Abstract
The recB268::Tn10 mutation was introduced into the HfrH strain of Escherichia coli. Compared with recB F− and recB F+ cells, the viability of this mutant strain was much lower. Compared with wild-type HfrH, the recB derivative donated much shorter fragments of its chromosome to the recipient. It is suggested that the recB gene product (i.e., RecBCD enzyme) participates in Hfr transfer.
Classical Hfr strains of Escherichia coli contain the prototypal conjugative plasmid F integrated into the chromosome. During exponential growth of Hfr cells, F replicates passively as a cluster of bacterial genes (8). Bacterial growth should therefore be unaffected by its presence. Experiments with HfrH cells carrying the gam+ plasmid pSF117 (9), however, gave surprising results. The rate of multiplication of these cells, which synthesize the Gam protein of phage λ, was drastically reduced (14). Gam protein does not exert such an effect on F+ cells, in which F behaves as an autonomous, cytoplasmic element. Apparently, under certain circumstances the integration of F into the chromosome may be deleterious to its host.
Since Gam protein interacts with the RecB subunit of the RecBCD enzyme (14), we wanted to determine whether mutation of recB is also detrimental to the growth of Hfr cells. For this purpose, an Hfr strain carrying recB268::Tn10 was constructed by transducing strain 3000 HfrH (13) with phage P1vir grown on N3071 (12). Tetracycline-resistant (Tcr) transductants were selected on TP plates (14) supplemented with tetracycline (10 μg/ml). They formed tiny colonies after 3 to 4 days of incubation at 37°C. The recB genotype of these Tcr transductants was confirmed by their sensitivity to UV light and by their ability to support the growth of a gene 2 mutant of phage T4 (see reference 18). The transductants retained the Hfr character (for experimental procedures, see reference 15); interestingly, F+ recB segregants could not be detected despite their growth advantage over the recB Hfr strain (14; also see below). However, when the recB derivatives of HfrH were restreaked on TP agar, some large-colony variants appeared among the progeny. Although these variants were both UV-resistant and recombination-proficient, they carried the original recB mutation (i.e., they retained the ability to propagate T4 2).
We also attempted to introduce recB268::Tn10 into the Hfr strains BW113 and B8 (13). Just as in the case of recB HfrH, the Tcr transductants of BW113 and B8 gave rise to minute colonies after 3 to 4 days of incubation at 37°C. They segregated large-colony variants so rapidly that the growth experiment with these strains could not be done. Further studies were therefore limited only to the recB derivative of strain 3000 HfrH, which we designated IRB101.
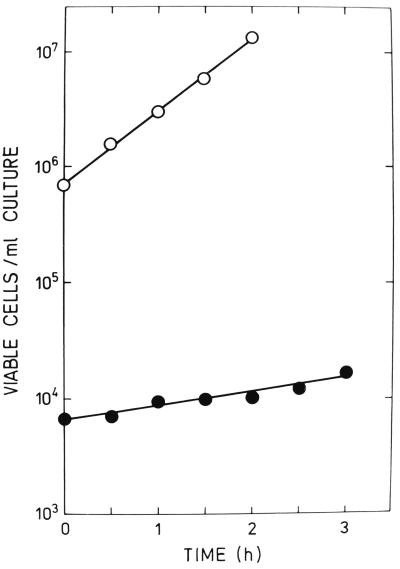
A small colony of IRB101 was inoculated into 10 ml of TP medium. After the culture had grown to saturation (≈107 viable cells/ml), it was diluted 1:50 in fresh TP and incubated for 5 h. The suspension was then diluted again (1:25 in TP). This was zero time of the growth experiment (see reference 6). During a 3-h period, samples were taken periodically and plated for colonies. All incubations were at 37°C. As a control, the growth experiment with the 3000 HfrH parent was also carried out.
The results presented in Fig. 1 show that cells of IRB101 grew extremely slowly. Their generation time (135 min) was four to five times longer than that of the parental strain (28 to 29 min). Large-colony variants interfered, however, with the results of some of the growth experiments. Although their numbers were always negligible at the beginning of these experiments, they represented a significant fraction of the total colony count after 3 h of incubation. (We were able to count large-colony variants separately. Their generation time was about 50 min; i.e., it was intermediate between those of 3000 HfrH and IRB101).
FIG. 1.
Growth curves in TP medium of HfrH cells. Symbols: ○, rec+ HfrH (strain 3000); ●, recB HfrH (strain IRB101).
Poor growth of strain IRB101 is probably due to the frequent appearance of non-colony-forming cells (“lethal sectors” [10]). These non-colony-forming (nonviable) cells are not necessarily metabolically inert (1). Like nonviable Gam-expressing HfrH bacteria, they might participate in conjugative DNA transfer.
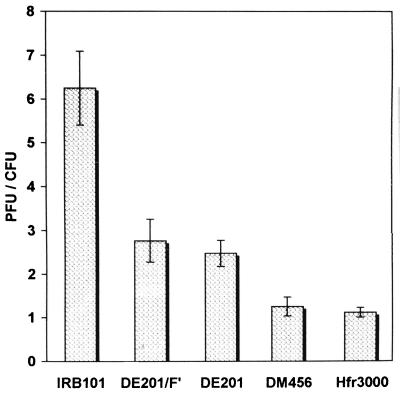
We used a simple method to assess the size of the population of nonviable but metabolically active bacteria (see reference 19). The cells were made lysogenic by the λcI857ind phage mutant. The λcI857ind lysogens are colony formers at 30°C and plaque formers at 42°C. The ratio of the number of PFU (at 42°C) to the number of CFU (at 30°C) is normally about 1.0. If this ratio significantly exceeds the unit, it indicates the presence of nonviable cells. Such nonviable cells are, however, metabolically active because they can support phage growth.
Figure 2 shows that an extremely high proportion of the metabolically active cells of IRB101 failed to form colonies. Less than one-sixth (≈15%) of these bacteria were colony formers. In F− bacteria, mutation of recB did not produce such a dramatic effect. About 40% of the metabolically active cells of strain DE201, a recB268::Tn10 (12) derivative of DM456 (16), were able to give rise to colonies. Of particular interest is that the presence of Fts114lac::Tn5 (21) in the cytoplasm of DE201 (strain DE201/F′) had little, if any, effect on viability. It therefore seems that only the integrated F interferes with the growth of recB cells. As expected, wild-type bacteria formed the same numbers of colonies and plaques, irrespective of whether they were F− (strain DM456) or Hfr (strain 3000).
FIG. 2.
Effects of the recB268::Tn10 mutation in λcI857ind lysogens on formation of plaques and colonies. The ratio of the number of PFU to the number of CFU is presented. Each value is the mean of five independent measurements ± standard deviation. For details, see the text.
We next performed Hfr conjugations, in which 3000 HfrH and IRB101 were used as donors. Both strains transferred chromosomal markers in the following order: thr+, leu+, pro+, pur+, his+. N3105, which is mutant for these loci, was used as a rec+ F− recipient. Strain N3105 (kindly provided by R. G. Lloyd, Nottingham, United Kingdom) is a purE85::Tn10 derivative of AB1157 (for other markers, see reference 2).
Hfr matings were carried out at 37°C in TP medium. The ratio of donor to recipient cells was 1:10. This ratio was determined on the basis of colony count (strains 3000 HfrH and N3105) or on the basis of the calculated number of all metabolically active cells (strain IRB101). Matings were for 80 min; this period was long enough to allow complete transfer of the most distal marker used in our studies. After 80 min, chromosome transfer was interrupted by vigorous agitation of the mating mixtures. The cells were then plated on recombinant-selective synthetic media (15). (Streptomycin at 100 μg/ml in the plate was used to counterselect the Hfr parent). Recombinants inheriting the various donor markers were scored after 48 h of incubation at 37°C.
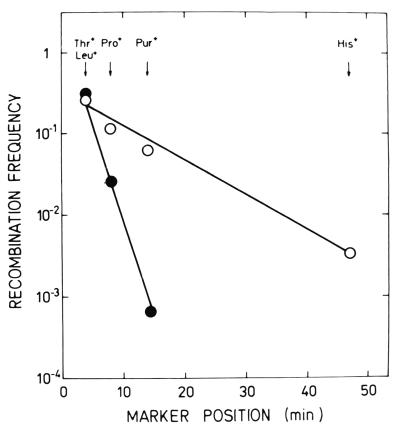
It should be stressed that the frequencies with which donor markers appear among the recombinants are always inversely related to their distances from the origin of transfer (e.g., see reference 22). Hence, there is a characteristic gradient of transmission of these markers, which coincides with their order on the Hfr chromosome. Figure 3 shows the curves illustrating such gradients obtained in matings with strains 3000 HfrH and IRB101. The slopes of these curves, which are supposed to reflect random breakage of the chromosome during transfer, should be the same (22). Contrary to expectations, the curve obtained with IRB101 was three to four times steeper than that obtained with the corresponding rec+ donor. Seemingly, the recB mutation greatly increases the probability of chromosome breakage during DNA transfer.
FIG. 3.
Effects of the recB268::Tn10 mutation in an Hfr strain on the transfer gradient. Symbols: ○, 3000 × N3105 (HfrH × F−); ●, IRB101 × N3105 (recB HfrH × F−).
The extrapolate number, another characteristic of the gradient, was also altered by the recB mutation. To determine this number, the curve representing the gradient of transmission is extended back to intercept the ordinate. The value thus obtained reflects the efficiencies with which an Hfr donor forms a mating pair with a recipient and with which DNA transfer is initiated (see reference 22). In matings with strain IRB101, this value was above 1.0; i.e., it was significantly higher than that obtained in the control cross (a value of ≈0.3). Perhaps recB HfrH is multipiliated and therefore forms mating aggregates rather than mating pairs. Another possibility is that the size of the population of metabolically active recB HfrH bacteria was underestimated. If this size is larger, the extrapolate number is lower. Even so, our results strongly suggest that the recB mutation does not affect the formation of mating pairs/aggregates and the initiation of DNA transfer. Similarly, in experiments with DE201/F′, neither mating pair formation nor F′ selftransmission was inhibited by this mutation (data not shown).
We also performed experiments on zygotic induction. Zygotic induction occurs when a chromosomal segment of the lysogenic Hfr donor carrying an inducible prophage enters the nonlysogenic F− recipient. In the case of HfrH (λ), the prophage is located ≈20 min from the integrated F. Upon its entry into the zygote, it is induced and begins vegetative growth. When plated with indicator bacteria, the zygotes therefore give rise to infectious centers. In this way, the frequency of zygotic induction following the transfer of λ from lysogenic rec+ HfrH (strain 3000) and recB HfrH (strain IRB101) to the F− recipient, N3105, was determined. Matings were for 45 min at 37°C. As expected, a large proportion (nearly 50%) of the rec+ donors transferred λ and formed infectious centers. However, only 0.1% of the total number of metabolically active recB HfrH cells were able to transfer the prophage to the recipient. This finding confirms the results presented in Fig. 3. Again, it may be inferred that the recB gene product (i.e., RecBCD enzyme) plays an essential role in Hfr transfer.
Until now, as far as we know, there have been no reports on the specific effects of recB on Hfr cells. It is noteworthy that one of the early recB Hfr constructs (strain JC5412 [25]) became a better growing, UV-resistant type in the course of subculturing (26). This strain was later shown to be a suppressed revertant carrying an sbcA mutation (3). We do not know whether large-colony variants observed in our experiments belong to the same class of mutants. It is clear, however, that the low viability of recB Hfr cells facilitates the detection and isolation of the various types of such fast-growing mutants.
The low level of viability of recB HfrH cells can easily be interpreted. It is known that the transfer (tra) genes of F are permanently derepressed. The relaxase specified by the traI gene (TraI protein) is therefore permanently present within F+ and Hfr bacteria. This protein introduces a site- and strand-specific nick at the origin of transfer (oriT) on F. The reaction is reversible and the interrupted DNA strand can be resealed. In fact, there is an equilibrium between nicked and ligated states (27; for a recent review, see reference 5). Certainly, the replisome may encounter the integrated F in its nicked state during replication of the Hfr chromosome. According to the current hypothesis (11), a nick in the template (parental) strand leads to collapse of the replication fork. The collapsed fork can be repaired by the RecBCD recombinational pathway (11). Obviously, recB HfrH bacteria are unable to repair such a lesion and, therefore, cell death ensues. This interpretation was further supported by an experiment in which recB268::Tn10 was introduced into strain 122-1. Strain 122-1 (kindly provided by E. L. Zechner, Graz, Austria) is an HfrH derivative with a deletion of the arcA locus. Due to arcA, the HfrH strain becomes deficient in the oriT-dependent nicking reaction (27). As may be expected, the growth rate of the recB arcA HfrH double mutant was therefore about the same as that of the recB F+ cells (data not shown).
The question of whether the low viability of recB HfrH donors can account for the altered DNA transfer described in the present report arises. One might assume that the premature cessation of Hfr transfer occurs after the last energy reserves of dying cells have been depleted. This assumption seems unlikely for the following reasons. (i) The low viability of Gam-expressing HfrH bacteria does not interfere with DNA transfer. (Evidently, Gam protein does not convert a recB+ cell to a RecB− phenotype [17, 20, 24].) (ii) We also examined the donor ability of large-colony revertants of strain IRB101. With respect to DNA transfer, the behavior of these revertants (which carry the original recB mutation) was similar to that of the parental recB Hfr donor (data not shown). It may therefore be concluded that RecBCD enzyme (most likely, its helicase activity) participates in Hfr transfer.
Clearly, the necessary prerequisite for Hfr transfer is the formation of mating pairs. The DNA strand nicked at oriT on the integrated F then begins to enter the recipient. TraI protein, which possesses both the relaxase and helicase activities, catalyzes the displacement of this strand. F selftransmission depends exclusively on the activity of TraI (5; but see reference 23). Host helicases are not involved in this process (5) although RecBCD enzyme can occasionally enter the plasmid in the cytoplasm (7). During Hfr transfer, however, TraI might normally be replaced by the RecBCD helicase. This notion challenges the widely held belief that the bacterial host plays only a passive role in the dissemination of its genes.
Hfr transfer is usually regarded as “F self-transmission, in which the bacterial chromosome is inserted into F” (4). Our studies modify this long-standing tenet. The F-encoded functions are indeed indispensable for mating pair formation and chromosome mobilization. However, the F conjugative system seems unsuited for transfer of the huge bacterial chromosome. Apparently, this transfer is largely dependent on the host-encoded RecBCD helicase.
Acknowledgments
This research was supported by the Croatian Ministry of Science (grant 00981001).
REFERENCES
- 1.Ankenbauer A G. Reassessing forty years of genetic doctrine: retrotransfer and conjugation. Genetics. 1997;145:543–549. doi: 10.1093/genetics/145.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann B J. Derivatives and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W C, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 2460–2488. [Google Scholar]
- 3.Barbour S D, Nagaishi H, Templin A, Clark A J. Biochemical and genetic studies of recombination proficiency in Escherichia coli. II. Rec+ revertants caused by indirect suppression of Rec− mutations. Proc Natl Acad Sci USA. 1970;67:128–135. doi: 10.1073/pnas.67.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broda P. Plasmids. W. H. Oxford, England: Freeman and Company; 1979. [Google Scholar]
- 5.Byrd D R, Matson S W. Nicking by transesterification: the reaction catalysed by a relaxase. Mol Microbiol. 1997;25:1011–1022. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- 6.Capaldo-Kimball F, Barbour S D. Involvement of recombination genes in growth and viability of Escherichia coli K-12. J Bacteriol. 1971;106:204–212. doi: 10.1128/jb.106.1.204-212.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter J F, Porter R D. traY and traI are required for oriT-dependent enhanced recombination between lac-containing plasmids and λplac5. J Bacteriol. 1991;173:1027–1034. doi: 10.1128/jb.173.3.1027-1034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chander M, Silver L, Roth Y, Caro L. Chromosome replication in an Hfr strain of Escherichia coli. J Mol Biol. 1976;104:517–523. doi: 10.1016/0022-2836(76)90285-0. [DOI] [PubMed] [Google Scholar]
- 9.Friedman S A, Hays J B. Selective inhibition of Escherichia coli RecBC activities by plasmid-encoded GamS function of phage lambda. Gene. 1986;43:255–263. doi: 10.1016/0378-1119(86)90214-3. [DOI] [PubMed] [Google Scholar]
- 10.Haefner K. Spontaneous lethal sectoring, a further feature of Escherichia coli strains deficient in the function of rec and uvr genes. J Bacteriol. 1968;96:652–659. doi: 10.1128/jb.96.3.652-659.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 12.Lloyd R G, Benson F E. Genetic analysis of conjugational recombination in Escherichia coli K-12 strains deficient in RecBCD enzyme. J Gen Microbiol. 1987;133:2531–2538. doi: 10.1099/00221287-133-9-2531. [DOI] [PubMed] [Google Scholar]
- 13.Low K B. Hfr strains of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W C, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 2402–2405. [Google Scholar]
- 14.Maršić N, Roje S, Stojiljković I, Salaj-Šmic E, Trgovčević Ž. In vivo studies on the interaction of RecBCD enzyme and λ Gam protein. J Bacteriol. 1993;175:4738–4743. doi: 10.1128/jb.175.15.4738-4743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 16.Mount D W. Isolation and genetic analysis of a strain of Escherichia coli K-12 with an amber recA mutation. J Bacteriol. 1971;109:388–389. doi: 10.1128/jb.107.1.388-389.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy K C. λ Gam protein inhibits the helicase and χ-stimulated recombination activities of Escherichia coli RecBCD enzyme. J Bacteriol. 1991;173:5808–5821. doi: 10.1128/jb.173.18.5808-5821.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver D B, Goldberg E B. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J Mol Biol. 1977;116:877–881. doi: 10.1016/0022-2836(77)90276-5. [DOI] [PubMed] [Google Scholar]
- 19.Petranović D, Petranović M, Salaj-Šmic E, Trgovčević Ž. A simple method for estimating the biological activity of irradiated Escherichia coli DNA by the use of the prophage. Int J Radiat Biol. 1976;29:187–190. doi: 10.1080/09553007614550201. [DOI] [PubMed] [Google Scholar]
- 20.Salaj-Šmic E, Maršić N, Trgovčević Ž, Lloyd R G. Modulation of EcoKI restriction in vivo: role of the λ Gam protein and plasmid metabolism. J Bacteriol. 1997;179:1852–1856. doi: 10.1128/jb.179.6.1852-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sansonetti P J, Kopecko D J, Formal S B. Shigella sonnei plasmids: evidence that a large plasmid is necessary for virulence. Infect Immun. 1981;34:75–83. doi: 10.1128/iai.34.1.75-83.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith G R. Conjugational recombination in E. coli: myths and mechanisms. Cell. 1991;64:19–27. doi: 10.1016/0092-8674(91)90205-d. [DOI] [PubMed] [Google Scholar]
- 23.Traxler B A, Minkley E G. Evidence that DNA helicase I and oriT site-specific nicking are both functions of the F TraI protein. J Mol Biol. 1988;204:205–209. doi: 10.1016/0022-2836(88)90609-2. [DOI] [PubMed] [Google Scholar]
- 24.Trgovčević Ž, Rupp W D. Interaction of bacterial and lambda phage recombination systems in the X-ray sensitivity of Escherichia coli K-12. Proc Natl Acad Sci USA. 1974;71:503–506. doi: 10.1073/pnas.71.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willetts N S, Clark A J, Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969;97:244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willetts N S, Mount D W. Genetic analysis of recombination-deficient mutants of Escherichia coli K-12 carrying rec mutations cotransducible with thyA. J Bacteriol. 1969;100:923–934. doi: 10.1128/jb.100.2.923-934.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zechner E L, Prüger H, Grohman E L, Espinosa M, Högenauer G. Specific cleavage of chromosome and plasmid DNA strands in gram-positive and gram-negative bacteria can be detected with nucleotide resolution. Proc Natl Acad Sci USA. 1997;94:7435–7440. doi: 10.1073/pnas.94.14.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]