Significance
We identify the interferon-γ inducible protein 16 (IFI16)-dependent STING signaling pathway as the crucial immune cascade conferring anti-HER2 trastuzumab resistance. Importantly, we provide an actionable epigenetic approach that combines EZH2 and histone deacetylase inhibitors that promotes IFI16-mediated CXCL10/11 signaling for the complete tumor eradication through increased CD8+ T cells infiltration and long-term T cell memory in HER2+ breast cancer.
Keywords: HER2-targeted therapy, epigenetic approach, HER2+ breast cancer, anti-HER2 resistance
Abstract
Relapse to anti-HER2 monoclonal antibody (mAb) therapies, such as trastuzumab in HER2+ breast cancer (BC), is associated with residual disease progression due to resistance to therapy. Here, we identify interferon-γ inducible protein 16 (IFI16)-dependent STING signaling as a significant determinant of trastuzumab responses in HER2+ BC. We show that down-regulation of immune-regulated genes (IRG) is specifically associated with poor survival of HER2+, but not other BC subtypes. Among IRG, IFI16 is identified as a direct target of EZH2, the underexpression of which leads to deficient STING activation and downstream CXCL10/11 expression in response to trastuzumab treatment. Dual inhibition of EZH2 and histone deacetylase (HDAC) significantly activates IFI16-dependent immune responses to trastuzumab. Notably, a combination of a novel histone methylation inhibitor with an HDAC inhibitor induces complete tumor eradication and long-term T cell memory in a HER2+ BC mouse model. Our findings demonstrate an epigenetic regulatory mechanism suppressing the expression of the IFI16-CXCL10/11 signaling pathway that provides a survival advantage to HER2+ BC to confer resistance to trastuzumab treatment.
The addition of an anti-HER2 monoclonal antibody (mAb) to chemotherapy-based neoadjuvant treatment (NAT) has significantly improved the pathological complete response rate (pCR) in patients with HER2+ breast cancer (BC), once thought to be untreatable. However, the challenges of treating HER2+ BC remain as up to 50% of patients develop the progressive disease by either low responsiveness or acquired resistance after treatment (1–3). The poor clinical outcomes have been associated with a lack of complete tumor response resulting in residual disease remaining, which may arise from clones resistant to anti-HER2 mAb (1, 3). Indeed, it seems that anti-HER2–based NAT can significantly improve survival only in patients with a pCR, but not in patients with remaining residual tumors after NAT (4, 5). Although various levels of HER2 expression in the cancer are associated with anti-HER2 pathologic response (6, 7), it remains unclear if additional molecular heterogeneity in HER2+ tumors contributes to anti-HER2 response. These limitations highlight the necessity of understanding anti-HER2 resistance mechanisms and developing a potent and durable therapeutic strategy to cure and eliminate HER2+ tumors.
Among issues relevant to anti-HER2 mAb resistance is the insensitivity of the tumor intrinsic signaling response to HER2-targeted mAb and the insufficient infiltration of tumor leukocytes (TILs). Indeed, more than 50% of HER2+ BC displayed limited TILs, showing a feature of “cold tumors” that are characterized by deficient interferon (IFN) signaling in the tumor immune microenvironment, which responds poorly to immunotherapy (8). It has been shown that the higher infiltration of IFN-secreting natural killer (NK) and CD8+ T cells improves the tumor responses to HER2-targeted mAb by mediating direct cytotoxicity to HER2+ BC cells (9–11). Particularly in NK cells, antibody-dependent cellular cytotoxicity (ADCC) mediated by anti-HER2 mAb induces direct cytolytic killing of HER2+ tumor cells through secretion of cytotoxic granules and proinflammatory cytokines, such as IFN-γ. Subsequently, these tumor-suppressive substances indirectly enhance the activation of antigen-specific T cell immunity (12). Activation of IFN response through epigenetic modulation has been shown to improve anti-HER2 mAb treatment in an animal model (13), suggesting a role of IFN signaling in anti-HER2 mAb immunotherapy.
On the other hand, induction of memory immunity, especially CD8+ memory T cells, is vital in combating residual disease by inducing durable antitumor immune responses in HER2+ patients (14, 15). Recently, an animal model treated with infused NK cells and trastuzumab showed that tumor infiltration of effector memory T (Tem) cells resulted in long-term survival (16). Interestingly, a HER2-specific chimeric antigen receptor T cell platform focused on engineering central memory T cells (Tcm) showed a positive correlation with metastatic tumor eradication and prolonged survival in HER2+ disease (17). Hence, it is reasonable to propose that activating the immune-related signaling pathway and the induction of TILs interplaying between innate (NK cell-mediated ADCC) and adaptive immunity (memory T cell induction) are valuable strategies to enhance effective anti-HER2 mAb immunotherapy.
Recently, activating STING signaling has emerged as a therapeutic approach to modulate cancer immunotherapy. The STING pathway has been shown to regulate immune surveillance by activating NK and T cells (18–21). The activation of the STING cascade promotes the cross-presentation of dendritic cells and CD8+ T cells through IFN and CXCL10 production (22–24). STING signaling can be activated by two primary cytoplasmic DNA sensors, cyclic GMP-AMP synthase (cGAS) and IFN-γ inducible protein 16 (IFI16) (25). Both cGAS and IFI16 are capable of initiating STING cascades, which results in the secretion of various cytokines and chemokines that play essential roles in enforcing immune cell activities (26, 27). However, compared to cGAS, whose role in cancer immunity has been studied extensively, little is known about IFI16. Moreover, cGAS silencing by DNA methylation is linked to the attenuation of cancer immunity (28, 29), while the inactivation of IFI16 in cancer, particularly BC, has not been previously reported, to our knowledge.
Here, we uncover a significant role of IFI16 in mediating the immune response to trastuzumab treatment in HER2+ BC. We demonstrate that IFI16 is down-regulated in HER2+ BC cells through cooperative histone modifications involving EZH2 and histone deacetylase (HDAC), which induces deficient STING-CXCL10/11 immune signaling associated with resistance to anti-HER2 mAb treatment. We show that epigenetic reactivation of IFI16-STING-CXCL10/11 elicited a potent immune cytotoxicity and memory response that lead to durable tumor suppression by anti-HER2 mAb therapy in both in vitro and mouse models. These findings show that the IFI16-mediated STING pathway is a crucial determinant of anti-HER2 mAb immunotherapeutic response. The results support a treatment strategy for achieving complete and durable tumor remissions and long-term immune memory in HER2-targeted BC.
Results
Down-Regulation of Immune Regulated Genes Is Associated with Metastatic Relapse Following Trastuzumab Treatment and Poor Prognosis in HER2+ BC Patients.
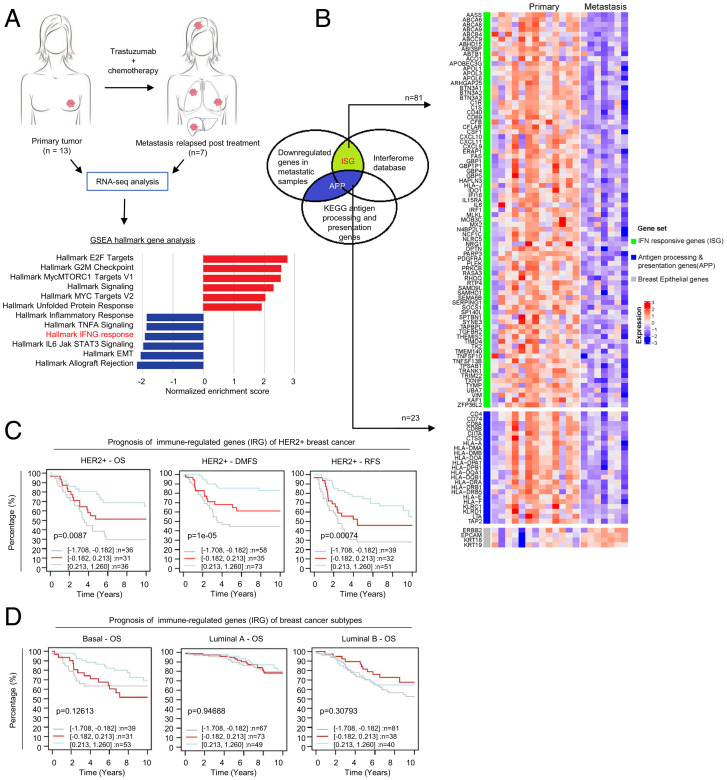
To investigate the molecular basis of cancer immunity associated with anti-HER2 mAb resistance, we performed RNA-sequencing (RNA-seq) analysis in treatment-naïve primary HER2+ BC tumors (n = 13) and distant metastasis in HER2+ tumors relapsed posttrastuzumab treatment (n = 7). Altered gene expression could distinguish most metastases from the primary BC tumors (SI Appendix, Fig. S1A), suggesting substantial transcriptional changes in metastasis compared to primary BC tumors. Among genes that exhibited altered expression (with a log fold-change cutoff > 1, P < 0.05), the IFN-γ response pathway was found to be one of the main pathways down-regulated in metastasis compared to primary BC tumors (Fig. 1A). Consistently, RNA-seq analysis using immune-cell gene signatures for profiling the microenvironment of solid tumors (Imsig) (30) showed that relapsed metastasis samples exhibited loss of infiltrating immune cells that include NK and T cells, but not macrophages and monocytes, compared to treatment-naïve primary BC tumors (SI Appendix, Fig. S1B). Among the 1,791 genes down-regulated in metastasis samples, 81 were identified as IFN-responsive genes (ISG) by the Interferome database (www.interferome.org). Another 23 genes were identified as antigen presentation and processing genes (APP) in the Kyoto Encyclopedia of Genes and Genomes (KEGG) APP genes database (https://www.gsea-msigdb.org/gsea/index.jsp). In total, we identified 104 immune-regulated genes (IRG), whose expression was down-regulated in metastases relapsed from initial trastuzumab treatment (Fig. 1B). RT-PCR analysis of representative IRG, including the key IFN signaling transcriptional factor IRF1, STING pathway regulator IFI16, chemotaxis regulators CXCL10 and CXCL11, as well as human leukocyte antigen encoding major histocompatibility complex (MHC) class I and II, HLA-A and HLA-DRA, further validated their down-regulation in metastasis compared to the primary BC tumors (SI Appendix, Fig. S1C). Of interest, hallmark EMT was found to be one of the top down-regulated pathways in metastatic tumors. Among them, mesenchymal markers but not epithelial markers were down-regulated in metastasis, which might be associated with a MET phenotype required for metastatic cell colonization and proliferation (SI Appendix, Fig. S1D).
Fig. 1.
IRG signature is correlated to clinical trastuzumab resistance in HER2+ BC patients. (A) Illustration showing sample preparation for RNA-seq analysis. Gene set enrichment analysis shows IFN-γ pathway as one of the top gene pathways down-regulated in relapse-metastasis samples (n = 7) compared to primary HER2+ tumors (n = 13). (B) Heatmap showing 104 IRG comprised of 81 ISG, and 23 APP genes that were differentially expressed in metastatic tumors compared to primary tumors (log fold cutoff >1, P < 0.05). (C) Lower expression of IRG is strongly associated with poorer overall survival (OS), distant metastasis-free survival (DMFS), and recurrence-free survival (RFS) in HER2+ BC. Data were collected from GOBO analysis (co.bmc.lu.se/gobo/). Datasets were stratified into three quantiles based on IRG expression (lower, medium, and upper quartile) and censored for 10-y follow-up. Log-rank P values are shown as −log10(P value). (D) Kaplan–Meier plots showing prognostic values of IRG for OS in basal, luminal A and luminal B BC subtypes.
To correlate IRG down-regulation with clinical outcomes of BC, we used the public database GOBO (co.bmc.lu.se/gobo/gsa.pl) to evaluate their prognostic values in BC. Interestingly, down-regulation of the IRG gene signatures were found to be significantly associated with poor survival only in HER2+ BC but not in other subtypes of BC, including basal, luminal A, and luminal B (Figs. 1 C and D and SI Appendix, Fig. S1E). Together, these findings from the clinical sample analysis suggest that these IRG, including those involved in IFN response and antigen presentation, are associated with HER2+ BC progression and resistance to trastuzumab treatment.
Epigenetic Drug Combinations Targeting Histone Modifications Induce Robust Reactivation of IRG in HER2+ BC Cells.
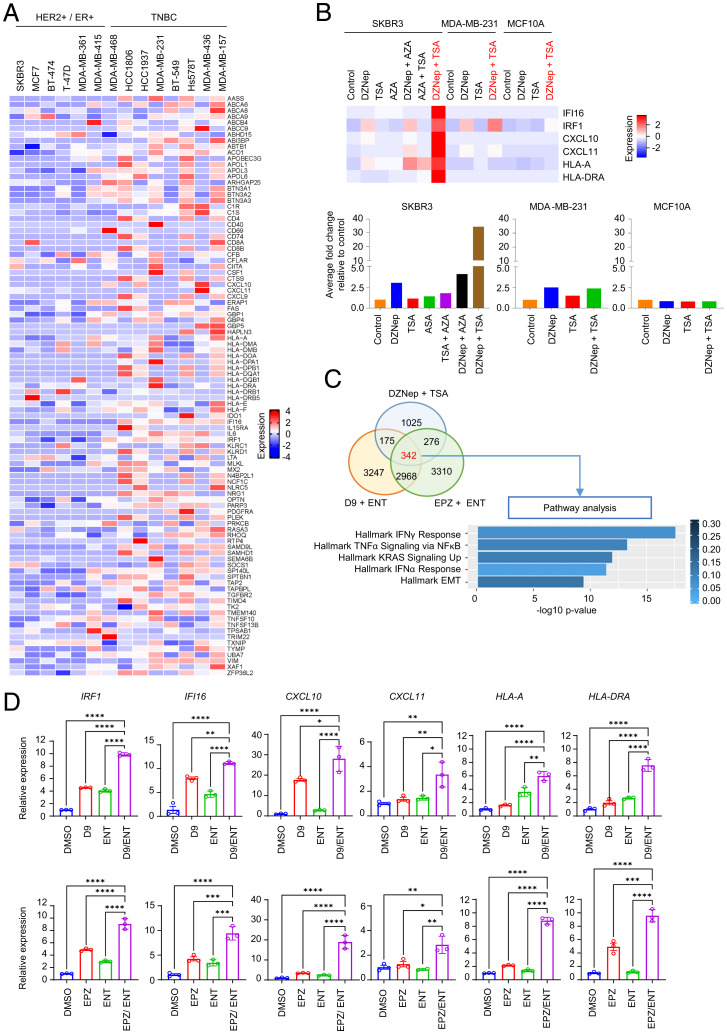
We next sought to characterize the heterogeneity of the IRG expression in various BC cell lines. Transcriptome analysis of our in-house BC cell line gene-expression database indicated the down-regulation of IRG is mainly in estrogen receptor (ER)+ or HER2+ BC subtype cell lines—including SKBR3, MCF7, BT-474, T-47D, MDA-MB-361, and MDA-MB-415—when compared to triple-negative BC subtype (TNBC) cell lines comprised of MDA-MB-468, HCC1806, HCC1937, MDA-MB-231, BT-549, Hs578T, MDA-MB-436, and MDA-MB-157 (Fig. 2A). We previously reported an association of EZH2-mediated epigenetic silencing of IFN signaling with resistance to IFN-γ–mediated cytotoxicity in prostate cancer (31), and have also shown that combined inhibition of EZH2 and HDAC can lead to robust synergistic induction of EZH2-target gene expression (32, 33). To investigate whether EZH2 is involved in suppressing IRG in HER2+ BC, we performed an EZH2 knockdown in the HER2+ cell line SKBR3 and found that it restored the expression of the representative IRG (SI Appendix, Fig. S2A). Moreover, a combination of EZH2 inhibitor DZNep with HDAC inhibitor Trichostatin A (TSA) resulted in synergistic induction of IRG in SKBR3, but not in the TNBC cell line MDA-MB-231 or in the noncancerous breast epithelial cell line MCF10A (Fig. 2B and SI Appendix, Table S1). In contrast, DNA methylation inhibitor 5-Azacytidine (AZA) alone or combined with TSA had only a modest effect on the IRG (Fig. 2B).
Fig. 2.
Epigenetic modulation targeting histone modification induces a robust IRG response in HER2+ cell lines. (A) Heatmap displaying 140 IRG down-regulated in HER2+ and ER+ BC cell lines compared to TNBC cell lines as determined by microarray analysis. (B) Heatmap of microarray analysis showing the expression profiles of six representative IRG in different BC subtypes (HER2+, SKBR3; TNBC, MDA-MB-231; and normal, MCF10A) treated with different combinations of various epigenetic inhibitors (histone methylation inhibitor, DZNep; HDAC inhibitor, TSA; and DNA methylation inhibitor, AZA). The average fold-change of six representative IRG are plotted to bar graphs at the bottom. (C) Venn diagram of microarray analysis showing the number of genes induced by different combinatorial inhibitors of EZH2 (DZNep, D9, and EPZ) and HDAC (TSA and ENT) in SKBR3 cells. Pathway analysis of the 342 up-regulated genes reveals IFN-γ response as top gene pathway induced by combinatorial inhibitors of EZH2 and HDAC. (D) RT-PCR showing the induction of 6 representative IRG in SKBR3 treated with D9 (Upper), EPZ (Lower), and ENT alone or in combination. GAPDH was used as a reference gene for normalization. Data are expressed as means ± SEM of three technical replicates, representing three independent experiments. P values were calculated with one-way ANOVA with Tukey’s multiple comparison test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant.
To further the validation of IRG induction by combining EZH2 and HDAC inhibitors, we also treated the SKBR3 cells with a specific EZH2 inhibitor EPZ-6438 (EPZ) and HDAC inhibitor Entinostat (ENT), both approved for clinical use. In addition, we also tested a second-generation HMT inhibitor D9, that we developed in-house, which is a DZNep analog that can induce EZH2 depletion in nanomolar concentrations (SI Appendix, Fig. S2B) with an improved safety profile in vivo (34, 35). Gene-expression analysis identified a set of 342 genes that were commonly induced by the three combinations (Fig. 2C). Intriguingly, IFN-γ response was identified as the primary pathway up-regulated by these treatments (Fig. 2C). RT-PCR analysis further validated the synergistic and specific induction of representative IRG in HER2+ cell lines SKBR3, MDA-MB-361, and BT-474 (Figs. 2D and SI Appendix, Fig. S2C), but not in TNBC MDA-MB-231 cells and MDA-MB-436 (SI Appendix, Fig. S2D). It is noteworthy that 100 nM D9 treatment combined with 250 nM ENT was sufficient to induce a robust IRG expression comparable to 500 nM EPZ with 250 nM ENT (Fig. 2D). These findings demonstrated that IRG are the top genes targeted by EZH2 and HDAC in HER2+ BC, and a coinhibition strategy is necessary to boost their expression. Of note, the cell line difference in response to EZH2 and HDAC inhibitors is not simply due to different EZH2 levels in these cell lines, as shown in (SI Appendix, Fig. S2E).
Combined Inhibition of EZH2 and HDAC Suppresses HER2+ Tumor Growth in an Immune-Dependent Manner.
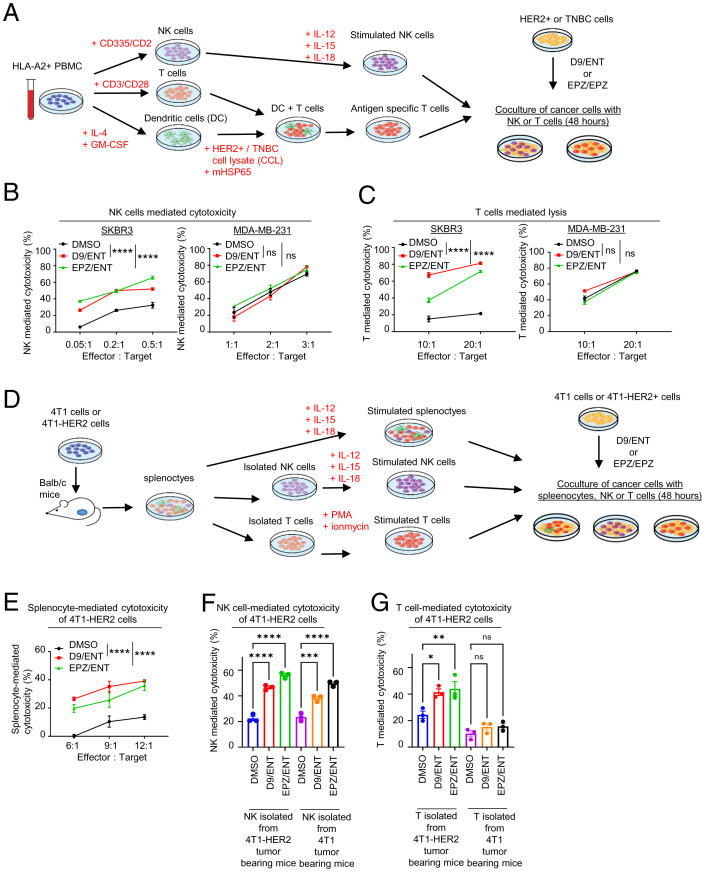
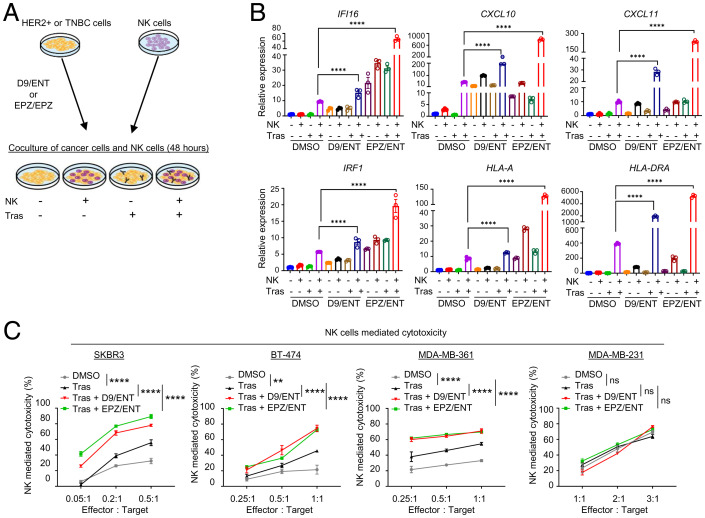
Cancer cells can escape immune surveillance through intrinsic deficiency in response to IFN-γ secreted from immune effector T and NK cells (36). Given that IRG are the top genes susceptible to combined EZH2 and HDAC inhibitor treatment, we assessed whether this modulation would increase anticancer immunity through increased NK or T cell-mediated killing of HER2+ BC. We isolated NK and T cells from peripheral blood mononuclear cells (PBMCs) of HLA-A2+ healthy human donors (to match HLA-A2+ SKBR3), followed by NK activation and expansion with interleukin (IL)-12/15/18 stimulation or T cell activation with dendritic cells prestimulated with antigens of HER2+ cell lysates, respectively (procedure illustrated in Fig. 3A). Cocultures of NK with BC cells expressing a luciferase reporter for viability measurement showed that D9/ENT or EPZ/ENT pretreatment of HER2+ BC cell lines SKBR3, BT-474, and MDA-MB-361 markedly enhanced susceptibility to NK-mediated killing (Figs. 3B and SI Appendix, Fig. S3A). Similar results were also observed in antigen-stimulated T cells (Fig. 3C). In contrast, the epigenetic combination did not enhance NK or T-mediated killing in the TNBC cell line MDA-MB-231 (Fig. 3 B and C). RT-PCR analysis of HER2+ cells consistently showed that D9/ENT or EPZ/ENT combinations markedly potentiated NK-stimulated IRG expression (SI Appendix, Fig. S3B), but not in MDA-MB-231 cells (SI Appendix, Fig. S3C). The findings support our hypothesis that epigenetic treatment may harness immunosurveillance toward HER2+ BC cells through enhanced immune response by NK and T effector cells.
Fig. 3.
Combination of EZH2 and HDAC inhibitors induce NK and T cell-mediated cytotoxicity in HER2+ cells. (A) Schematic overview of coculture experiment of PBMC expanded NK and T cells with epigenetic inhibitor-treated BC cell lines. (B) Representative data showing D9/ENT or EPZ/ENT boosts NK-mediated cytotoxicity and (C) dendritic cell-primed specific CD8+ T-mediated cytotoxicity of SKBR3 and MDA-MB-231 cells. Data are expressed as means ± SEM of three technical replicates, representative of three independent experiments. P values were calculated with two-way ANOVA with Sidak’s multiple comparison test, ****P < 0.0001, ns, not significant. (D) Schematic overview of coculture experiment of mouse immune cells and epigenetic inhibitors treated mouse cancer cells. (E) Representative lysis data showing D9/ENT or EPZ/ENT boosts the 4T1-HER2 cell lysis by splenocytes, and (F) NK cells and (G) T cells isolated from 4T1-HER2 tumor-bearing Balb/c mouse. Data are expressed as means ± SEM of three technical replicates, representing two independent experiments. P values were calculated with one-way ANOVA with Tukey’s multiple comparison test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant.
To evaluate the above hypothesis in vivo, we overexpressed HER2 in 4T1 mouse mammary cancer cells, referred to as 4T1-HER2 cells (SI Appendix, Fig. S3D). Previous studies have demonstrated that the 4T1-HER2 model may mimic human HER2+ tumors in vivo (37, 38). We first showed that 4T1-HER2 cells also exhibited reduced IRG expression compared to the parental 4T1 cells, including the mouse homolog of IFI16, IFI204, and mouse MHC class I and II, H2-Kd and I-Ad (SI Appendix, Fig. S3E). To determine the in vitro cellular cytotoxicity of the mouse immune cells toward 4T1-HER2 cells, we inoculated 4T1 or 4T1-HER2 cells into immune-competent Balb/c mice to form xenograft tumors, and splenocytes were isolated from the corresponding mouse spleen (Fig. 3D). Like human HER2+ BC cells, 4T1-HER2 cells pretreated with D9/ENT or EPZ/ENT were more vulnerable to splenocyte-mediated killing (Fig. 3E). Similar results were observed in stimulated NK cells isolated from splenocytes of 4T1-HER2–bearing and 4T1-bearing tumors (Fig. 3F). On the other hand, D9/ENT or EPZ/ENT increased the lysis of T cells isolated from splenocytes of 4T1-HER2–bearing tumors but not 4T1-bearing tumors (Fig. 3G). Consistently, combined D9/ENT or EPZ/ENT markedly enhanced splenocyte-stimulated IRG expression in 4T1-HER2 cells (SI Appendix, Fig. S3F).
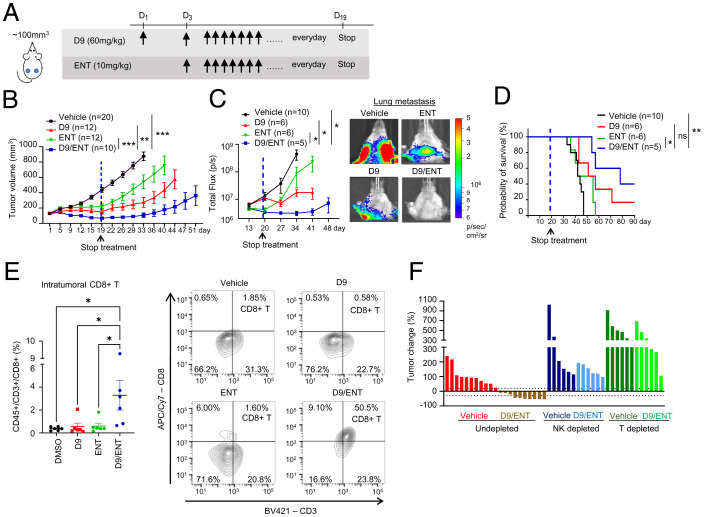
Next, we investigated the efficacy of D9 (60 mg/kg), EPZ (100 mg/kg), and ENT (10 mg/kg) alone or in combination in suppressing 4T1-HER2 tumor growth in vivo (treatment dosage and schedule as illustrated in Fig. 4A). Indeed, combined D9/ENT treatment administered daily was much more robust in suppressing 4T1-HER2 tumor growth than single-agent treatment (Fig. 4B). The combinatory treatment also halted lung metastasis (Fig. 4C) and enhanced the survival probability of these mice (Fig. 4D). EPZ combination with ENT also showed a tumor-suppressive effect, though the efficacy was less strong than the D9/ENT combination (SI Appendix, Fig. S4). In agreement with the tumor response, xenograft tumors treated with D9/ENT exhibited more infiltrating CD8+ T cells than single agent-treated tumors (Fig. 4E). A significant finding was that depletion of NK cells or T cells in mice abrogated the D9/ENT or EPZ/ENT-induced tumor regression (Fig. 4F), validating the role of antitumor immunity in the therapeutic efficacy of the epigenetic combination treatments. Collectively, data from both in vitro and in vivo studies demonstrated that a combination of EZH2 and HDAC inhibitor treatment effectively suppressed HER2+ tumor growth in an immune-dependent manner.
Fig. 4.
Combination of EZH2 and HDAC inhibitors suppresses tumor growth and metastasis in an immune-dependent manner. (A) Illustration of treatment dosage and schedule. Balb/c mice were treated with vehicle, 60 mg/kg D9, 10 mg/kg ENT, or combination of D9/ENT every day. (B) 4T1-HER2 primary tumor growth of Balb/c mice treated with vehicle, D9, ENT, or combination of D9/ENT. Blue dotted line indicates the day when the treatment was stopped. The number of subjects (n) per group were indicated in the graph. Data are expressed as means ± SEM of n number of samples. P values were calculated with Mann–Whitney unpaired t test, **P < 0.01, ***P < 0.001, ns, not significant. (C) Combination of D9 and ENT significantly reduced 4T1-HER2 lung metastasis compared to vehicle, D9 and ENT (Left). Representative bioluminescent imaging showing lung metastasis of 4T1-HER2 tumors on day 34 (Right). The number of subjects (n) per group were indicated in the graph. Data are expressed as means ± SEM of n number of samples. P value were calculated with Mann–Whitney unpaired t test, *P < 0.05. (D) Improved survival probability of 4T1-HER2 bearing mice treated with D9/ENT compared to vehicle and single treatment of D9 and ENT. The number of subjects (n) per group were indicated in the graph. P value were calculated with Logrank Mantel–Cox test, *P < 0.05, **P < 0.01, ns, not significant. (E) Percentage of tumor infiltrating CD8+ T cells (CD45+/CD3+/CD8+ population in whole tumor) assessed by flow cytometry following treatment at the end of experiments (Left). Each dot represents one individual tumor. Representative flow cytometric plots of CD3+/CD8+ T cells for each treatment group were shown (Right). Data are expressed as means ± SEM of six representative 4T1-HER2 tumors per group. P values were calculated with one-way ANOVA with Tukey’s multiple comparisons test, *P < 0.05. (F) Percentage of tumor change showing depletion of NK cells or T cells in Balb/c mice abolished the therapeutic effect of D9/ENT. Percentage of tumor change was calculated based on tumor volume on day 15 from the baseline of tumors before treatment. Each bar represents one individual tumor.
The Combination of EZH2 and HDAC Inhibitors Further Enhances Anti-HER2 Immunotherapy.
Trastuzumab binds to the Fcγ receptor on NK cells to elicit ADCC-mediated killing of HER2+ BC cells (39). We next investigated whether the epigenetic combination also potentiates trastuzumab-mediated ADCC efficacy by pretreating HER2+ BC cells with combined D9/ENT or EPZ/ENT before coculturing with NK cells in the presence of trastuzumab (Fig. 5A). RT-PCR analysis showed that NK and trastuzumab-stimulated SKBR3 cells elicited the IRG response, which was further enhanced by combined D9/ENT or EPZ/ENT treatment, but not in MDA-MB-231 cells (Fig. 5B and SI Appendix, Fig. S5A). Corresponding to the IRG boosting, D9/ENT or EPZ/ENT, when combined with trastuzumab, also markedly potentiated NK killing of HER2+ BC cells, including SKBR3, BT-474, and MDA-MB-361 cells, but not the TNBC line MDA-MB-231(Fig. 5C). Collectively, these data suggest that a combination of EZH2 and HDAC inhibitors leads to a robust boost of IRG-related immune responses by trastuzumab.
Fig. 5.
Combination of EZH2 and HDAC inhibitors enhances anti-HER2 immune response in vitro. (A) Illustration showing coculture experiments of NK cells and epigenetic inhibitors treated cancer cells in the presence of trastuzumab (Tras). (B) Representative RT-PCR showing that the D9/ENT and EPZ/ENT have a strong synergistic effect to boost NK and trastuzumab (Tras)-induced IRG in HER+ SKBR3 cells. GAPDH was used as a reference gene for normalization. Data are expressed as means ± SEM of three technical replicates, representative of three independent experiments. P values were calculated with two-way ANOVA with Sidak’s multiple comparisons test, ****P < 0.0001, ns not significant. (C) D9/ENT or EPZ/ENT combinations plus trastuzumab boost NK cells cytotoxicity toward HER2+ cells (SKBR3, BT-474, and MDA-MDA-MB-361) but not TNBC (MDA-MB-231). Data are expressed as means ± SEM of three technical replicates, representative of two independent experiments. P values were calculated with two-way ANOVA with Sidak’s multiple comparisons test, **P < 0.01, ****P < 0.0001, ns, not significant.
To reproduce the above results in a mouse model, we used a mouse monoclonal anti-HER2 mAb, clone 7.16.4 (referred to as α-ErbB2 mAb). This α-ErbB2 mAb binds to the trastuzumab binding epitope of human ErbB2 (40) and can be recognized by mouse immune cells to elicit an attack of mouse tumor cells overexpressing the human HER2 (40, 41). As in the human system, mouse splenocytes treated with α-ErbB2 mAb showed an increase in the splenocyte-mediated killing of 4T1-HER2, but not 4T1 cells, upon treatment in combination with D9/ENT or EPZ/ENT (SI Appendix, Fig. S5B).
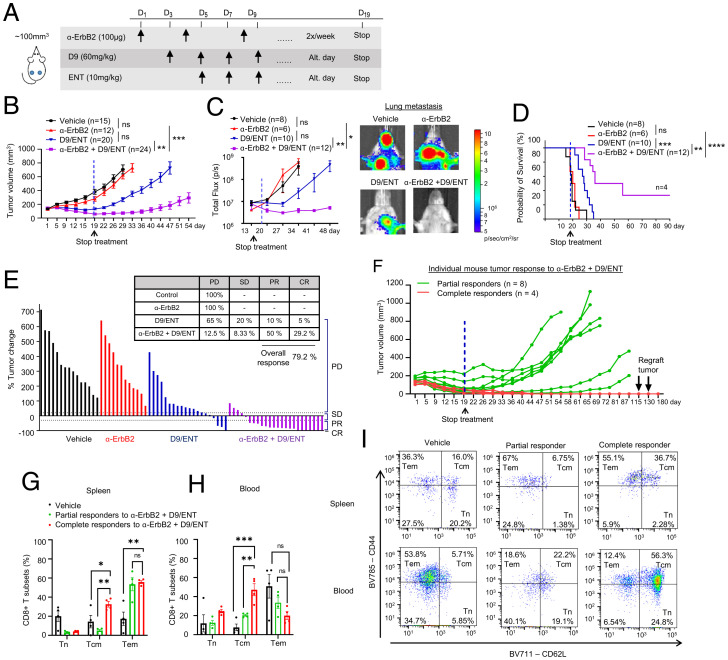
To investigate the effect of combined D9/ENT or EPZ/ENT in enhancing α-ErbB2 mAb efficacy in vivo, we first treated 4T1-HER2 tumor-bearing mice with α-ErbB2 mAb, followed by treatments with or without D9/ENT combination on alternate days instead of every day (Fig. 6A). In this treatment condition, 4T1-HER2 tumors were resistant to α-ErbB2 mAb treatment. Although both D9/ENT and α-ErbB2+D9/ENT combinations effectively suppressed tumor growth, the triple combination resulted in markedly prolonged suppression of tumor growth and metastasis even after the termination of the drug administration (Fig. 6 B and C). This treatment effect was translated into a significant survival benefit (Fig. 6D). Using response evaluation criteria in solid tumors (RECIST) to measure tumor response, α-ErbB2+D9/ENT triple combination significantly improved 4T1-HER2 tumor eradication by achieving an overall response rate of 79.2% compared to 15% in D9/ENT-treated tumors (Fig. 6E). In particular, we observed complete tumor eradication in 30% of α-ErbB2+D9/ENT-treated mice (n = 4), which remained tumor-free for up to 180 d, at which point the mice were killed for analysis (Fig. 6F). EPZ/ENT also showed a combinatorial effect with α-ErbB2 mAb (SI Appendix, Fig. S6 A–C), but the overall tumor response was only 29.6% (SI Appendix, Fig. S6D).
Fig. 6.
Combination of EZH2 and HDAC inhibitors sensitizes anti-HER2 therapy to induce complete tumor eradication and long-term immune memory. (A) Illustration of treatment dosage and schedule. Balb/c mice were treated with vehicle, 100 μg α-ErbB2, 60 mg/kg D9, and 10 mg/kg ENT, or α-ErbB2+D9/ENT. (B) Tumor volume along treatment in 4T1-HER2 tumor bearing Balb/c mice model which were treated with vehicle, α-ErbB2, D9, and ENT, or α-ErbB2+D9/ENT. Blue dotted line indicates the day when the treatment was stopped. The treatment schedule is indicated at the top of the plot. The number of subjects (n) per group are indicated in the graph. Data are expressed as means ± SEM of n number of samples. P value were calculated with Mann–Whitney unpaired t test, **P < 0.01, ***P < 0.001, ns, not significant. (C) Combination of D9 and ENT enhances α-ErbB2 treatment in reducing 4T1-HER2 lung metastasis compared to vehicle, α-ErbB2 and D9/ENT treatment (Left). Representative bioluminescent imaging showing lung metastasis of 4T1-HER2 tumors on day 34 (Right). The number of subjects (n) per group were indicated in the graph. Data are expressed as means ± SEM of n number of samples. P values were calculated with Mann–Whitney unpaired t test, *P < 0.05, **P < 0.01, ns, not significant. (D) Prolonged overall survival of 4T1-HER2–bearing mice treated with α-ErbB2+D9/ENT compared to vehicle, α-ErbB2, or D9/ENT. The number of subjects (n) per group were indicated in the graph. P values were calculated with Logrank Mantel–Cox test, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant. (E) Percentage of tumor change showing the therapeutic effect of vehicle, α-ErbB2, D9/ENT, or combination. The percentage of tumor change was calculated based on day 22 from the baseline of tumors before treatment. Tumor volume changes were analyzed based on RECIST criteria to categorize the tumor response into progressive disease (PD). stable disease (SD), progressive repression (PR), and complete response (CR). Each bar represents one individual tumor. (F) Individual mouse tumors response (average volume of two tumors from left and right mammary fat pads) treated with α-ErbB2+D9/ENT. n represents the number of mice. Partial responders are defined as tumors that undergo relapse after treatment was stopped. Complete responders are tumors that completely disappear after treatment and continuously reject tumor growth. 4T1-HER2 tumors were reinoculated twice on day 115 and 130 to complete responders. (G) Flow cytometry analysis showing T cells compartment in spleen and (H) blood comparing between 4T1-HER2–bearing mice (vehicle) and mice achieving partial or complete response after α-ErbB2+D9/ENT treatment. CD8+ T cell subset population are defined by CD44−CD62L+ naïve T cell (Tn), CD44+CD62L− Tem and CD44+CD62L+ Tcm populations. Blood and spleen were collected at the end of the experiment. Data are expressed as means ± SEM of four 4T1-HER2 spleen or blood samples in each group. Each dot represents one individual sample. P values were calculated with two-way ANOVA with Tukey’s multiple comparisons test, *P < 0.05, **P < 0.01, ***P < 0.001, ns not significant. (I) Representative flow cytometry plots showing CD8+ T cell subset population for spleen (Upper) and blood (Lower) samples.
Two attempts at reengraftment of the 4T1-HER2 cells in the four tumor-free α-ErbB2+D9/ENT-treated mice after 100 d of initial tumor engraftment failed to form tumors, suggesting that these mice had developed a durable immune memory resulting in effective tumor rejection (Fig. 6F). Analysis of the splenocytes of these four α-ErbB2+D9/ENT “cured” tumor-free mice showed a significant expansion of Tcm (CD44+CD62L+ population) and Tem (CD44+CD62L−) of CD8+ T cells when compared with splenocytes from untreated or partial responders in α-ErbB2+D9/ENT-treated mice (Fig. 6 G and I). Analysis of blood PBMCs of these mice also revealed a significant increase of CD8+ Tcm cells (Fig. 6 H and I). These findings indicate that combined EZH2/HDAC inhibitor, in particular D9/ENT, can potentiate the efficacy of the anti-HER2 mAb to achieve robust and durable tumor suppression. Notably, the mice tolerated the triple combination with no overt signs of toxicity and only suffered a modest weight loss (SI Appendix, Fig. S6E).
IFI16 Is a Direct Target of EZH2 that Regulates Trastuzumab-Stimulated STING Signaling and CXCL10/11 Expression.
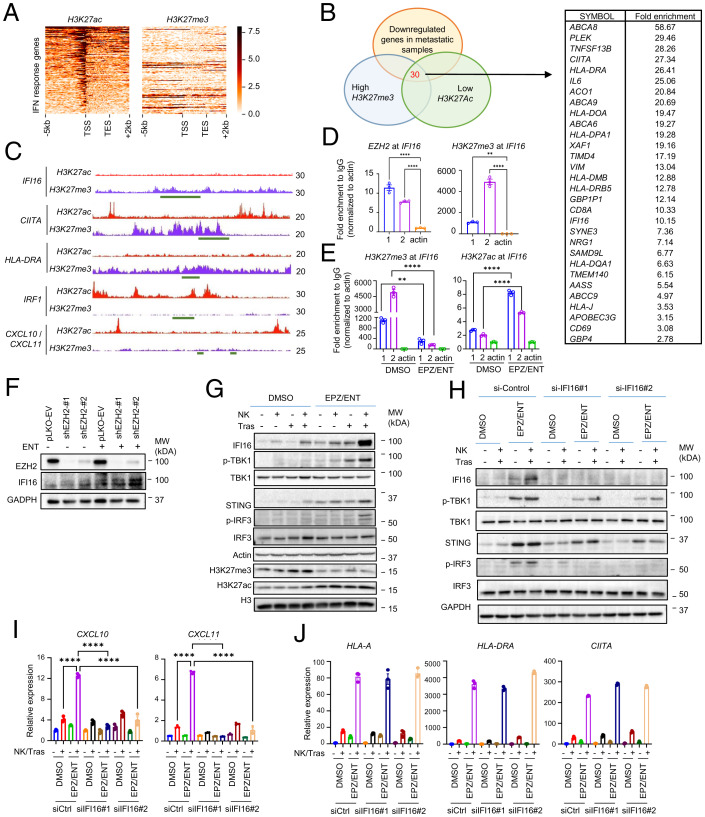
The synergistic induction of IRG by combined EZH2/HDAC inhibitors suggests that these genes are coregulated by both EZH2 and HDACs. EZH2-associated increases in H3K27me3 and HDAC-associated decreases in H3K27ac are hallmarks of repressive chromatin modifications. To identify whether H3K27me3 and H3K27ac are involved in IRG expression, we performed chromatin immunoprecipitation-sequencing (ChIP-seq) analysis of H3K27me3 and H3K27ac in SKBR3 cells and compared these genes with the down-regulated genes in metastatic relapsed HER2+ BC tumors. ChIP-seq analysis revealed only a small subset of genes showing high enrichment in H3K27me3, but low in H3K27ac (Fig. 7A). Comparing these H3K27me3hi/H3K27aclow genes with IRG down-regulated in metastasis identified a set of 30 genes as potential direct targets of EZH2 (Fig. 7B). Of these, genes involved in antigen presentation, such as MHC-II class HLA-DRA and class II MHC transactivator CITTA, have been previously reported as EZH2 targets (21, 42). IFI16, a STING pathway regulator, has not been previously shown to be an EZH2 target. The other IRG—including IRF1, CCL10, and CXCL11—did not show apparent H3K27me3 enrichment and thus are unlikely to be direct targets of EZH2 (Fig. 7C).
Fig. 7.
IFI16 is a direct target of EZH2 and critical for activation of STING signaling and downstream CXCL10 and CXCL11. (A) ChIP-seq analysis of SKBR3 cells showing H3K27ac and H3K27me3 histone mark enrichment of IFN response gene set near individual transcription start site (−5 kbp to +2 kbp) genome wide. (B) Venn diagram showing the overlapping of H3K27me3hi/H3K27aclow IRG to identify potential direct target of EZH2. (C) ChIP-seq analysis showing H3K27me enrichment and H3K27ac reduction at IFI16, CIITA, and HLA-DRA promoter but not at IRF1, CXCL10, and CXCL11. (D) ChIP-PCR analysis of EZH2 and H3K27me enrichment on IFI16 promoter in SKBR3 cells, using two pairs of ChIP primers flanking the promoter. Data are expressed as means ± SEM of three technical replicates. P values were calculated with one-way ANOVA with Tukey’s multiple comparisons test, **P < 0.01, ****P < 0.0001, ns, not significant. (E) ChIP-PCR analysis showing EPZ/ENT abolished H3K27me3 enrichment and induced H3K27ac activation at IFI16 in SKBR3 cells, using two pairs of ChIP primers flanking the promoter. Data are expressed as means ± SEM of three technical replicates. P values were calculated with one way ANOVA with Tukey’s multiple comparisons test, **P < 0.01, ****P < 0.0001, ns, not significant. (F) Knockdown of EZH2 in SKBR3 cells induces IFI16, which can be further enhanced by ENT. (G) EPZ/ENT treatment exhibits synergic effects with NK cells and trastuzumab (Tras) in SKBR3 cells in inducing the expression of IFI16, p-TBK1, p-IRF3, and STING. (H) Knockdown of IFI16 in SKBR3 cells abolished the synergic effects of EPZ/ENT treatment, NK cells and trastuzumab in STING signaling pathway. All Western blot images are representative of two independent experiments. (I) RT-PCR showing knockdown of IFI16 abolished the CXCL10 and CXCL11 induction in SKBR3 cocultured with NK cells and trastuzumab. Data are expressed as means ± SEM of three technical replicates, representative of two independent experiments. P values were calculated two-way ANOVA with Sidak’s multiple comparisons test, ****P < 0.0001, ns, not significant. (J) RT-PCR showing expression of HLA-A, HLA-DRA, and CIITA were not affected by IFI16 knockdown. Data are expressed as means ± SEM of three technical replicates, representative of two independent experiments.
Recent studies have indicated an important role of STING activation in the efficacy of immunotherapy (43, 44). Given the emerging role of IFI16 in regulating STING signaling, we investigated the epigenetic regulation of IFI16 and its functional role in the anti-HER2 mAb responses. ChIP-PCR confirmed the enrichment of EZH2 and H3K27me3 at IFI16 (Fig. 7D). Consistent with the synergistic induction of IFI16, combined EPZ/ENT treatment resulted in depletion of H3K27me3, while H3K27ac at the IFI16 locus was induced (Fig. 7E). Moreover, the knockdown of EZH2 in SKBR3 increased the expression of IFI16, which was further enhanced by adding ENT (Fig. 7F). These findings demonstrate that IFI16 is a direct target of EZH2, and combined inhibition of EZH2 and HDAC can maximize its induction.
Western blot analysis further confirmed that combined EPZ/ENT or D9/ENT boosted the induction of IFI16 protein expression and the downstream STING signaling hallmarks p-TBK1, p-IRF3, and STING in SKBR3 and MDA-MB-361 cells in the presence of NK and trastuzumab (Fig. 7G and SI Appendix, S7 A and B). Similar results were seen in 4T1-HER2, but not 4T1, cells (SI Appendix, Fig. S7 C and D). Knockdown of IFI16 abolished the induction of the STING signaling cascade (Fig. 7H), confirming the role of IFI16 in regulating STING signaling in response to NK and trastuzumab stimulation.
Interestingly, IFI16 knockdown also abrogated NK and trastuzumab-stimulated induction of CXCL10 and CXCL11 expression in HER2+ cells (Fig. 7I). CXCL10 and CXCL11 have been previously shown to be downstream of the STING pathway (45, 46). Indeed, STING knockdown also reduced the NK and trastuzumab-elicited CXCL10 and CXCL11 induction in SKBR3 and 4T1-HER2 cell lines (SI Appendix, Figs. S7 E and F), indicating that IFI16-STING may control the expression of the two critical chemoattractants that act to recruit T cells to the tumor in response to trastuzumab treatment. Significantly, IFI16 knockdown did not affect the MHC-I/II class genes, such as HLA-A, HLA-DRA, and CITTA (Fig. 7J), suggesting that antigen presentation might not be IFI16-dependent. FACS analysis of surface markers of MHC class I molecules, HLA-ABC and MHC class II molecules, HLA-DR, DQ, and DP in SKBR3 cells also indicated that trastuzumab+ D9/ENT and trastuzumab+ EPZ/ENT were able to induce surface expression of MHC class I and II molecules on BC cells (SI Appendix, Fig. S7G), but IFI16 depletion did not change their expression (SI Appendix, Fig. S7 H and I). Collectively, these findings suggest that IFI16 is a crucial regulator of STING signaling in the anti-HER2 mAb immune response that may act as a functional node to integrate innate and adaptive immunity in HER2+ BC. Loss of IFI16-CXCL10/11 expression may compromise the anti-HER2 mAb therapeutic immune response.
IFI16-CXCL10/11 Blockade Is Sufficient to Abolish the Tumor Response to the Combination Treatments.
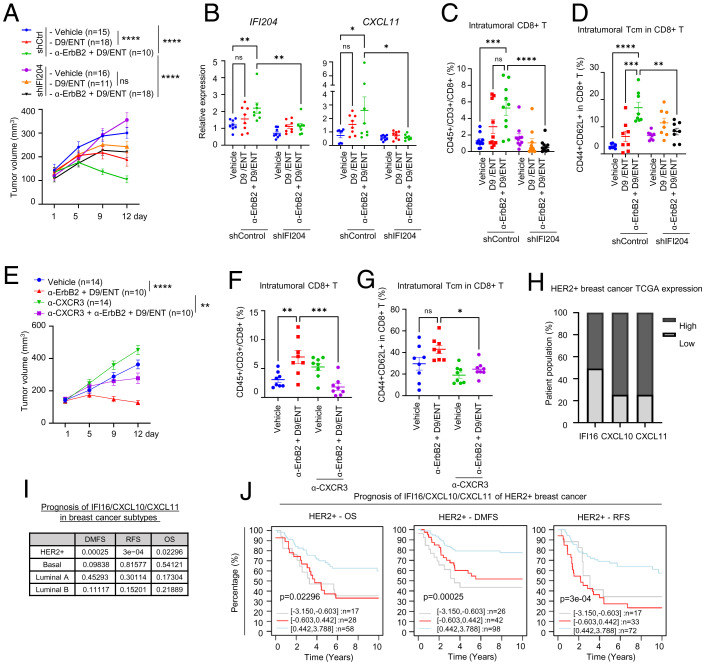
To ascertain the role of IFI16 and CXCL10/11 in the α-ErbB2+D9/ENT combinatorial therapeutic effect, we assessed the impact of IFI204 (mouse homolog of IFI16) knockdown in the 4T1-HER2 mouse model. We first conducted an in vitro experiment to confirm the effect of IFI204 depletion and found that IFI204 knockdown in 4T1-HER2 cells resulted in a loss of splenocyte-mediated killing of 4T1-HER2 cells in the presence of α-ErbB2+D9/ENT (SI Appendix, Fig. S8A). When 4T1-HER2 cells expressing short-hairpin RNA (shRNA)-control and shRNA-IFI204 were engrafted in Balb/c mice, tumors with IFI204 depletion were no longer responsive to the α-ErbB2+D9/ENT treatment (Fig. 8A), confirming the indispensable role of IFI204 in the combination treatment. The α-ErbB2+D9/ENT-induced tumor-infiltrating CD8+ T cells, intratumoral CD8+ Tcm cells, and splenic CD8+ Tcm cells, as well as intratumoral CXCL11, were also blunted in IFI204 knockdown tumors (Fig. 8 B–D and SI Appendix, Fig. S8C). In contrast, MHC class I and II genes, H2-Kd and I-Ad, were not affected in tumors with IFI204 knockdown (SI Appendix, Fig. S8B).
Fig. 8.
Depletion of IFI204 (IFI16 analog in mice) is sufficient to rescue the combination effect (A) Tumor volume along treatment in 4T1-HER2 control (shCtrl) and IFI204 knockdown (shIFI204) tumor-bearing Balb/c mice model were treated with vehicle, 60 mg/kg D9 and 10 mg/kg ENT, 100 μg α-ErbB2+D9/ENT on a treatment regime as indicated in Fig. 6. The number of subjects (n) per group were indicated in the graph. Mice were killed on day 12 for tumor analysis. Data are expressed as means ± SEM of indicated number of samples per group. P value were calculated based on endpoints with Mann–Whitney unpaired t test, ****P < 0.0001. (B) RT-PCR showing expression levels of IFI204 and CXCL11 of 4T1-HER2 tumors. Data are expressed as means ± SEM of eight tumors from each group. P values were calculated two-way ANOVA with Sidak’s multiple comparisons test, *P < 0.05, **P < 0.01, ns not significant. (C) Percentage of tumor infiltrating CD8+ T cells (CD45+/CD3+/CD8+) assessed by flow cytometry. Data are expressed as means ± SEM of 10 tumors from each group. P values were calculated with two-way ANOVA with Sidak’s multiple comparisons test, ***P < 0.001, ****P < 0.0001, ns not significant. (D) Percentage of intratumoral Tcm (CD44+CD62L+) in CD8+ T cells assessed by flow cytometry. Data are expressed as means ± SEM of eight tumors from each group. P values were calculated with two-way ANOVA with Sidak’s multiple comparisons test, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns not significant. (E) Tumor volume along treatment in 4T1-HER2 tumor-bearing Balb/c mice model, which were treated with vehicle or α-ErbB2+D9/ENT on a similar treatment regime as indicated in Fig. 6. CXCR3 inhibitor (α-CXCR3) was treated twice per week at 100 μg. The number of subjects (n) per group were indicated in the graph. Mice were killed on day 12 for tumor analysis. Data are expressed as means ± SEM of n number of samples. P values were calculated based on endpoints with Mann–Whitney unpaired t test. **P < 0.01, ****P < 0.0001, ns not significant. (F) Percentage of tumor infiltrating CD8+ T cells (CD45+/CD3+/CD8+) assessed by flow cytometry. Data are expressed as means ± SEM of eight tumors from each group. P values were calculated with two-way ANOVA with Sidak’s multiple comparisons test. **P < 0.01, ***P < 0.001, ns not significant. (G) Percentage of intratumoral Tcm (CD44+CD62L+) in CD8+ T cells assessed by flow cytometry. Data are expressed as means ± SEM of eight tumors from each group. P values were calculated with two-way ANOVA with Sidak’s multiple comparisons test. *P < 0.05, ns not significant. (H) Percentage of HER2+ BC patients with high (expression >1-fold) and low expression (expression <1-fold) of IFI16, CXCL10 and CXCL11. Data were extracted from TCGA dataset. (I) Summary of clinical prognosis of IFI16/CXCL10/CXCL11 in various BC subtypes, collected from GOBO analysis (co.bmc.lu.se/gobo/). Logrank P values are shown as −log10(P value). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns not significant. (J) Kaplan–Meier plots of IFI16/CXCL10/CXCL11 for HER2+ BC collected from GOBO analysis. Datasets were stratified into three quantiles based on IRG expression (lower, medium, and upper quartile) and censored for 10-y follow-up. Logrank P values are shown as −log10(P value).
To further validate the role of CXCL10/11 in anti-HER2 mAb tumor eradication, we also tested whether blockade of CXCR3, the receptor of CXCL10/11 ligands on T cells, could abolish the efficacy of α-ErbB2+D9/ENT treatment. As in IFI204 knockdown, adding an anti-CXCR3 mAb to α-ErbB2+D9/ENT treatment regimen also rescued the tumor suppression effect (Fig. 8E) and inhibited the infiltrating CD8+ T cells, intratumoral CD8+ Tcm cells, and splenic CD8+ Tcm cells (Fig. 8 F and G and SI Appendix, Fig. S8D). Collectively, these data demonstrated that IFI16-regulated CXCL10/11 is crucially associated with the efficacy of α-ErbB2+D9/ENT treatment.
The role of IFI16-CXCL10/11 in T cell memory induction was further validated in vitro. In the coculture assay consisting of NK, T cells and SKBR3 cells, we showed that D9/ENT pretreatment of SKBR3 cells increased the proportion of Tcm (CD45RO+CD62L+) among CD8+ T cells in the presence of NK cells and trastuzumab (SI Appendix, Fig. S8E), but this induction was blocked by both IFI16 knockdown and anti-CXCR3 mAb (SI Appendix, Fig. S8 F and G). These findings indicate that the IFI16-CXCL10/11 axis contributes to the development of T cell memory. Analysis of The Cancer Genome Atlas (TCGA) data show that down-regulation of IFI16-CXCL10/11 expression occurs in 49% of HER2+ BC (Fig. 8H), suggesting that these tumors may harness resistance to trastuzumab. Prognosis analysis revealed that low expression of IFI16/CXCL10/11 was strongly associated with poor overall survival, recurrence-free survival, and distant metastasis-free survival in HER2+, but not other subtypes of BC patients (Fig. 8 I and J).
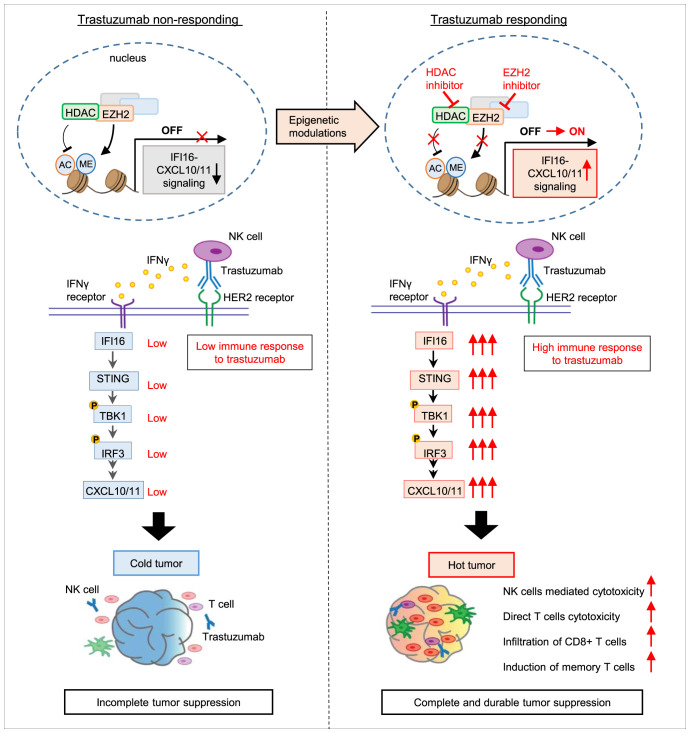
Finally, we validated the association of EZH2/HDAC, IFI16, and CD8 T cells infiltration using our clinical samples of HER2+ tumors. Analysis using CIBERSORTx (https://cibersortx.stanford.edu/) showed a negative correlation of EZH2/HDAC1 to -3 with infiltrated immune cells, including activated NK cells and CD8 T cells (SI Appendix, Fig. S9A), supporting that high expression of EZH2 and HDAC is associated with cold tumor. Correlation analysis also revealed that EZH2/HDAC 1 to -3 expression is negatively correlated with IFI16 expression (SI Appendix, Fig. S9B). Collectively, as depicted in Fig. 9, our data support a model in which down-regulation of IFI16-CXCL10/11 by EZH2 and HDACs is associated with a “cold immunosuppressive” tumor microenvironment that contributes to anti-HER2 mAb immune resistances. Epigenetic reactivation of the IFI16-STING-CXCL10/11 pathway by inhibiting EZH2/HDACs can convert the cold tumor to a “hot tumor” microenvironment, leading to increased NK-mediated ADCC and infiltration of CD8+ T cells, thus sensitizing anti-HER2 response. Our findings suggest that the EZH2/HDAC cotargeting could be a practical therapeutic approach to target trastuzumab resistance and achieve long-term durable tumor suppression.
Fig. 9.
Model illustrating a role of IFI16-CXCL10/11 inactivation in trastuzuamb resistance and reactivation strategy. Model illustrating the mechanisms by which EZH2 and HDAC induces inactivation of IFI16-CXCL10/11 signaling. Treatment with EZH2 and HDAC inhibitors, D9 or EPZ and ENT, reactivates IFI16-directed immune responses, thereby sensitizing the trastuzumab antitumor responses in HER2+ BC patients. The reactivation of IFI16-CXCL10/11 converts immunosuppressive cold tumor to immunocompetent hot tumor as shown by improved CD8+ T cell infiltration and increased memory T cells population within HER2+ tumors.
Discussion
In the present study, we identified an epigenetic regulatory mechanism suppressing the expression of the IFI16-CXCL10/11 signaling pathway that provides a survival advantage to HER2+ BC cells to confer resistance to trastuzumab treatment. We present a combination epigenetic treatment approach that induces robust activation of IFI16-CXCL10/11 signaling, which could enable trastuzumab to induce complete tumor eradication and long-term T cell memory in vivo.
Recent clinical trials suggest that patients who achieved pCR had significantly better long-term survival without cancer remission than those who did not (4, 5). The inadequate clinical tumor response to anti-HER2 mAb treatment has been linked to a lack of TILs (9–11), suggesting that the antitumor immunogenicity plays a vital role in mediating the therapeutic activity of anti-HER2 mAb therapies. Moreover, trastuzumab resistance still develops even in some tumors exhibiting a relatively higher number of TILs, suggesting the existence of complex immune escape mechanisms (47). In our model, the combinatorial effect of EZH2 and HDAC inhibitors in tumor suppression was lost in mice depleted of NK or T cells, confirming the vital role of both innate and adaptive immunity in the therapeutic response, which was further supported by increased infiltrating CD8+ T cells in tumors treated with epigenetic drugs and HER2-targeted mAb. Moreover, one-third of the mice treated with α-ErbB2 mAb and EZH2/HDAC inhibitors exhibited a complete tumor response and tumor-free survival for up to 180 d. The re-engraftment of HER2+ BC cells into these mice led to tumor rejection, indicating the presence of specific immune memory. Accordingly, the spleen and blood in these mice showed higher levels of systemic CD8+ Tcm than those without treatment. Together, our findings uncovered an epigenetic mechanism leading to immune deficiency as an important factor contributing to resistance to anti-HER2 mAb immunotherapy.
Our study revealed that a substantial set of HER2+ BC tumors exhibit down-regulation of IRG, including genes involved in IFN response (IFI16, IRF1), T cell recruitment (CXCL10, CXCL11), and antigen presentation (MHC-I/II class genes HLA-A, HLA-DRA, and CIITA). The epigenetic mechanism leading to deficiency of IFN signaling has been reported in multiple cancers and associated with immune evasion and resistance to immunotherapy (48). Our ChIP-seq and ChIP-PCR analysis identified IF116, a key DNA sensor that triggers a STING-dependent type I IFN activation, as a direct target of EZH2-H3K27me3. IFI16 has been implicated as a tumor suppressor to induce cancer cell death or promote tumor immune evasion through modulating immune cell activity (43, 44, 49). Moreover, the emergence of the STING pathway in antitumor responses through the infiltration of lymphocytes via IFN signaling comprises a promising direction for cancer immunotherapy. Interestingly, it has been recently reported that the recruitment of AKT by HER2 disrupts DNA sensing and prevents STING-mediated antitumor immunity (50). Our findings thus add an epigenetic mechanism mediating STING pathway inactivation in cancer. In contrast, ChIP-PCR and ChIP-seq did not identify the H3K27me3 mark at CXCL10 and CXCL11 locus, contrary to the previous finding on the direct suppression of EZH2 in CXCLs in ovarian cancer (51). Instead, epigenetic inhibitor-induced CXCL10 and CXCL11 is more likely a result of an indirect effect of IFI16 induction, as IFI16 has been reported to induce CXCL10 expression, and type I IFN primes IFN-γ–mediated immune responses (26, 52). Of note, we only observed a modest IRG induction by a DNA methyltransferase inhibitor, suggesting that histone chromatin modifications are the primary mechanism governing the down-regulation of IRG in HER2+ BC, though the aberrant expression of DNA methyltransferases and HDAC have been previously reported to correlate with HER2+ BC proliferation, differentiation, and metastasis (53–55). These data support our observations that inhibition of these epigenetic dysregulations could reverse the inactivation of the IFI16-mediated STING pathway, suggesting the relevance of poor IFI16-STING responses to anti-HER2 immune resistance.
We found that the down-regulation of IRG expression and its restoration by EZH2/HDAC inhibitor is limited to ER+/HER2+ tumors but not in TNBC lines. We have previously shown that while EZH2 acts canonically as a gene repressor in ER+ BC but acts noncanonically as a gene activator in TNBC (56). Hence, the canonical gene silencing activity of EZH2 might power the specificity of down-regulation of EZH2-mediated IRG in HER2+ BC, while this canonical gene silencing activity is insubstantial in TNBC.
Currently, HER2 expression is the only biomarker for clinical decisions on trastuzumab treatment. However, a high rate of trastuzumab resistance and relapse (at least 50%) highlights the need to identify a biomarker predictive of the clinical outcome of trastuzumab treatment. Analysis of a cohort of HER2+ BC patients from TCGA database revealed a comparable number of patients with down-regulation of IFI16 (49.2%), CXCL10 (25.4%), and CXCL11 (25.4%). Consistent with our findings, the high proportion of trastuzumab resistance might arise from HER2+ BC tumors having low IFI16, CXCL10, and CXCL11 expression, indicating that IFI16-CXCL10/11 could potentially serve as biomarkers to predict anti-HER2 mAb resistance. Interestingly, recent data show that EZH2 inhibition can boost an antiviral type I IFN response that sensitizes anti-HER2 mAb treatment (13). Our study demonstrates that combined inhibition of EZH2 and HDAC is necessary for robust IRG activation and immunotherapeutic responses, including IFI16-CXCL10/11. These results support the possibility of IFI16-CXCL10/11 as a biomarker determining anti-HER2 mAb resistance and evaluating the clinical efficacy of our proposed combination drug regime.
The present work demonstrates that in addition to modulating intrinsic HER2 downstream signaling, EZH2 also suppresses immune responsive pathways to limit tumor immunity against HER2+ BC. We have previously reported that the EZH2 induces the inactivation of PP2A tumor suppressor complex, thus leading to sustained mTOR/RSK signaling to promote resistance to anti-HER2 therapy (57). Without the reactivation of the IRG expression, particularly the IFI16/STING/CXCLs cascade, the trastuzumab-mediated NK or T cells lysis is limited. Induction of IFI16/STING/CXCLs cascade by EZH2/HDAC inhibitors revitalizes the trastuzumab responses to ADCC-mediated killing, allowing NK and T cell immune response in sensitizing the anti-HER2 treatment in BC. Thus, the EZH2-mediated epigenetic mechanism can engage both tumor intrinsic and extrinsic mechanisms to enable resistance to anti-HER2 therapy.
Of notice, our combinatorial epigenetic treatment also increases the expression of tumor antigen presentation machinery MHC class I and II related molecules, as represented by HLA-A, HLA-DRA, and CIITA. The availability of tumor antigens favors antigen presentation to T cells and subsequently facilitates the development of tumor-specific T cell immunity (58). Our combinatorial epigenetic treatment could increase the expression of MHC class I and II related molecules, as represented by HLA-A, HLA-DRA, and CIITA. Although we cannot exclude the roles of antigen presentation in the combination drug effects, we noticed that blockade of IFI16-CXCL10/11 signaling was sufficient to eliminate the impact of the epigenetic drug in vitro and in vivo, suggesting that activation of the IFI16 signaling cascade is indispensable for an effective anti-HER2 mAb response. We propose that epigenetic inactivation of IFI16 modifies both NK/ADCC-mediated innate immunity and subsequently the adaptive immune response, which involves T cell activation and memory immunity induction. We demonstrated that the T cell-dependence of the epigenetic treatment is also associated with IFI16-dependent expression of CXCL10 and CXCL11 as the tumor infiltration of CD8+ T cells were suspended in IFI204 (IFI16) knockdown tumors or in mice treated with anti-CXCR3 mAb, which inhibits T cell CXCR3 receptor binding to CXCL10 and CXCL11. The abrogation of the epigenetic inhibitor-activated antitumor effect indicates the importance of resensitizing the tumor microenvironment to provide an immunocompetent niche for responding to antitumor immunity.
This study shows that the compound we developed, D9, can induce complete tumor regression in 30% of mice when combined with ENT and α-ErbB2 mAb, which translated to long-term tumor-free survival. This observation has significant translational implications as it is known that in the clinic, the pCR with no residual disease after anti-HER2 mAb-based NAT correlates with patient survival benefits. Although EPZ plus ENT and α-ErbB2 can elicit a similar combination effect, it did not yield a complete and durable tumor eradication in our model for reasons that remain to be defined. However, EZH2 is known to have both catalytic and noncatalytic tumor-promoting effects in cancers, including BC (56, 59). While EPZ is an EZH2 catalytic inhibitor, DZNep is known to act as an EZH2 degrader (60), which has a powerful antitumor effect targeting both catalytic and noncatalytic activities of EZH2 (61, 62). The DZNep analog D9 has a similar capacity to deplete EZH2 with an improved safety profile in mice (34). However, whether the more potent efficacy of D9/ENT is related to its ability to target the additional noncatalytic function of EZH2 remains to be determined. Nevertheless, the translational value of D9 and the combination strategy warrant further investigation.
Overall, our study establishes the importance of IFI16 activation to both innate and adaptive immune-mediated responses in anti-HER2 mAb treatment. We also provide a clinically applicable strategy to improve current antitumor therapy for HER2+ BC patients, potentially targeting residual or refractory metastatic disease to improve clinical outcomes.
Methods and Materials
Human Samples.
All human biological samples were collected with informed consent. The study was approved by the Ethics Committee of Tan Tock Seng Hospital (Singapore), the Ethics Committee of the Region of Southern Denmark (Denmark), and the Ethics Committee of Saint John’s Cancer Institute (United States).
Animal Models.
All in vivo experiments were conducted in compliance with animal protocols approved by the Institutional Animal Care and Use Committee of the A*STAR-Biopolis Institutional Animal Care and Use Committee of Singapore. 4T1-HER2-luc (5 × 104 cells) were injected into both mammary fat pads (left and right) of 8- to 12-wk-old female Balb/C mice (strain name: BALB/cAnNTac) obtained from The Jackson Laboratory. Mice were randomized to different treatment groups and monitored for tumor growth twice per week. Two different treatment schedules and dosages were carried out. For the epigenetic drug treatment-only experiments, 60 mg/kg D9 or 100 mg/kg EPZ were administrated daily (starting from day 1), followed by daily administration of 10 mg/kg ENT 2 d posttreatment of D9 (day 3). For experiments involving α-ErbB2, α-ErbB2 was administered twice per week at an initial loading dose of 200 μg on day 1, followed by 100 μg. Next, 60 mg/kg D9 or 50 mg/kg EPZ plus 10 mg/kg ENT (D9/ENT) were given on alternate days in sequential order in which D9 or EPZ treatment began on day 3, while ENT treatment began on day 5. Control mice received an equivalent volume of vehicle and the isotype-matched control antibody. All treatments were given via intraperitoneal injection. Mice were weighed three times per week and doses were adjusted to body weight. Lung metastasis was evaluated by injected 100 μL of luciferin via intraperitoneal injection and measured using the IVIS imaging system (Xenogen). For the in vivo NK cell depletion experiment, 100 μg of antiasialo-GM1 (Life Technologies; Cat. no. 16-6507-39) was administered to mice 1 d before tumor inoculation and 1 d before treatment. For T cell depletion, 250 μg of 2.43 depletion (Bioxcell; Cat. no. BP0061) and 500 μg GK1.5 depletion antibodies (Bioxcell; Cat. No. BE0003-1) were used. For the CXCR3 inhibition experiment, anti-mouse CXCR3 (CD183) antibody (Bioxcell; Cat. no. BE0249) was administrated twice per week at 100 μg from day 1. The detailed protocols are described in SI Appendix.
In Vitro Cell Line Drug Treatments.
The following epigenetic drug treatment schedules were performed in all in vitro drug treatments unless stated otherwise. For single-drug treatment, cancer cells were subjected to 500 nM EPZ-6438, EPZ (Glpbio Technology; Cat. no. GC14062) and 100 nM D9 for 5 d, or 10 nM TSA (Sigma-Aldrich; Cat. no. T8552), 500 nM AZA (Sigma-Aldrich; Cat. no. A3656), and 250 nM ENT (Medchemexpress; Cat. no. HY-12163) for 2 d. For combination drug treatment, 1 µM DZNep (Sigma-Aldrich; Cat. no. 120964-45-6), 500 nM EPZ, or 100 nM D9 was added to cancer cells for 3 d before the addition of 250 nM ENT or 10 nM TSA for an additional 2 d. All epigenetic drugs were prepared in DMSO for all in vitro experiments. Trastuzumab was obtained from Roche. Anti-human/rat HER2, clone 7.16.4, α-ErbB2 (Cat. no. BE0277) was acquired from Bioxcell.
Coculture Systems.
Drug-treated cancer cells were seeded in 96-well plates for 1 d with epigenetic drugs to form monolayers. Culture media with drugs were then removed from cancer cells. For activated immune cells: NK, T, or splenocytes, were subsequently added to the cancer cells with or without anti-HER2 antibody for 48 h. Cancer cells were subsequently used for cell viability, protein expression, or gene-expression analyses. Cocultured T cells were subjected to flow cytometry analysis. The detailed protocols are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Mei Yee Aau, Poulina Phangrestu, Michelle Patricia, and Ee Ching Oh for assistance with the experiments; and M. K. Occhipinti for editorial assistance This study was supported by the Agency for Science, Technology, and Research of Singapore (A*STAR) and the Ministry of Health’s National Medical Research Council Open Fund Individual Research Grants NMRC/OFIRG/0023/2016 (to Q.Y.) and NMRC/CIRG/2021 (to S.C.L. and Q.Y.), and A*STAR/CDF/2021 (to L.-T.O. and K.S.M.Y.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201376119/-/DCSupplemental.
Data Availability
RNA-seq data for primary and metastatic HER2+ breast tumors in Fig. 1A were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE191230)(63). Uncropped Western blots, along with molecular weight standards, are included in SI Appendix.
References
- 1.Luque-Cabal M., García-Teijido P., Fernández-Pérez Y., Sánchez-Lorenzo L., Palacio-Vázquez I., Mechanisms behind the resistance to trastuzumab in HER2-amplified breast cancer and strategies to overcome it. Clin. Med. Insights Oncol. 10 (suppl. 1), 21–30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pohlmann P. R., Mayer I. A., Mernaugh R., Resistance to trastuzumab in breast cancer. Clin. Cancer Res. 15, 7479–7491 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swain S. M., et al. , Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA study): Overall survival results from a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 14, 461–471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huober J., et al. , Survival outcomes of the NeoALTTO study (BIG 1-06): Updated results of a randomised multicenter phase III neoadjuvant clinical trial in patients with HER2-positive primary breast cancer. Eur. J. Cancer 118, 169–177 (2019). [DOI] [PubMed] [Google Scholar]
- 5.LeVasseur N., et al. , Impact of pathologic complete response on survival after neoadjuvant chemotherapy in early-stage breast cancer: A population-based analysis. J. Cancer Res. Clin. Oncol. 146, 529–536 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurvitz S. A., et al. , Neoadjuvant trastuzumab, pertuzumab, and chemotherapy versus trastuzumab emtansine plus pertuzumab in patients with HER2-positive breast cancer (KRISTINE): A randomised, open-label, multicentre, phase 3 trial. Lancet Oncol. 19, 115–126 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Filho O. M., et al. , Impact of HER2 heterogeneity on treatment response of early-stage HER2-positive breast cancer: Phase II neoadjuvant clinical trial of T-DM1 combined with pertuzumab. Cancer Discov. 11, 2474–2487 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorsson V., et al. ; Cancer Genome Atlas Research Network, The immune landscape of cancer. Immunity 48, 812–830.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denkert C., et al. , Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 19, 40–50 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Ignatiadis M., et al. , Tumor-infiltrating lymphocytes in patients receiving trastuzumab/pertuzumab-based chemotherapy: A TRYPHAENA substudy. J. Natl. Cancer Inst. 111, 69–77 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salgado R., et al. , Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with lapatinib and trastuzumab: A secondary analysis of the NeoALTTO trial. JAMA Oncol. 1, 448–454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muntasell A., et al. , Interplay between natural killer cells and anti-HER2 antibodies: Perspectives for breast cancer immunotherapy. Front. Immunol. 8, 1544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirukawa A., et al. , Reduction of global H3K27me3 enhances HER2/ErbB2 targeted therapy. Cell Rep. 29, 249–257.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Kuznetsova M., et al. , Cytotoxic activity and memory T cell subset distribution of in vitro-stimulated CD8+ T cells specific for HER2/neu epitopes. Front. Immunol. 10, 1017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byrne A., et al. , Tissue-resident memory T cells in breast cancer control and immunotherapy responses. Nat. Rev. Clin. Oncol. 17, 341–348 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Szöőr Á., et al. , Trastuzumab derived HER2-specific CARs for the treatment of trastuzumab-resistant breast cancer: CAR T cells penetrate and eradicate tumors that are not accessible to antibodies. Cancer Lett. 484, 1–8 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Murad J. P., et al. , 204. HER2-specific chimeric antigen receptor T cells for the treatment of breast-to-brain metastasis. Mol. Ther. 24 (suppl 1), S79–S80 (2016). [Google Scholar]
- 18.Lam A. R., et al. , RAE1 ligands for the NKG2D receptor are regulated by STING-dependent DNA sensor pathways in lymphoma. Cancer Res. 74, 2193–2203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parkes E. E., et al. , Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J. Natl. Cancer Inst. 109, djw199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yum S., Li M., Frankel A. E., Chen Z. J., Roles of the cGAS-STING pathway in cancer immunosurveillance and immunotherapy. Annu. Rev. Cancer Biol. 1, 323–344 (2019). [Google Scholar]
- 21.Morel K. L., et al. , EZH2 inhibition activates a dsRNA-STING-interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nat. Can. 2, 444–456 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diamond M. S., et al. , Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 208, 1989–2003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H., et al. , Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563, 131–136 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Woo S. R. Sr, et al. , STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Naour J., Zitvogel L., Galluzzi L., Vacchelli E., Kroemer G., Trial watch: STING agonists in cancer therapy. OncoImmunology 9, 1777624 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jønsson K. L., et al. , IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 8, 14391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunphy G., et al. , Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol. Cell 71, 745–760.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emran A. A., et al. , Targeting DNA methylation and EZH2 activity to overcome melanoma resistance to immunotherapy. Trends Immunol. 40, 328–344 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Li T., et al. , Antitumor activity of cGAMP via stimulation of cGAS-cGAMP-STING-IRF3 mediated innate immune response. Sci. Rep. 6, 19049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nirmal A. J., et al. , Immune cell gene signatures for profiling the microenvironment of solid tumors. Cancer Immunol. Res. 6, 1388–1400 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Wee Z. N., et al. , EZH2-mediated inactivation of IFN-γ-JAK-STAT1 signaling is an effective therapeutic target in MYC-driven prostate cancer. Cell Rep. 8, 204–216 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Sun F., et al. , Combinatorial pharmacologic approaches target EZH2-mediated gene repression in breast cancer cells. Mol. Cancer Ther. 8, 3191–3202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X., et al. , CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS One 4, e5011 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang X., et al. , Functional characterization of D9, a novel deazaneplanocin A (DZNep) analog, in targeting acute myeloid leukemia (AML). PLoS One 10, e0122983 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tam E. K., et al. , 3-Deazaneplanocin A and neplanocin A analogues and their effects on apoptotic cell death. ChemMedChem 10, 173–182 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Ivashkiv L. B., IFNγ: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 18, 545–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang F., et al. , Combating HER2-overexpressing breast cancer through induction of calreticulin exposure by Tras-Permut CrossMab. OncoImmunology 4, e994391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P., et al. , The third generation anti-HER2 chimeric antigen receptor mouse T cells alone or together with anti-PD1 antibody inhibits the growth of mouse breast tumor cells expressing HER2 in vitro and in immune competent mice. Front. Oncol. 10, 1143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandó P., Rivero S. G., Rizzo M. M., Pinkasz M., Levy E. M., Targeting ADCC: A different approach to HER2 breast cancer in the immunotherapy era. Breast 60, 15–25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H., et al. , Shared antigenic epitopes and pathobiological functions of anti-p185(her2/neu) monoclonal antibodies. Exp. Mol. Pathol. 67, 15–25 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Drebin J. A., Link V. C., Weinberg R. A., Greene M. I., Inhibition of tumor growth by a monoclonal antibody reactive with an oncogene-encoded tumor antigen. Proc. Natl. Acad. Sci. U.S.A. 83, 9129–9133 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Truax A. D., Thakkar M., Greer S. F., Dysregulated recruitment of the histone methyltransferase EZH2 to the class II transactivator (CIITA) promoter IV in breast cancer cells. PLoS One 7, e36013 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu J., et al. , STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med. 7, 283ra52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang M., et al. , cGAS-STING, an important pathway in cancer immunotherapy. J. Hematol. Oncol. 13, 81 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corrales L., et al. , Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohkuri T., et al. , Intratumoral administration of cGAMP transiently accumulates potent macrophages for anti-tumor immunity at a mouse tumor site. Cancer Immunol. Immunother. 66, 705–716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vonderheide R. H., Domchek S. M., Clark A. S., Immunotherapy for breast cancer: What are we missing? Clin. Cancer Res. 23, 2640–2646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manguso R. T., et al. , In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon J., Bakhoum S. F., The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 10, 26–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu S., et al. , HER2 recruits AKT1 to disrupt STING signalling and suppress antiviral defence and antitumour immunity. Nat. Cell Biol. 21, 1027–1040 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Peng D., et al. , Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527, 249–253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Almine J. F., et al. , IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat. Commun. 8, 14392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y., et al. , Comparative epigenetic analyses reveal distinct patterns of oncogenic pathways activation in breast cancer subtypes. Hum. Mol. Genet. 23, 5378–5393 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Terada K., et al. , Association between frequent CpG island methylation and HER2 amplification in human breast cancers. Carcinogenesis 30, 466–471 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Singla H., et al. , Recent advances in HER2 positive breast cancer epigenetics: Susceptibility and therapeutic strategies. Eur. J. Med. Chem. 142, 316–327 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Lee S. T., et al. , Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol. Cell 43, 798–810 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Bao Y., et al. , EZH2-mediated PP2A inactivation confers resistance to HER2-targeted breast cancer therapy. Nat Commun. 11, 5878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griguolo G., Pascual T., Dieci M. V., Guarneri V., Prat A., Interaction of host immunity with HER2-targeted treatment and tumor heterogeneity in HER2-positive breast cancer. J. Immunother. Cancer 7, 90 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahara S., et al. , HIFI-α activation underlies a functional switch in the paradoxical role of Ezh2/PRC2 in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 113, E3735–E3744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan J., et al. , Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 21, 1050–1063 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koyen A. E., et al. , EZH2 has a non-catalytic and PRC2-independent role in stabilizing DDB2 to promote nucleotide excision repair. Oncogene 39, 4798–4813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan J., et al. , EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood 121, 4512–4520 (2013). [DOI] [PubMed] [Google Scholar]
- 63.L.-T Ong, H. Ditzel, Q. Yu, RNA-seq analysis on human primary breast cancer tumor and distant metastasis tumor. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE191230. Deposited 19 December 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data for primary and metastatic HER2+ breast tumors in Fig. 1A were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE191230)(63). Uncropped Western blots, along with molecular weight standards, are included in SI Appendix.