Significance
Using ribosome profiling of cilia regeneration in green algae Chlamydomonas, we uncovered that ciliary genes undergo an early transcriptional activation and late translational suppression, the pattern of which guided us to identify serine palmitoyltransferase (SPT) as an essential regulator for ciliogenesis. We found that SPT-product ceramides interact with intraflagellar transport particle proteins, connecting the ciliary membrane with the axoneme, a bundle of microtubules and associated proteins forming the core of cilia. SPT mutations from patients with hereditary sensory neuropathy disrupted cilia, which could be rescued by sphingolipid supplementation, implying an effective therapeutic strategy.
Keywords: cilia, sphingolipid, ribosome profiling, cryo-electron tomography
Abstract
Cilium formation and regeneration requires new protein synthesis, but the underlying cytosolic translational reprogramming remains largely unknown. Using ribosome footprinting, we performed global translatome profiling during cilia regeneration in Chlamydomonas and uncovered that flagellar genes undergo an early transcriptional activation but late translational repression. This pattern guided our identification of sphingolipid metabolism enzymes, including serine palmitoyltransferase (SPT), as essential regulators for ciliogenesis. Cryo-electron tomography showed that ceramide loss abnormally increased the membrane-axoneme distance and generated bulged cilia. We found that ceramides interact with intraflagellar transport (IFT) particle proteins that IFT motors transport along axoneme microtubules (MTs), suggesting that ceramide–IFT particle–IFT motor–MT interactions connect the ciliary membrane with the axoneme to form rod-shaped cilia. SPT-deficient vertebrate cells were defective in ciliogenesis, and SPT mutations from patients with hereditary sensory neuropathy disrupted cilia, which could be restored by sphingolipid supplementation. These results reveal a conserved role of sphingolipid in cilium formation and link compromised sphingolipid production with ciliopathies.
Cilia are conserved organelles that power cell motility (where they are often termed flagella) and perceive environmental signals (1–3). Defects in cilium formation or function cause at least 35 types of diseases, collectively termed ciliopathies, ranging from primary ciliary dyskinesia to situs in versus (1). Cilium regeneration has received additional attention at this challenging time of the COVID-19 pandemic. A temporary loss of smell or anosmia resulting from olfactory cilia dysfunction becomes evident as a common symptom of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A study using early transmission electron microscopy of the nasal mucosa from a coronavirus-infected patient revealed that olfactory cilia loss represented one of the most striking ultrastructural abnormalities (4). The observed intermediate and short cilia in the recovering patients implied that cilia regeneration might correlate with smell restoration (5).
Like other regeneration processes, cilium regeneration requires the signaling pathways and protein machinery that build the organelle during cilium formation (2, 3, 6). Systematic surveys have identified more than 1,000 flagella-associated proteins that assemble cilia and perform ciliary motility and signaling (7, 8). A pioneering study with Chlamydomonas described that cycloheximide, which blocks eukaryotic translational elongation, inhibited flagellar regeneration after amputation, indicating that new protein synthesis in the cytoplasm is vital for flagellar regeneration (9). Although ciliogenesis relies on the cytoplasmic protein machinery, we know little about how nonciliary proteins act outside cilia to promote cilium formation or regeneration.
Cilia consist of a specialized membrane surrounding an axoneme composed of doublet microtubules (MTs) that originate from the basal body (10–12). The axoneme protrudes from the plasma membrane and elongates for 5 to 10 μm with a diameter of ∼300 nm. Cilia, or flagella, are evolutionarily conserved organelles that develop in diverse organisms ranging from Chlamydomonas to mammals (6, 13). In addition to Chlamydomonas, other well-established cilia models include the Caenorhabditis elegans amphid and phasmid cilia, mouse inner medullary collecting duct (IMCD3) epithelial cells, and zebrafish lateral line neuromasts and olfactory placode (14–16). Previous research has been dedicated to understanding how membrane lipids regulate ciliary signaling (2, 11, 17). Still, it remains a mystery regarding how membrane lipids interact with ciliary proteins to form and maintain cilia with rod-like morphology. The sphingolipid ceramide localizes in the Chlamydomonas flagella and the primary cilia of murine ependymal cells, and pharmacological inhibition of ceramide synthesis blocks flagellar regeneration (12, 18, 19). These studies postulate that ceramides regulate ciliogenesis by activating cytosolic glycogen synthase kinase-3 or preventing activation of HDAC6 via cytosolic aPKC and AurA, arguing that the contribution of ceramide to ciliogenesis is independent of the ciliary membrane (18).
Because protein synthesis is restricted to the cytoplasm, ciliogenesis requires the selective import and transport of ciliary proteins out to the tip by intraflagellar transport (IFT), a MT-based transport event. The MT plus-end motor protein kinesin-2 drives the anterograde movement of IFT particles, delivering ciliary precursors to the distal tip to elongate cilia, and the cytoplasmic dynein-2 mediates retrograde IFT, recycling the anterograde IFT machinery and ciliary turnover products (2, 6, 17). IFT particles move between the ciliary membrane and axoneme, but their association with the ciliary membrane is unclear.
Early transcription studies reveal the transcriptional pattern of ciliogenesis (8, 20–23), but RNA sequencing (RNA-Seq) is inadequate to describe which messenger RNAs (mRNAs) are translated into proteins. By contrast, ribosome footprint sequencing (Ribo-Seq) measures translation. Ribo-Seq studies have been conducted in Chlamydomonas (24–26). However, cells with intact cilia rather than cells that regenerate cilia were used. Thus, it remains unknown how nascent proteins are translated to reassemble cilia. By performing global translatome profiling of Chlamydomonas flagellar regeneration, we identified enzymes catalyzing sphingolipid biosynthesis as crucial regulators for cilium formation. We showed that ciliary ceramides interact with IFT particles that IFT motors transport along axoneme MTs, revealing a motile lipid–protein complex that connects the ciliary membrane with the axoneme and is essential for cilium formation and morphology.
Results
Translatome Profiling during Chlamydomonas Flagellar Regeneration.
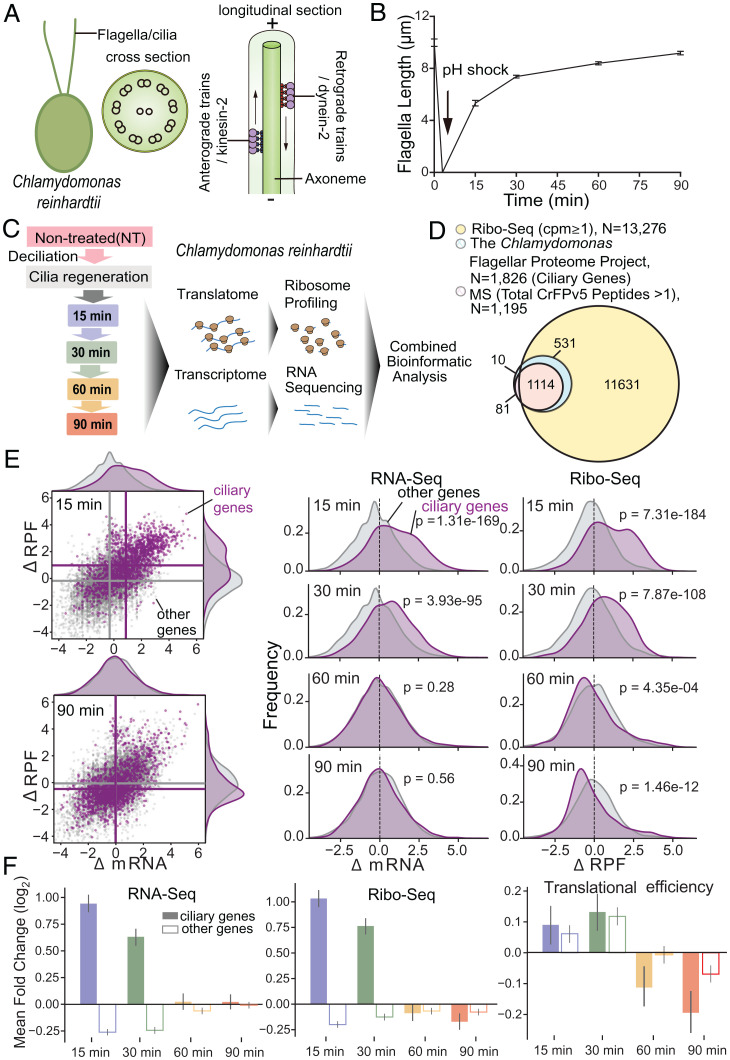
Chlamydomonas cells form flagella that are virtually identical to human motile cilia. The axoneme of flagella usually contains one pair of singlet MTs surrounded by nine outer doublet MTs organized in a circle, and the selective import and transport of ciliary proteins out to the tip is mediated by IFT (Fig. 1A). To monitor the global translational changes during flagellar regeneration, we collected the cells of the synchronized Chlamydomonas before deflagellation (nontreated [NT]) and during regeneration at 15, 30, 60, and 90 min to generate libraries for Ribo-Seq and RNA-Seq (Fig. 1B and C). Our RNA-Seq and Ribo-Seq measurements show high reproducibility (SI Appendix, Fig. S1A). Samples during flagellar regeneration are separated from controls on principal component analysis plots (SI Appendix, Fig. S1B). Sequenced ribosomal footprints display a three-nucleotide periodicity, indicating that they are protected by actively translating ribosomes (SI Appendix, Fig. S1C). The correlation coefficient for the overall transcriptional and translational levels in cilia regeneration was about 0.57 (SI Appendix, Fig. S1D). This suggests that similar to yeast, mammalian cells, and plants (27–29), changes at the transcriptional and translational levels during cilia regeneration in Chlamydomonas are not fully correlated. We also noticed that ribosome biogenesis-related genes were translationally up-regulated immediately during cilia regeneration (SI Appendix, Fig. S1E). Cycloheximide inhibited cilia regeneration after amputation, indicating that new protein synthesis is vital for cilia regeneration (9). Thus, during cilia regeneration, the ribosome-related genes are translationally up-regulated before translating other proteins, which agrees with the notion that the synthesis of the translational apparatus is regulated at the translational level (30). From Ribo-Seq data, we inferred protein synthesis for 13,276 genes, ∼11 times more than ciliary proteins previously identified by mass spectrometry (MS) from the purified flagella that comprise only a subset of the cellular proteome (7) (Fig. 1D and Dataset S1). Several early ribosome profiling studies from Chlamydomonas (24–26) provided valuable information for understanding microRNA or chloroplasts but not cilia, likely because these studies prepared Chlamydomonas with intact flagella rather than ones with regenerating cilia.
Fig. 1.
Flagellar gene transcription and translation during regeneration. (A) The structure of Chlamydomonas flagella. The cross-section shows one central pair of MTs surrounded by nine outer MT doublets organized in a circle which is termed axoneme. The longitude section shows the selective import and transport of ciliary proteins out to the tip are mediated by intraflagellar transport (IFT), which is a molecular motor-driven process. As eukaryotic flagella are essentially identical organelles to cilia, the terms cilia and flagella are often used interchangeably. (B) The kinetics of flagellar regeneration in a population of C. reinhardtii. (C) Schematic overview of the experimental approach. The samples used for quantification in B are the same for ribosome profiling. (D) The Venn diagram shows the overlap between ciliary genes [from Chlamydomonas Flagellar Proteome (7), chlamyfp.org] and detected genes by Ribo-Seq or MS. (E) Right: Scatter plots show log2-fold change of RPFs (y-axis) and mRNA (x-axis) at 15 and 90 min after flagellar regeneration (30- and 60-min data are shown in SI Appendix, Fig. S2). Ciliary genes are in purple; other genes are in gray. Mean values per group are indicated as lines. Left: Kernel density estimate (KDE) plots show the distribution of log2-fold change of mRNA and RPFs at 15, 30, 60, and 90 min after flagellar regeneration. (F) The mean log2-fold change (P value is shown in SI Appendix, Fig. S2) of RNA-Seq (Left), Ribo-Seq (Middle), and translational efficiency (Right) at 15 (blue), 30 (green), 60 (orange), and 90 (red) min after flagellar regeneration for ciliary genes (solid bars) and detected genes other than ciliary genes (hollow bars).
Early Transcriptional Activation and Late Translational Repression of Flagellar Genes during Regeneration.
To dissect the translational and transcriptional changes during flagellar regeneration, we compared the ribosome profile, RNA level, and translational efficiency (the ratio of normalized Ribo-Seq and RNA-Seq reads per transcript) of postdeflagellation (15, 30, 60, and 90 min) with those before deflagellation (NT). The flagellar genes which are annotated from the Chlamydomonas Flagellar Proteome Project (chlamyfp.org) in the Chlamydomonas flagellar proteome study with frequent updates (7) were used for the flagellar gene database in our study. We found that the annotated flagellar genes’ RNA levels are most activated at 15 min postdeflagellation (SI Appendix, Fig. S2A and Dataset S2), which is consistent with early transcriptome studies (8, 20–22). If transcriptional activation is the primary driver for flagellar regeneration, an increase in ribosome-protected fragments (RPFs) should parallel RNA up-regulation in ciliary genes. Conversely, if translational activation is the leading cause, the RPF increase should be more prominent than RNA. We observed significant RNA activation of ciliary genes compared to other genes at 15 and 30 min postdeflagellation, a rapid phase of flagellar regeneration (Fig. 1E and F and SI Appendix, Fig. S2B and C), with corresponding increases in RPFs. Intriguingly, at 60 and 90 min after amputation, the slow step of flagellar regeneration, we detected a statistically significant decrease in translational efficiency without an apparent reduction in corresponding RNA levels (Fig. 1E and F and SI Appendix, Fig. S2B and C). These results indicate that ciliary genes undergo transcriptional activation in parallel with active translation in the early rapid flagella regeneration phase, followed by late translational repression.
To further analyze ciliary gene transcription and translation, we clustered them into three categories using a hierarchical clustering algorithm based on changes of mRNA and RPFs (Fig. 2A and SI Appendix, Fig. S3A). We observed that 61.2% of ciliary genes were transcriptionally activated at 15 and 30 min but were translationally repressed at 60 and 90 min (Fig. 2B and C and SI Appendix, Fig. S3B), including genes that encode outer arm dynein, inner arm dynein, radial spokes, central pair apparatus, dynein regulatory complex, and IFT (Fig. 2D and Dataset S2). Cluster I revealed a characteristic transcriptional and translational pattern of ciliary gene during regeneration. The other ciliary genes encoding components in the centriole, membrane, and transition zone partially fell into the other two clusters: Cluster II accounted for 17.5%, which were not transcriptionally induced and were only translationally activated during cilia regeneration, whereas cluster III accounted for 21.3%, which were transcriptionally activated but translationally repressed (Fig. 2B–D and Dataset S2).
Fig. 2.
Ciliary gene clusters and novel ciliary regulators. (A) A global view of fold changes of mRNA and RPFs of ciliary genes at 15, 30, 60, and 90 min after flagellar regeneration. Shown is the result of hierarchical clustering (SI Appendix, Fig. S3A) of both mRNA and RPF data. Columns are genes, and rows are time points. The total signal is normalized per column to allow the comparison of patterns. (B) Pie charts of different categories of ciliary genes: 61.2% of the genes are transcriptionally activated at 15 and 30 min but translationally repressed at 60 and 90 min (cluster I), 17.5% of the genes sustain translationally repressed all the time (cluster II), and 21.3% of the genes maintain transcriptionally repressed but translationally activated after flagellar regeneration (cluster III). (C) The timing of the mean log2-fold change of RNA-Seq (green, Left axis), Ribo-Seq (gray, Left axis), and transcriptional efficiency (TE; blue, Right axis) is plotted for three ciliary cluster genes. (D) Histograms show the frequency distribution of various ciliary genes in three clusters. The sum of the three bars of each ciliary gene class is equal to 1. (E) The timing of the mean log2-fold change of RNA-Seq (green, Left axis), Ribo-Seq (gray, Left axis), and transcriptional efficiency (TE, blue, Right axis) is plotted for genes with a similar regulation pattern to ciliary genes cluster I. (F) The Venn diagram shows the overlap between genes in (E) with orthologs in C. elegans and genes in E with orthologs in Homo sapiens. A schematic overview of the screening strategy is on the right.
Sphingolipids Regulate Ciliogenesis in C. elegans.
We sought to identify novel regulators of ciliogenesis from genes that display the early transcriptional activation and late translational repression during flagellar regeneration. We found that 135 genes exhibit this pattern, but their ciliary roles have not been well documented (Fig. 2E, SI Appendix, Fig. S3C, and Dataset S3). Next, we searched for their homologs in C. elegans and humans and identified 32 as evolutionarily conserved (Fig. 2F and Dataset S3). The accessibility of the C. elegans mutant animals carrying mutations of the corresponding homologs facilitated us to determine their roles in ciliogenesis rapidly. Dye-filling defects (Dyf) indicate the failure of C. elegans to contact with their environment and suggest aberrant ciliary structure or function (31). By performing a visual screen for ciliary defects, we uncovered that 5 out of 18 examined mutant strains were Dyf, and four encode enzymes crucial for sphingolipid metabolism (SI Appendix, Fig. S3D). Next, we conducted a systematic survey of sphingolipid biosynthesis genes in C. elegans ciliogenesis. Out of 18 examined viable C. elegans mutant animals in sphingolipid metabolism (32), 15 developed the Dyf phenotype, albeit at the mild penetrance, which might prevent the previous forward genetic screens from isolating them (Fig. 3A for genes labeled in red). Nevertheless, by introducing a green fluorescent protein (GFP)–tagged ciliary marker IFT52/OSM-6::GFP into 13 mutant strains, we found that these Dyf animals indeed developed markedly shortened cilia (Fig. 3B and C).
Fig. 3.
Sphingolipid metabolism genes regulate ciliogenesis. (A) Schematic of the C. elegans orthologs of the sphingolipid metabolism pathway. Mutations of genes in red but not in blue cause ciliary defects. Genes in gray have not been examined due to the lack of mutant strains. (B) Amphid (Top, the C. elegans head sensory organ that contains chemosensory neurons forming sensory cilia at the dendritic endings) and phasmid (Bottom, C. elegans tail sensory organ) cilia in WT and sphingolipid-related mutants were labeled with IFT52/OSM-6::GFP. Arrowheads indicate the ciliary base and transition zone; arrows denote the junctions between the middle segment (m.s.) and the distal segment (d.s.) of sensory cilia. (Scale bar, 5 μm.) (C) Quantifications of the animals showing defects in cilium length (mean ± SD) in WT and mutant animals (n = 95 to 199 cilia in three independent experiments) (Left) and dye-filling assay (n = 250 to 350 animals in three independent experiments) (Right). Statistical significance was calculated by the Student’s t test using the means of biological replicates: *P < 0.05; **P < 0.01; ***P < 0.001; and N.S., not significant. (D) Total ceramide levels of SPT mutants relative to the WT animals are shown in columns. Errors bars represent the SD in three replicates. Statistical significance was calculated by the Student’s t test using the means of biological replicates: ***P < 0.001.
We focused on serine palmitoyltransferase (SPT), which catalyzes the first and rate-limiting reaction of de novo sphingolipid biosynthesis by condensing L-serine and palmitoyl-coenzyme A to 3-oxosphinganine (33). The C. elegans SPT holoenzyme contains the common SPTL-1 subunit and SPTL-2 or its isoform SPTL-3 (32, 34). We performed mRNA-Seq of the C. elegans sptl-1(ok1693), sptl-2(ok2753), and sptl-3(ok1927) mutant alleles to characterize their RNA defects caused by molecular lesions. In agreement with removing the 5′-untranslated region of sptl-1, the ok1693 allele reduced the sptl-1 mRNA level, indicating a weak loss-of-function allele (SI Appendix, Fig. S3E). The lesions in the sptl-2(ok2753) and sptl-3(ok1927) mutants almost entirely truncate their aminotransferase motifs, which suggest that they may be severe loss-of-function alleles (SI Appendix, Fig. S3E). In line with the redundant biochemical activity between SPTL-1/-2 and SPTL-1/-3 (32, 34), we successfully generated the viable sptl-1; sptl-2 or sptl-1; sptl-3 double mutant animals but failed to create the sptl-2; sptl-3 double mutants. Because sphingolipids are essential for embryonic development (35), the viable SPT mutant alleles are either partial loss-of-function (sptl-1) or have functional redundancy (sptl-2 and sptl-3), resulting in mild ciliary defects (Fig. 3C).
We performed electrospray ionization MS in C. elegans SPT mutants to quantify the total ceramide levels, a crucial product from sphingolipid metabolism (36). The ceramide amounts in sptl-1 and sptl-2 single-mutant or sptl-1; sptl-2 double-mutant animals were significantly lower than wild-type (WT) animals (Fig. 3D). To determine the ciliary ceramide level, we used immunofluorescence with anticeramide antibodies to stain the WT and sptl-2 mutant animals whose cilia were marked with GFP-tagged IFT-52/OSM-6. While we detected the ceramide in all the examined WT cilia, 31% of sptl-2 mutant animals did not show an apparent ceramide signal within cilia (SI Appendix, Fig. S3F, n = 15 or 16 for WT or sptl-2 animals, respectively). These results collectively indicate that sphingolipid metabolic enzymes regulate ciliogenesis and suggest an involvement of ceramides in this process.
Sphingolipids Regulate Ciliogenesis in Chlamydomonas.
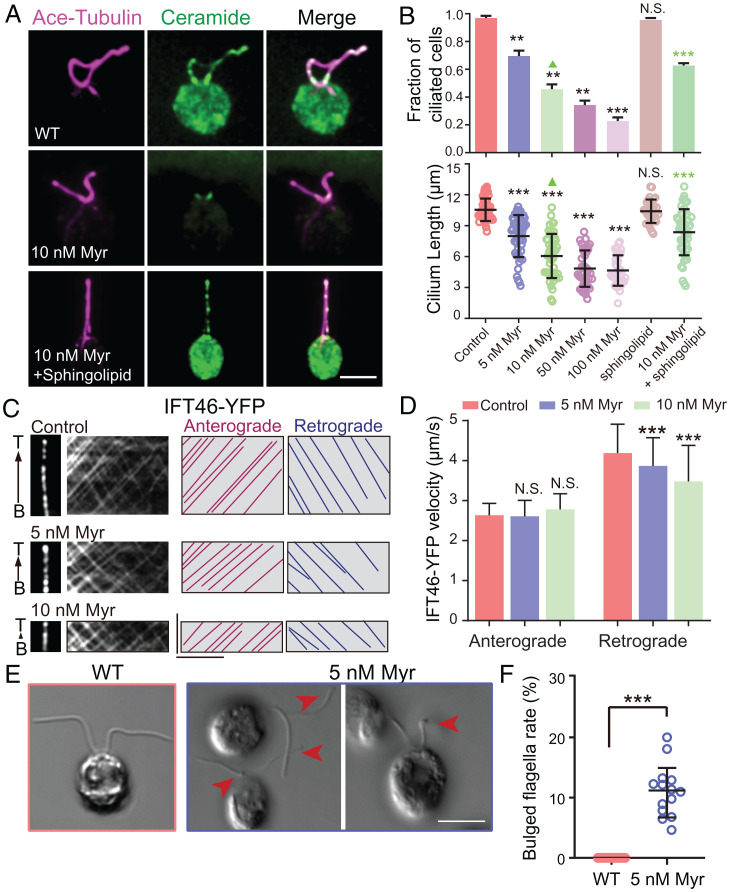
To address how the ciliary ceramides regulate cilium formation mechanistically, we aimed to perform cryo-electron tomography (cryo-ET) and biochemical experiments, for which Chlamydomonas flagella are advantageous. Immunofluorescence with anticeramide antibodies detected the ciliary localization of ceramides, which was effectively reduced by treating cells with the de novo sphingolipid biosynthesis inhibitor (Fig. 4A and SI Appendix, Fig. S4A). Myriocin reduced the fraction of ciliated cells and cilium length in a dose depedent manner and sphingolipid mixture partially restored them (Fig. 4B). Following yellow fluorescent protein (YFP)–labeled IFT46 motility in myriocin-treated algae cells, we found that the velocities of retrograde but not anterograde IFT were reduced in a dose-dependent manner (Fig. 4C and D), indicating that ciliary ceramides specifically promote IFT-dynein motility. Sphingolipid biosynthesis enzyme genes undergo a similar translational pattern to ciliary genes (SI Appendix, Fig. S4B), and myriocin treatment blocked ciliogenesis (Fig. 4B) and caused defective phototaxis (SI Appendix, Fig. S4C). In agreement with early studies (18), these findings demonstrate the crucial roles of ceramides in Chlamydomonas flagellar assembly and function.
Fig. 4.
Sphingolipids regulate ciliogenesis in Chlamydomonas. (A) Colocalization of ceramide and acetylated tubulin immunofluorescence in cilia of Chlamydomonas cells treated with ceramide biosynthesis inhibitors (0 to 10 nM myriocin) with or without exogenous 2 μM sphingolipid mixture [sphingolipid mix contains sphingosine (C17 base), sphinganine (C17 base), sphingosine-1-P (C17 base), sphinganine-1-P (C17 base), lactosyl(β) C12 ceramide, 12:0 sphingomyelin, glucosyl(β) C12 ceramide, 12:0 ceramide, 1and 2:0 ceramide-1-P) (magenta, acetylated tubulin; green, ceramide)]. (Scale bar, 5 μm.) (B) Quantifications in Chlamydomonas cells treated with ceramide biosynthesis inhibitors (0 to 100 nM myriocin) with or without exogenous 2 μM sphingolipid mixture. n = 32 to 52 cells from three independent experiments. Values are the mean ± SD for combined cell populations. Statistical significance was calculated by the Student’s t test using the means of biological replicates: **P < 0.01, ***P < 0.001, and N.S., not significant. (C) Kymographs show the anterograde and retrograde movement of YFP-tagged IFT46. Representative particle traces are marked with lines. (Scale bars represent, 5 μm [horizontal] and 5 s [vertical].) (D) Quantification of IFT velocities. N = 75 to 180 tracks from flagella in 20 to 35 alga cells from three independent experiments were used for statistical analysis. Values are the mean ± SD for combined cell populations. Statistical significance was calculated by the Student’s t test using the means of biological replicates: N.S., not significant; ***P < 0.001. (E) WT, 5 nM myriocin treated algae cells were imaged by differential interference contrast microscopy. Arrows indicate flagellar bulges. (Scale bar, 5 μm.) (F) Quantification of bulged flagella. n = 14 fields from three independent experiments were used for statistical analysis. Values are the mean ± SD. Statistical significance was calculated by the Student’s t test: ***P < 0.001.
Ceramide Loss Increased the Membrane-Axoneme Distance and Caused Bulged Cilia.
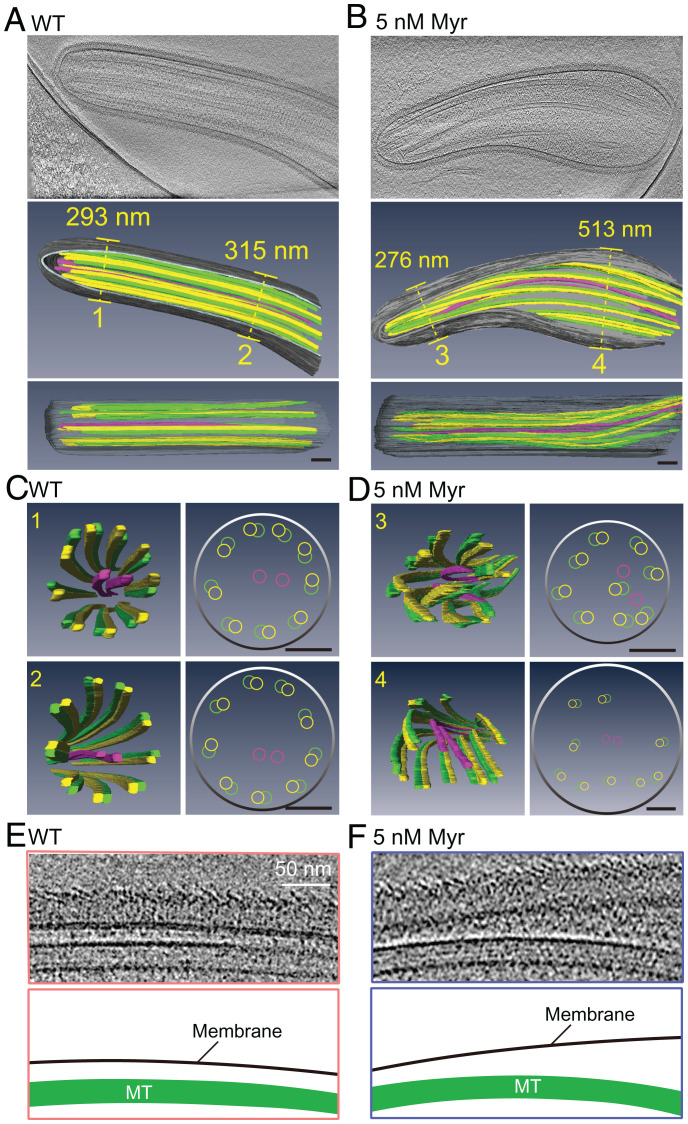
To examine the flagella structure, we employed differential interference contrast microscopy to show that myriocin treatment caused algae cells to form short flagella with bulges (Fig. 4E and F). Using a fluorescence lipid dye, Dioc6(3), we confirmed short and bulged flagella formation in myriocin-treated cells (SI Appendix, Fig. S5A and B). The purified flagella from myriocin-treated algae became more bulbous at room temperature for 90 min than WT (SI Appendix, Fig. S5C and D). The 300-nm thickness of the Chlamydomonas flagella allows structural analysis of flagella in close-to-native conditions using cryo-ET (37). We observed 8 out of 18 examined flagella from myriocin-treated cells developing bulbous flagella (Fig. 5A and B and SI Appendix, Fig. S5E), consistent with light microscopy results. At the ultrastructural level, nine doublets symmetrically distribute and surround two singlet MTs in WT flagella (Fig. 5C and Movie S1); however, myriocin treatment caused doublet MTs to spiral into the axonemal lumen (Fig. 5D and Movie S2), revealing an impaired nine-fold symmetry of axonemal MTs. In the bulbous flagella, the distance between the plasma membrane and axonemal MTs abnormally increased to more than 500 nm (Fig. 5E and F, SI Appendix, Fig. S5F, and Movies S1 and S2), which indicates that ceramide loss impaired the connection between the ciliary membrane and axonemal MTs.
Fig. 5.
Ceramide loss disrupted the membrane–axoneme interaction. (A–D) Longitudinal (A and B) and cross-section (C and D) cryo-ET analysis of the cilia in WT and cell treated by 5 Nm Myr. Representative images of flagella longitudinal sections in A and B and cross-sections in C and D (Movies S1 and S2). (Scale bars, 100 nm.) (E and F) Magnified views of longitudinal section cryo-ET micrographs and the model below (Movies S1 and S2). (Scale bar, 50 nm.)
Ceramides Bind to IFT Particles.
To understand whether and how the ciliary membrane interacts with axonemal MTs via ceramides, we performed immunoprecipitation experiments with biotin-linked ceramide immobilized streptavidin agarose beads and used biotin immobilized streptavidin agarose beads as a control according to the manufacturer’s instructions (Fig. 6A). Using the lysate from purified Chlamydomonas flagella, we isolated potential ceramide binding proteins and subjected them to MS analysis (Fig. 6A and SI Appendix, Fig. S5G). We found that ceramide beads but not the control beads pulled down IFT particle components (Fig. 6B, SI Appendix, Fig. S5H, and Dataset S4). We confirmed the ceramide–IFT particle interaction using Western blots (Fig. 6C). Given that the IFT motor protein transports IFT particles along axonemal MTs, we propose that the ceramide–IFT particle–IFT motor–MT association forms a physical connection between the ciliary membrane and axoneme (Fig. 6D). Although such a lipid–protein complex is mobile and undergoes dynamic changes at the ciliary base and tip, invariant IFT speeds and high frequency of IFT events (2) in both anterograde and retrograde directions allow the complex to be constant along the entire flagella, thereby stabilizing the ciliary membrane–axoneme connection.
Fig. 6.
Ceramides bind to IFT particles. (A) Schematics isolating ceramide binding proteins by immunoprecipitation using biotinylated ceramide immobilized on streptavidin agarose beads for MS and Western blot (WB) analysis, the biotin immobilized streptavidin agarose beads were used as control beads. (B) Mass spectrometric analysis of potential ceramide binding proteins by biotin-linked ceramide streptavidin agarose beads using the lysate from purified Chlamydomonas flagella. The plot compares proteins coprecipitated with control beads (y-axis) and ceramide beads (x-axis), and the colorful dots represent IFT proteins (Dataset S4 and SI Appendix, Fig. S5H). (C) Protein lysate from purified Chlamydomonas flagella, followed by SDS–polyacrylamide gel electrophoresis and immunoblotting using antibodies against IFT121, IFT122, and IFT88. The results are consistent in three replicates. (D) Model depicting the role of the ceramide–IFT particle–IFT motor–MT association in maintaining a rod-shaped cilium structure. The loss of ciliary ceramide caused short cilia with bulges.
Roles of Ceramide in Mouse and Zebrafish Cilia.
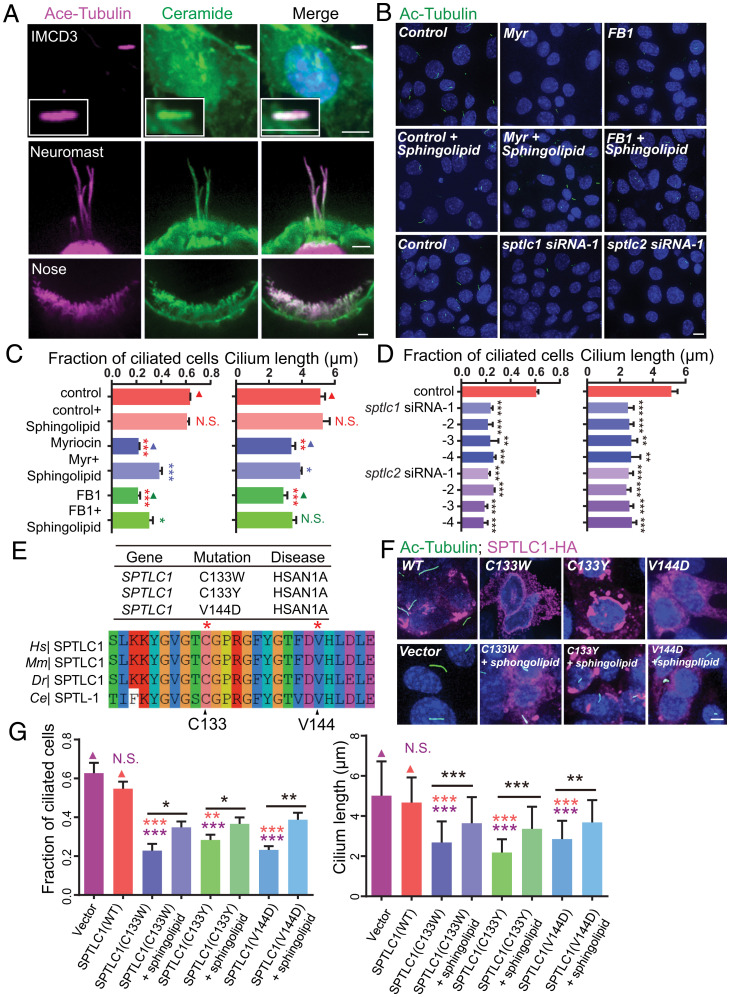
Cilia are evolutionarily conserved organelles. Model organisms, including Chlamydomonas reinhardtii, C. elegans to mammalian cells, and zebrafish, have been widely used to study cilium formation and model ciliopathies (14–16). To test whether ceramide has a conserved role in mammals, we examined whether ceramides enter cilia in mouse IMCD3 cells and zebrafish lateral line neuromasts and olfactory placode. Immunofluorescence with anticeramide antibodies detected the ciliary localization of ceramides in mouse IMCD3 cells and the zebrafish olfactory placode and lateral line neuromasts (Fig. 7A). By blocking ceramide generation with myriocin or ceramide synthase inhibitor fumonisin B1 (FB1), we showed that ceramide reduction inhibited cilium formation in the zebrafish olfactory placode and dramatically shortened cilium length in the lateral line neuromasts (SI Appendix, Fig. S6A–C). Likewise, incubating IMCD3 cells with either drug significantly reduced the fraction of ciliated cells or truncated cilia, which was partially restored by supplementing the medium with the sphingolipid mix containing ceramide (Fig. 7B and C). Consistent with the pharmacological results, small interfering RNA silencing of sptlc1 and sptlc2 genes in IMCD3 cells reduced ciliation and cilium length (Fig. 7B and D and SI Appendix, Fig. S6D). These results reveal that ceramide plays a conserved role in cilium formation in vertebrates.
Fig. 7.
Roles of ceramide in mouse and zebrafish cilia. (A) Colocalization of ceramide and acetylated tubulin immunofluorescence in cilia of mouse IMCD3 epithelial cells and zebrafish lateral line neuromasts (rosette-like sensory organs that detect directional water flow) and olfactory placode (nose, the lateral edge of the anterior neural plate) (magenta, acetylated tubulin; green, ceramide; blue, DAPI). (Scale bar, 5 μm.) (B) Representative immunocytochemistry images in IMCD3 cells treated with ceramide biosynthesis inhibitors (5 μM myriocin, 10 μM FB1) with or without exogenous 10 μM sphingolipid mixture [sphingolipid mix contains sphingosine (C17 base), sphinganine (C17 base), sphingosine-1-P (C17 base), sphinganine-1-P (C17 base), lactosyl(β) C12 ceramide, 12:0 sphingomyelin, glucosyl(β) C12 ceramide, 12:0 ceramide, and 12:0 ceramide-1-P] or transferred with sptlc1 or sptlc2 siRNA (green, acetylated tubulin; blue, DAPI) for 48 h. (Scale bar, 5 μm.) (C) Quantifications in IMCD3 cells treated with ceramide biosynthesis inhibitors (5 μM myriocin, 10 μM FB1) with or without exogenous 10 μM sphingolipid mixture. n = 161 to 179 cells from three independent experiments. Values are the mean ± SD combined cell populations. Statistical significance was calculated by the Student’s t test using the means of biological replicates: *P < 0.05; **P < 0.01; ***P < 0.001; and N.S., not significant. (D) Quantifications in IMCD3 cells transferred with sptlc1 or sptlc2 siRNA. n = 115 to 200 cells from three independent experiments. Values are the mean ± SD combined cell populations. Statistical significance was calculated by the Student’s t test using the means of biological replicates: **P < 0.01 and ***P < 0.001. (E) Alignment of sequences flanking the dominant human diseases-associated residue C133 and V144 in SPTLC1. (F) Representative immunocytochemistry images in IMCD3 cells transfected with SPTLC1 related mutants (green, acetylated tubulin; magenta, SPTLC1; blue, DAPI). (Scale bar, 5 μm.) (G) Quantifications in IMCD3 cells transfected with SPTLC1-related mutants. n = 90 to 120 cells from three independent experiments. Values are the mean ± SD combined cell populations. Statistical significance was calculated by the Student’s t test using the means of biological replicates: *P < 0.05; **P < 0.01; ***P < 0.001; and N.S., not significant.
Human SPT Mutations Affect Cilium Formation.
Multiple mutations in the human SPTLC1 subunit of SPT lead to hereditary sensory neuropathy type I (HSAN1), a common degenerative disorder of peripheral sensory neurons (38, 39). We wondered whether HSAN1-associated mutations in SPTLC1 affected cilium assembly. We studied three missense mutations (C133W, C133Y, and V144D) in SPTLC1 that occurred on conserved amino acid residues (Fig. 7E). SPTLC1 mutation inheritance mode is autosomal dominant, and transgenic mice overexpressing the C133W mutation developed an HSAN1-like phenotype (40, 41). To assay the effects of these mutations on ciliogenesis, we transfected the SPTLC1 mutants to IMCD3 cells. The transfected cells strikingly reduced the ciliation and cilia length, which could be rescued by supplemented sphingolipids (Fig. 7F and G). To examine the impact of these mutations in C. elegans, we introduced the C133W and C133Y variations in the sptl-1 gene’s corresponding location. By expressing them under the control of a ciliated neuron-specific promoter, we found that both SPTL-1 mutations reduced the ciliary length, whereas the transformation of the WT sptl-1 gene at the same DNA concentration did not cause any ciliary defects (SI Appendix, Fig. S6E and F). These results show that HSAN1-associated mutations in SPT disrupt cilium formation in mammalian cells and C. elegans.
Discussion
In summary, systematic translatome analyses of flagellar regeneration guided us to discover the function of sphingolipid metabolism enzymes in ciliogenesis. Focusing on SPT genes, this study provides an example of how global translatome profiling facilitates the discovery of novel ciliary regulators acting from the cytoplasm. We revealed an evolutionarily conserved role of ceramides in ciliogenesis and that abnormal sphingolipid production by mutations in patients with hereditary sensory neuropathy (HSN) disrupted ciliogenesis, linking ceramides to ciliary disorders. We provide evidence that the interaction between ciliary ceramides and IFT particles moved by IFT motor proteins on axonemal MTs connects the ciliary membrane with axonemes, crucial for generating and maintaining cilia with the characteristic rod-like morphology.
Compared to other systematic approaches such as proteome or transcriptome datasets (7, 20), Ribo-Seq results provide large-scale protein translational information about the previously uncharacterized cytoplasmic events during cilium formation and regeneration. Ribo-Seq data illustrate the translational status for 92% of the annotated protein-coding genes in the Chlamydomonas genome during flagellar regeneration (Dataset S1). The number of genes in the Ribo-Seq dataset is 11 times greater than that from MS-based ciliary protein identification (Fig. 1D and Dataset S1). The higher throughput is likely because some abundant ciliary proteins (e.g., tubulin or axonemal dynein) hinder the detection of lower-expressed proteins by MS and because many proteins that are vital for cilium formation do not necessarily localize in cilia. Future studies can be fruitful by characterizing the ciliary roles of other genes that undergo similar transcriptional and translational changes during flagellar regeneration.
We know relatively little about pathways outside cilia to promote cilium assembly compared to extensive ciliary protein studies. Considering that cilium regeneration starts from a cell that loses its cilium under stress or disease conditions, the most upstream is that the cell senses the ciliary loss or dysfunction and communicates with the nuclei to turn on the transcription program to recover cilia. Other intracellular processes involve translation regulation of ciliary proteins and biosynthesis of ciliary lipids. From this point of view, cytoplasmic regulators of cilium formation and regeneration are perhaps equivalently vital to well-characterized ciliary proteins. We speculate that some cytosolic molecules may not be required for ciliogenesis during development but may promote the restoration of cilium structure and function during patient recovery from diseases such as SARS-CoV-2 infection.
We identified enzymes in the sphingolipid metabolism as a set of essential regulators for ciliogenesis. The ciliary lipids are known for their function in ciliary signaling transduction (11, 17), but their contribution to building the ciliary structure was unclear. The chemical perturbation of sphingolipid biosynthesis inhibited flagellar regeneration in Chlamydomonas and suggested that sphingolipids may regulate membrane polarity at the early stage of cilium initiation (18). Our work shows that impaired sphingolipid metabolism reduces the ciliary ceramide level, which increases the ciliary membrane and axoneme distance and shortens cilia with bulges, revealing a direct contribution of sphingolipids to ciliogenesis. The loss of ciliary ceramides reduced IFT-dynein but not IFT-kinesin motility, suggesting that the ciliary lipid composition might more easily influence dynein motility. Mechanistic studies addressing the impact of ciliary ceramides on dynein motility will require new tools for visualizing lipids, like seeing proteins, in a living cilium.
The HSN patient mutations significantly reduce cilia length and number in mammalian cells, implying that HSN disease mutations in SPT might be unidentified molecular lesions that cause ciliary disorders. The causative link between impaired sphingolipid metabolism and ciliopathies still awaits further investigation. Several studies have recently identified mutations in a ceramide kinase-like gene leading to autosomal recessive retinitis pigmentosa in patients, mirroring the eye dysfunction of ciliopathies (42). A congenital pansensory neuropathy and progressive retinitis pigmentosa were reported in the same patient (42). Interestingly, mice expressing the SPTLC1(C133W) mutation had a reduced sperm count and motility (40, 41). While abnormal sphingolipid composition can lead to axonal neuropathy, the defective ciliary structure or function in HSAN1 neurons might be an alternative cause for axonal neuropathy. Cilia have not been linked to this disease, but abnormal cilia formation has been shown to disrupt axonal projection (43).
Along with our results, these data might offer a glimpse into the potential role of sphingolipid metabolism in ciliopathies. Further massive sequencing efforts of the human genome and disease modeling and functional studies in model organisms will be required to establish the connection between sphingolipid metabolism and ciliary diseases. The exogenous addition of sphingolipids partially restored ciliogenesis in cells with impaired sphingolipid generation (Figs. 4A and B and 7B, C, F, and G), and serine supplementation significantly improved the sperm count and motility in SPTLC1 (C133W) mutant mice (40, 41). These findings demonstrate the proof-of-principle prototypes in which ciliary defects can be restored by manipulating lipid generation, holding great promise for curing ciliopathies and accelerating cilia regeneration.
Materials and Methods
Strains and Genetics.
SI Appendix, Table S1 summarizes the strains used in this study. All the plasmids and primers are listed in SI Appendix, Table S2. All the animal experiments were performed following governmental and institutional guidelines.
C. reinhardtii (WT strain CC-125) was grown in Tris-acetate-phosphate (TAP) growth medium with a light/dark cycle of 14:10 h at 20 °C. Chlamydomonas cells were harvested and resuspended in 10 mM Hepes buffer for 2 h and then were deflagellated by adding acetic acid to the TAP growth medium to pH 4.5 for 30 s. Sodium hydroxide was rapidly added to neutralize the pH, and cells were further incubated in the TAP medium (44) and then harvested at 0, 15, 30, and 90 min.
Danio rerio WT strain Tübingen embryos were raised at 28.5 °C. Fish maintenance followed the institutional animal care and use committee protocol, with the approval of the Tsinghua University Animal Care and Use Committee. Whole-mount immunostaining in zebrafish was carried out based on the standard protocol using the following primary antibodies: antiacetylated α-tubulin antibody (3971, Cell Signaling Technology, 1:500 dilution), anticeramide antibody (C8104, Sigma, 1:100 dilution). The 24-hours post fertilization WT embryos were treated with 25 μM myriocin and 250 μM FB1 or dimethyl sulfoxide; after 4 d, they were collected for the whole-mount immunostaining analysis.
C. elegans strains were raised with Escherichia coli strain OP50 on nematode growth medium (NGM) plates at 20 °C. Adult hermaphrodite worms were used in live-cell imaging experiments and Dyf assays. Worms at mixed stages were collected to perform lipid MS and RNA-Seq. All animal experiments were performed following governmental and institutional guidelines.
IMCD3 cells were maintained in DMEM/F12 (Invitrogen) containing 10% fetal bovine serum, 0.3 mg/mL glutamine (Sigma-Aldrich), 100 U/mL penicillin (Invitrogen), and 100 U/mL streptomycin (Invitrogen) in a 5% CO2, 37 °C incubator until reaching confluency.
Measurement of Flagellar Length and Flagellated Cells.
Chlamydomonas cells were fixed with 0.5% Lugol’s solution at room temperature and imaged with an Axio Observer Z1 microscope (Carl Zeiss MicroImaging) equipped with a 100× 1.45 numerical aperture (NA) objective and an EM charge coupled device(CCD) camera (Andor iXon + DU-897DC00-# BV-500). Images were acquired with an exposure time of 200 ms by μManager (https://micro-manager.org/) and processed with ImageJ software. At least 100 cells were counted for each condition in biological triplicates.
Young-adult C. elegans hermaphrodites were anesthetized with 0.1 mmol/L levamisole, mounted on 3% agarose pads at 20 °C, and imaged with an Axio Observer Z1 microscope (Carl Zeiss MicroImaging) equipped with a 100× 1.45 NA objective, an EM CCD camera (Andor iXon + DU-897DC00-# BV-500), and the 488- and 568-nm lines of a Sapphire CW CDRH USB Laser System with a spinning-disk confocal scan head (Yokogawa CSU-X1 Spinning Disk Unit). Time-lapse images were acquired with an exposure time of 200 ms at 0-s intervals by μManager and processed with ImageJ software. The identical parameters were used to image the WT and mutant animals.
Phototaxis Assays.
Chlamydomonas cells were resuspended with phototaxis assay buffer (5 mM Hepes, pH 7.4, 1 mM KCl, 0.3 mM CaCl2, and 0.2 mM ethylene glycol tetraacetic acid) and were activated by red light (λ = 620 nm) for 5 to 15 min before the assays. Two-milliliter cell suspensions (∼107 cells/mL) were transferred to 35-mm Petri dishes and were illuminated from a defined side with a green light (λ = 525 nm, ∼1 μmol photon·m−2·s−1 for 30 min). All phototaxis experiments were performed 8 h after the light phase began.
Immunofluorescence Staining.
Chlamydomonas or IMCD3 cells were fixed with methanol at −20 °C for 20 min or 4% paraformaldehyde/phosphate-buffered saline (PBS) for 15 min at room temperature, respectively. Cells were washed three times with PBS and permeabilized by incubation with 0.5% Triton X-100 in PBS for 15 min. IMCD3 cells were incubated on ice for 30 min to depolymerize cytoskeletal MTs before fixation. Nonspecific binding sites were blocked with 4% bovine serum albumin (BSA) for 1 h at 37 °C. Cells were incubated with primary antibodies at 4 °C overnight. The following primary antibodies were used for immunofluorescence: antiacetylated-tubulin rabbit IgG (1:1,000, catalog [Cat.] #ab179484, Abcam) and anticeramide mouse IgG (1:100, Cat. #C8104-50TST, Sigma-Aldrich). Dylight 488 goat anti-rabbit IgG (1:500, Cat. #A23220-1, Abbkine), Dylight 549 goat anti-rabbit IgG (1:500, Cat. #A23320-1, Abbkine ), Dylight 488 goat anti-mouse IgG (1:500, Cat. #A23210-1, Abbkine), and Dylight 549 goat anti-mouse IgG (1:500, Cat. #A23310-1, Abbkine) were diluted 1:500 in 4% BSA/PBS and cells were incubated for 1 h at 37 °C. DAPI (Sigma-Aldrich) was used to visualize the nuclei.
Dyf Assay.
Young-adult worms were collected randomly into 200 μL dye solution (DiI 1,1′-dioctadecyl-3, 3, 3′, 3′-tetramethylindo-carbocyanine perchlorate, Sigma) at working concentration (20 mg/mL), followed by 30-min incubation in the dark at room temperature. Worms were transferred to NGM plates seeded with OP50. After 1 h, we examined dye uptake using a fluorescence stereoscope. Each strain was examined for at least 100 worms in three independent experiments. We examined the mutant alleles of genes pmk-3(ok169), kgb-1(um3); kgb-2(km16), sek-1(km4), rom-2(ok996), scl-1(ok1185), shw-3(ok1884), sel-12(ok2078), trxr-2(ok2267), sms-5(ok2498), F15D4.4(ok3200), cca-1(ok3442), drh-1(ok3495), Y55F3AR.2(ok1737), sms-3(ok3540), spl-2(ok490), and hyl-1(ok976), and they did not show any dye-filing defect. The mutant alleles of genes C30F12.2(ok3505), sms-1(ok2399), sptl-1(ok1693), sptl-2(ok2753), sptl-3(ok1927), hyl-2(ok1766), lagr-1(gk327), lagr-1(gk331), asah-2(ok564), asah-1(tm495), sphk-1(ok1097), cerk-1 (ok1252), asm-1(tm5023), asm-2(tm3746), asm-3(ok1744), sms-2(tm2757), and ttm-5(tm6585) showed a dye-filing defect with penetrance ranging from 2.6 to 10% from three independent experiments. The WT animals did not show any dye-filling defects (n > 300). The strain information is provided in SI Appendix, Table S1.
Lipid Analysis.
Unsynchronized WT Bristol N2 or sphingolipid-related mutant strains were collected and washed three times with M9 buffer. Worm pellets were subjected to liquid nitrogen three times and thawing followed by fragmentation by 0.5-mm-diameter glass beads using FastPrep-24 (MP Biomedicals). Total lipids were extracted using a mixture of chloroform and methanol (2:1; vol/vol) at 48 °C overnight and then back-extracted with chloroform. The lower organic phase was collected after centrifugation, washed with the artificial upper phase (chloroform/methanol/H2O; 3:48:47) once, and dried under nitrogen gas. Sphingolipidomics analysis by liquid chromatography–tandem MS was performed at the lipidomics facility of Tsinghua University in China. The ceramide composition was normalized to lipid phosphate.
Image Processing and Analysis.
We used ImageJ software (https://rsbweb.nih.gov/ij/) to process images and generate kymographs. The ciliary marker YFP::IFT46 was used to measure IFT speeds. Separated anterograde and retrograde kymographs were generated with the KymographClear toolset plugin in ImageJ (www.nat.vu.nl/erwinp/downloads.html) by manually drawing lines along cilia at the width of 3 pixels. Complete tracks of individual IFT particles were manually selected and carefully drawn with the KymographClear toolset.
Flagella Isolation.
Flagella were isolated from Chlamydomonas cells using the pH shock method as described previously (45) with a few modifications. Briefly, WT cells were grown in 8 L of TAP medium until the cell density reached ∼5 × 106 cell/mL and the algae cells were harvested and resuspended in 500 mL of 10 mM Hepes buffer for 2 h. The algae cells were then resuspended with 100 mL of 10 mM Hepes containing 5% sucrose. The pH of the suspension was quickly lowered to 4.5 for about 30 s by the addition of 0.5 mM acetic acid while stirring with a magnetic stirrer. Then 0.5 mM KOH was added to the suspension to raise the pH to 7.0. The suspension was then transferred to 50-mL polycarbonate tubes for about 35 mL suspension per tube. Then 10 mL of 25% sucrose in 10 mM Hepes was injected into the underlayer of the tubes. Centrifugation at 2,500 × g for 10 min at 4 °C sedimented the cell body through 25% sucrose. The flagella were in the supernatant which contained 5% sucrose. The supernatant was transferred to a new 50-mL tube and underlaid again with 25% sucrose to remove any remaining cell bodies. The 5% sucrose layer was then transferred to a new 50-mL tube and centrifuged at 10,000 × g for 10 min. The pellets were resuspended in HMDEK buffer [20 mM Hepes, pH 7.2, 5 mM MgCl2, 1 mM ethylene diamine tetraacetic acid (EDTA), and 1 mM dithiothreitol (DTT) containing a complete protease inhibitor mixture from Roche]. For flagellar fractionation, about 100 μL flagella (50 μg/μL) was first incubated with 0.5% igepal for 30 min and centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was collected as the membrane plus matrix fraction for the affinity purification assay.
Labeling Flagella with Fluorescent Dye Dioc6(3).
Flagella were stained using a previously described protocol (46) with a few modifications. The algae cells were grown in TAP medium until the cell density reached ∼5 × 106 cell/mL and they were harvested and resuspended in TAP medium to a final cell concentration of ∼1 × 107 cell/mL Chlamydomonas cells were labeled with Dioc6(3) (0.05 μg/mL, Invitrogen) for 1 min at room temperature and imaged with an Axio Observer Z1 microscope (Carl Zeiss MicroImaging) equipped with the 488- and 568-nm lines of a Sapphire CW CDRH USB Laser System with a spinning-disk confocal scan head (CSU-X1 Spinning Disk Unit, Yokogawa).
Affinity Purification Using Ceramide-Coated Beads.
The ceramide pull-down assay was performed as per the manufacturer’s instructions. Briefly, the exact amount of flagellar lysates was incubated with 100 μL slurry of ceramide-coated beads (P-BCer, biotin-linked ceramide immobilized streptavidin agarose beads, Echelon Bioscience) at 4 °C for 3 h under rotary agitation. We chose the biotin immobilized streptavidin agarose beads (P-B000, Echelon Bioscience) as the control according to the manufacturer’s instructions, although a different membrane component can be the better control. Igepal was added to the supernatant at 0.5% of final concentration. Beads were washed four times with ceramide binding-washing buffer containing 0.5% igepal. The bound proteins were eluted by a 2× sodium dodecyl sulfate (SDS) sample buffer and then subjected to SDS–polyacrylamide gel electrophoresis either for immune blotting or proteomics analysis.
Cell Transfection.
Plasmids were transfected into IMCD3 cells using Lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions. The control scrambled plasmids and targets were bought from the mouse short hairpin RNA library from Sigma. During the cultivation period, cells were also incubated with inhibitors affecting ceramide biosynthesis (20 μM FB1, 5 μM Myrocin), sphingolipid mixture (LM-6005-1Ea, Avanti), or combinations of these reagents. Cells were shifted from 10% serum to 0.5% serum for 48 h to induce ciliogenesis before fixation.
Cryosample Preparation.
Two-hundred-mesh grids were glow discharged. Cryo-EM grids were prepared with a Leica EM GP (47). A drop of 4 μL cultured cells was added to the grid and blotted with filter paper for 6 s, followed by an immediate plunge-frozen in liquid ethane.
Cryo-ET Data Collection.
We collected the cryo-ET data using the Titan Krios microscope (Thermo Fisher Scientific) operated at a voltage of 300 KV and equipped with Cs corrector, GIF Quantum energy filter (Gatan), and K3 Summit direct electron detector (Gatan). The tilt series were recorded from 68° to −68° with SerialEM software (48). The recording state was at a nominal magnification of 33,000× in counting mode with a pixel size of 2.656 Å⋅pixel−1. Each stack was exposed for 2.4 s with an exposure time of 0.3 s per frame and recorded as a movie of eight frames, resulting in the total dose rate of ∼2 electrons per Å2 for each stack. GIF was set to a slit width of 20 eV. The defocus ranged from −5 to −8 μm. The MotionCor2 program was used to correct the beam-induced motion (49).
Cryo-ET Reconstruction and Three-Dimensional Model Reconstruction.
Tilt series were aligned with the patch-tracking method in IMOD software (50). Three-dimensional models were reconstructed with Amira 5.2.2 software.
Ribosome Profiling and RNA-Seq Libraries Processing.
Ribo-Seq libraries were performed using a previously published protocol (51) with a few modifications. Briefly, Chlamydomonas cells were flash-frozen and stored in liquid nitrogen. The sample pellets were crushed into powder in a mortar filled with liquid nitrogen. The powdered samples were lysed in 400 μL of lysis buffer containing 20 mM Tris⋅HCl (pH 7.4) (AM9850G, Invitrogen), 150 mM NaCl (AM9760G, Invitrogen), 5 mM MgCl2 (AM9530G, Invitrogen), 1 mM DTT (43816, Sigma-Aldrich), 100 μg/mL cycloheximide (C4859, Sigma-Aldrich), 1% Triton X-100 (T8787, Sigma-Aldrich), and 25 U/mL Turbo DNase I (AM2238, Invitrogen) at 4 °C for 10 min. Then 200 μL of the clarified supernatant was used to purify the RPFs, and 100 μL of the supernatant was used for the total RNA-Seq in parallel.
For the RPFs, 15 U RNase I (Epicentre, N6901K) was added to 200 μL of the lysate, and the lysate was incubated with gentle mixing at room temperature for 45 min. To terminate digestion, we added 10 μL SUPERase In RNase Inhibitor (AM2696, Life Technologies) to the lysate and chilled it on ice. RPFs were purified by Sephacryl S400 spin column chromatography (27514001, GE Healthcare) and extracted with TRIzol (15596018, Invitrogen) following the manufacturer’s protocol. The rRNA was depleted using the RiboZero kit (MRZPL1224, Illumina). The 25- to 35-nt fragments were excised from the 15% urea gel and eluted in 400 μL RNA gel extraction buffer (300 mM ammonium acetate [AM9070G, Invitrogen], 0.25% vol/vol of 10% SDS [AM9823, Invitrogen], and 1 mM EDTA [AM9260G, Invitrogen]) followed by isopropanol (I9030, Sigma-Aldrich) precipitation. The RNA pellets were resuspended in 10 μL of 10 mM Tris, pH 8.0, and incubated with T4 polynucleotide kinase (M0201S, New England Biolabs) at 37 °C for 1 h to dephosphorylate. The dephosphorylated RNA was ligated with preadenylated linker using T4 RNA ligase 2 (truncated K227Q, M0351S, New England Biolabs) at 22 °C for 3 h. The ligation products were reverse transcript by SuperScript III at 50 °C for 30 min. The reverse transcription products were added with 1 M NaOH and incubated at 98 °C for 20 min to hydrolyze RNA. The reverse transcription bands were excised from 15% urea gel and eluted in 400 μL DNA gel extraction buffer (300 mM sodium chloride [AM9070G, Invitrogen]), 10 mM Tris [AM9855G, pH 8.0, Invitrogen], and 1 mM EDTA [AM9260G, Invitrogen]) followed by isopropanol (I9030, Sigma-Aldrich) precipitation. The purified reverse transcription products were circularized by CircLigase II (CL9021K, Epicentre) at 60 °C for 1 h. Deep sequencing libraries were generated from the circularized products. The amplified PCR products were excised from 8% polyacrylamide nondenaturing gels and eluted in 400 μL DNA gel extraction buffer, followed by isopropanol precipitation. The purified amplificated PCR products were sequenced on an Illumina HiSeq platform.
Total RNA from the Chlamydomonas cells was isolated with TRIzol (15596018, Invitrogen) from 100 μL clarified lysate, and the rRNA was depleted using the RiboZero kit (MRZPL1224, Illumina). The rRNA-depleted total RNA was fragmented at 95 °C for 20 min. Deep sequencing libraries were generated in parallel with Ribo-Seq and sequenced on an Illumina HiSeq platform. Total RNA from C. elegans was extracted from mixed-stage worms with TRIzol. The RNA quality was assessed on the Agilent Bioanalyzer 2100 system. The samples with an RNA integrity number > 6 were processed; 50 to 500 ng of total RNA was used for library preparation using the KAPA RNA HyperPrep Kit (KAPA Biosystems). Library samples were sequenced on an Illumina HiSeq platform.
RNA-Seq and Ribo-Seq Data Analyses.
Ribosome profiling data were processed using the established methods (51, 52). The 5′ base of the adapter-trimmed Ribo-Seq reads was removed, which is usually an artifact of reverse transcription. For removing abundant contamination from digested ribosomal RNA (rRNA) present in the libraries, the reads aligned to a collection of rRNA sequences obtained from GenBank and University of California at Santa Cruz using Bowtie2 were discarded.
For C. reinhardtii data, both Ribo-Seq and RNA-Seq reads were aligned to the Chlamydomonas v5.5 genome (53), using Tophat (54) guided by Chlamydomonas annotation, JGI v5.5 (Phytozome 10), allowing for two mismatches per read and unique alignment only. For C. elegans data, RNA-Seq reads were aligned to ce11/WBcel235 genome assembly using STAR_2.5.1b (55) and an index with splice junction information from the Ensemble 94 transcriptome annotation.
Read assignment was achieved by HT-Seq (56). Ribo-Seq was calculated for all genes that passed reads per million reads mapped (CPM) ≥1 in the coding sequence (CDS) threshold in two biological replicates. Differential expression analysis for transcriptomes and translatomes was achieved using the Bioconductor package edgeR, based on the negative binomial distributions (57). Transcriptome fold changes for expressed genes were determined using reads assigned to exons by the gene. Translatome fold changes for protein-coding genes were measured using reads assigned to CDS by the gene. Translational efficiency is the ratio of normalized Ribo-Seq and RNA-Seq reads per transcript and log2-fold change of translational efficiency was calculated for each gene by normalizing log2-fold change of RPFs to mRNA as reported (51). The nucleotide resolution of the coverage around start and stop codons, and the 3-nt periodicity of Ribo-Seq data were analyzed by RiboCode (58). Pearson correlations were used to test for coregulation between transcriptome and translatome among all translated genes using CPM. Gene Ontology enrichment analysis was performed with the Gene Ontology Resource website (geneontology.org/) with Fisher’s exact test (59, 60).
Ciliary Gene Clustering.
Hierarchical clustering with Euclidean distance followed by single-linkage algorithms was used for classifying ciliary genes, which was annotated from the Chlamydomonas Flagellar Proteome Project (7) (chlamyfp.org). Z-scores were used to normalize fold changes of RPFs or mRNA. Z-scores are: z = (x − mean)/std, which ensures that each row (column) has a mean of 0 and a variance of 1. Hierarchical clustering and visualization were done using clustermap function from the Python package seaborn (https://seaborn.pydata.org/). All statistical analyses were carried out using custom-written Python scripts and the R statistical language.
Similar Gene Selection and Ortholog Finding.
We screened genes with a similar regulatory pattern to ciliary genes cluster I from the whole genome by setting the following thresholds and logic operations: Transcription or translation is up-regulated at least 0.8 log2-fold change at 15 or 30 min and decrease at 60 and 90 min after flagellar regeneration, and translational efficiency is down-regulated at 60 and 90 min compared to 15 or 30 min. Orthologs were obtained from OrthoDB(v10) database (61) (https://www.orthodb.org) by searching with the gene UniProtKB/TrEMBL ID.
Quantification and Statistical Analysis.
Two-tailed Student’s t test (Figs. 3 C and D, 4 B, D, and F, and 7 C, D, and G, and SI Appendix, Figs. S5 B and D and S6B) was used to examine significant differences between groups as indicated in the figure legends. For each group, quantification was represented by the mean value ± SD. Statistical significance was determined when the P value was lower than 0.05 (N.S. indicates not significant). N represents the number of animals used for Dyf assay (Fig. 3C), the number of animals or cells used for cilium length quantification (Figs. 3C, 4B, 7 C, D, and G and SI Appendix, Fig. S6B), the number of animals or cells used for cilium fraction quantification (Figs. 4B and 7C, D, and G), the number of algae used for bulged flagella rate quantification (Fig. 4F and SI Appendix, Fig. S5B and C), or the number of IFT particles used for velocity quantification (Fig. 4D).
Supplementary Material
Acknowledgments
We thank Profs. Z. Lu, X. Yang, and W. Qian for their advice on bioinformatics. We thank Prof Pan J. for providing some Chlamydomonas strains and primary antibodies. We thank the Caenorhabditis Genetics Center (CGC) and the National Bioresource Project of Japan for providing some strains, the Lipidomics Facility at the Center for Biomedical Analysis of Tsinghua University for sample analysis, the Protein Chemistry Facility at the Center for Biomedical Analysis of Tsinghua University for sample analyses, and the Tsinghua University Cryo-EM Facility of China National Center for Protein Sciences (Beijing) for tomography data collection. This work was supported by the National Natural Science Foundation of China (Grants 31991191, 31730052, 31525015, 31561130153, 31671444, and 31871352), the National Basic Research Program of China (973 Program; Grant 2017YFA0102900), and the Newton Advanced Fellowship (Grant NA140490) from the Royal Society.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.F.-L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201096119/-/DCSupplemental.
Data Availability
Ribo-Seq and RNA-Seq data were deposited in Gene Expression Omnibus (accession no. GSE149844). All other study data are included in the article and/or supporting information.
References
- 1.Reiter J. F., Leroux M. R., Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18, 533–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishikawa H., Marshall W. F., Intraflagellar transport and ciliary dynamics. Cold Spring Harb. Perspect. Biol. 9, a021998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breslow D. K., Holland A. J., Mechanism and regulation of centriole and cilium biogenesis. Annu. Rev. Biochem. 88, 691–724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afzelius B. A., Ultrastructure of human nasal epithelium during an episode of coronavirus infection. Virchows Arch. 424, 295–300 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rautiainen M., Nuutinen J., Kiukaanniemi H., Collan Y., Ultrastructural changes in human nasal cilia caused by the common cold and recovery of ciliated epithelium. Ann. Otol. Rhinol. Laryngol. 101, 982–987 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Khan S., Scholey J. M., Assembly, functions and evolution of archaella, flagella and cilia. Curr. Biol. 28, R278–R292 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Pazour G. J., Agrin N., Leszyk J., Witman G. B., Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stolc V., Samanta M. P., Tongprasit W., Marshall W. F., Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. U.S.A. 102, 3703–3707 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbaum J. L., Moulder J. E., Ringo D. L., Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J. Cell Biol. 41, 600–619 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatsu F., A phosphoinositide code for primary cilia. Dev. Cell 34, 379–380 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Garcia G. III, Raleigh D. R., Reiter J. F., How the ciliary membrane is organized inside-out to communicate outside-in. Curr. Biol. 28, R421–R434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaiser F., Huebecker M., Wachten D., Sphingolipids controlling ciliary and microvillar function. FEBS Lett. 594, 3652–3667 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Snell W. J., Pan J., Wang Q., Cilia and flagella revealed: From flagellar assembly in Chlamydomonas to human obesity disorders. Cell 117, 693–697 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Inglis P. N., Ou G., Leroux M. R., Scholey J. M., “The sensory cilia of Caenorhabditis elegans.” In: WormBook: The Online Review of C. elegans biology (WormBook, Pasadena, CA, 2007), pp. 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Z., Zhang X., Jia S., Yelick P. C., Zhao C., Zebrafish as a model for human ciliopathies. J. Genet. Genomics 43, 107–120 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa H., Thompson J., Yates J. R. III, Marshall W. F., Proteomic analysis of mammalian primary cilia. Curr. Biol. 22, 414–419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nachury M. V., Mick D. U., Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Biol. 20, 389–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong J. N., et al. , Regulation of Chlamydomonas flagella and ependymal cell motile cilia by ceramide-mediated translocation of GSK3. Mol. Biol. Cell 26, 4451–4465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Q., et al. , Characterization of an apical ceramide-enriched compartment regulating ciliogenesis. Mol. Biol. Cell 23, 3156–3166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albee A. J., et al. , Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3 (Bethesda) 3, 979–991 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chamberlain K. L., Miller S. H., Keller L. R., Gene expression profiling of flagellar disassembly in Chlamydomonas reinhardtii. Genetics 179, 7–19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silflow C. D., et al. , Expression of flagellar protein genes during flagellar regeneration in Chlamydomonas. Cold Spring Harb. Symp. Quant. Biol. 46, 157–169 (1982). [DOI] [PubMed] [Google Scholar]
- 23.Zones J. M., Blaby I. K., Merchant S. S., Umen J. G., High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27, 2743–2769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trösch R., et al. , Commonalities and differences of chloroplast translation in a green alga and land plants. Nat. Plants 4, 564–575 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Chung B. Y., et al. , The use of duplex-specific nuclease in ribosome profiling and a user-friendly software package for Ribo-seq data analysis. RNA 21, 1731–1745 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung B. Y. W., Deery M. J., Groen A. J., Howard J., Baulcombe D. C., Endogenous miRNA in the green alga Chlamydomonas regulates gene expression through CDS-targeting. Nat. Plants 3, 787–794 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Z., et al. , Pervasive, coordinated protein-level changes driven by transcript isoform switching during meiosis. Cell 172, 910–923.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Z., et al. , Ribosome profiling reveals translational regulation of mammalian cells in response to hypoxic stress. BMC Genomics 18, 638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu G., et al. , Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545, 487–490 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyuhas O., Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 267, 6321–6330 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G., Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117, 456–487 (1986). [DOI] [PubMed] [Google Scholar]
- 32.Cui M., et al. , Starvation-induced stress response is critically impacted by ceramide levels in Caenorhabditis elegans. Genetics 205, 775–785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hannun Y. A., Obeid L. M., Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 175–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng X., et al. , Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans. Science 322, 110–115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seamen E., Blanchette J. M., Han M., P-type ATPase TAT-2 negatively regulates monomethyl branched-chain fatty acid mediated function in post-embryonic growth and development in C. elegans. PLoS Genet. 5, e1000589 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breslow D. K., Sphingolipid homeostasis in the endoplasmic reticulum and beyond. Cold Spring Harb. Perspect. Biol. 5, a013326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan M. A., Diener D. R., Stepanek L., Pigino G., The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat. Cell Biol. 20, 1250–1255 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Dawkins J. L., Hulme D. J., Brahmbhatt S. B., Auer-Grumbach M., Nicholson G. A., Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat. Genet. 27, 309–312 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Bejaoui K., et al. , SPTLC1 is mutated in hereditary sensory neuropathy, type 1. Nat. Genet. 27, 261–262 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Eichler F. S., et al. , Overexpression of the wild-type SPT1 subunit lowers desoxysphingolipid levels and rescues the phenotype of HSAN1. J. Neurosci. 29, 14646–14651 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garofalo K., et al. , Oral L-serine supplementation reduces production of neurotoxic deoxysphingolipids in mice and humans with hereditary sensory autonomic neuropathy type 1. J. Clin. Invest. 121, 4735–4745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuson M., Marfany G., Gonzàlez-Duarte R., Mutation of CERKL, a novel human ceramide kinase gene, causes autosomal recessive retinitis pigmentosa (RP26). Am. J. Hum. Genet. 74, 128–138 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee C. H., et al. , Primary cilia mediate early life programming of adiposity through lysosomal regulation in the developing mouse hypothalamus. Nat. Commun. 11, 5772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson N. F., Lefebvre P. A., Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot. Cell 3, 1307–1319 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Craige B., Brown J. M., Witman G. B., Isolation of Chlamydomonas flagella. Curr. Protoc. Cell Biol. Chapter 3, Unit 3.41–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright R., Fluorescence labeling of flagellar membranes. Methods Cell Biol. 47, 413–418 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Li M., Ma J., Li X., Sui S. F., In situ cryo-ET structure of phycobilisome-photosystem II supercomplex from red alga. eLife 10, e69635 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Ingolia N. T., Brar G. A., Rouskin S., McGeachy A. M., Weissman J. S., The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brar G. A., et al. , High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merchant S. S., et al. , The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCarthy D. J., Chen Y., Smyth G. K., Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao Z., et al. , De novo annotation and characterization of the translatome with ribosome profiling data. Nucleic Acids Res. 46, e61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashburner M., et al. ; The Gene Ontology Consortium, Gene Ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gene Ontology C.; Gene Ontology Consortium, The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 49, D325–D334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kriventseva E. V., et al. , OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47, D807–D811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Ribo-Seq and RNA-Seq data were deposited in Gene Expression Omnibus (accession no. GSE149844). All other study data are included in the article and/or supporting information.