Significance
In our work, we address poorly understood mechanisms underlying root-to-shoot communication that lead to a rapid readjustment of shoot growth and development after nitrate provision. Applying a combination of genetics, cell and developmental biology approaches, grafting experiments, and hormonal analytics, we identified a molecular framework orchestrating shoot development with a root nitrate sensory system. We demonstrate that in response to nitrate supply, NLP7, a master regulator of nitrate response in plants, promotes the expression of cytokinin biosynthesis and facilitates translocation of this plant hormone to shoots. There, cytokinin response factors (CRFs) as direct regulators of PINs, stimulate the transport of auxin, and thereby promote shoot growth and development.
Keywords: nitrate, plant development, macronutrient
Abstract
Mineral nutrition is one of the key environmental factors determining plant development and growth. Nitrate is the major form of macronutrient nitrogen that plants take up from the soil. Fluctuating availability or deficiency of this element severely limits plant growth and negatively affects crop production in the agricultural system. To cope with the heterogeneity of nitrate distribution in soil, plants evolved a complex regulatory mechanism that allows rapid adjustment of physiological and developmental processes to the status of this nutrient. The root, as a major exploitation organ that controls the uptake of nitrate to the plant body, acts as a regulatory hub that, according to nitrate availability, coordinates the growth and development of other plant organs. Here, we identified a regulatory framework, where cytokinin response factors (CRFs) play a central role as a molecular readout of the nitrate status in roots to guide shoot adaptive developmental response. We show that nitrate-driven activation of NLP7, a master regulator of nitrate response in plants, fine tunes biosynthesis of cytokinin in roots and its translocation to shoots where it enhances expression of CRFs. CRFs, through direct transcriptional regulation of PIN auxin transporters, promote the flow of auxin and thereby stimulate the development of shoot organs.
Nitrogen, a building block of organic macromolecules such as nucleic acids and proteins, constitutes one of the essential chemical elements determining the growth and development of all organisms (1). Plants, as sessile organisms, rely on absorption of this macroelement from the soil, where it is available in different forms such as nitrate, ammonium, urea, or amino acids. While nitrate represents one of the major inorganic forms of nitrogen in aerobic soil, its availability might dramatically fluctuate in both time and space (2). To face these constraints, plants developed a wide range of adaptive mechanisms triggered by sensing systems to balance nitrate uptake with internal homeostasis (3, 4). At the molecular level, balancing nutrients acquisition with the plant’s requirements implies a close communication between pathways controlling uptake, distribution, and homeostasis of nutrients and the pathways coordinating plant growth and development. Over the last few years, mechanisms of nitrate acquisition, sensing, and signaling have been dissected and key components including the nitrate transceptor NPF6.3/NRT1.1/CHL1 (5–8) and transcriptional regulators of the nitrate response such as NLP6/7, TCP20, LBD37/38/39, SPL9, and TGA1/TGA4 identified. They mediate primary nitrate responses including feedback on the expression of nitrate transport and assimilation genes (9–15).
Flexible adjustment of developmental programs is an essential part of plant adaptation to nitrate availability and a key foraging strategy to optimize its uptake and overcome temporal deficiency of this nutrient. Plant responses to nitrate availability can occur locally, confined to the root organ where nitrate is sensed and absorbed and systemically, involving regulatory signals that elicit responses in distant plant organs (16). The root–shoot–root systemic signaling theme appears to be central in communicating N status and in the coordination of root and shoot organ growth and development according to resources of nitrate (17–20). Mobile signaling molecules such as hormones and peptides have been identified as key components of systemic regulatory pathways (19–23). Among them, cytokinin has been proposed as a principal hormonal integrator, adjusting shoot development including apical meristem activity and leaf expansion to nitrate status sensed by the root system (20, 24–29). While cytokinin’s role as an important component of the systemic nitrate response pathway is firmly established, little is known about the downstream effectors that coordinate the early shoot adaptive response to nitrate. Likewise, the role of auxin and polar auxin transport (PAT), as a core hormonal machinery coordinating plant growth and developmental processes, has been associated with plant adaptation to nitrate (9, 30). However, insights into molecular mechanisms underlying regulation of PAT by nitrate and the physiological role of its components in plant adjustment to nitrate availability are scarce.
Here, we identified a molecular framework that integrates inputs of nitrate and cytokinin regulatory pathways to effectively adjust early shoot development according to the nitrate availability sensed by the root system. We demonstrate that nitrate, through NLP7-mediated signaling in roots, enhances biosynthesis and transport of cytokinins to shoots. Cytokinin response factors (CRFs), encoding APETALA2/ERF transcription factors, provide a readout of cytokinin levels in shoots, and through a fine-tuning expression of PIN auxin efflux carriers steer the flow of auxin, thereby promoting the growth of shoot organs.
Results
Developmental Adaptation to Nitrate Involves Adjustment of PIN Auxin Transporter Expression in the Shoot.
Plants use nitrate (NO3−) and ammonium (NH4+) as major sources of inorganic nitrogen, although their preference for different forms might differ. Fluctuations in both concentrations and the form of nitrogen available in the soil have prominent effects on plant growth and development. To explore how Arabidopsis responds and adapts to different forms of nitrogen, seedlings were grown on ammonium succinate (AS) as an exclusive nitrogen source for 7 d and afterward transferred to media containing either AS or nitrate (day of transfer is indicated as D0). The growth of seedlings was monitored for 1, 4, and 14 d after transfer (DAT). As previously reported, nitrate promoted the growth of primary roots (PRs) and lateral roots (LRs), but not the density of LRs when compared to roots on media supplied with AS (SI Appendix, Fig. S1A–C) (31–33). To examine developmental responses of shoot organs, the surface area of cotyledons and fresh weight of shoots 1 and 14 DAT to nitrate, respectively, were quantified. Embryonic leaves of seedlings 1 DAT on medium supplied with nitrate expanded significantly more when compared to those supplemented with AS (Fig. 1A and B). Consistently, 14 DAT fresh weight of shoots was significantly higher in seedlings supplied with nitrate than with AS (SI Appendix, Fig. S1D). Hence, consistently with reports, nitrate provided to seedlings triggers overall modulation of plant development, including root and shoot organs . At the molecular level, the transfer of seedlings from AS to nitrate-supplemented medium resulted in rapid up-regulation (within 1 h) of the early nitrate responsive genes, including HRS1 and TGA1 in roots (9, 35, 36) (SI Appendix, Fig. S1E).
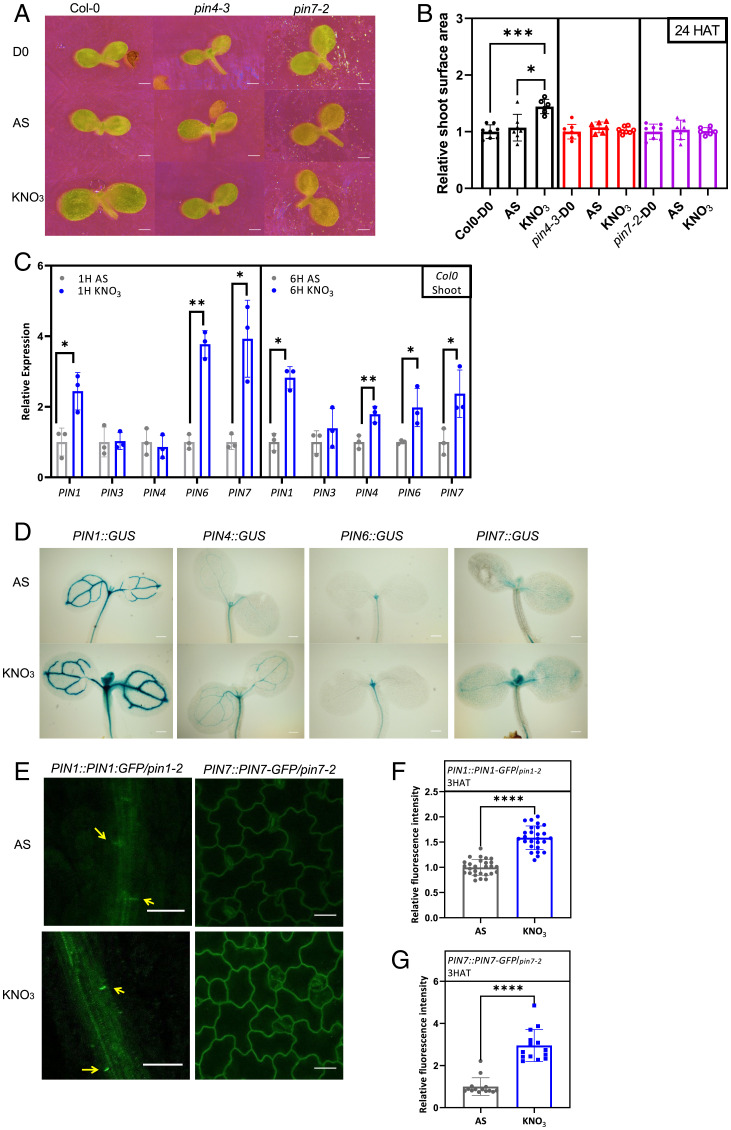
Fig. 1.
Modulation of PIN expression is part of nitrate-mediated transcriptional reprogramming. (A and B) Imaging (A) and surface area quantification (B) of cotyledons from 7-d-old wild-type (Col 0), pin4-3, and pin7-2 seedlings grown on AS (0.5 mM) (D0), and 24 HAT to either ammonium (AS)- or nitrate (5 mM KNO3)-containing media. Relative size was quantified as mean surface area of cotyledons on AS or KNO3 24 HAT compared to D0 (n = 5 to 8 seedlings per treatment). The experiment was repeated three times; the result from one representative experiment is presented. Significant differences were determined by one-way ANOVA followed by a significant Tukey’s multiple comparison test; *P < 0.05, ***P < 0.001. (C) qRT-PCR expression analysis of PIN genes normalized to UBQ10 (AT4G05320) in Col 0 shoots, 1 and 6 HAT to AS- or KNO3-containing media. All qRT-PCR reactions were carried out with biological and technical triplicates. Statistical difference was determined by Student’s t test; *P < 0.05, **P < 0.01. (D) Expression analyses of PIN::GUS reporters in shoots of 7-d-old seedlings, 6 HAT to AS and KNO3. (E–G) Monitoring (E) and quantification of PIN1::PIN1-GFP (F) and PIN7::PIN7-GFP (G) membrane signal in vasculature (yellow arrows) and adaxial epidermal cells of cotyledons, respectively, 3 HAT on AS- or KNO3-supplemented media. PIN-GFP signal measured in n = 2 epidermal cells, and at least two cells of vasculature in six to seven seedlings per treatment. Significant differences were determined by Student’s t test; ****P < 0.0001 (F and G). (Scale bars in A, 400 μm; D, 200 μm; and E, 24 μm.)
Auxin and its graded distribution play an instructive function in all plant growth and developmental processes (37–40). Thus, we asked whether and/or how the auxin transport mediated through PIN auxin efflux carriers contributes to the developmental adaptation of plant organs to nitrate provision. qRT-PCR analyses revealed relatively modest alterations of PIN transcription in roots after transfer of seedlings to nitrate-supplemented medium. Among tested PINs, PIN3 expression was significantly increased in roots 1 and 6 h after seedlings were transferred on nitrate when compared to AS-containing medium, while PIN7 transcription was attenuated 6 hours after transfer (HAT) (SI Appendix, Fig. S1F). Intriguingly, provision of nitrate triggered a profound response in shoots, in which transcription of several PIN genes, including PIN1, PIN4, PIN6, and PIN7, was enhanced 1 HAT and/or 6 HAT to nitrate when compared to AS (Fig. 1C). Analysis of PIN::GUS reporter lines corroborated the qRT-PCR results (Fig. 1D and SI Appendix, Fig. S1G). The nitrate-promoted expression of PIN genes correlated with the abundance of PIN proteins quantified in PIN1::PIN1-GFP and PIN7::PIN7-GFP reporter lines. Significantly higher PIN1-GFP and PIN7-GFP signals at the plasma membrane (PM) of vasculature and epidermal cells at the adaxial side of cotyledons, respectively, were detected in seedlings provided by nitrate than by AS (Fig. 1E–G).
Hence, early phases of seedling adaptation to nitrate provision might be accompanied by adjustment of polar auxin transport in shoots. To examine whether PIN-mediated auxin transport is essential for shoot growth enhancement triggered by nitrate, mutants in PIN genes were analyzed. To dissect alterations in adaptive responses to nitrate provision from general developmental defects that potentially might be caused by the lack of the respective PIN function, growth response was calculated as a difference in cotyledon size at D0 versus D1. Unlike wild type, no significant increase in the size of cotyledon surface area 1 DAT to nitrate when compared to AS was detected in the pin4 and pin7 mutants (Fig. 1A and B). Consistently, 14 DAT to nitrate, shoot fresh weight of pin mutants was significantly lower when compared to wild-type control (SI Appendix, Fig. S1H–J). Intriguingly, no pronounced shoot developmental defects in either pin4 or pin7 mutants have been reported previously (41–44), presumably because of the functional redundancies among PINs. Hence, to examine whether functionally redundant PINs mitigate deficiency in shoot growth at optimal nutrient conditions more efficiently than under nitrate deficiency, we performed an additional set of experiments. We grew wild-type and pin mutants for 21 d on 0.5×Murashige and Skoog (MS) medium. In parallel, to obtain a direct comparison, wild type and mutants were grown for 7 d on a medium supplemented with AS and transferred to nitrate for 14 d. Whereas no significant difference in shoot fresh weight was detected between wild-type and pin4 and pin7 grown on the MS medium, shoot fresh weight gain was significantly lower in pin4 and pin7 than in wild type when seedlings were transferred to nitrate (SI Appendix, Fig. S1K and L). This suggests that intact PIN-mediated transport is essential for plants to recover from the stress caused by the initial growth under suboptimal nitrogen supply (AS), and effectively adjust their development to nitrate provision.
Cytokinin Response Factors Mediate Shoot Developmental Adaptations to Nitrate.
Up-regulation of the PIN transcription in shoots after the provision of nitrate motivated us to screen for upstream regulators involved in this response. CRFs, which have been identified as direct transcriptional regulators of PINs (45), were detected among the early nitrate responsive genes in several transcriptome profiling data (9, 14, 29). In agreement with these reports, qRT-PCR analyses confirmed up-regulation of CRF2 and CRF6 in shoots provided with nitrate, when compared to AS (Fig. 2A and B), whereas in roots no significant changes could be detected (SI Appendix, Fig. S2A). Consistently with qRT-PCR, staining of CRFs::GUS reporters corroborated nitrate responsive expression of CRFs in shoots (Fig. 2C).
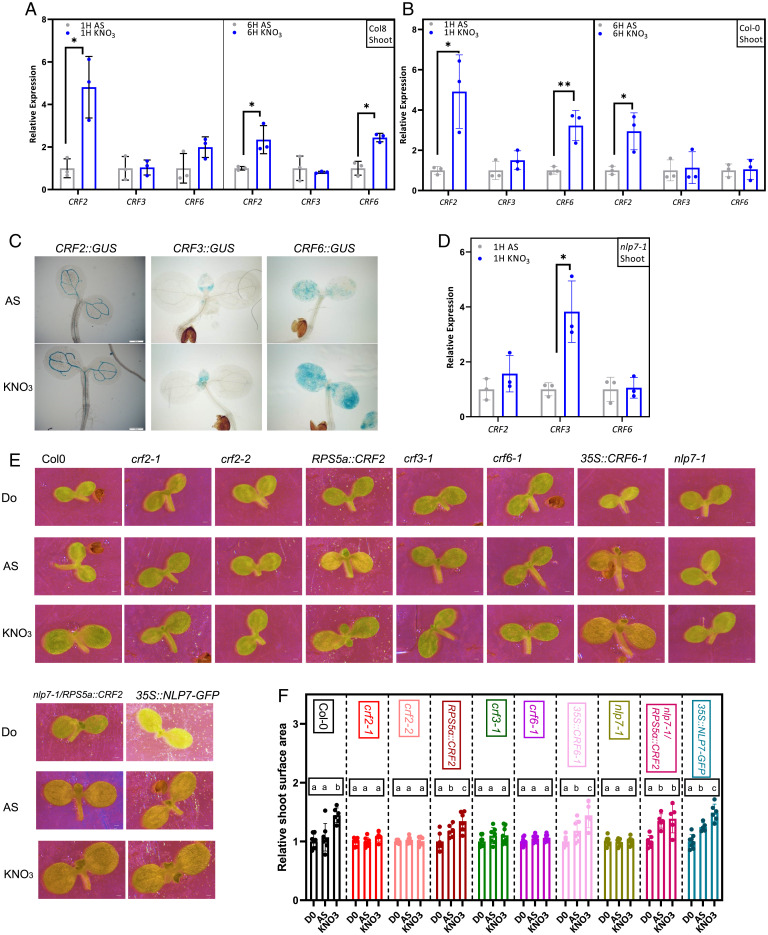
Fig. 2.
Cytokinin response factors mediate shoot developmental adaptations to nitrate. (A–D) Expression analyses of CRFs using qRT-PCR (A, B, and D) and CRF::GUS reporters (C) in wild-type (Col 0) (A), (Col 8) (B), and nlp7 (D) shoots of 7-d-old seedlings 1 HAT (A–D) and 6 HAT (A and B) to AS and KNO3. Expression of CRF genes normalized to UBQ10 (AT4G05320) 1 HAT and 6 HAT on AS- or KNO3-containing media. All qRT-PCR reactions were carried out with biological and technical triplicates. Statistical difference was determined by Student’s t test; *P < 0.05, **P < 0.01. (E and F) Imaging (E) and surface area quantification (F) of cotyledons from 7-d-old wild type (Col 0), mutants, and CRF overexpressors grown on AS (D0), and 24 HAT to AS-or KNO3-containing media. Relative size was quantified as mean surface area of cotyledons on AS or KNO3 24 HAT compared to D0 (n = 5 to 8 seedlings per treatment). The experiment was repeated twice; the result from one representative experiment is presented. Significant differences were determined by one-way ANOVA followed by Tukey’s multiple comparison test; lowercase letters indicate significant differences of at least *P < 0.05. (Scale bars in C , 200 μm and E, 100 μm.)
NLP7, a NIN-like transcription factor, is a master regulator of plant adaptation to nitrate. Nitrate triggers nuclear retention of NLP7 protein, where it activates transcriptional responses (SI Appendix, Fig. S2B and C) (10, 12). To examine whether nitrate-promoted expression of CRFs is dependent on NLP7-mediated signaling we analyzed the nlp7 loss-of-function mutant. Unlike wild type, neither CRF2 nor CRF6 transcription was enhanced in shoots or roots of nlp7 seedlings 1 HAT to nitrate, when compared to AS (Fig. 2D and SI Appendix, Fig. S2D). These data suggest that transcriptional modulation of CRFs is part of the specific plant adaptive responses to nitrate mediated by the core nitrate signaling machinery (9, 14, 46).
To further explore the function of CRFs in nitrate-regulated plant growth, seedlings with the modulated activity of CRFs were examined. Similar to nlp7 mutants, which are defective in the shoot growth response to nitrate provision (Fig. 2E and F and SI Appendix, Fig. S2E and F) (10, 47), loss of CRF function affected seedling sensitivity to nitrate provision. The nitrate-promoting effects on cotyledon expansion 1 DAT were reduced in multiple crf2, crf3, crf6, and single crf2, crf3, and crf6 mutants when compared to wild type (Fig. 2E and F). Shoot fresh weight 14 DAT was strongly reduced in crf6 and crf2, crf3, crf6 mutants when compared to wild type (SI Appendix, Fig. S2G and H). In contrast to the attenuated response of the crf mutants to nitrate, seedlings expressing CRF2 and CRF6 driven by constitutive RPS5a or 35S promoter, respectively, displayed enhanced expansion of cotyledons and shoot growth irrespective of nitrate provision (Fig. 2E and F and SI Appendix, Fig. S2I and J). In addition, RPS5a::CRF2 introduced into the nlp7 background resulted in enhanced cotyledon expansion and shoot growth when compared to nlp7 supplemented with either AS or nitrate (Fig. 2E and F and SI Appendix, Fig. S2I and J).
These data strongly support the role of CRFs as molecular components of the pathway that, downstream of NLP7-mediated signaling, regulate plant growth adaptation to nitrate provision.
NLP7 Fine Tunes Expression of PINs and Shoot Developmental Adaptation to Nitrate through CRFs.
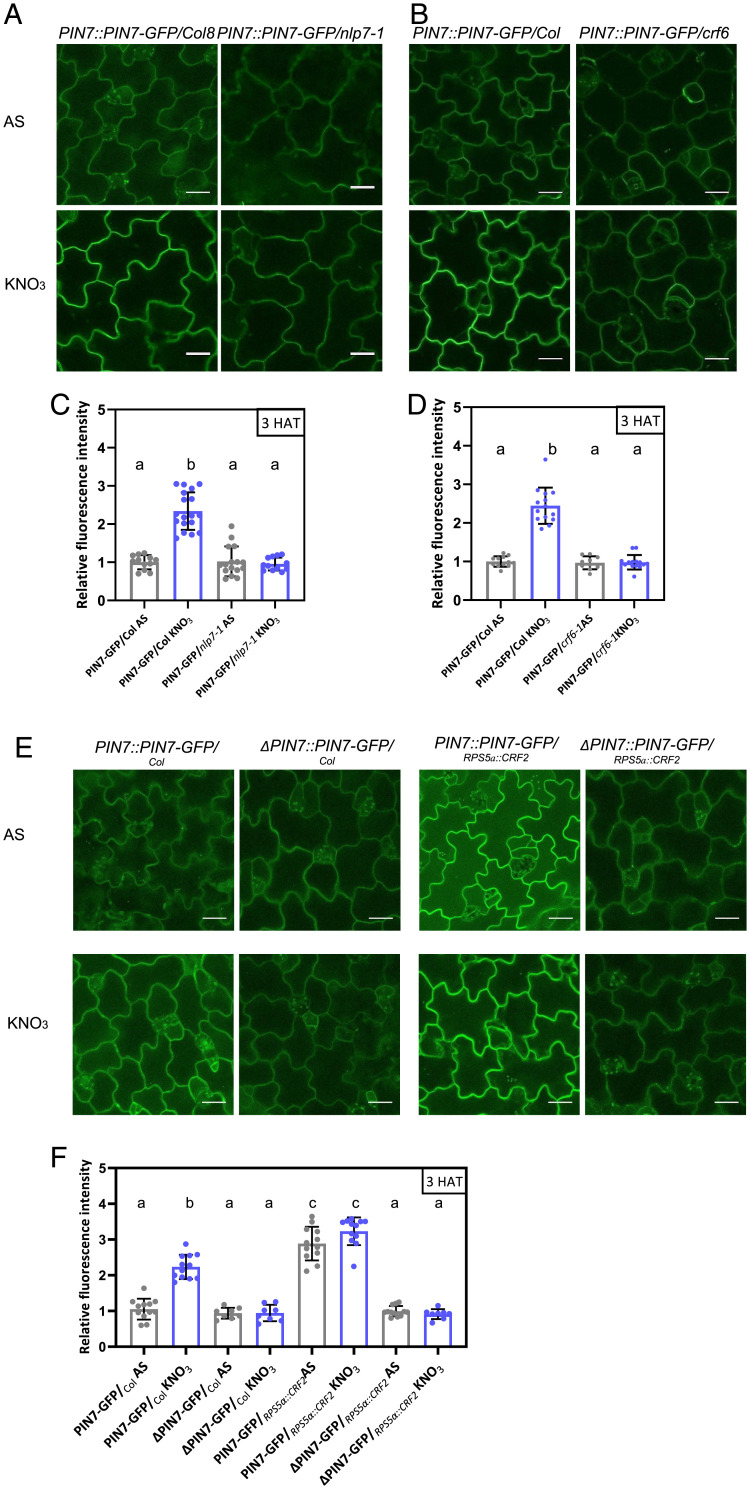
To examine whether nitrate promotes the expression of PINs in shoots through NLP7-mediated regulation of CRFs, crf and nlp7 mutants were examined closely. Unlike wild type, in which supply of nitrate led to a significant increase in shoot expression of several PINs, in nlp7 and crf2 mutants, transcription of PINs was largely unaltered 1 HAT to nitrate when compared to AS (SI Appendix, Fig. S3A and B compared to Fig. 1C). PIN7-GFP and PIN1-GFP signals at the PM of epidermal and vascular cells in cotyledons in nlp7, crf6, and crf2 mutants, respectively, remained unaffected after transfer to nitrate, thus further corroborating the role of NLP7 and CRFs in the adjustment of PIN-mediated transport during shoot response to nitrate provision (Fig. 3A–D and SI Appendix, Fig. S3C–G). Conversely, in RPS5a::CRF2 seedlings, an elevated PIN7-GFP abundance at the PM of epidermal cells in cotyledons was detected irrespectively of the nitrogen source. The strength of the PIN7-GFP signal in cotyledons of the RPS5a::CRF2 seedlings was significantly higher than that detected in wild-type seedlings supplemented with nitrate (Fig. 3E and F). CRFs control PIN transcription through binding to cytokinin response elements (PCRE) in their promoters (45). Notably, nitrate provision did not enhance the expression of PIN7-GFP driven by the ΔPIN7 promoter, which lacks the CRF binding domain in cotyledons of wild-type as well as RPS5a::CRF2 seedlings (Fig. 3E and F). In line with the role of CRF-mediated regulation of PINs in developmental adaptation of shoots to nitrate provision, neither ΔPIN7::PIN7:GFP nor ΔPIN1::PIN1:GFP was able to recover cotyledon growth of pin7 and pin1 mutants in response to nitrate provision (SI Appendix, Fig. S3H and I).
Fig. 3.
NLP7 fine tunes expression of PINs and shoot developmental adaptation to nitrate through CRFs. (A–F) Imaging (A, B, and E) and quantification (C, D, and F) of PIN7-GFP membrane signal in adaxial epidermal cells of cotyledons 3 HAT to medium supplemented with AS or KNO3 in nlp7-1 (A and C), crf6-1 (B and D), and PIN7::PIN7-GFP, Col 0 and ΔPIN7::PIN7-GFP,RPS5a::CRF2 (E) lines. Different lowercase letters indicate significant difference at one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05). PIN7-GFP signal measured in n = 2 epidermal cells, in at least five seedlings per treatment. Experiments were repeated at least twice; the result from one experiment is presented (C, D, and F). (Scale bars in A, B, and E, 24 μm.)
To gain further insights into the mechanism underlying nitrate-promoting effect on the cotyledon expansion, we examined growth at the cellular level. For these purposes, we observed pavement cells at the adaxial side of cotyledons at D0 and D1 after transfer to either AS- or nitrate-supplemented medium. Pavement cells of wild-type seedlings supplied with nitrate displayed a pronounced jigsaw puzzle–like cell pattern and expanded significantly more when compared to those supplemented with AS. Mutants in NLP7, CRF2, PIN4, and PIN7 were severely affected in expansion and patterning of pavement cells in response to nitrate, when compared to wild type (SI Appendix, Fig. S4).
Altogether, these results support a role of the NLP7-CRF-PIN module in shoot adaptation to nitrate and indicate that adjustment of the PIN-mediated auxin transport is important for the auxin-driven expansion and patterning of pavement cells.
NLP7-Mediated Signaling in Roots Controls Cytokinin Levels in Shoots.
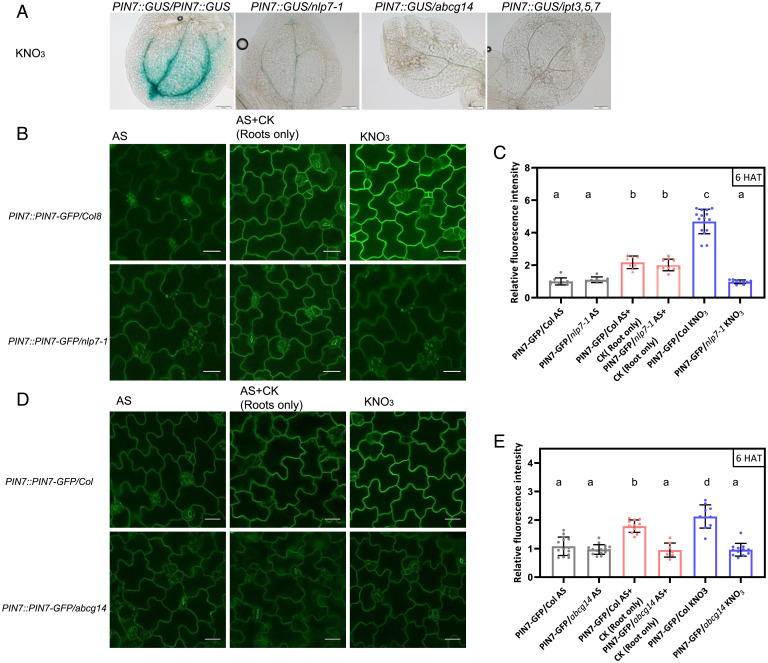
Our analyses indicated that enhanced growth of shoots in response to nitrate is dependent on NLP7/CRF-mediated regulation of PIN-directed auxin transport. Intriguingly, while the root is the principal organ for nitrate uptake, significant alterations in the transcription of PINs by nitrate supply were detected in shoots. This raised a question as to whether adjustment of PIN-mediated auxin transport and shoot growth in response to nitrate might involve some mobile signal translocated from roots. NLP7 is a core molecular component of the nitrate signaling pathway, whose loss of activity severely affects shoot growth in response to nitrate (10, 47) (Fig. 2E and F and SI Appendix, Fig. S2E and F). To examine whether proper uptake and sensing of nitrate in roots can recover shoot development of nlp7, we implemented grafting experiments. Roots and shoots of 7-d-old wild-type and nlp7-1 seedlings germinated on media supplemented with AS were separated, and using a sealing tube, various grafting combinations were performed. Grafted seedlings were transferred to nitrate-containing medium and monitored for 14 d. As expected, shoots of Col 8 wild-type grafted on Col 8 roots (ColR/ColS) displayed enhanced development and bigger size when compared to shoots of nlp7 grafted on the mutant roots (nlp7R/nlp7S). Notably, the partial rescue of the nlp7 shoot development was detected in grafts with Col 8 roots, while Col 8 shoots grafted on roots of nlp7 mutant displayed retarded development when compared to ColR/ColS grafts (SI Appendix, Fig. S5A). This suggested that nitrate, in an NLP7-dependent manner, promotes translocation of some mobile signal from roots to above-ground organs where it coordinates shoot development.
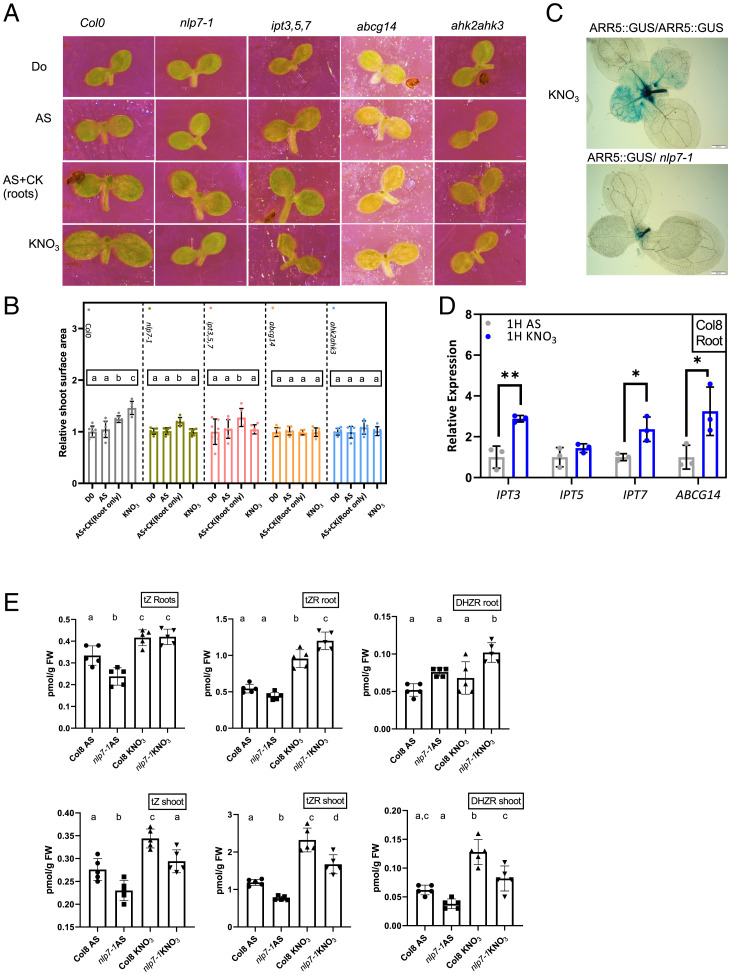
Cytokinin is an established positive hormonal regulator of shoot growth, strongly associated with adaptive responses to nitrate (20–22, 48). Mutants defective either in cytokinin perception (ahk2 and ahk3), long-distance root-to-shoot cytokinin transport (abcg14), or biosynthesis (ipt3,5,7) are strongly affected in shoot growth adaptation to nitrate provision (Fig. 4A and B and SI Appendix, Fig. S5B and C). Analysis of the early cytokinin response gene ARR5 revealed that 6 HAT of seedlings from AS to nitrate-containing medium expression of ARR5::GUS reporter in shoots is enhanced (SI Appendix, Fig. S5D). To examine whether nitrate promotes the expression of ARR5 in an organ-autonomous manner or through the root-derived hormonal signal, the ARR5::GUS shoots were grafted on roots of ARR5::GUS as well as on the roots of abcg14 and ipt3,5,7 mutants affected in cytokinin long-distance transport and biosynthesis, respectively. ARR5::GUS reporter expression was stronger in shoots grafted on wild-type roots when compared to shoots grafted on either abcg14 or ipt3,5,7 roots (SI Appendix, Fig. S5E). Consistently with previous studies (20–22), these results suggest that also in our system cytokinin acts as a mobile signal that is transported to shoots, upon transfer of seedlings to nitrate. Recently, measurements of cytokinins in the nlp7,nlp6 seedlings revealed reduced levels of the trans-Zeatin (tZ) type of cytokinins when compared to wild type after nitrate supply hinting at their role in regulation of cytokinin biosynthesis (41). To explore whether a reduced amount of cytokinin transported from roots to shoots might underlie the retarded development of nlp7 shoots despite sufficient nitrate availability, ARR5::GUS shoots were grafted on nlp7 roots. Expression of ARR5::GUS remained low in shoots grafted on roots of the nlp7 mutants when compared to those grafted on control roots (Fig. 4C). These results suggest that NLP7 signaling activated by nitrate in roots is involved in the control of cytokinin translocation to shoots where it promotes their growth.
Fig. 4.
NLP7-mediated signaling in roots controls cytokinin levels in shoots. (A and B) Imaging (A) and surface area quantification (B) of cotyledons from 7-d-old wild type (Col 0), nlp7-1, ipt3,5,7, abcg14, and ahk2ahk3 seedlings grown on AS (D0), and 24 HAT to either AS, AS supplemented with cytokinin (CK, 100 nM 6-benzylaminopurine), and KNO3-containing media. Relative size was quantified as mean cotyledon surface area on AS, AS plus CK, or KNO3 24 HAT compared to D0 (n = 6 to 8 seedlings per treatment). The xperiment was repeated at least twice; results from one representative experiment are presented. Significant differences were determined by one-way ANOVA followed by Tukey’s multiple comparison test; different lowercase letters indicate P < 0.05 (B). (C) Representative images of ARR5::GUS expression in shoots of 7-d-old seedlings grafted on Col 0 or nlp7-1 roots. Grafted seedlings after 72 h of healing on AS medium were then transferred to KNO3 for 24 h. (D) qRT-PCR analysis of IPT genes and ABCG14 expression normalized to UBQ10 (AT4G05320) in Col 8 roots 1 HAT to AS- or KNO3-containing media. All qRT-PCR reactions were carried out with biological and technical triplicates. Statistical difference was determined with a t test; *P < 0.05,**P < 0.01 . (E) Quantification of tZ, tZR, and DHZR in roots and shoots of Col 8 and nlp7-1 seedlings 6 HAT to AS- or KNO3-containing medium. Different lowercase letters indicate significant one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05) (n = 5 biological replica/genotype/treatment). (Scale bars in A, 100 μm and C, 200 μm.)
NLP7-Mediated Nitrate Signaling Controls Cytokinin Levels in Shoot.
Weaker ARR5::GUS expression in shoots grafted on nlp7 roots when compared to those on control roots indicates that NLP7-mediated nitrate signaling in roots plays a critical role in regulating cytokinin provision to the shoot.
In agreement with previous reports (20–22, 29, 41, 49, 50), qRT-PCR analyses showed that in wild-type seedlings, root expression of IPT3, IPT7, and ABCG14 is enhanced 1 HAT to nitrate. In shoots, no major changes in expression of either cytokinin biosynthesis or transport genes were observed 1 HAT to nitrate, relative to AS transfer, in line with a note that cytokinin acts as a mobile signal to mediate root-to-shoot communication (Fig. 4D and SI Appendix, Fig. S4F). Unlike wild type, no increase in IPT3 and ABCG14 expression by nitrate was detected in nlp7 roots or shoots, supporting NLP7 function in fine tuning cytokinin biosynthesis and transport in roots supplied with nitrate (SI Appendix, Fig. S4G and H).
To further investigate the role of NLP7 in cytokinin biosynthesis and/or transport to shoots, we examined whether the provision of cytokinins locally to roots can recover retarded shoot development in the nlp7 mutant. To validate the experimental setup we first monitored cytokinin effects on shoot recovery in ipt3,5,7, abgc14, and ahk2,ahk3 mutant. Cytokinin applied to roots promoted shoot growth in the ipt3,5,7 mutant defective in cytokinin biosynthesis, while no enhancement could be detected in abcg14 or ahk2,ahk3 mutants in transport and perception, respectively. Notably, cytokinin applied to roots promoted shoot growth in nlp7, thus hinting at the role of NLP7 in the regulation of root-derived cytokinins to coordinate shoot growth (Fig. 4A and B).
In summary, NLP7-mediated regulation of cytokinin biosynthesis and transport derived from roots is an important part of the shoot developmental adaptation to nitrate provision.
Nitrate-Dependent Cytokinin Biosynthesis and Distribution Are Affected in nlp7.
To examine the role of NLP7 in the adjustment of cytokinin levels to nitrate provision to shoots we performed hormonal analyses in wild type and nlp7 mutant 6 HAT to either AS- or nitrate-supplemented medium. In agreement with previous reports (20, 41, 49, 50) we found that provision of nitrate resulted in significant increase of biologically active cytokinin tZ, as well as its transport form transzeatin riboside (tZR) and dihydrozeatin riboside (DHZR) in both roots and shoots of wild-type seedlings, when compared to those supplemented with AS (Fig. 4E). Provision of nitrate also enhanced accumulation of isopentenyladenosine-5′-monophosphate (iPRMP) and isopentenyladenosine (iPR), but not isopentenyladenine (iP) in shoots when compared to those on AS (SI Appendix, Fig. S4I and Table S1). Levels of cis-zeatin (cZ) and its derivatives were not altered by nitrate provision, indicating that the pathway encompassing cytokinin synthesis from the tRNA-IPT pathway is not significantly contributing to the adjustment of the cytokinin pool to nitrate availability (SI Appendix, Table S1). Likewise, levels of biologically inactive or storage forms of cytokinins such as O-, N7-, N9-glucosides remained largely unaffected by nitrate provision, suggesting that increased levels of active cytokinins after transfer to nitrate do not originate from the conversion of their storage counterparts (SI Appendix, Table S1). Overall, measurements of cytokinin derivatives in wild-type seedlings are consistent with previous reports demonstrating that provision of nitrate is accompanied by enhanced biosynthesis of cytokinins, and in particular tZ types of cytokinins, which are translocated to shoots via the xylem (20, 49, 51).
Unlike wild type, transfer of nlp7 to nitrate resulted in significantly lower accumulation of tZ, tZR, and DHZR in shoots (Fig. 4E). Noteworthy, levels of tZR and DHZR in roots of nlp7 mutants 6 HAT to nitrate were higher than in wild-type roots, suggesting that their transport from roots to shoots might also be affected in the mutant background (Fig. 4E). Surprisingly, in nlp7 mutants supplemented with AS, increased levels of iP in both roots and shoots were detected when compared to wild type, hinting at activation of a mechanism compensating for low levels of tZ cytokinins in the nlp7 mutant (SI Appendix, Fig. S4I and Table S1). Provision of nitrate led to reduction of the iP forms in nlp7 presumably due to their conversion of tZ triggered by nitrate. No dramatic alterations in amounts of cytokinin conjugates in nlp7 mutant on AS versus nitrate could be detected, in line with the assumption that modulation of cytokinin levels in response to nitrate mediated by NLP7 does not involve reactivation of storage forms of cytokinins (SI Appendix, Table S1). Overall, these data support the previously established role of cytokinin as signaling molecule involved in plant adaptation to nitrate availability (20–22, 26, 41, 49, 52) and show that NLP7, in addition to fine tuning cytokinin metabolism (41), might also play an important role in its distribution according to nitrate levels.
Adjustment of PIN Expression and Shoot Growth by Nitrate Is Coordinated by Cytokinin Derived from Roots.
To test whether shoot expression of PIN genes in seedlings transferred to nitrate is dependent on cytokinin transported from roots, we grafted shoots of the PIN7::GUS reporter line on roots of wild type, ipt3,5,7, or abcg14, respectively. Expression of PIN7::GUS in grafts on control roots was stronger when compared to shoots grafted on ipt3,5,7, abcg14, or nlp7 roots, in line with the assumption that cytokinin is a mobile signal produced in roots supplied with nitrate and transported to shoots where it might activate expression of PINs (Fig. 5A). Consistently, expression of PIN7::PIN7-GFP was enhanced in cotyledons of seedlings that were transferred to media supplemented with AS and had cytokinin applied locally to roots. In the nlp7 mutant, the cytokinin treatment enhanced PIN7-GFP signal in epidermal cells of cotyledons (Fig. 5B and C), whereas in the abcg14 mutant no enhancement of PIN7-GFP abundance was observed under this condition (Fig. 5D and E).
Fig. 5.
Nitrate-dependent fine tuning of PIN expression in shoots is driven by cytokinin translocated from roots. (A) Representative images of PIN7::GUS expression in shoots grafted on roots of PIN7::GUS, nlp7, abcg14, or ipt3,5,7 roots. Grafted seedlings after 48 h of healing on AS, then transferred to KNO3-containing medium for 24 h. (B–E) Imaging (B and D) and quantification (C and E) of PIN7-GFP membrane signal in adaxial epidermal cells of cotyledons 6 HAT on AS, AS plus cytokinin (CK, 100 nM 6-benzylaminopurine), or KNO3-supplemented media in nlp7-1 (B and C) and abcg14 (D and E). PIN7-GFP signal was measured in n = 2 epidermal cells, four different seedlings per treatment. Experiments were repeated at least twice; results from one experiment are presented. Different lowercase letters indicate significant one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05) (C and E). (Scale bars in A, 100 μm and B and D, 24 μm.)
Enhanced shoot growth was observed in PIN1::PIN1-GFP/pin1 and PIN7::PIN7-GFP/pin7 seedlings provided with AS and with cytokinin applied to roots. In contrast, PIN-GFP driven by promoters lacking elements recognized by CRFs, such ΔPIN1 and ΔPIN7, were unable to recover pin1 and pin7 shoot growth when cytokinin was added to AS-containing medium (SI Appendix, Fig. S3G and H). These data support a model where modulation of PIN expression, driven by cytokinins translocated from roots, play an important role in controlling shoot growth adaptation to nitrate.
Discussion
Over the last few years, nitrate-dependent regulation of several components of PAT, including Aux/LAX, PINs, and ABCBs efflux transporter has been reported (29, 33, 53–55). However, insights into molecular mechanisms underlying regulation of PAT by nitrate and its function in plant developmental adaptations to nitrate availability and nitrogen status are scarce. Interestingly, although the root is the primary organ for nitrate uptake and sensing, we detected a profound nitrate-promoted transcription reprogramming of PINs in shoot organs, pointing at the role of the PIN-mediated auxin transport in shoot adaptive responses to nitrate provision. Consistently with this assumption, interference with the activity of PIN genes significantly attenuated stimulatory effects of nitrate on shoot development when compared to wild-type seedlings.
CRFs, a subgroup of transcription factors from the AP2/ERF family, emerged as potential molecular components underlying convergence of nitrate signaling and the PAT from the overlap between nitrate responsive transcriptomes (9, 12, 14, 29, 56) and transcriptional regulators of PIN genes identified by the yeast one-hybrid screen (45). Nitrate-sensitive transcription of CRF genes in shoots, deficiency of the respective mutants to respond to nitrate provision by enhanced shoot growth, and promoting effects of CRF2 overexpression on shoot growth irrespective of nitrate supply, collectively, support a role of these transcription factors in the regulation of shoot developmental responses to nitrate. Importantly, nitrate-stimulated expression of PINs is attenuated in crf mutants, or when the PIN promoters are deprived of the cis-elements targeted by CRFs in the wild type as well as RPS5a::CRF2 background, pointing at CRF-PINs as a regulatory module coordinating shoot developmental adaptation to nitrate.
The CRF-mediated regulation of PIN expression in shoots needs to be tightly coordinated with processes such as nitrate absorption, distribution, and signaling occurring primarily in the root. Cytokinins have been identified as one of the principal long-distance signaling molecules that mediate root-to-shoot communication about the nitrate status (20–22, 26, 49). Nitrate provision has been reported to induce expression of cytokinin biosynthesis genes, such as IPT3, and root-to-shoot translocation of cytokinins (21, 22, 29, 41, 49). Our expression analysis and measurements of cytokinins revealing up-regulation of cytokinin biosynthesis and transport genes after transfer to nitrate in comparison with AS, and enhanced accumulation of cytokinin derivatives such as tZ, tZR, and DHZR, are fully consistent with these reports (20, 43, 49). In light of these findings, CRFs as transcription factors whose expression is stimulated by cytokinins (46), are plausible candidates for regulators that, in response to cytokinins translocated to shoots, adjust the activity of the PAT.
However, a question arises about the molecular components and pathways underlying adjustment of cytokinins to nitrate availability. Our analyses of the mutant lacking functional NLP7, a master regulator of the nitrate response, revealed that deficient shoot response of nlp7 to nitrate provision can be partially recovered by either its grafting on wild-type roots, cytokinin supply to roots, or expression of CRF2 under a constitutive promoter. Altogether, these observations suggest that NLP7 impinges on the regulation of cytokinin metabolism in roots or transport of the hormone to shoots. In line with these assumptions, expression of the cytokinin biosynthetic gene IPT3 in roots is stimulated by nitrate in an NLP7-dependent manner. Interestingly, loss of NLP7 did not significantly interfere with nitrate-induced transcription of IPT7, thus hinting at potential redundancies in the regulation of the cytokinin biosynthetic genes by NLP7 homologs (12, 56). Furthermore, enhanced transcription of ABCG14 in response to nitrate might be part of a positive feedback loop to reinforce the transport of cytokinins to the shoot that seems to be dependent on NLP7 function. Analyses of cytokinins in nlp7 mutants when compared to wild type further strongly support the role of NLP7 in the adjustment of cytokinin biosynthesis and distribution according to nitrate availability. Thus, our findings extend the previously recognized link between nitrate and cytokinins with NLP7 as a molecular component (20–22, 41), which converges both regulatory pathways and coordinate distribution of this hormone between roots and shoots.
Recently, chromatin immunoprecipitation sequencing (ChIP-seq) experiments predicted several CRF promoters to be directly bound by NLP7 upon its translocation into the nucleus (12, 56). Additionally, the PIN7 promoter has been detected among the potential genes directly regulated by NLP7 in the protoplast system (56, 57). Thus, alternatively, NLP7 might regulate PIN7 locally in tissues and organs where their expression coincides. Our findings do not rule out a contribution of such a direct interaction in addition to NLP7-mediated enhancement of cytokinin activity and transport from root to shoot, which might provide an efficient way for the robust CRF-PIN activation in the early phases of the shoot adaptation to nitrate.
Altogether, our study reveals a molecular framework orchestrating shoot developmental processes with a root nitrate sensory system. In response to nitrate supply, NLP7 promotes the expression of cytokinin biosynthesis and facilitates cytokinin translocation to shoots. There, CRFs as direct regulators of PINs, stimulate the transport of auxin and thereby promote shoot growth and development.
Materials and Methods
Plant Material.
All Arabidopsis thaliana lines used in this study are in the Columbia ecotype, detailed information of which is provided in SI Appendix, Materials and Methods.
Growth Conditions.
Seeds of A. thaliana sterilization and seedling growth conditions used in this study and external hormonal treatment with cytokinin (100 nM 6-benzylaminopurine [BAP]) are detailed in SI Appendix, Materials and Methods.
Phenotype Analyses of Primary Roots, Lateral Roots, and Cotyledon Surface Area.
Seven-day-old seedlings grown on 0.5 mM AS containing medium were transferred on media supplemented with either 0.5 mM AS, or 5 mM KNO3. Seedlings were imaged using an Epson Perfection V700 flatbed scanner on respective time points for primary root length. For lateral root density and cotyledon surface area measurements an Olympus BX53 microscope and the Leica EZ4HD were used, respectively. Detailed descriptions are provided in SI Appendix, Materials and Methods.
Pavement Cell Imaging and Cell Size Measurements.
For observation of cotyledon pavement cells, the adaxial side of the cotyledon surface was placed on melted 2% (wt/vol) low-melt agar containing 0.01% (wt/vol) bromophenol blue on a glass slide according to Horiguchi et al. (58). For details refer to SI Appendix, Materials and Methods.
Shoot Fresh Weight Measurement.
Seven-day-old seedlings grown on AS were transferred to KNO3 containing medium and grown for 14 d vertically under the previously reported growth conditions. The experiments consisted of three biological replicates, each replicate containing eight seedlings. On the 14th day the shoots were excised and weighed on a VWR analytical balance (series no. IT1301517). Presented are average shoot fresh weights per seedling. Significant differences were calculated by Tukey’s test following a one-way ANOVA. Details are in SI Appendix, Materials and Methods.
GUS (β-Glucuronidase) Staining.
GUS expression was analyzed in 7-d-old seedlings transferred for 6 h on AS or KNO3 supplemented media. Seedlings were incubated in staining buffer containing 1 mM of ferro- and ferricyanide, 150 mM sodium phosphate buffer (pH 7) and 1 mg/mL of X-Gluc (dissolved in Dimethyl sulfoxide (DMSO)) at 37 °C for 4 to 24 h (time adjusted to the strength of GUS expression in individual lines). For more details refer to SI Appendix, Materials and Methods.
qRT-PCR Analysis.
Total RNA was extracted from excised 7-d-old seedling’s roots and shoots 1 and 6 HAT to AS- or KNO3-containing plates using the RNeasy Plant Mini kit from (Qiagen) according to the manufacturer’s protocol. A total of 1 and 0.5 µg of RNA was used to synthesize cDNA (shoot and root, respectively) using the iScriptTM cDNA synthesis kit (Bio-Rad). The analysis was carried out on a LightCycler 480 II (SW1.5.1 version; Roche Diagnostics) with the SYBR Green I Master kit (Roche Diagnostics) according to the manufacturer’s instructions. All PCR reactions were carried out with three biological and technical triplicates. The levels of expression of each gene were first measured relative to AT4G05320 (UBQ10) and then to respective mock treatment (AS). For more details refer to SI Appendix, Materials and Methods.
Grafting.
The 7-d-old seedlings were grafted according to the hypocotyl-grafting procedure provided in ref. 59. For detailed description of grafting refer to SI Appendix, Materials and Methods.
Imaging and Image Analysis of PIN-GFP Expression.
The 7-d-old seedlings were transferred onto the AS- or KNO3-containing media for 3 h, then mounted onto slides in a droplet of water and imaged with a Zeiss, LSM800 vertical confocal microscope equipped with a 20×/0.8 Plan-Apochromat M27 objective. Fluorescence signals for GFP (excitation 488 nm, emission 507 nm) and chlorophyll A signal for autofluorescence (excitation 640 nm, emission 645 to 700 nm) to verify the authenticity of membrane-bound signal were detected. Maximum intensity Z-stack projections of confocal images of at least four cotyledons from four different seedlings per treatment were used. To monitor PIN7::PIN7-GFP expression pattern, cotyledons in their middle part were imaged. The membrane PIN-GFP signal of two epidermal cells at the adaxial side per cotyledon was measured using the segmented line function to mark the cell membrane to obtain a quantification of mean gray value using Fiji (v. 152). Significant differences were calculated by Tukey’s test following one-way ANOVA. Experiments were conducted two to three times; representative images and quantification from one experiment are shown.
For monitoring PIN1::PIN1-GFP, a clearing method using Clearsee solution was utilized to diminish chlorophyll autofluorescence and to enhance deep imaging and optical sectioning of the vasculature extending between the petiole and the first vasculature branching junction in the cotyledon. The clearing process was performed according to manufacturer protocols (60). Maximum intensity Z-stack projection images of at least four different cotyledons from four different seedlings per treatment were obtained. The plasma membrane of cells within the vasculature were marked using the segmented line function in Fiji (v. 152) and a mean gray value as a quantification of PIN1-GFP intensity was obtained. Experiments were repeated a total of two to three times; representative images and quantification from one repetition are shown. Significant differences were calculated by Tukey’s test following a one-way ANOVA.
Measurements of Endogenous Cytokinins.
Quantification of cytokinin metabolites was performed according to the method described in ref. 61, including modifications described in ref. 62. For more information refer to SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge Hana Semeradova, Juan Carlos Montesinos, Nicola Cavallari, Marçal Gallemí, Kaori Tabata, Andrej Hurný, and Sascha Waidmann for sharing materials; and Marina Borges Osorio for critical reading of the manuscript. Work in the E. Benková laboratory was supported by the Austrian Science Fund (FWF01_I1774S) to K.O., R.A., and E. Benková. We acknowledge the Bioimaging Facility and Life Science Facilities of the Institute of Science and Technology Austria. We give sincere thanks to Hana Martínková and Petra Amakorová for their help with cytokinin analyses. This work was funded by the Czech Science Foundation (Project No. 19-00973S).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122460119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Frink C. R., Waggoner P. E., Ausubel J. H., Nitrogen fertilizer: Retrospect and prospect. Proc. Natl. Acad. Sci. U.S.A. 96, 1175–1180 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford N. M., Forde B. G., Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book 1, e0011 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiba T., Krapp A., Plant nitrogen acquisition under low availability: Regulation of uptake and root architecture. Plant Cell Physiol. 57, 707–714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Brien J. A., et al. , Nitrate transport, sensing, and responses in plants. Mol. Plant 9, 837–856 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Bouguyon E., et al. , Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 1, 15015 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Ho C.-H., Lin S.-H., Hu H.-C., Tsay Y.-F., CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Krouk G., et al. , Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Tsay Y. F., Schroeder J. I., Feldmann K. A., Crawford N. M., The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Alvarez J. M., et al. , Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80, 1–13 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Castaings L., et al. , The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57, 426–435 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Guan P., et al. , Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U.S.A. 114, 2419–2424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchive C., et al. , Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4, 1713 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Rubin G., Tohge T., Matsuda F., Saito K., Scheible W.-R., Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21, 3567–3584 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varala K., et al. , Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc. Natl. Acad. Sci. U.S.A. 115, 6494–6499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J.-W., Czech B., Weigel D., miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Ruffel S., Gojon A., Systemic nutrient signalling: On the road for nitrate. Nat. Plants 3, 17040 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Chen X., et al. , Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Ohkubo Y., Tanaka M., Tabata R., Ogawa-Ohnishi M., Matsubayashi Y., Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 3, 17029 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Ota R., Ohkubo Y., Yamashita Y., Ogawa-Ohnishi M., Matsubayashi Y., Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat. Commun. 11, 641 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poitout A., et al. , responses to systemic nitrogen signaling in Arabidopsis roots involve trans-zeatin in shoots. Plant Cell 30, 1243–1257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruffel S., et al. , Nitrogen economics of root foraging: Transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc. Natl. Acad. Sci. U.S.A. 108, 18524–18529 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruffel S., Poitout A., Krouk G., Coruzzi G. M., Lacombe B., Long-distance nitrate signaling displays cytokinin dependent and independent branches. J. Integr. Plant Biol. 58, 226–229 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Tabata R., et al. , Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Bouguyon E., Gojon A., Nacry P., Nitrate sensing and signaling in plants. Semin. Cell Dev. Biol. 23, 648–654 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Krouk G., Hormones and nitrate: A two-way connection. Plant Mol. Biol. 91, 599–606 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Landrein B., et al. , Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc. Natl. Acad. Sci. U.S.A. 115, 1382–1387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Z. H., Hirakawa T., Yamaguchi N., Ito T., The roles of plant hormones and their interactions with regulatory genes in determining meristem activity. Int. J. Mol. Sci. 20, 4065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray J. A. H., Jones A., Godin C., Traas J., Systems analysis of shoot apical meristem growth and development: Integrating hormonal and mechanical signaling. Plant Cell 24, 3907–3919 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R., et al. , Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 136, 2512–2522 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vidal E. A., Gutiérrez R. A., A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 11, 521–529 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Little D. Y., et al. , The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. U.S.A. 102, 13693–13698 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.López-Bucio J., Cruz-Ramírez A., Herrera-Estrella L., The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6, 280–287 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Ötvös K., et al. , Modulation of plant root growth by nitrogen source-defined regulation of polar auxin transport. EMBO J. 40, e106862 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier M., Liu Y., Lay-Pruitt K. S., Takahashi H., von Wirén N., Auxin-mediated root branching is determined by the form of available nitrogen. Nat. Plants 6, 1136–1145 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Medici A., et al. , AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 6, 6274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gan Y., Bernreiter A., Filleur S., Abram B., Forde B. G., Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 53, 1003–1016 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Benková E., et al. , Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K., Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Friml J., et al. , Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Reinhardt D., Vascular patterning: More than just auxin? Curr. Biol. 13, R485–R487 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Maeda Y., et al. , A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 9, 1376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guenot B., et al. , Pin1-independent leaf initiation in Arabidopsis. Plant Physiol. 159, 1501–1510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawchuk M. G., Edgar A., Scarpella E., Patterning of leaf vein networks by convergent auxin transport pathways. PLoS Genet. 9, e1003294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett T., et al. , Connective auxin transport in the shoot facilitates communication between shoot apices. PLoS Biol. 14, e1002446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Šimášková M., et al. , Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat. Commun. 6, 8717 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Raines T., et al. , The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 85, 134–147 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Yu L.-H., et al. , Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 6, 27795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakakibara H., Cytokinin biosynthesis and transport for systemic nitrogen signaling. Plant J. 105, 421–430 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Takei K., et al. , AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 45, 1053–1062 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Kiba T., Takei K., Kojima M., Sakakibara H., Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell 27, 452–461 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Kudo T., Kiba T., Sakakibara H., Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52, 53–60 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Naulin P. A., et al. , Nitrate Induction of Primary Root Growth Requires Cytokinin Signaling in Arabidopsis thaliana. Plant Cell Physiol. 61, 342–352 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Gutiérrez R. A., et al. , Insights into the genomic nitrate response using genetics and the Sungear Software System. J. Exp. Bot. 58, 2359–2367 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Ma W., et al. , Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 78, 70–79 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Maghiaoui A., et al. , The Arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J. Exp. Bot. 71, 4480–4494 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Alvarez J. M., et al. , Transient genome-wide interactions of the master transcription factor NLP7 initiate a rapid nitrogen-response cascade. Nat. Commun. 11, 1157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu K. H., et al. , Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545, 311–316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horiguchi G., Fujikura U., Ferjani A., Ishikawa N., Tsukaya H., Large-scale histological analysis of leaf mutants using two simple leaf observation methods: Identification of novel genetic pathways governing the size and shape of leaves. Plant J. 48, 638–644 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Turnbull C. G. N., Booker J. P., Leyser H. M. O., Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 32, 255–262 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Kurihara D., Mizuta Y., Sato Y., Higashiyama T., ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Svačinová J., et al. , A new approach for cytokinin isolation from Arabidopsis tissues using miniaturized purification: Pipette tip solid-phase extraction. Plant Methods 8, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Antoniadi I., et al. , Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. Plant Cell 27, 1955–1967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.