Comparisons were with control or mock, unless otherwise indicated by a line between two samples.
Supplementary Material
Acknowledgments
We thank the Roca lab members, especially Donald Sim and Eling Goh, for discussions. We thank Kyoka Ikemori and Hy Q. Nguyen for help with U1 experiments and R scripts, respectively. We acknowledge Su I-Hsin and group members for sharing trackers. This work was funded by the NTU Integrated Medical Biological and Environmental Life Sciences NIM/02/2017 (to X.R. and D.L.), Academic Research Fund Tier 1 (RG22/21) from Singapore’s Ministry of Education (to X.R.), NRF2019-NRF-ISF003-3104 from the National Research Foundation of Singapore (to X.R.), and the OFIRG17nov084 grant from Singapore’s National Medical Research Council (to D.L.).
Footnotes
The authors declare no competing interest. Nanyang Technological University (NTU) has applied for a patent, Singapore Provisional Application No. 10202107930U related to the therapeutic modulation of ZAPL/S with ZAP ssASOs and regulation of cellular transcripts by ZAPL/S.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121453119/-/DCSupplemental.
Data Availability
We deposited the raw fastq and the processed files in the National Center for Biotechnology Information’s Gene Expression Omnibus (42) and are accessible through GEO Series accession number GSE203529 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203529).(43)
References
- 1.Gao G., Guo X., Goff S. P., Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297, 1703–1706 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Bick M. J., et al. , Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 77, 11555–11562 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müller S., et al. , Inhibition of filovirus replication by the zinc finger antiviral protein. J. Virol. 81, 2391–2400 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiu H. P., et al. , Inhibition of Japanese encephalitis virus infection by the host zinc-finger antiviral protein. PLoS Pathog. 14, e1007166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y., et al. , Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 15834–15839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo X., Carroll J.-W. N., Macdonald M. R., Goff S. P., Gao G., The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 78, 12781–12787 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerns J. A., Emerman M., Malik H. S., Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 4, 0150–0158 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charron G., Li M. M. H., MacDonald M. R., Hang H. C., Prenylome profiling reveals S-farnesylation is crucial for membrane targeting and antiviral activity of ZAP long-isoform. Proc. Natl. Acad. Sci. U.S.A. 110, 11085–11090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang N., et al. , Viral induction of the zinc finger antiviral protein is IRF3-dependent but NF-kappaB-independent. J. Biol. Chem. 285, 6080–6090 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregersen L. H., et al. , SCAF4 and SCAF8, mRNA anti-terminator proteins. Cell 177, 1797–1813.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwerk J., et al. , RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nat. Immunol. 20, 1610–1620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y. T., et al. , Human cytomegalovirus evades ZAP detection by suppressing CpG dinucleotides in the major immediate early 1 gene. PLoS Pathog. 16, e1008844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M. M. H., et al. , Characterization of novel splice variants of zinc finger antiviral protein (ZAP). J. Virol. 93, e00715-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S., et al. , The SARS-CoV-2 RNA interactome. Mol. Cell 81, 2838–2850.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmer M. M., et al. , The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting. Nat. Commun. 12, 7193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayakawa S., et al. , ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat. Immunol. 12, 37–44 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Todorova T., Bock F. J., Chang P., PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat. Commun. 5, 5362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Perez A. C., et al. , The zinc finger antiviral protein ZAP restricts human cytomegalovirus and selectively binds and destabilizes viral UL4/UL5 transcripts. MBio 12, 1–23 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hetz C., Zhang K., Kaufman R. J., Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21, 421–438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeisberg M., Neilson E. G., Biomarkers for epithelial-mesenchymal transitions. J. Clin. Invest. 119, 1429–1437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roca X., Krainer A. R., Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat. Struct. Mol. Biol. 16, 176–182 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y., et al. , POSTAR2: Deciphering the post-transcriptional regulatory logics. Nucleic Acids Res. 47 (D1), D203–D211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhullar D., Jalodia R., Kalia M., Vrati S., Cytoplasmic translocation of polypyrimidine tract-binding protein and its binding to viral RNA during Japanese encephalitis virus infection inhibits virus replication. PLoS One 9, e114931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cammas A., et al. , Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol. Biol. Cell 18, 5048–5059 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoggins J. W., et al. , A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Todorova T., Bock F., Chang P., PARP13 and RNA regulation in immunity and cancer. Trends Mol. Med. 21, 373–384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dermit M., et al. , Subcellular mRNA localization regulates ribosome biogenesis in migrating cells. Dev. Cell 55, 298–313.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jan C. H., Williams C. C., Weissman J. S., Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346, 1257521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujimoto M., et al. , The HSF1-PARP13-PARP1 complex facilitates DNA repair and promotes mammary tumorigenesis. Nat. Commun. 8, 1638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inada M., Izawa G., Kobayashi W., Ozawa M., 293 cells express both epithelial as well as mesenchymal cell adhesion molecules. Int. J. Mol. Med. 37, 1521–1527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aiello N. M., et al. , EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell 45, 681–695.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gläsker S., Töller M., Kümmerer B. M., The alternate triad motif of the poly(ADP-ribose) polymerase-like domain of the human zinc finger antiviral protein is essential for its antiviral activity. J. Gen. Virol. 95, 816–822 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Ly P. T., Tang S. J., Roca X., Alternative polyadenylation expands the mRNA isoform repertoire of human CD46. Gene 625, 21–30 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Vorlová S., et al. , Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol. Cell 43, 927–939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodier J. L., Pereira G. C., Cheung L. E., Rose R. J., H. H. Kazazian, Jr., The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet. 11, e1005252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S., et al. , Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nat. Struct. Mol. Biol. 19, 430–435 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Lee H., et al. , Zinc-finger antiviral protein mediates retinoic acid inducible gene I-like receptor-independent antiviral response to murine leukemia virus. Proc. Natl. Acad. Sci. U.S.A. 110, 12379–12384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leung A. K. L., et al. , Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42, 489–499 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., et al. , Association of low zinc finger antiviral protein expression with progression and poor survival of patients with hepatocellular carcinoma. Cell. Physiol. Biochem. 49, 1007–1018 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Huang W., et al. , ZC3HAV1 promotes the proliferation and metastasis via regulating KRAS in pancreatic cancer. Aging (Albany NY) 13, 18482–18497 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weston S., et al. , The SKI complex is a broad-spectrum, host-directed antiviral drug target for coronaviruses, influenza, and filoviruses. Proc. Natl. Acad. Sci. U.S.A. 117, 30687–30698 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.P. T. Ly, & X. Roca, National Center for Biotechnology Information’s Gene Expression Omnibus. 21 May 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We deposited the raw fastq and the processed files in the National Center for Biotechnology Information’s Gene Expression Omnibus (42) and are accessible through GEO Series accession number GSE203529 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203529).(43)