Significance
“Rare somatic mutations” are present in a small fraction of DNA molecules at as low as one in 107 to 108 bp in normal tissues, and are extremely difficult to detect. EcoSeq was developed here by introducing a strong genome size reduction into duplex sequencing, and had a high efficiency, in addition to high sensitivity and accuracy. Accordingly, EcoSeq revealed that high levels of somatic mutations were accumulated in normal-appearing blood cells, a surrogate for bone marrow stem cells, of pediatric sarcoma patients with prior chemotherapy. EcoSeq is capable of analyzing DNA samples potentially accumulating somatic mutations, such as tissues exposed to inflammation, smoking, and extreme cell turnover, and will enable assessment of past exposure and future cancer risk.
Keywords: normal tissue, somatic mutation, duplex sequencing, next-generation sequencing, chemotherapy
Abstract
Somatic mutations are accumulated in normal human tissues with aging and exposure to carcinogens. If we can accurately count any passenger mutations in any single DNA molecule, since their quantity is much larger than driver mutations, we can sensitively detect mutation accumulation in polyclonal normal tissues. Duplex sequencing, which tags both DNA strands in one DNA molecule, enables accurate count of such mutations, but requires a very large number of sequencing reads for each single sample of human-genome size. Here, we reduced the genome size to 1/90 using the BamHI restriction enzyme and established a cost-effective pipeline. The enzymatically cleaved and optimal sequencing (EcoSeq) method was able to count somatic mutations in a single DNA molecule with a sensitivity of as low as 3 × 10−8 per base pair (bp), as assessed by measuring artificially prepared mutations. Taking advantages of EcoSeq, we analyzed normal peripheral blood cells of pediatric sarcoma patients who received chemotherapy (n = 10) and those who did not (n = 10). The former had a mutation frequency of 31.2 ± 13.4 × 10−8 per base pair while the latter had 9.0 ± 4.5 × 10−8 per base pair (P < 0.001). The increase in mutation frequency was confirmed by analysis of the same patients before and after chemotherapy, and increased mutation frequencies persisted 46 to 64 mo after chemotherapy, indicating that the mutation accumulation constitutes a risk of secondary leukemia. EcoSeq has the potential to reveal accumulation of somatic mutations and exposure to environmental factors in any DNA samples and will contribute to cancer risk estimation.
Accumulation of mutations and aberrant DNA methylation in so-called “normal” tissues constitutes cancer risk (1, 2), and their accurate measurement is important to assess future risk of cancer development and past exposure to various environmental carcinogenic factors (3–12). Normal tissues often refer to tissues without any tumors, but may contain small expanded clonal patches with or without histological clonal expansion (3, 7, 8, 10, 11). Mutations involved in such small clonal expansion are driver genes suitable for clonal growth in a specific ecosystem and often present in specific positions of specific genes in multiple clones. At the same time, there are far more mutations not involved in clonal expansion, namely passenger mutations (2, 13). Detection of passenger mutations in polyclonal normal tissues is challenging because they are extremely rare (one per 106 to 108 bp) (12, 14, 15), present only in one clone, and scattered around the genome.
To detect rare somatic mutations, small-size samples with a small number of clonal patches (3, 7, 8, 10, 11) and organoids or cloned cells derived from a single cell (4, 9, 16) were used. The use of a small number of clones enables sequencing of multiple cells from a clone, namely multiple DNA molecules with the same mutations, which enables distinction of a mutation from a sequencing error. However, most of these methods require meticulous technique, and live cells are essential to prepare organoids and cloned cells. Alternatively, sequencing accuracy is enhanced by various methods (5, 6, 12, 17–22). Among these, duplex sequencing can distinguish single-strand DNA damage from real mutations by tagging both DNA strands in individual DNA molecules and enables accurate detection of a mutation in a single DNA molecule (17, 18). However, duplex sequencing needs a large number of sequencing reads to assemble reads with the same molecular barcodes. To address this issue, NanoSeq was developed by combining duplex sequencing and reduced representation sequencing (23, 24), which reduces the genome size to 1/2 to 1/3 using a restriction enzyme, HpyCH4III (12). However, NanoSeq still needs 300 M paired-end (PE) reads to analyze a single mammalian sample and also additional sequencing data to exclude single nucleotide polymorphisms (SNPs).
On the application side of rare somatic mutations in normal tissues, estimation of the risk of secondary malignancy in pediatric patients is very important. It is known that survivors of childhood cancer have a three to six times increased risk of developing a secondary malignancy, associated with prior chemotherapy and radiation therapy (25, 26). Especially, platinum-based drugs, alkylating agents, and topoisomerase II inhibitors are associated with the risk of therapy-related myeloid neoplasms (27–31). However, there are no methods to estimate the risk of an individual for therapy-related myeloid neoplasms that may develop after a few years. If accumulation levels of somatic mutations in normal blood cells, a surrogate for bone marrow stem cells, can be measured and are correlated with the risk of secondary malignancy, future risk estimation, and adoption of less mutagenic therapy may become possible.
In this study, to count rare somatic mutations in DNA samples at a relatively low cost, we improved the duplex sequencing by reducing genomic regions to ∼1/90 using the BamHI restriction enzyme. Using the method, we analyzed peripheral blood cells of pediatric sarcoma patients who received chemotherapy to reveal accumulation of somatic mutations and the presence of mutational signatures reflecting therapeutic agents.
Results
Reduction of Analyzed Genomic Regions by BamHI Digestion and Size Selection.
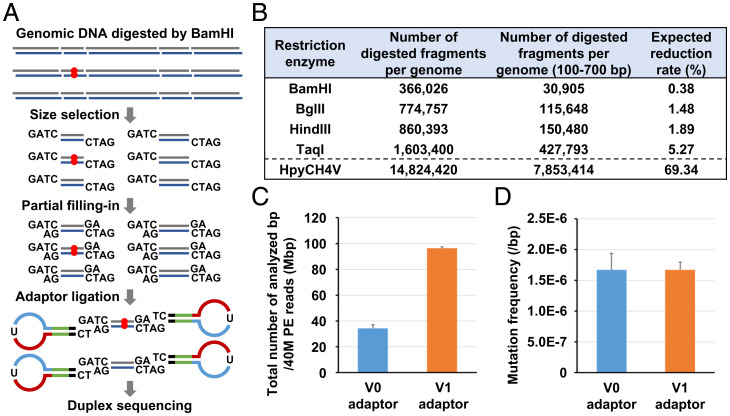
To reduce the large number of sequencing reads needed for duplex sequencing of a mammalian genome, analyzed genomic regions were reduced by restriction digestion and selection of fragments with 100 to 700 bp (Fig. 1A and SI Appendix, Fig. S1A). This reduction was expected to increase coverage depth, enabling distinguishment of SNPs from somatic mutations and sparing the need for whole genome sequencing. We compared three six-base-cutting restriction enzymes (BamHI, BglII, and HindIII) used for representational difference analysis (32), and the most frequently used four-base-cutting restriction enzyme TaqI (Fig. 1B). HpyCH4V was also added because it was used in NanoSeq (12). The expected reduction rate of genomic regions was highest (0.38%) when using BamHI.
Fig. 1.
Reduction of analyzed genomic regions by EcoSeq. (A) Schema of genomic region reduction and adaptor ligation of EcoSeq. Analyzed genomic regions are reduced by digestion using the BamHI restriction enzyme and size selection. Partial filling-in using dATP and dGTP enabled specific ligation to a 5′-TC-tailed adaptor with a sticky end, and excluded illegitimate inserts. Red circle represents a mutation present in a single DNA molecule. (B) Expected reduction rate using five restriction enzymes. Enzymatically digested fragments of 100 to 700 bp were expected to be used to prepare the EcoSeq library. A reduction rate was calculated as genomic regions covered by digested fragments of 100 to 700 bp within the entire genome. BamHI was expected to have the highest reduction rate and adopted for EcoSeq. (C) Total number of analyzed base pairs from 40 M PE reads by EcoSeq. Ligation using a 5′-TC-tailed adaptor from 100 ng genomic DNA (v1 adaptor, n = 3) showed a larger number of analyzed base pairs than normal 3′-dA-tailed adaptor from 500 ng genomic DNA (v0 adaptor, n = 4) using the same cell line sample. Error bar represents the SD. (D) Mutation frequency of EcoSeq library prepared by v0 and v1 adaptors. Mutation frequencies of the same cell line sample were consistent between v1 adaptor (n = 3) v0 adaptor (n = 4). Error bar represents the SD.
Also, to improve the low ligation efficiency of duplex sequencing, partial filling in with deoxyadenosine triphosphate (dATP) and deoxyguanosine triphosphate (dGTP) was introduced, which enabled specific ligation to a 5′-TC-tailed adaptor (adaptor) with a sticky end (33) (Fig. 1A and SI Appendix, Fig S1A). In comparison to the original ligation using a 3′-dA-tailed v1 adaptor (v0 adaptor), a larger number of sequencing reads with BamHI-digested ends were mapped, resulting in a larger number of base pairs analyzed (Fig. 1C and SI Appendix, Fig S1B). In the following experiments, the analyses were performed using either adaptor because mutation frequencies and the spectrum of the same cell line sample were consistent between the v0 and v1 adaptors (Fig. 1D and SI Appendix, Fig S1C). Duplex sequencing with strong reduction of analyzed genomic regions was designated EcoSeq.
Successful Detection of Rare Mutations Artificially Prepared.
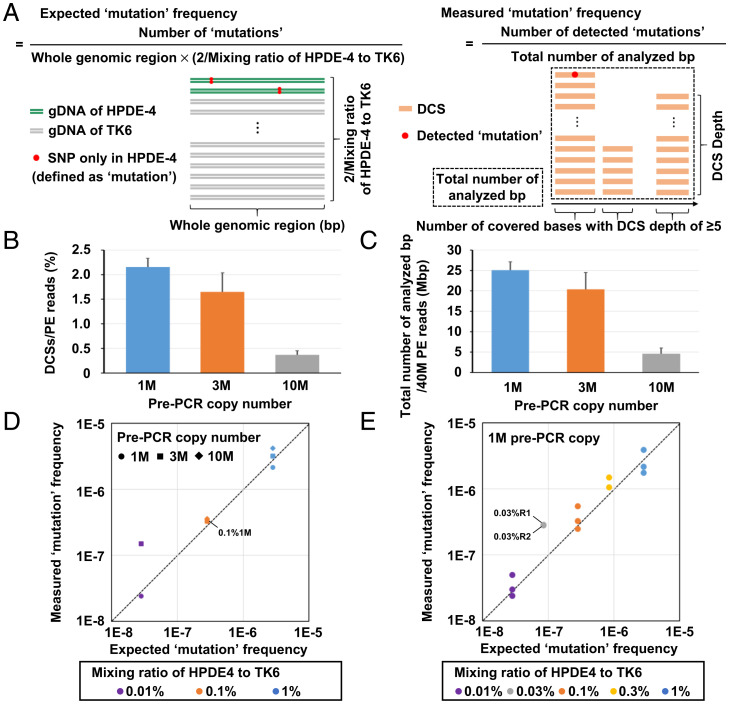
To confirm the ability of EcoSeq to detect rare somatic mutations, we prepared model DNA samples with very low mutation frequencies. A small amount of genomic DNA (gDNA) of a normal cell line HPDE-4 was mixed into gDNA of another normal cell line TK6, and SNPs present only in HPDE-4 were defined as artificial rare “mutations” (Fig. 2A). We calculated the expected mutation frequency based upon the mixing ratio. We also calculated measured mutation frequency as the number of detected mutations divided by the total number of analyzed base pairs. At the same time, we compared three pre-PCR copy numbers (1 M, 3 M, and 10 M pre-PCR copy number) to decide an optimal pre-PCR copy number under the conditions of 40 M PE reads per sample. Although 10 M copies may enable us to analyze a large number of DNA fragments, the high diversity may result in low efficiency to create duplex consensus sequences (DCSs). In contrast, although 1 M copies allow us to analyze only a limited number of DNA fragments, the low diversity may result in high efficiency to create DCSs. As a result, in 1 M pre-PCR copy number, we were able to create the largest number of DCSs per PE read and analyze the largest number of base pairs by 40 M PE reads (Fig. 2B and C and Dataset S1). Regarding the number of sequencing reads used for a single-strand consensus sequence (SSCS) assembly, 1 M pre-PCR copy number showed the highest efficiency to create SSCSs with a peak at 9 to 11 reads (SI Appendix, Fig. S2A), in accordance with a previous report (18). To assemble SSCS and DCS, EcoSeq used sequencing reads with completely identical unique molecular identifiers (UMIs) only. The Hamming distance between any pairs of UMIs showed a distribution at zero or ≥10, with 4% of reads showing in-between values (SI Appendix, Fig. S2B). Thus, the number of sequencing errors in UMIs, which show in-between Hamming distances, was limited, and the loss of discarding the error UMIs was considered negligible.
Fig. 2.
Detection of artificially prepared rare mutations. (A) Schema of expected and measured mutation frequencies using model DNA samples. A small amount of HPDE-4 gDNA was mixed into TK6 gDNA, and SNPs present only in HPDE-4 were defined as mutations (artificially prepared mutations). (B) Efficiency in DCS creation. The percentage of DCSs created from PE reads using three pre-PCR copy numbers (1 M, 3 M, and 10 M; n = 3) was analyzed. 1 M pre-PCR copy number showed the highest efficiency to create DCSs. Error bar represents the SD. (C) Total number of analyzed base pairs from 40 M PE reads by EcoSeq. Total number of analyzed base pairs from 40 M PE reads using three pre-PCR copy numbers (1 M, 3 M, and 10 M; n = 3) was compared. 1 M pre-PCR copy number allowed the largest total number of analyzed base pairs. Error bar represents the SD. (D) Accordance between the measured and expected mutation frequencies. The libraries with three mixing ratios (1, 0.1, and 0.01%) prepared from three pre-PCR copy numbers (1 M, 3 M, and 10 M) were analyzed. High accordance was shown in all three mixing ratios prepared from 1 M pre-PCR copy number. No mutations were detected with a mixing ratio of 0.01% and 10 M pre-PCR copy number. (E) Accordance with two additional mixing ratios. The libraries with five mixing ratios (1, 0.3, 0.1, 0.03, and 0.01%) prepared from optimal pre-PCR copy number (1 M) were analyzed in triplicates (1, 0.1, and 0.01%) or duplicates (0.3 and 0.03%). High accordance was shown in all five mixing ratios.
From the three mixing ratios (1, 0.1, and 0.01%) and the number of HPDE-4–specific SNPs, expected mutation frequencies were calculated as 2.8 × 10−6, 2.8 × 10−7, and 2.8 × 10−8 per base pair, respectively. Importantly, measured mutation frequencies were in accordance with the expected mutation frequencies in 1 M pre-PCR copy number, regardless of mixing ratios (Fig. 2D and SI Appendix, Table S1). In the conditions of optimal copy number (1 M), libraries with five mixing ratios (1, 0.3, 0.1, 0.03, and 0.01%) were analyzed in triplicate (1, 0.1, and 0.01%) or duplicate (0.3 and 0.03%), and high accordance was confirmed (Fig. 2E and SI Appendix, Table S1). These results showed that EcoSeq is able to detect rare mutations accurately at a frequency of as low as 3 × 10−8 per base pair.
Unexpectedly Detected but Real Mutations Present in Background Cells.
In the analysis of the model DNA samples, we unexpectedly detected mutations other than SNPs in HPDE-4 (background mutations). The average background mutation frequency in 13 samples was 12.4 ± 2.3 × 10−7 per base pair under the conditions of 1 M pre-PCR copy number (SI Appendix, Fig. S3A and Table S1). These model DNA samples were prepared from TK6 and HPDE-4 gDNA, and these cell lines had been cultured with long-term passages before their DNA purification. Thus, we hypothesized that the background mutations might be real mutations with small clonal populations induced in the TK6 cells after long-term culture. Indeed, mutational spectra of these background mutations showed a 22% signature associated with cell culture (34) (SI Appendix, Fig. S3B).
To confirm that the background mutations were derived from the long-term cell culture, we conducted EcoSeq of TK6 cell clones after cloning, in addition to the parental TK6 used for the model DNA. The mutation frequency in a single TK6 clone was 1.4 × 10−7 per base pair and was lower than that in the parental TK6 cells (9.3 × 10−7 per base pair) (SI Appendix, Fig. S3A and Table S1). To confirm the presence of mutations in the TK6 clones, we conducted Sanger sequencing of 30 independent TK6 clones for 11 mutations randomly selected from the 502 background mutations detected in the 13 samples (SI Appendix, Fig. S3 A and C and Table S2). In one of the 30 TK6 clones, one of the 11 mutations was detected, and it was not archived in the common SNP database (NCBI dbSNP build 153). This result also supported that the background mutations were not errors but real mutations present in small clonal populations.
Detection of Rare Mutations Induced by a Mutagen.
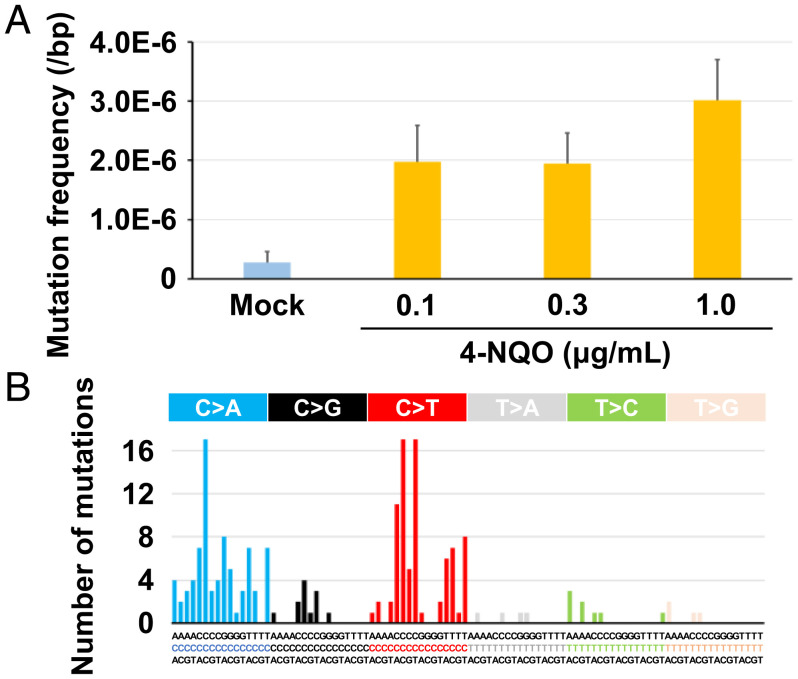
We further verified the sensitivity of EcoSeq by detecting rare mutations induced by a mutagen in a cell culture. A 293FT clone was treated with 4-NQO, a mutagen that induces specific mutations containing G:C to A:T transitions and G:C to T:A transversions (35, 36), for three doses (0.1, 0.3, and 1 µg/mL) (SI Appendix, Fig. S4A). EcoSeq showed that the mutation frequency was elevated from 2.8 × 10−7 per base pair in mock-treated cells to 30.1 × 10−7 per base pair in 4-NQO–treated cells (Fig. 3A, and SI Appendix, Table S3). In addition, the mutations in cells treated with 4-NQO showed the increases of G:C to A:T transitions and G:C to T:A transversions (Fig. 3B). These results showed that EcoSeq was able to detect rare mutations induced by mutagen treatment at a mutation frequency of ≤10−6 per base pair.
Fig. 3.
Detection of rare mutations induced by a mutagen. (A) Mutation frequencies in cells treated with 4-nitroquinoline 1-oxide (4-NQO) for three doses (0.1, 0.3, and 1.0 µg/mL). Mutation induction by 4-NQO was successfully detected in 293FT cells after cloning. Error bar represents 95% confidence interval (CI). (B) The mutational signatures in cells treated with 4-NQO. The signatures associated with 4-NQO treatment including G:C to A:T transitions and G:C to T:A transversions were observed.
Mutation Accumulation in Blood Cells of Pediatric Patients with Chemotherapy.
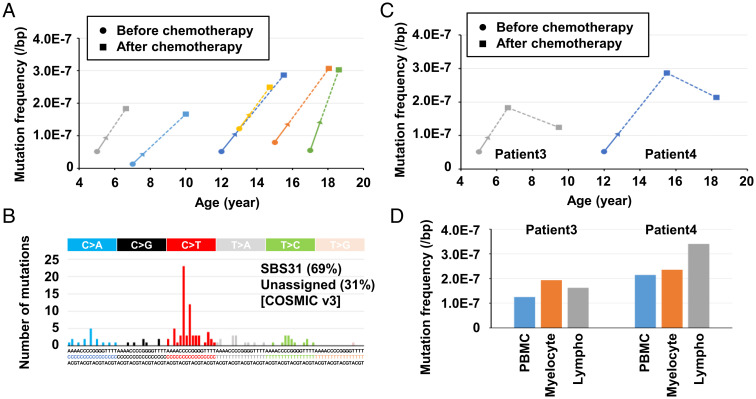
EcoSeq was applied to a long-standing biological question of mutation accumulation after chemotherapy. We quantified somatic mutations in peripheral blood cells of pediatric sarcoma patients who received chemotherapy (n = 10) and those who did not (n = 10) (SI Appendix, Table S4). The mutation frequency of patients with chemotherapy was 31.2 ± 13.4 × 10−8 per base pair, and was significantly higher than that of patients without (9.0 ± 4.5 × 10−8 per base pair) (P < 0.001) (Fig. 4A, and SI Appendix, Table S5). Mutational signatures in the patients with chemotherapy showed an association of 43% with prior platinum-based chemotherapy (SBS35 in COSMIC v3 signatures) (37) (Fig. 4B). Among the 10 patients with chemotherapy, six patients who received platinum-based drugs tended to show a higher mutation frequency than the other four patients who received other drugs (3.7 ± 1.4 × 10−7 per base pair vs. 2.2 ± 0.3 × 10−7 per base pair, P = 0.085) (Fig. 4C and SI Appendix, Table S5). In contrast, between patients treated with and without radiation therapy, no difference in mutation frequency was observed (P = 0.385) (SI Appendix, Fig. S5A and Table S5). It is noteworthy that the mutation frequency in patients without chemotherapy was correlated with age (Fig. 4D and SI Appendix, Table S5). These results showed that mutations have already accumulated in normal peripheral blood cells depending upon age, and chemotherapy can increase the mutation frequency.
Fig. 4.
Mutation accumulation in blood cells with chemotherapy. (A) Mutation frequencies of normal peripheral blood cells in pediatric sarcoma patients who received chemotherapy (n = 10) and those who did not (n = 10). Somatic mutations were significantly accumulated in patients with chemotherapy at a level of 10−7 per base pair (P < 0.001). Error bar represents the SD. (B) The mutational signatures in patients with chemotherapy. The signature associated with prior platinum-based chemotherapy (SBS35 in COSMIC v3 signatures) was observed. (C) Mutation frequencies in patients who received platinum-based drugs (n = 6) and those who received other drugs (n = 4). Somatic mutations tended to be accumulated in patients with platinum-based drugs (P = 0.085). Error bar represents the SD. (D) Correlation between mutation frequency and age. Mutation frequencies in patients without chemotherapy (blue dot) were correlated with age.
We further quantified mutation accumulation before and 12 to 31 mo after chemotherapy in peripheral blood cells of six pediatric sarcoma patients (SI Appendix, Table S6). All six patients showed much higher levels of somatic mutations after chemotherapy with a mutation signature of prior platinum-based chemotherapy (SBS31 in COSMIC v3 signatures) (Fig. 5A and B and SI Appendix, Table S7). Mutation accumulation after 46 to 64 mo was further analyzed in two of them, and the increased levels slightly decreased but persisted (Fig. 5C). Finally, the differences of mutation accumulation between myelocytes and lymphocytes were also analyzed in the two patients. No clear differences were observed (Fig. 5D and SI Appendix, Fig. S6 A and B). These results showed that substantial fractions of mutations in peripheral blood cells induced by chemotherapy persist for years after chemotherapy.
Fig. 5.
Mutation accumulation at multiple time points and in different cell types. (A) Mutation accumulation at two time points of the same pediatric sarcoma patients. Peripheral blood cell samples before and 12 to 31 mo after chemotherapy were analyzed in six patients. Blood cells after chemotherapy showed 2.1 to 12.5 times higher mutation frequencies than those before chemotherapy. Arrows represent the chemotherapy periods, and the dotted lines represent the chemotherapy-free period. (B) The mutational signatures in patients after chemotherapy. The signature associated with prior platinum-based chemotherapy (SBS31 in COSMIC v3 signatures) constituted 69%. (C) Mutation accumulation at three time points of the same pediatric sarcoma patients. Increased mutation levels after chemotherapy persisted even after 46 to 64 mo. (D) Mutation accumulation by chemotherapy in myelocytes and lymphocytes. No differences of mutation accumulation were observed. PBMC, peripheral blood mononuclear cells; Lympho, lymphocytes.
Discussion
EcoSeq achieved a high efficiency (low sequencing cost) by a strong reduction of analyzed genomic regions, in addition to the high accuracy (distinction of single-stranded premutagenic lesions and sequence errors) and high sensitivity (detection limit of 3 × 10−8 bp) of duplex sequencing. EcoSeq allows simultaneous analysis of multiple gDNA samples and detection of mutations present in polyclonal normal tissues. While NanoSeq combined duplex sequencing with a four-base-cutting restriction enzyme HpyCH4V and reduced genomic regions to 1/2 to 1/3 to reflect mutations in the whole human genome as much as possible (12), EcoSeq introduced a stronger reduction using a six-base-cutting restriction enzyme BamHI, reducing genomic regions to ∼1/90, and decreased the number of sequencing reads to analyze a sample to 40 M PE reads (Fig. 1C). Analysis of a number of normal tissues, such as tissues exposed to inflammatory bowel disease and interstitial pneumonia, by EcoSeq will clarify the potential accumulation of somatic mutations and mutational signatures reflecting past exposure to various environmental carcinogenic factors. Age-dependent accumulation of somatic mutations was recently documented (4, 5, 7–9, 11, 12, 16), and confirmed in this study.
The high sensitivity and accuracy of EcoSeq was confirmed using model DNA samples with artificially prepared rare mutations. In this experiment, we were able to detect artificial mutations at 3 × 10−8 per base pair from 40 M PE reads (Fig. 2D and E and SI Appendix, Table S1). In addition, mutation induction in cells treated with 4-NQO was successfully detected (Fig. 3A and SI Appendix, Table S3), and the mutation frequency of mock-treated cells was 2.8 × 10−7 per base pair. Furthermore, the mutation frequency of pediatric blood cells without chemotherapy was 9.0 ± 4.5 × 10−8 per base pair (Fig. 4A and SI Appendix, Table S5), and basal mutation frequency in normal tissues that had hardly been exposed was estimated at ∼1 × 10−7 per base pair. This frequency was in accordance with previous reports in which normal tissues or organoids were analyzed (4, 12).
Reproducibility of EcoSeq was high because mutation frequencies of the same DNA sample were 1.67 × 10−6 ± 1.22 × 10−7 (±7.3%) using the v0 adaptor and 1.67 × 10−6 ± 2.70 × 10−7 (±16%) using the v1 adaptor (Fig. 1D and SI Appendix, Table S3) and mutational spectra were similar (SI Appendix, Fig. S1C). Since EcoSeq counts a somatic mutation on a single DNA molecule, mutations detected in replicate experiments using the same DNA solution are different because analyzed DNA molecules are not identical. Nevertheless, mutation frequencies and spectra were consistent, potentially reflecting the nature of the DNA source, such as age and exposure to carcinogens, and relatively homogenous distribution of mutations in the genome.
Library preparation, sequencing, and informatics were optimized for the following four issues. First, partial filling-in in the v1 system increased the total number of base pairs effectively analyzed from 34 M with 500 ng of genomic DNA (v0) to 96 M in 100 ng (v1). The original duplex sequencing and our v0 system utilized T tailing for adaptor ligation, and even illegitimate DNA fragments could be ligated. Second, a loop adaptor was introduced to prevent double-strand DNA that constituted a UMI from denaturation during storage (Fig. 1A and SI Appendix, Figs. S1A and S7 A and B). Third, the pre-PCR copy numbers, reflecting the diversity of libraries, were optimized under the conditions of 40 M PE reads by quantifying 1 M pre-PCR copy that showed the highest efficiency (Fig. 2B and C). Fourth, SNPs were excluded by using information from sequencing reads that failed to assemble into SSCS or DCS, in addition to the final DCS. Further exclusion of common SNPs based upon a SNP database had little effect on the results (SI Appendix, Fig. S4B and Table S3), showing that the exclusion method was successful and that it eliminated the need for additional whole-genome sequencing of the same sample to detect SNPs. In contrast with SNPs, clonal mutations in a minor cell population of <2.5% (100% ÷ 40) could be detected as a mutation because the DCS depth of EcoSeq is about 40.
Chemotherapy was shown to have significantly induced somatic mutations in normal peripheral blood cells, likely reflecting those in hematopoietic stem cells in bone marrow (Fig. 4A and SI Appendix, Table S5). In addition, chemotherapy enriched the mutational signature associated with prior platinum-based chemotherapy (Fig. 4B). Importantly, mutation frequencies in peripheral blood cells increased 2.1 to 12.5 times after chemotherapy, and remained high 46 to 64 mo after chemotherapy (Fig. 5A–C and SI Appendix, Table S7). In adult secondary acute myeloid leukemia (AML) after platinum-based chemotherapy, a significant increase of mutation frequencies and platinum-related signatures are reported to be present compared to primary AML (31). Thus, somatic mutations in secondary AML are considered to originate from somatic mutations that had already accumulated in normal blood cells. The finding in secondary AML indicates that quantification of somatic mutations in normal blood cells may be useful to estimate the risk of therapy-related myeloid neoplasms.
There were no differences in mutation accumulation between myelocytes and lymphocytes after chemotherapy. We anticipated that myelocytes might accumulate a larger number of somatic mutations than lymphocytes since myeloid tumors are 2.3 times more frequent than lymphoid leukemia as therapy-related leukemia (38). It was considered that we measured the frequency of point mutations, and that frequencies of gene rearrangement could be different. Alternatively, the sizes of genomic regions that could drive tumorigenesis or the number of driver mutations required for tumorigenesis could be different between myeloid and lymphoid tumors.
Some limitations exist in this study. First, the mutation frequency and mutational signatures in analyzed regions by EcoSeq and those in the entire genome may not be completely in accordance. Indeed, a G+C content in EcoSeq libraries (∼48%) was higher than the average G+C content in a human genome (∼41%) (39) (Dataset S1), which may affect somatic mutation frequencies (40). In silico digestion also showed higher G+C content and proportion of coding regions in EcoSeq libraries (SI Appendix, Table S8). However, mutational spectra of nontreated pediatric peripheral blood cells observed in EcoSeq were similar to those of granulocytes from healthy donors observed in NanoSeq (SI Appendix, Fig. S5B).
Second, there are no data on whether rare somatic mutations also accumulate in hematopoietic stem cells in bone marrow, as in peripheral blood cells. This could happen if hematopoietic stem cells are more resistant to mutation induction than peripheral blood cells, and peripheral blood cells persist for a long time. However, mutation accumulation by chemotherapy in peripheral blood cells persisted even after a >12-mo chemotherapy-free period (Fig. 5A and C), strongly indicating that hematopoietic stem cells also accumulated mutations as peripheral blood. Third, mutations in circulating tumor cells (CTCs) might have been detected using EcoSeq. However, in general, the number of mutations per megabase pairs in various cancers are reported as ≤100 (41) and CTCs from metastatic tumors in 7.5 mL whole blood (≤5 × 107 leukocytes) are reported as ≤500 (42, 43), suggesting that the mutation frequency of CTCs is expected to be at ≤1 × 10−9 per base pair. Thus, CTCs are expected to have little effect on the results here.
Materials and Methods
A more detailed method is available in SI Appendix. The shell script and R script for the data processing are available at the GitHub repository (https://github.com/EpigeneticField/EcoSeq).
Sample Preparation.
gDNAs of cell lines were purified by the phenol/chloroform method (TK6 and HPDE-4 cells) or a QIAamp DNA Mini Kit (Qiagen) (293FT cells), and that of human peripheral blood cells were extracted by a FlexiGene DNA Kit (Qiagen) or a QIAamp DNA Mini Kit. A part of gDNA of human blood cells was stored in the National Cancer Center Biobank (Tokyo, Japan). This study was approved by the Institutional Review Board of the National Cancer Center (approval No. 2017-454, and 2018-024), and all the specimens were obtained with written informed consents.
EcoSeq Library Preparation, Sequencing, and Data Processing.
A duplex-loop adaptor was prepared from a 94-mer oligonucleotide with UMI by filling-in, restriction digestion, and 3′-dT tailing (v0 adaptor) or 5′-TC tailing (v1 adaptor) (SI Appendix, Fig. S7 A and B and Table S8). A total of 500 ng (v0 adaptor) or 100 ng (v1 adaptor) gDNA was digested by BamHI, size-selected, end-repaired or partial filling-in, and ligated to the adaptor. After cutting at a dU-base, real-time PCR was performed to measure 1 M pre-PCR copy number as a template for PCR (SI Appendix, Table S9). A sequencing library was prepared by PCR, and sequenced using HiSeq X Ten (Illumina) or NovaSEq. 6000 (Illumina) to achieve 40 M PE reads per sample at 150-bp PE sequencing.
A flowchart of the data processing is shown in SI Appendix, Fig. S8. Three or more sequencing reads containing identical UMI sequences were merged into a SSCS. Two SSCSs were merged into a DCS. Mutation calling was performed by using the mapped DCS reads. The mutation frequency was calculated by dividing the number of detected mutations by the total number of analyzed base pairs. The difference in mutation frequencies was assessed by the Student’s t test, and a P value of <0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments
We are grateful to Kazuki Tanimura (Department of Pediatric Oncology, National Cancer Center Hospital, Japan) for collecting clinical information. This work was supported by the National Cancer Center Biobank, Japan. The study was supported by Japan Agency for Medical Research and Development (AMED) under JP21gm1310006, JP21ck0106552, and JP21bk0104097; and by Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) Grant Nos. JP21H02773 (T.U.), JP19K07744 (N.I.), and JP19K07745 (S.Y.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123241119/-/DCSupplemental.
Data Availability
Algorithms and computer code data have been deposited in GitHub (Available at https://github.com/EpigeneticField/EcoSeq). Sequencing data of EcoSeq libraries have been deposited with links to DRA accession number DRA014481 in the DDBJ Sequence Read Archive (Available at https://ddbj.nig.ac.jp/resource/sra-submission/DRA014481)(45).
References
- 1.Yang Z., et al. , Correlation of an epigenetic mitotic clock with cancer risk. Genome Biol. 17, 205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ushijima T., Clark S. J., Tan P., Mapping genomic and epigenomic evolution in cancer ecosystems. Science 373, 1474–1479 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Martincorena I., et al. , Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blokzijl F., et al. , Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoang M. L., et al. , Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 113, 9846–9851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamashita S., et al. , A novel method to quantify base substitution mutations at the 10−6 per bp level in DNA samples. Cancer Lett. 403, 152–158 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Martincorena I., et al. , Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokoyama A., et al. , Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 565, 312–317 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Franco I., et al. , Whole genome DNA sequencing provides an atlas of somatic mutagenesis in healthy human cells and identifies a tumor-prone cell type. Genome Biol. 20, 285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R., et al. , Macroscopic somatic clonal expansion in morphologically normal human urothelium. Science 370, 82–89 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Yoshida K., et al. , Tobacco smoking and somatic mutations in human bronchial epithelium. Nature 578, 266–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abascal F., et al. , Somatic mutation landscapes at single-molecule resolution. Nature 593, 405–410 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Takeshima H., Ushijima T., Accumulation of genetic and epigenetic alterations in normal cells and cancer risk. NPJ Precis. Oncol. 3, 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gossen J. A., et al. , Efficient rescue of integrated shuttle vectors from transgenic mice: a model for studying mutations in vivo. Proc. Natl. Acad. Sci. U.S.A. 86, 7971–7975 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohler S. W., et al. , The use of transgenic mice for short-term, in vivo mutagenicity testing. Genet. Anal. Tech. Appl. 7, 212–218 (1990). [DOI] [PubMed] [Google Scholar]
- 16.Franco I., et al. , Somatic mutagenesis in satellite cells associates with human skeletal muscle aging. Nat. Commun. 9, 800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt M. W., et al. , Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 14508–14513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy S. R., et al. , Detecting ultralow-frequency mutations by Duplex Sequencing. Nat. Protoc. 9, 2586–2606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sloan D. B., Broz A. K., Sharbrough J., Wu Z., Detecting Rare Mutations and DNA Damage with Sequencing-Based Methods. Trends Biotechnol. 36, 729–740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pel J., et al. , Duplex Proximity Sequencing (Pro-Seq): A method to improve DNA sequencing accuracy without the cost of molecular barcoding redundancy. PLoS One 13, e0204265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salk J. J., Schmitt M. W., Loeb L. A., Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 19, 269–285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumura S., et al. , Genome-wide somatic mutation analysis via Hawk-Seq™ reveals mutation profiles associated with chemical mutagens. Arch. Toxicol. 93, 2689–2701 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Van Tassell C. P., et al. , SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat. Methods 5, 247–252 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Luca F., Hudson R. R., Witonsky D. B., Di Rienzo A., A reduced representation approach to population genetic analyses and applications to human evolution. Genome Res. 21, 1087–1098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhatia S., Sklar C., Second cancers in survivors of childhood cancer. Nat. Rev. Cancer 2, 124–132 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Reulen R. C., et al. ; British Childhood Cancer Survivor Study Steering Group, Long-term risks of subsequent primary neoplasms among survivors of childhood cancer. JAMA 305, 2311–2319 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Vardiman J. W., Harris N. L., Brunning R. D., The World Health Organization (WHO) classification of the myeloid neoplasms. Blood 100, 2292–2302 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Larson R. A., Etiology and management of therapy-related myeloid leukemia. Hematology (Am. Soc. Hematol. Educ. Program) 2007, 453–459 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Bhatia S., Therapy-related myelodysplasia and acute myeloid leukemia. Semin. Oncol. 40, 666–675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNerney M. E., Godley L. A., Le Beau M. M., Therapy-related myeloid neoplasms: when genetics and environment collide. Nat. Rev. Cancer 17, 513–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pich O., et al. ; O. G3 (Bethesda)Pich et al, The evolution of hematopoietic cells under cancer therapy. Nat. Commun. 12, 4803 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lisitsyn N., Lisitsyn N., Wigler M., Cloning the differences between two complex genomes. Science 259, 946–951 (1993). [DOI] [PubMed] [Google Scholar]
- 33.Zabarovsky E. R., Allikmets R. L., An improved technique for the efficient construction of gene libraries by partial filling-in of cohesive ends. Gene 42, 119–123 (1986). [DOI] [PubMed] [Google Scholar]
- 34.Kucab J. E., et al. , A Compendium of Mutational Signatures of Environmental Agents. Cell 177, 821–836.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prakash L., Stewart J. W., Sherman F., Specific induction of transitions and transversions of G-C base pairs by 4-nitroquinoline-1-oxide in iso-1-cytochrome c mutants of yeast. J. Mol. Biol. 85, 51–65 (1974). [DOI] [PubMed] [Google Scholar]
- 36.Downes D. J., et al. , Characterization of the mutagenic spectrum of 4-nitroquinoline 1-oxide (4-NQO) in Aspergillus nidulans by whole genome sequencing. G3 (Bethesda) 4, 2483–2492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tate J. G., et al. , COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 47 (D1), D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neglia J. P., et al. , Second malignant neoplasms in five-year survivors of childhood cancer: childhood cancer survivor study. J. Natl. Cancer Inst. 93, 618–629 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Lander E. S., et al. ; International Human Genome Sequencing Consortium, Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Lawrence M. S., et al. , Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexandrov L. B., et al. ; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain, Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allard W. J., et al. , Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 10, 6897–6904 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Corces-Zimmerman M. R., Majeti R., Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia 28, 2276–2282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamashita S., Liu Y. Y., Data from "EcoSeq". GitHub, Available at https://github.com/EpigeneticField/EcoSeq. Deposited 26 May 2022.
- 45.Ueda S., Data from "DRA014481" DDBJ Sequence Read Archive, Available at https://ddbj.nig.ac.jp/resource/sra-submission/DRA014481. Deposited 12 July 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Algorithms and computer code data have been deposited in GitHub (Available at https://github.com/EpigeneticField/EcoSeq). Sequencing data of EcoSeq libraries have been deposited with links to DRA accession number DRA014481 in the DDBJ Sequence Read Archive (Available at https://ddbj.nig.ac.jp/resource/sra-submission/DRA014481)(45).