Significance
The acidic environment is critical for the proper function of many intracellular organelles. It is also associated with various diseases such as ischemia, cancer, and inflammation. In response to acidic pH, the proton-activated chloride (PAC) channel permeates chloride ions across membranes and plays an important role in endosomal acidification. It is also involved in acid-induced cell death and ischemic brain injury. However, how proton binding leads to the opening of this newly identified family of ion channels remains unknown. In this study, we identified several critical protonation sites and intersubunit interactions that work together to determine PAC pH sensitivity. Our work reveals a distinct pH-sensing mechanism and is relevant to the development of inhibitors targeting PAC in diseases associated with acidosis.
Keywords: proton-activated chloride channel, PAC, TMEM206, pH sensitivity, allosteric regulation
Abstract
In response to acidic pH, the widely expressed proton-activated chloride (PAC) channel opens and conducts anions across cellular membranes. By doing so, PAC plays an important role in both cellular physiology (endosome acidification) and diseases associated with tissue acidosis (acid-induced cell death). Despite the available structural information, how proton binding in the extracellular domain (ECD) leads to PAC channel opening remains largely unknown. Here, through comprehensive mutagenesis and electrophysiological studies, we identified several critical titratable residues, including two histidine residues (H130 and H131) and an aspartic acid residue (D269) at the distal end of the ECD, together with the previously characterized H98 at the transmembrane domain–ECD interface, as potential pH sensors for human PAC. Mutations of these residues resulted in significant changes in pH sensitivity. Some combined mutants also exhibited large basal PAC channel activities at neutral pH. By combining molecular dynamics simulations with structural and functional analysis, we further found that the β12 strand at the intersubunit interface and the associated “joint region” connecting the upper and lower ECDs allosterically regulate the proton-dependent PAC activation. Our studies suggest a distinct pH-sensing and gating mechanism of this new family of ion channels sensitive to acidic environment.
Opened by extracellular acidic pH, the proton-activated Cl− (PAC) channel activity has been observed in a wide range of mammalian cells (1–8). Through unbiased RNA interference (RNAi) screens, we and others recently identified a novel gene, PACC1 (also known as TMEM206), as the molecular identity of the PAC channel (9, 10). Tissue acidosis is one of the pathological features in many diseases, including ischemia, cancer, infection, and inflammation. We showed that PAC is involved in acid-induced neuronal cell death in vitro and ischemic brain damage in mice (9, 11, 12). In addition to its pathological role, we also found a surprising physiological function for the PAC channel in regulating endosomal acidification (13). PAC traffics from the plasma membrane to endosomes, where it functions as a low pH sensor, preventing hyperacidification by releasing Cl− from the lumen (13, 14).
Without obvious sequence homology with other membrane proteins, PAC represents a new family of Cl− channels (9, 10). We recently solved two cryo-electron microscopy (cryo-EM) structures of human PAC in a pH 8 resting closed state and a pH 4 proton-bound nonconducting state (15). Indeed, the structures revealed a trimeric channel architecture that is different from known Cl− channels. Each PAC subunit consists of a transmembrane domain (TMD) with two membrane-spanning helices (TM1 and TM2) and a large extracellular domain (ECD). TM2 lines the ion-conducting pore and the positively charged lysine 319 residue in TM2 determines the anion selectivity of the channel. The hand-like shape of ECD is composed of a palm, a finger, and a β-ball domain, all formed by β-strands, and a thumb domain consisting of two short α-helices (Fig. 1A). Surprisingly, the ECD of PAC shows remarkable structural similarities to the acid-sensitive ion channel (ASIC) and the epithelial sodium channel, despite very limited sequence similarity and opposite ion selectivity (15–17). However, PAC lacks several peripheral pH-sensing helices found in the ECD of ASIC (15, 18) and has a continuous TM2 helix instead of a two-segment TM2 structure that is critical for the gating of the ASIC channel (15, 19). These key distinctions suggest that the PAC channel has a novel pH-sensing and gating mechanism. The cryo-EM structure of pufferfish PAC at neutral pH is very similar to that of the human homolog (15, 20), indicating conservation among the PAC family members across vertebrate species.
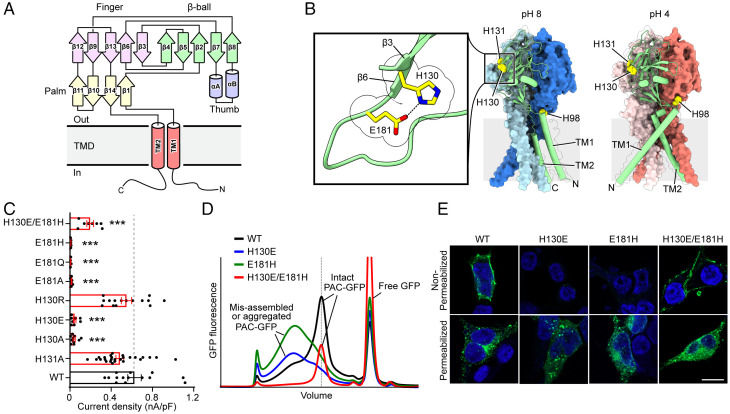
Fig. 1.
H130–E181 interaction is essential for PAC channel formation and function. (A) Domain organization of PAC. Secondary structure elements that constitute the palm, finger, thumb, and β-ball domains are grouped and labeled (15). (B) Cryo-EM structures of the trimeric structure of pH 8–PAC and pH 4–PAC viewed parallel to the membrane (Protein Data Bank ID: 7JNA and 7JNC) (15). The green subunit is shown as a cartoon and the other two subunits are shown in surface representation. H98 in the ECD-TM1 interface, and H130, E181, and H131 in the extracellular domain are highlighted (yellow). Insert: Close-up view of the side-chain interaction between H130 and E181 on the β3 and β6 strands, respectively (highlighted in yellow and shown in stick representation). (C) Current densities of wild-type PAC and mutants of H130 and E181. Data are the mean ± SEM of the pH 4.6–induced currents at +100 mV. ***P < 0.001, one-way ANOVA with Bonferroni post hoc test. (D) The FSEC profile of GFP-tagged wild-type PAC (black) and mutants of H130E (blue), E181H (green), and H130E/E181H (red) solubilized using glyco-diosgenin detergent. The expected position of intact GFP-tagged PAC protein is indicated in a dotted vertical line. The peak positions of both H130E and E181H single mutants, but not the double mutant H130E/E181H, are broader and shifted compared to the wild-type, suggesting that single mutants H130E and E181H interfere with the proper assembly of PAC. A Superose 6 Increase 10/300 GL column was used. (E) Cell surface immunostaining of HEK293T PAC KO transiently transfected with wild-type PAC-FLAG277 (FLAG tag at position 277), H130E, E181H, or H130E/E181H cDNA constructs. The cells were either permeabilized to allow intracellular access to FLAG antibody (Permeabilized) or live-labeled to enhance only plasma membrane staining of PAC-FLAG277(Non-Permeabilized). (Scale bar, 10 µm.)
When the pH drops from 8 to 4, PAC undergoes dramatic conformational changes in the TMD and ECD–TMD interface (15). TM1 of the TMD rotates and switches its interaction partner from the cognate TM2 to the TM2 in the adjacent subunit. In the ECD–TMD interface, the histidine 98 (H98) residue is decoupled from its resting position and inserted into an acidic pocket comprising E107, D109, and E250. The importance of the interaction between H98 and the acidic pocket is further supported by functional analysis, in which mutations of H98 and E107 altered the pH sensitivity of PAC (15). However, these mutants were still largely pH sensitive. Thus, similar to ASIC, the pH sensing of PAC is likely also determined by additional titratable residues in the ECD. Here, we performed comprehensive mutagenesis and functional studies and identified additional titratable residues, including H130, H131, and D269, which determine PAC pH sensitivity. Combined with structural analysis and molecular dynamics (MD) simulations, we found that enhanced intersubunit interactions between the β12 strand and the associated joint region connecting the upper and lower ECDs is another critical element in facilitating the pH-dependent conformational changes of PAC.
Results
Interaction between H130 and E181 Is Essential for PAC Channel Assembly and Function.
Given the range of pH sensitivity of the PAC channel, histidine residues are prime candidates for pH sensing because the pKa value (∼6) of the imidazole side chain is close to the pH of half-maximal activation (pH50) value (∼5) of the channel (9). In addition to the previously characterized H98, there are two conserved adjacent histidine residues (H130 and H131) in the β3 strand of the PAC extracellular finger domain (Fig. 1A and B and SI Appendix, Fig. S1). To examine their role in PAC channel function, we performed site-directed mutagenesis and whole-cell patch-clamp recordings on PAC knockout (KO) HEK293T cells transfected with mutant cDNAs. Substitution of H130, but not H131, with alanine resulted in a loss of channel activity (Fig. 1C), indicating the importance of H130 in PAC function. Replacing H130 with the negatively charged glutamic acid (H130E) also led to a dead mutant (Fig. 1C). To understand why this mutant is nonfunctional, we first performed fluorescence size-exclusion chromatography (FSEC) to analyze channel expression and assembly. Instead of a sharp peak observed for wild-type PAC protein, the green fluorescent protein (GFP)–tagged H130E mutant exhibited a much broader and shifted peak (Fig. 1D), suggesting misfolding or misassembly of the mutant PAC protein. Consistent with the recording data and biochemical analysis, live labeling experiments also showed a lack of expression of the H130E mutant on the cell plasma membrane (Fig. 1E). In the PAC cryo-EM structures, H130 is in close contact with E181 on the β6 strand of the finger domain in the same subunit (Fig. 1B and SI Appendix, Fig. S2A). We hypothesized that the side chain of H130 forms a hydrogen bond with the side chain of E181 and that this interaction is important for proper folding and/or assembly of the channel. Indeed, replacing E181 with alanine, glutamine, or histidine residues all produced dead mutants (Fig. 1C), and expression analysis of E181H revealed defects in the PAC assembly profile and cell surface localization (Fig. 1D and E). In contrast, replacing H130 with a positively charged residue (H130R), which presumably does not abolish the interaction with E181, preserved the channel activity (Fig. 1C). To further examine the potential interaction between H130 and E181, we generated a double mutant, H130E/E181H, by swapping these two critical residues. While both single mutants, H130E and E181H, were not properly expressed and were dead, the double mutant was functional with biochemical profile and plasma membrane expression similar to the wild-type PAC (Fig. 1C–E). Together, these data suggest that the intrasubunit interaction between H130 and E181 in the distal finger domain is critical for a properly folded ECD, which in turn is important for the PAC channel assembly and its trafficking to the plasma membrane.
Three Conserved Histidine Residues Are Key Determinants of PAC pH Sensing.
To determine the role of conserved histidine residues (H98, H130, and H131) in PAC pH sensing, we first examined the pH sensitivity of mutants in which only one of the three histidine residues was replaced with arginine, which is constitutively protonated at physiological pH (Fig. 2A). As shown previously (15), H98R mutation resulted in a modestly increased pH sensitivity (Fig. 2A and B). This substitution likely facilitates the repositioning of the protonated side chain at position 98 from its resting position to the acidic pocket in the palm domain, which is a major conformational change of PAC induced by low pH (15). While the H131R mutant had a pH50 similar to that of the wild type, the H130R mutant exhibited an increased pH sensitivity, shifting the pH50 to a less acidic value by ∼0.2 units (Fig. 2A and B). Given the potential intrasubunit interaction between H130 and E181, the increased pH sensitivity of the H130R mutant suggests that strengthening the interaction between protonated H130 and E181 in the distal ECD may promote the pH-induced PAC channel activation. Notably, the swapping mutant H130E/E181H exhibited a reduced pH sensitivity compared to the wild-type PAC channel (Fig. 2A and B), suggesting that the location of the histidine residue may affect its pH-sensing ability because its side-chain pKa is influenced by the local environment.
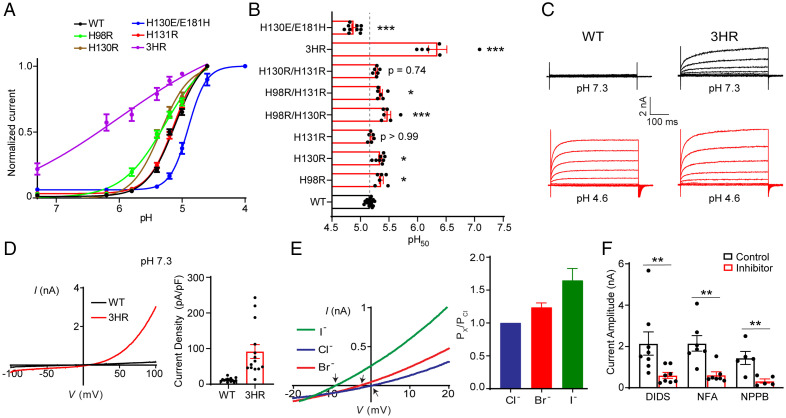
Fig. 2.
H98, H130, and H131 are key determinants of PAC pH sensing. (A) pH dose-response curve of wild-type PAC, H98R, H130R, H131R, H130E/E181H, and H98R/H130R/H131R (3HR). The currents are normalized to the pH 4.6– or 4.0–induced currents [n = 14 (wild-type PAC); n = 6 (H98R); n = 8 (H130R); n = 6 (H131R); n = 10 (H130E/E181H); n = 6 (3HR)]. The normalized data are fitted to the Hill equation unconstrained, allowing both the top and bottom to be defined by the maximum and minimum normalized currents. Data are the mean ± SEM of the normalized currents at +100 mV. (B) pH50 values estimated from the pH dose-response curves in (A). *P < 0.05; ***P < 0.001, one-way ANOVA with Bonferroni post hoc test. (C) Representative whole-cell currents of wild-type PAC (Left) and 3HR mutant (Right) at pH 7.3 (black) and 4.6 (red) monitored by voltage-step protocol. (D) Left: Representative whole-cell currents of wild-type PAC (black trace) and 3HR mutant (red trace) at pH 7.3 monitored by voltage-ramp protocol. Right: Current densities (mean ± SEM) of wild-type PAC and 3HR mutant at pH 7.3 and +100 mV. (E) Left: Representative I to V relationship of 3HR mutant recorded in extracellular equimolar I− (green trace), Br− (red trace), or Cl− (blue trace) pH 7.3 solution. Arrows indicate the reversal potentials. Right: Anion permeability ratio of 3HR mutant (n = 7 cells). Data represent mean ± SEM. (F) Quantification of the inhibition of the 3HR basal currents at neutral pH by three PAC inhibitors: 100 µM 4,4′-diisothiocyanatostilbene-2,2′-disulfonate (DIDS), 100 µM niflumic acid (NFA), and 100 µM 5-nitro-2-(3-phenylpropylamino)-benzoic acid (NPPB). Current amplitudes (mean ± SEM) at +100 mV (control) before inhibitors were applied. **P < 0.01; two-tailed Student’s t test.
Although single histidine replacements with arginine increased the pH sensitivity, none of them produced basal PAC currents at neutral pH. To test whether the three histidine residues cooperatively regulate PAC pH sensing and channel activation, we generated double and triple histidine substitutions with arginine. Two double mutants (H98R/H130R and H98R/H131R) showed significant changes in the pH50 compared to wild-type PAC (Fig. 2B), yet they retained similar pH sensitivities as the single H98R mutant.
Interestingly, the triple mutation (3HR) not only markedly increased the pH sensitivity (shifting the pH50 by ∼1.2 units to a less acidic value) but also gave rise to large basal currents at pH 7.3 and pH 8.0 (Fig. 2A–D and SI Appendix, Fig. S2B). These data suggest that protonation of the three histidine residues synergistically regulates the pH sensitivity and channel activation of PAC. The basal currents mediated by the 3HR mutant shared similar characteristics to the wild-type PAC currents, including time-dependent facilitation at positive membrane voltages (Fig. 2C), strong outward rectification (Fig. 2D), and I− > Br− > Cl− permeability sequence (Fig. 2E) (9). They were also blocked by several nonspecific PAC inhibitors, including 4,4′-diisothiocyanatostilbene-2,2′-disulfonate, niflumic acid, and 5-nitro-2-(3-phenylpropylamino)-benzoic acid (Fig. 2F and SI Appendix, Fig. S2G). Therefore, the biophysical properties and pharmacological profile of the 3HR-mediated basal currents at neutral pH match those of the wild-type PAC currents activated by acidic pH. Together, these results demonstrate that H98 at the TM1–ECD interface and H130 and H131 in the distal finger domain of the ECD are key regulators of PAC pH sensitivity and channel activation.
We next tested whether replacing one or more of the three histidine residues with glutamine, a residue that cannot be protonated, results in a loss-of-function phenotype. Surprisingly, the H98Q mutant showed an increased pH sensitivity (SI Appendix, Fig. S2C and D). This is unexpected because a glutamine residue would be less attracted by the acidic pocket than a protonated histidine residue. A similar result was previously observed for the H98A mutant (15). It is likely that these mutations at position 98 introduced additional conformational changes at the TMD–ECD interface rather than simple side-chain substitutions, which renders the channel more sensitive to protons. The two single mutants, H130Q and H131Q, exhibited a pH sensitivity similar to that of the wild-type PAC channel (SI Appendix, Fig. S2D), whereas the double (H130Q/H131Q) and triple (3HQ) mutants abolished acid-induced PAC currents because they failed to localize to the plasma membrane and exhibited defects in channel assembly (SI Appendix, Fig. S2C–E).
The Joint Region Is an Allosteric Transducer of pH-Dependent Conformational Changes.
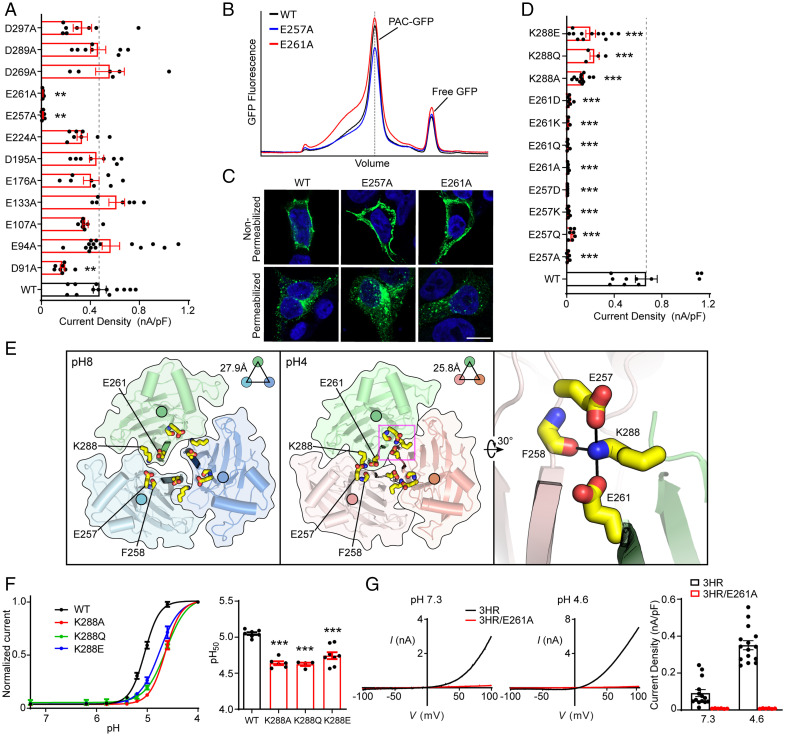
The residual pH sensitivity of the 3HR mutant indicates the existence of additional structural elements that contribute to PAC pH sensing. We focused our search for these elements on acidic residues (aspartic acid and glutamic acid) because the predicted pKa of many of them are above pH 4, which puts them in the pH range for PAC channel activation (15). We performed an alanine scanning screen on 12 acidic residues conserved from fish to humans (SI Appendix, Fig. S1), excluding E181, which we have already characterized in Fig. 1. While most mutants were functional (which will be discussed in the next section), the E257A and E261A mutants lost their acid-induced PAC channel activities (Fig. 3A), even when the pH was dropped to 4 (SI Appendix, Fig. S3A). Unlike other dead mutants described earlier in this work, the lack of channel activity was not due to defects in protein folding/assembly and cell surface expression (Fig. 3B and C). Moreover, neutralizing or reversing the charges by replacing E257 and E261 with glutamine or lysine residues also resulted in nonfunctional channels (Fig. 3D). The ECD of PAC has a loose intersubunit interface with obvious gaps between subunits at pH 8, which are mostly filled at pH 4 (Fig. 3E). This iris-like rotation of the ECD creates extensive interactions between the β12 strands of the three PAC subunits. E257 and E261 are located at the linker between β11 and β12, and the lower β12 strand, respectively (SI Appendix, Fig. S1). Due to their location at the intersubunit interface, E257 and E261 were previously proposed as potential pH sensors (20). However, we consider it unlikely because neutralizing the negative charges with glutamine led to a loss of channel function rather than an increased pH sensitivity (Fig. 3D). In addition, substitutions with aspartic acid, which also carries a negative charge, resulted in dead channels too (Fig. 3D), suggesting that the precise side-chain length of residues at the location is critical for channel function.
Fig. 3.
The joint region is an essential allosteric transducer for pH-dependent PAC activation. (A) Current densities of wild-type and mutant PAC in the ECD acidic residue screen. Data are the mean ± SEM of pH 4.6–induced currents at +100 mV. **P < 0.01; one-way ANOVA with Bonferroni post hoc test. (B) The FSEC profile of GFP-tagged wild-type PAC (black), E257A (blue), and E261A (red) solubilized using glyco-diosgenin detergent. The expected position of intact GFP-tagged PAC protein is indicated in a dotted vertical line. (C) Cell surface immunostaining of HEK293T PAC KO transiently transfected with wild-type PAC-FLAG277 (the same representative image as in Fig. 1E), E257A, or E261A cDNA constructs. (Scale bar, 10 µm.) (D) Current densities of wild-type PAC and various mutations on E257, E261, and K288 residues. Data are the mean ± SEM of the pH 4.6–induced currents at +100 mV. ***P < 0.001, one-way ANOVA with Bonferroni post hoc test. (E) Top view of the pH 8–PAC (Left) and pH 4–PAC (Middle) structures depicting the pH-dependent conformational change of the intersubunit interface. E257, F258, E261, and K288 are highlighted in yellow sticks. A close-up view of the intersubunit interactions involving E257, F258, E261, and K288 in the pH 4 state is shown on the Right. (F) Left: pH dose-response curve of wild-type PAC, K288A, K288Q, and K288E. The currents are normalized to the pH 4.0–induced currents [n = 7 (wild-type PAC); n = 6 (K288A); n = 4 (K288Q); n = 8 (K288E)]. The normalized data are fitted to the Hill equation unconstrained. Data are the mean ± SEM of the normalized currents at +100 mV. Right: pH50 values (mean ± SEM) estimated from the pH dose-response curves. ***P < 0.001, one-way ANOVA with Bonferroni post hoc test. (G) Left two panels: Representative whole-cell currents of 3HR (black trace) and 3HR/E261A (red trace) mutants induced by extracellular pH 7.3 (Left) and 4.6 (Middle) solutions, monitored by voltage-ramp protocols. Right: Current densities (mean ± SEM) at +100 mV shown on the Left.
In the pH 4–PAC structure, in addition to being directly involved in intersubunit contact (e.g., through potential interactions with the side chain of W230 and backbone of Q260), the side chain of E261 forms a salt bridge interaction with K288 located between the β13 and β14 strands of the same subunit, which in turn forms additional polar interactions with the side chain of E257 and the backbone of F258 of the adjacent subunit (Fig. 3E). We thus hypothesized that K288 is also critical in mediating intersubunit interactions at low pH, thereby promoting the proton-induced contraction of the ECD. Indeed, substitution of K288 with alanine, glutamine, or glutamate—which is expected to disrupt the intersubunit interaction (Fig. 3E)—all resulted in markedly reduced PAC currents (Fig. 3D) and a decreased pH sensitivity (Fig. 3F). These data suggest that maintaining a tight intersubunit interaction at low pH is essential for PAC channel activation.
E257, E261, and K288 are all strategically located at the intersubunit interface while linking the β sheets of the upper finger domain (β12, β13) with the lower palm domain (β11, β14) (SI Appendix, Fig. S1). We referred to the region between the finger and palm domains as the “joint region,” which undergoes a significant conformational change when the pH drops from 8 to 4. Specifically, while E261 and E257 are solvent exposed in the structure at pH 8, both form critical interactions at the intersubunit interface in the structure at pH 4 (Fig. 3E). Therefore, we hypothesized that the joint region is poised to transduce pH-dependent conformational changes in the upper finger domain to the lower palm domain and TMDs, thus allosterically controlling channel gating. To test this hypothesis, we introduced the E261A substitution into the 3HR mutant. The combined 3HR/E261A mutant not only lost its basal activity at neutral pH, but also was nonfunctional at acidic pH (Fig. 3G). The loss of activity was not due to abnormal channel expression, as the mutant proteins were detected at the cell surface (SI Appendix, Fig. S3B). Similarly, the acid-induced currents were also greatly reduced in the 3HR/E257A mutant (SI Appendix, Fig. S3C). These data further support the important function of E261 and E257 in PAC activation and are consistent with an essential role of the joint region in the allosteric control of proton-dependent channel gating.
D269 Is Another Critical Regulator of PAC pH Sensitivity.
Next, we determined the pH sensitivity of the remaining 10 functional mutants from the acidic residue screen. Among them, three mutants (D91A, E94A, and E107A) exhibited a modest increase in the pH50 values (Fig. 4A). E107 is part of the acidic pocket, interacting with H98 in the pH 4–PAC structure (SI Appendix, Fig. S2A) (15). E94 and D91 are located in the TM1 helix close to the TMD–ECD interface where H98 is located. It is likely that mutations of these residues affect the interaction between H98 and the acidic pocket, which in turn leads to an increased pH sensitivity.
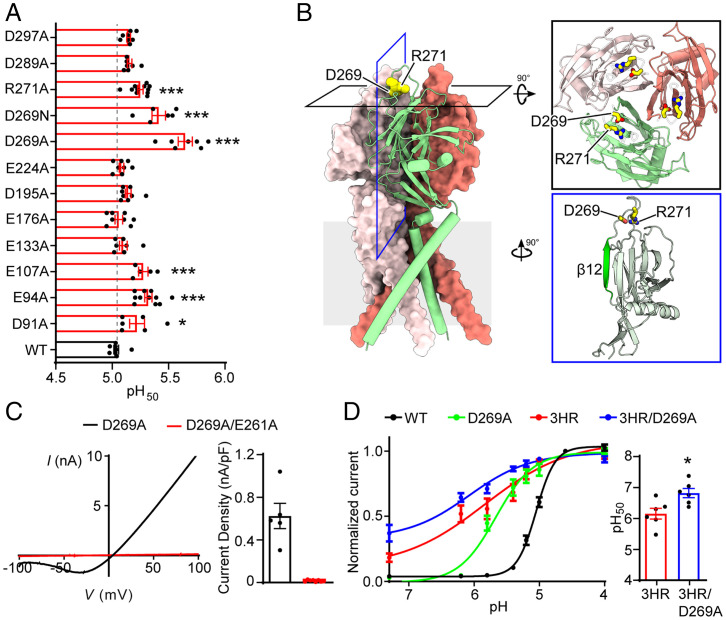
Fig. 4.
Screening of acidic residues in the ECD identifies D269 as another regulator of PAC pH sensitivity. (A) pH50 values (mean ± SEM) of wild-type PAC and mutants. *P < 0.05; ***P < 0.001, one-way ANOVA with Bonferroni post hoc test. (B) Cryo-EM structure models of pH 4–PAC structure viewed parallel to the membrane (Protein Data Bank ID: 7JNC). The green subunit is shown as a cartoon and the other two subunits are shown in surface representation. D269 and R271 are highlighted as yellow spheres. Inserts: Top view (black frame) and side view (blue frame) highlighting D269 and R271. The β12 strand is highlighted in green in the side view. (C) Left: Representative whole-cell currents of D269A (black) and D269A/E261A (red) mutants at pH 4.6, and monitored by voltage-ramp protocol. Right: pH 4.6–induced current densities (mean ± SEM) of D269A and D269A/E261A mutants at +100 mV. (D) Left: The pH dose-response curve of wild-type PAC (black), D269A (green), 3HR (red), and 3HR/D269A (blue). The currents are normalized to the pH 4.0–induced currents [n = 7 (wild-type PAC); n = 7 (D269A); n = 6 (3HR); n = 6 (3HR/D269A)]. The normalized data are fitted to the Hill equation unconstrained. Data are the mean ± SEM of the normalized currents at +100 mV. Right: pH50 values (mean ± SEM) of 3HR and 3HR/D269A mutants estimated from the pH dose-response curve in (D). *P < 0.05, two-tailed Student’s t test.
D269 is most interesting among all the tested acidic residues because although it is situated at the top of ECD, distal to the channel gate (Fig. 4B and SI Appendix, Fig. S1), the D269A mutant exhibited a dramatic increase in pH sensitivity. The pH50 was shifted by ∼0.6 units to a less acidic value (Fig. 4A), which was the largest change among all the single PAC mutations tested, suggesting that protonation of D269 at acidic pH may play an important role in pH-dependent channel activation. Consistently, neutralizing the charge by asparagine substitution (D269N) also resulted in an increased pH sensitivity (Fig. 4A). In the structural model of PAC at pH 4, D269 forms a salt bridge interaction with the adjacent R271. Replacing R271 with an alanine residue (R271A) also led to an increase in the pH50, although not as dramatic as the D269 mutation (Fig. 4A). These results indicate that D269 is involved in pH-dependent PAC activation, partially through its interaction with R271. Disrupting this interaction might affect PAC pH sensing by changing the conformation of the intersubunit interface, because D269 is only three residues away from the β12 strand that forms the intersubunit interface. Consistent with this hypothesis, introducing the key substitution at the β12 strand (E261A) into the D269A mutant resulted in a loss of PAC channel activity (Fig. 4C), although the double-mutant proteins were expressed and trafficked to the plasma membrane (SI Appendix, Fig. S3B). Together, our data identify the acidic residue D269 near the β12 strand as another important determinant of PAC pH sensing.
To test whether D269 and three conserved histidine residues synergistically regulate pH-dependent PAC channel activation, we combined all four mutations and generated a quadruple mutant (3HR/D269A). Whole-cell recordings revealed that the 3HR/D269A mutant mediated large basal currents at neutral pH, which had similar biophysical properties to the wild-type PAC currents (SI Appendix, Fig. S4A and B). More importantly, the quadruple mutant lost most of its pH sensitivity with an even higher pH50 compared to the 3HR mutant (Fig. 4D). These data suggest that three histidine residues (H98, H130, and H131) and D269 cooperatively determine PAC pH sensitivity and channel activation.
The Lower and Upper Parts of the β12 Strand Differentially Regulate PAC Channel Activity.
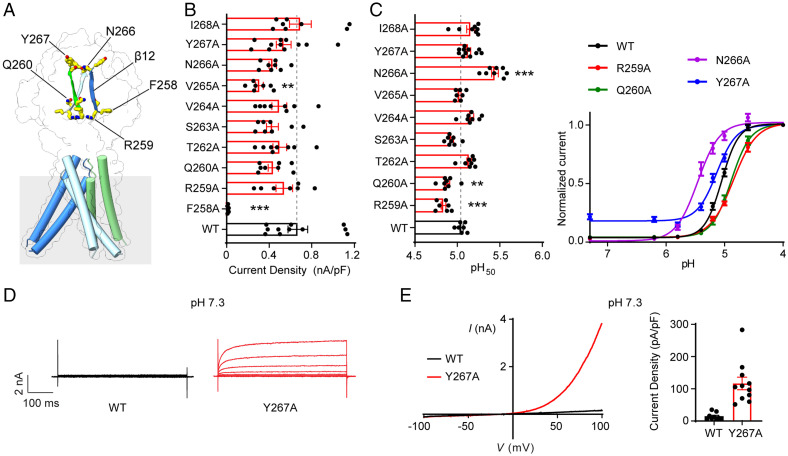
The β12 strand of the finger domain forms a main intersubunit interface at the ECD of the pH 4–PAC structure (15). The critical role of E261 within β12 and two other flanking residues (E257 and D269) in channel activation suggests an important function of the β12 strand. To further examine its role in PAC channel activation, we performed an unbiased alanine scanning screen (F258 to I268, excluding the already characterized E261) and assayed their channel activity and pH sensitivity (Fig. 5A and SI Appendix, Fig. S1). Interestingly, mutations of 5 out of 10 amino acids exhibited significant defects in PAC channel activity (Fig. 5B and C), highlighting the importance of the β12 strand. The mutants were clustered in the lower and upper parts of β12, respectively, with two different types of phenotypes. In the lower β12, like the adjacent E261 and E257, alanine mutation of F258 resulted in a loss of PAC channel function (Fig. 5B). The R259A and Q260A mutants were functional; however, both exhibited a reduced pH sensitivity (Fig. 5B and C) similar to mutations of K288 located in the joint region (Fig. 3F). These data reinforced our hypothesis that the lower β12 strand and the associated joint region between the finger and palm domains allosterically regulate the pH-dependent channel activation. In contrast to the mutations in the lower β12, the N266A mutant in the upper β12 had an increased pH sensitivity (Fig. 5C), similar to mutations of the adjacent D269 and R271 right above the β12 strand. In addition, the Y267A mutant mediated large basal currents at neutral pH with similar biophysical properties to the wild-type PAC currents (Fig. 5D and E). Together, these results indicate that different regions of the β12 strand play important but distinct roles in PAC channel activation. Because most of these residues are not titratable, they are likely involved in allosteric regulation of pH-induced conformational changes required for PAC channel gating.
Fig. 5.
The lower and upper parts of the β12 strand differentially regulate PAC channel activity. (A) The Cryo-EM structure of pH 8–PAC structure highlighting residues of the β12 strand, F258, R259, Q260, N266, and Y267. (B) pH 4.6–induced current densities (mean ± SEM) at +100 mV of wild-type PAC and alanine scanning mutants of the β12 strand. **P < 0.01; ***P < 0.001, one-way ANOVA with Bonferroni post hoc test. (C) Left: pH50 values (mean ± SEM) of wild-type PAC and β12 strand mutants estimated from their respective pH dose-response curves. Right: pH dose-response curve of wild-type PAC, R259A, Q260A, N266A, and Y267A. The currents are normalized to the pH 4.0–induced currents. The normalized data are fitted to the Hill equation unconstrained. Data are the mean ± SEM of the normalized currents at +100 mV. **P < 0.01; ***P < 0.001, one-way ANOVA with Bonferroni post hoc test. (D) Representative whole-cell currents of wild-type PAC (Left) and Y267A (Right) at pH 7.3 monitored by voltage-step protocol. (E) Left: Representative whole-cell currents of wild-type PAC (black trace) and Y267A (red trace) at pH 7.3 monitored by voltage-ramp protocol. Right: Current densities (mean ± SEM) of wild-type PAC and Y267A mutant at +100 mV induced by extracellular pH 7.3 or 4.6 solution.
MD Simulations Reveal a Mechanism of pH-Dependent Allosteric Communication in PAC.
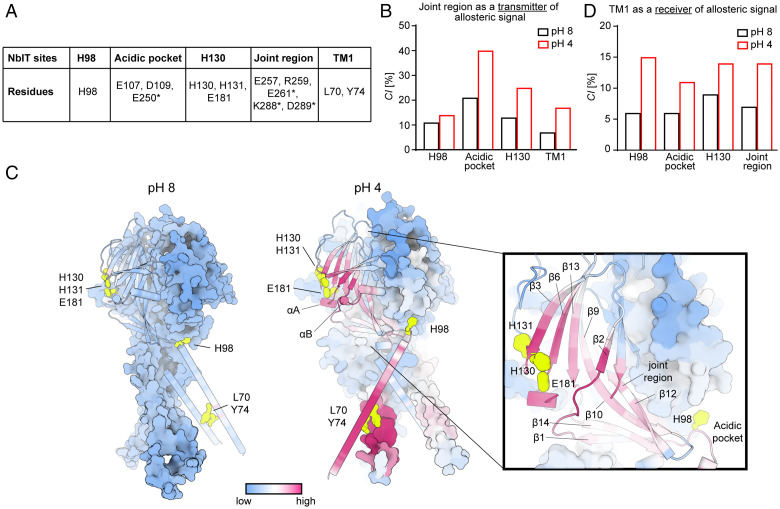
To specify in structural and quantitative terms the pH-dependent allosteric pathways involved in the functional pH sensitivity observed for the PAC channel, we simulated the dynamics of the molecular systems. Extensive MD simulations (36 replicas of 750 ns each, for a total sampling of 27 µs) of PAC were carried out at pH 8 and pH 4 using the equilibrium constant pH (ECpH) MD method (21, 22). ECpH MD achieves accurate prediction of conformational pH dependence in protein systems by utilizing a force-field modification protocol in which the protonation probability for each titratable site defines the system’s protonation state at a certain pH (Methods) (21, 22). This eliminates the need for a binary protonated/nonprotonated approximation used in the conventional MD approach. To reveal the allosteric communication between the activation of the pH-sensing region of the ion channel and the intracellular opening, the resulting ECpH-MD trajectories were analyzed with the N-body information theory (NbIT) method (23) (Methods). NbIT (23, 24) quantifies the allosteric communication between various regions of the protein in the “coordination information” term (CI) that expresses the extent of coordination of one region (identified as “receiver”) by another (identified as “transmitter”). The path of the allosteric communication (aka “the allosteric pathway”) is then defined by the set of residues that share the highest amount of information with the transmitter–receiver pair, as quantified by the “mutual coordination information” term (MCI) (see Methods and ref. 23 for a detailed description of the NbIT formalism).
The allosteric nature of pH-dependent conformational change observed in the PAC channel is encoded in its structure where the ECD is exposed to different pH levels and the TMD undergoes the pH-dependent conformational rearrangements. The conformational response to a pH change sensed in the ECD, and observed in the distal TMD, is thus likely to result from a switch in protonation state of one or more ECD residues. To reveal the allosteric pathway, the first step in the NbIT analysis was to identify potential transmitters of allosteric signal among the titratable residues of the PAC ECD at pH 4 that trigger the dramatic pH-dependent rotational movement of the receiver TM1 (details in Methods). Among titratable sites in ECD (transmitter), the largest CI values calculated in this step (SI Appendix, Fig. S5B) identified a prominent transmitter cluster, comprised of the residues E257, R259, and E261—part of the joint region (see cluster S9 in SI Appendix, Fig. S5A and quantification in SI Appendix, Fig. S5B).
The crucial role of the joint region as a key transmitter of allosteric signaling in the PAC protein at pH 4 (Fig. 6B) was further validated in the NbIT analysis of the mechanism of allosteric communication by values calculated for selected sites shown experimentally to be important for PAC pH sensitivity (Fig. 6A). Thus, the highest CI values for the joint region acting as the transmitter were toward the acidic pocket (40%) and H130/H131/E181 (25%) (Fig. 6B). Remarkably, when the same communication was quantified from the ECpH-MD trajectory at pH 8, the calculated CI values were dramatically decreased (a loss of 20% for the acidic pocket, and 12% for the H130 site) (Fig. 6B). These quantitative findings corroborate the experimental data by illuminating the underlying allosteric involvement of these regions in pH-dependent activation of PAC.
Fig. 6.
MD simulations and NbIT analysis reveal pH-dependent allosteric communication in PAC. (A) Definition of NbIT sites. Asterisks indicate residues from a neighboring PAC subunit. (B) CI values for the joint region (transmitter) coordination of other NbIT sites averaged over three subunits of PAC at pH 8 and 4. (C) The allosteric coordination channel between the H130/H131/E181 region and TM1 of PAC from NbIT analysis of ECpH-MD trajectories simulated at pH 8 (Left) and 4 (Right). Per-residue MCI values are represented on the structure with a uniform colormap ranging from low to high. (D) CI values for TM1 (receiver) coordination by other NbIT sites averaged over three subunits of PAC at pH 8 and 4.
To identify the specific allosteric communication path between the pH-dependent H130/H131/E181 (H130 site) (transmitter) and TM1 (receiver), we quantified MCI between the two sites and other residues of the protein from NbIT analysis of ECpH-MD trajectories simulated at pH 8 and pH 4 (Fig. 6C). The results show that in the pH 4–PAC state, the allosteric signal passes from β3 through β6 to the β5 to β6 and β1 to β2 loops (SI Appendix). SI Appendix, Fig. S6 identifies specific “trigger” residues along the pathway. At pH 4, the interactions among these residues support the propagation of the allosteric signal; but at pH 8, these interactions are disrupted, and the allosteric pathway fades away. The allosteric path then proceeds through the helical regions αA and αB located between β7 and β8 (SI Appendix, Fig. S1) where it connects to H98 and the acidic pocket through the β10 to β12 strands and the joint region (Fig. 6C). CI and MCI values calculated from the pH 8–PAC trajectories do not sustain such an allosteric interaction path (Fig. 6C). Consistent with our data from the functional experiments, the CI values for TM1 (receiver) with several regions of the protein involved in pH sensing (transmitters; Fig. 6D) were found to correspond to high levels of allosteric coordination at pH 4 and to be reduced at pH 8 to low-level nonspecific CI values (Fig. 6D). Notably, the biggest pH-dependent changes in allosteric coordination occurred for H98 and the joint region. Apart from demonstrating a high CI value for TM1 coordination by H98, which in itself is not surprising based on its association with the pH-dependent conformational change elucidated previously (15), the NbIT analysis unraveled the important allosteric communication between the joint region (transmitter) and TM1 (receiver) . Moreover, our results show that the joint region is an integral component of the long allosteric pathway connecting TM1 with the H130 site (Fig. 6C). This was brought to light by the finding that the CI values calculated for TM1 as the receiver with either the H130 site or the joint region alone (transmitters) were each 14% (Fig. 6D); when the two regions were included in a combined transmitter, the calculated CI was 28%. According to the mathematical formulation of NbIT, CI is a nonadditive function (specific details in Methods), and additivity occurs only if the individual regions used as a combined transmitter belong to the same allosteric pathway.
Consistent with the structural information, we observed in the ECpH trajectories a pH-dependent switch in orientation of the nonbonded interactions at the subunit interface in the regions of β12 (Ε261), β13 (K288, D289), and β11 (E257, R259) (SI Appendix, Fig. S7). In the joint region, this switch from intrasubunit at pH 8 to intersubunit at pH 4 results in a large decrease in transmitter CI toward the acidic pocket and H130 site at pH 8 (Fig. 6B) as well as in the fading away of the CI allosteric pathway between the H130 site and TM1 (Fig. 6C). The weakening of PAC-subunit interactions, and of the allosteric communication, at pH 8 (Fig. 6C) appears to relate to a ∼40 to 50% increase in mobility of the Cα atoms in the β strands of the ECD that was quantified with root-mean-square fluctuation analysis of the MD trajectories (SI Appendix, Table S1).
Discussion
Based on the cryo-EM structures of human PAC in a high-pH resting closed state and a low-pH proton-bound nonconducting (likely desensitized) state, we previously identified the flipping of H98 at the ECD–TMD interface from the resting position to the acidic pocket as an important element of the proton-induced activation of the PAC channel. However, the H98 mutants exhibited relatively small changes in pH sensitivity. Thus, the detailed molecular mechanism by which protons activate PAC remains to be addressed. Here, guided by the structural information, we employed a broad mutagenesis study together with MD simulations to investigate the pH-sensing and gating mechanism of the PAC channel. Although the large number of titratable residues that could serve as potential proton-binding sites in PAC are distributed throughout the ECD, our study indicates that some of them are important for its pH sensitivity. We identified two adjacent histidine residues (H130 and H131) at the β3 strand and an acidic residue D269 above the β12 strand, all in the finger domain of the ECD, as key determinants of PAC pH sensitivity. H130 is particularly interesting, in that it is required for proper PAC channel assembly and trafficking to the plasma membrane. Together with H98, these titratable residues account for most of the pH sensitivity of PAC. Furthermore, the basal currents at neutral pH mediated by the combined mutants (3HR and 3HR/D269A) exhibited biophysical characteristics similar to those of the wild-type PAC currents activated by low pH. Thus, these gain-of-function mutations specifically affect pH-dependent channel activation and do not disturb the ion-conducting pathway and other channel properties. Unlike H98, we did not observe marked pH-induced conformational changes of H130, H131, D269, and their interaction partners in the available PAC structures, including an activated conformation published when the present work was under review (25). Therefore, we cannot exclude the possibility that mutations of these residues indirectly change the pH sensitivity of the PAC channel. Nevertheless, acidic pH may induce subtle conformational changes at multiple protonation sites, which synergistically drive the inward contraction of the ECD (Fig. 7).
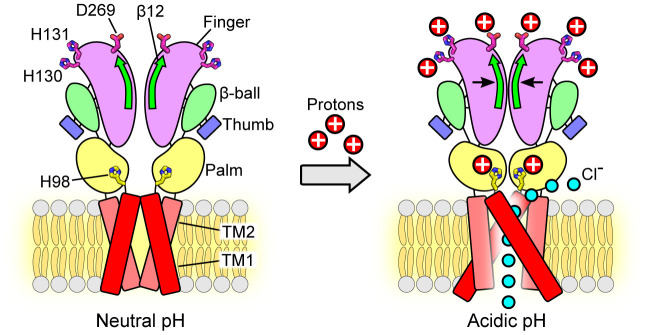
Fig. 7.
Schematic representation of the conformational changes during pH-dependent activation and allosteric gating of the PAC channel. Representative cartoon of PAC, depicting a dimer of the homotrimeric subunit, in the closed and open states. Each subunit is composed of two TMDs, N and C termini, and a large ECD. In the closed state, TM1 and TM2 run parallel and interact with each other. At pH 4, TM1 swings and interacts with TM2 of the adjacent subunit. The ECD contracts toward the pore axis, resulting in a shorter overall structure and a more compact ECD in comparison to that at pH 8. The joint region is at the intersubunit interface and connects the upper finger domain to the lower palm domain. The potential proton-binding sites (H130, H131, D269) are located at the top of the ECD, distal to the ion-conducting pore and channel gate. Protonation of these sites allosterically affects the state of channel gate leading to the PAC channel activation. The β12 strand and the joint region allosterically transduce the pH-dependent conformational changes from the peripheral proton-binding sites to the channel pore. Therefore, combining the available structural information, we propose a model in which protonation of residues in the extracellular finger domain induces the contraction of the ECD, which, together with the conformational change of H98 in the ECD–TMD interface, leads to the opening of the ion-conducting pore. The permeation of Cl− ions through the fenestration site is illustrated (15).
Because of the pH-induced ECD contraction, several acidic residues, including E249, E250, E257, D289, and D297, at the intersubunit interface move closer to potentially interact and were proposed as possible pH sensors (20, 25, 26). However, we did not observe significant changes in pH sensitivity for alanine substitutions of D289 and D297 (Fig. 4A). Even neutralizing three adjacent negative charges together (E249A/E250A/D251A) only resulted in a modest decrease rather than a predicted increase in the pH50 (SI Appendix, Fig. S8). Furthermore, all tested mutants of E257 eliminated PAC channel function, except for E257Q that exhibited a very small channel activity and a modest decrease in pH sensitivity (Fig. 3) (25). This decrease is unexpected because the E257Q mutant represents a constitutively protonated form of this putative pH sensor and, therefore, one would anticipate an increase in pH sensitivity. Together, these results suggest that the acidic amino acids at the intersubunit interface unlikely function as important pH sensors despite their conformational changes induced by low pH.
The candidate proton-binding sites we identified in this study are located at the top of the ECD, distal to the ion-conducting pore and channel gate. Thus, the effect of their protonation state is expected to allosterically affect the state of the channel gate leading to PAC activation. Indeed, during the screen for potential pH sensors, we serendipitously identified that the β12 strand at the intersubunit interface and the associated joint region connecting the upper finger domain to the lower palm domain play a key role in allosterically transducing pH-dependent conformational changes from the peripheral proton-binding sites to the channel pore. Our MD simulations also support a critical function of the joint region in connecting the distal protonation sites to the TMD and coordinating pH-dependent conformational changes. Moreover, the joint region exhibits allosteric coordination for other key regions of PAC, including the acidic pocket, indicating its involvement in the allosteric communications throughout the PAC protein. Based on the current study and the available structural information, we propose a model in which protonation of residues in the extracellular finger domain induces the contraction of the ECD, which, together with the conformational change of H98 in the ECD–TMD interface, leads to the opening of the ion-conducting pore (Fig. 7). Future studies using short-range distance measurement techniques, such as Förster resonance energy transfer and double electron-electron resonance spectroscopy, are required to probe the mechanistic details of our proposed allosteric model of pH-dependent allosteric conformational transitions in the ECD and ECD–TMD interface of PAC.
Notably, even the quadruple mutant (3HR/D269A) still retained some residual pH sensitivity, suggesting the existence of additional pH sensors. A large body of work on the ASIC channels in the last two decades also revealed many titratable residues (histidine and acidic amino acids) as potential proton sensors, which work together to drive pH-dependent conformational changes essential for both channel activation and desensitization (27). Although the specific residues and their location in the ECD are not conserved, the existence of multiple proton-binding sites underlying pH-dependent activation for PAC and ASIC appears to be a common feature shared by these two acid-sensitive channel families.
The elucidation of these structure-based allosteric models would enable a focused approach to the use of the PAC channel as a potential therapeutic target for diseases associated with tissue acidosis. Severe local acidosis causes tissue damage in many pathological conditions (28, 29), and when activated by acidic environment, the PAC channel is involved in acid-induced cell death and contributes to ischemic brain damage in a mouse experimental stroke model (9). The findings reported here are relevant to understand how acidic pH activates the PAC channel and to facilitate the development of inhibitors that target its proton-sensing and gating machinery.
Methods
Cell Culture.
PAC KO HEK293T cells, previously generated using CRISPR technology (9), were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a humidified 95% CO2 incubator.Plasmid DNA was transfected using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Cells were seeded on 12-mm-diameter poly-D-lysine–coated glass coverslips (BD) and were recorded or immunostained 1 d after transfection.
Constructs and Mutagenesis.
The coding sequence of human PAC (NP_060722) previously subcloned into pIRES2–enhanced green fluorescent protein (EGFP) vector (Clontech) using XhoI and EcoRI restriction enzyme sites (9) was used for whole-cell patch-clamp recording. For all immunostaining experiments, the human PAC (NP_060722) with FLAG tag (DYKDDDDK) at position 277 (PAC-FLAG277) previously subcloned into pcDNA5/FRT/TO plasmid using KpnI and EcoRV restriction sites was used (13). The pEG BacMam vector (15) containing PACC1 with a thrombin cutting site, EGFP, and 8×His tag in the C terminus was used for protein expression. Site-directed mutagenesis was introduced into plasmids using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) and confirmed by Sanger sequencing.
Immunostaining.
PAC KO HEK293T cells were transfected with FLAG-tagged human PAC cDNA and mutants. Twenty-four hours after transfection, cells were fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature (RT) and washed three times with phosphate-buffered saline (PBS). Cells were blocked in PBS containing 5% bovine serum albumin (BSA) and 0.2% Tween-20 for 30 min at RT. Cells were then permeabilized and incubated with primary antibody (mouse anti-FLAG, 1:500; Sigma F1804) for 2 h at RT in PBS containing 1% BSA and 0.2% Tween-20. Cells were then washed three times with PBS, followed by Alexa Fluor–conjugated secondary antibody (1:1,000; Thermo Fisher Scientific) incubation for 1 h at RT.
To stain the cell surface membranes, cells were incubated directly with the primary antibody (mouse anti-FLAG, 1:200) for 2 h at 4 °C in PBS containing Ca2+ and Mg2+ prior to fixing with 4% PFA. After fixation, cells were washed three times and then incubated with Alexa Fluor 488–conjugated secondary antibody (1:500) for 1 h at RT. Coverslips were then mounted on microscope slides with ProLong Gold Antifade Reagent (Invitrogen) and images were taken using a Zeiss LSM900 confocal microscope.
Protein Expression and FSEC Analysis.
Adherent human tsA201 cells transiently transfected with GFP-tagged PAC wild-type and mutant plasmids were grown in DMEM supplemented with 10% fetal bovine serum at 37 °C. After 24 h, 10 mM sodium butyrate was added, and the cells were moved to grow at 30 °C. The cells were harvested the next day and washed with Tris-buffered saline (TBS) buffer (20 mM Tris, pH 8, and 150 mM NaCl). PAC protein was extracted in TBS buffer containing 1% glyco-diosgenin detergent on ice for 30 min. The sample was then clarified by ultracentrifugation at 40,000 rpm and the supernatant was collected. The clarified sample was injected into a high-performance liquid chromatography machine connected with a Superpose 6 Increase 5/150 GL column or a Superose 6 10/300 GL column (GE Healthcare). The GFP fluorescence signal (excitation of 488 nm, emission of 507 nm) against the retention volume was detected.
Sequence Alignment.
PAC N-terminal sequence alignment was created using MAFFT software v7.427 (2019 March 29) (30). Protein sequences from different vertebrate species were obtained from UniProt: human PAC (Q9H813), rat PAC (Q66H28), mouse PAC (Q9D771), frog PAC (Q0V9Z3), zebrafish PAC (Q7SY31), and chicken ASIC1 (Q1XA76).
Electrophysiology.
PAC KO HEK293T cells were seeded on poly-D-lysine–coated coverslips and transiently transfected with wild-type or mutant PAC plasmids using Lipofectamine 2000 (Thermo Fisher). The cells were recorded ∼1 d after transfection. Whole-cell patch-clamp recordings were performed as described previously (15). The extracellular recording solution contained 145 mM NaCl, 10 mM Hepes, 2 mM MgCl2, 2 mM KCl, 1.5 mM CaCl2, and 10 mM glucose (300 to 310 mOsm/kg; pH 7.3, titrated with NaOH). Acidic extracellular solutions were made of the same ionic composition, buffered with 5 mM sodium citrate instead of Hepes, and pH was adjusted using citric acid. Solutions were applied locally using a gravity perfusion system with a small tip 100 to 200 µm away from the recording cell. Recording pipettes (2 to 4 MΩ) were pulled with a Model P-1000 multistep puller (Sutter Instruments) filled with intracellular solution containing 135 mM CsCl, 2 mM CaCl2, 1 mM MgCl2, 5 mM EGTA, 4 mM MgATP, and 10 mM Hepes (280 to 290 mOsm/kg; pH 7.2, titrated with CsOH).
For proton-activated Cl− current recordings, voltage-ramp pulses were applied every 3 s from −100 to +100 mV at a speed of 1 mV/ms and a holding potential of 0 mV. For the voltage-dependent activation, cells were held at −60 mV and voltage-step pulses (500-ms duration, 5-s interval) were applied from −100 to +100 mV in 20-mV increments. For measurement of pH sensitivity, all pH-induced currents were normalized to the maximal current at pH 4.6 or 4.0. GraphPad Prism was used to fit the normalized data to the Hill equation [Y = Bottom + (Top − Bottom)/(1 + 10^ ((Log pH50 − X) × HillSlope))] to estimate the pH50, where Top and Bottom are plateaus of the maximum and minimum normalized currents, respectively, and Y is the response variable. All normalized data were fitted to the pH dose-response curve unconstrained, allowing the Top and Bottom to be defined by the maximum and minimum normalized currents. To avoid the complication of channel desensitization, we returned pH-induced current recordings to baseline by perfusing cells with neutral pH solution before applying other acidic solutions.
For anionic selectivity experiments, minimal solutions contained 145 mM NaX, 10 mM Hepes (300 to 310 mOsm/kg; pH 7.3, titrated with NaOH) were bath applied, where X was Cl−, Br−, or I−. The recorded currents were used to generate I to V curves for reversal potential determination. The permeability ratios were calculated from shifts in the reversal potential using the Goldman–Hodgkin–Katz equation. All recordings were done with the MultiClamp 700B amplifier and 1550B digitizer (Molecular Devices) at RT. Current signals were filtered at 2 kHz and digitized at 10 kHz. Series resistance was compensated for at least 80%. All data were analyzed using Clampfit 10.7, and GraphPad Prism 8 software was used for all statistical analyses.
MD Simulations.
To generate initial input conformations for MD simulations, we assembled the molecular systems of PAC protein conformations at pH 4 and pH 8 (Protein Data Bank ID: 7JNC, and 7JNA, respectively) (15) in phosphatidylcholine (POPC) bilayers by using the Membrane Builder module on the CHARMM-GUI web server (31). The protein-membrane constructs were further solvated in 150 mM KCl explicit water solution. The assembled molecular systems were equilibrated with NAMD2.13 simulation software (32) following a standard six-step protocol provided by CHARMM-GUI (33, 34). The production phases of simulations were run with OpenMM 7.5.0 (35) software and CHARMM36 (36) force-field parameters for both wild-type PAC constructs. Simulations were performed under NPT conditions, with pressure control implemented with MonteCarloMembraneBarostate at 1 atm, with a temperature bath maintained at 310 K with the Langevin integrator of OpenMM. The electrostatic interactions were treated with the Potential Mesh Ewald (PME) protocol with cutoff set to 12 Å and switching distance set to 10 Å. The integration step size was 2 fs.
The all-atom MD simulations were performed under the selected pH conditions with the ECpH method described recently (21). ECpH is an equilibrium constant pH MD method for accurate prediction of conformational pH dependence in protein systems (22). The algorithm is based on a force-field modification protocol in which linear interpolation is applied between force-field representations of protonated and deprotonated titratable residues to scale their nonbonded interactions with the rest of the system. It takes advantage of a definition of protonation probability for each titratable site and yields a definition of the system in a given protonation state composed of numerous individual titratable sites at a certain pH. A major advantage of this procedure is that it eliminates the approximation of representing protonation states as a binary protonated/nonprotonated choice used in the conventional MD approach. Individual pKa values for the initial conformation of PAC at pH 4 were obtained with the PROPKA protocol (37). The production runs for each starting structure of the wild-type PAC consisted of 18 replicas for each pH value between 4 and 8. Each replica was run for 750 ns, resulting in 13.5 µs of sampling time for each pH value.
NbIT Analysis.
The NbIT analysis estimates the configurational entropy of an equilibrated protein state from the covariance matrix of atomic positions (21, 23, 38). The application of NbIT protocols to MD trajectories reveals the modes and extents of allosteric communication between various structural elements and regions of the protein using an information theory formalism that quantifies the allosteric communication in a complex molecular system by a CI term. The CI quantifies the extent of coordination of one region (identified as the receiver) by another (identified as the transmitter).
The path of the allosteric communication (allosteric pathway) between two protein regions is revealed from the evaluation of the MCI between the two communicating regions, and every other residue in the system. Residues that share the highest amount of information with the transmitter–receiver pair of regions are identified as the allosteric coordination channel (CC) connecting these regions in the allosteric pathway.
The CI for a pair of NbIT sites (transmitter A and receiver B) was evaluated according to equation 10.1 in the previous NbIT description in ref. 23:
where is a total correlation of site A and is a conditional total correlation between site A conditioning on site B. and are derived directly from configurational entropy () as following (equations 11.1 and 11.2 in ref. 23):
Hence, calculation of CI values relies on evaluation of configurational entropy (H) that is defined by the determinant of a covariance matrix of the positions of all atoms included in the NbIT site. By its definition, therefore, CI is a nonadditive function, because the difference between the sum of CI values of two sites [i.e., and the CI value of a combined site comes from the difference between a sum of two determinants of the two covariance matrices and the determinant of the combined matrix.
To identify the CC between sites A and B according to ref. 23, we evaluated the MCI shared between these sites and each residue () of the PAC trimer according to equation 13.1 in ref. 23:
The CI difference for a pair of NbIT sites was calculated as a difference between CI at pH 4 and at pH 8:
The NbIT analysis (23, 24) was implemented on the last ∼400 ns of equilibrated segments of ECpH-MD trajectories obtained for the wild-type PAC systems at pH 4 and 8. The covariance matrices were calculated from the motion of all the heavy atoms of the protein.
The first stage of NbIT analysis is used to establish the main regions of allosteric coordination in the system. To this end, we clustered all titratable residues of the PAC trimer into 16 arbitrary selected NbIT sites per subunit (16 × 3 = 48 total) based on their proximity (SI Appendix, Fig. S5A and B). NbIT sites were defined only by the heavy atoms of corresponding residues. From this calculation, we were able to identify 1) the regions of highest allosteric connectivity, i.e., the joint region (SI Appendix, Fig. S5A and B, site S9), and 2) the threshold values of CI levels based on the clustering procedure of 48 × 48 pairwise values of CI. To this end, we used a k-means algorithm to split all site pairs into three groups based on their CI values: a group of noncoordinated sites (below 7%), a group of sites with fair coordination (8 to 15%), and a group of highly coordinated sites (above ∼15%). Thus, the level of background noise for CI calculation in PAC protein was set to 7%.
For the main round of NbIT calculation, the number of sites was reduced to include only those that shared high allosteric communication supported by experimental data presented in this paper (Fig. 6B).
Data Analysis.
Data and statistical analyses were performed using Clampfit 10.7, GraphPad Prism 8, and Excel. Statistical analysis between two groups was performed using the two-tailed Student’s t test, unless reported otherwise. Multiple-group comparisons were performed using one-way ANOVA with the Bonferroni post hoc test. Nonsignificant P values were generally not reported, unless stated. All numeric data are shown as the mean ± SEM and numbers per group are represented in the bars or legends of each figure panel. The significance level was set at P < 0.05.
Supplementary Material
Acknowledgments
We thank Gavin Li and Nisha A. Rajakrishna for technical assistance and the Qiu lab for helpful discussions. J.O.-O. was supported by an American Heart Association (AHA) predoctoral fellowship (Grant 18PRE34060025). E.K. and H.W. are supported by NIH Grant R37 NS085318-07 and NSF Award BIGDATA: IA: Collaborative Research: In Situ Data Analytics for Next Generation Molecular Dynamics Workflows (NSF #1740990). Z.R. was supported by an AHA postdoctoral fellowship (Grant 20POST35120556).L.M. was supported by a predoctoral fellowship from Boehringer Ingelheim Fonds. W.L. was supported by NIH Grant R01NS112363. Z.Q. was supported by a McKnight Scholar Award, a Klingenstein-Simon Scholar Award, a Sloan Research Fellowship in Neuroscience, and NIH Grants R35GM124824 and R01NS118014. The computational work was performed using the following resources: the Oak Ridge Leadership Computing Facility (INCITE allocations BIP109) at the Oak Ridge National Laboratory, which is a Department of Energy Office of Science User Facility supported under Contract DE-AC0500OR22725; the RPI Artificial Intelligence Multiprocessing Optimized System (AiMOS) system; and the computational resources of the David A. Cofrin Center for Biomedical Information in the Institute for Computational Biomedicine at Weill Cornell Medicine.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200727119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Capurro V., et al. , Functional analysis of acid-activated Cl− channels: Properties and mechanisms of regulation. Biochim. Biophys. Acta 1848, 105–114 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Auzanneau C., Thoreau V., Kitzis A., Becq F., A novel voltage-dependent chloride current activated by extracellular acidic pH in cultured rat Sertoli cells. J. Biol. Chem. 278, 19230–19236 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Lambert S., Oberwinkler J., Characterization of a proton-activated, outwardly rectifying anion channel. J. Physiol. 567, 191–213 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato-Numata K., Numata T., Okada T., Okada Y., Acid-sensitive outwardly rectifying (ASOR) anion channels in human epithelial cells are highly sensitive to temperature and independent of ClC-3. Pflugers Arch. 465, 1535–1543 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Ma Z. Y., et al. , A proton-activated, outwardly rectifying chloride channel in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 371, 437–440 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto S., Ehara T., Acidic extracellular pH-activated outwardly rectifying chloride current in mammalian cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 290, H1905–H1914 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Kajiya H., et al. , Characteristics of ClC7 Cl- channels and their inhibition in mutant (G215R) associated with autosomal dominant osteopetrosis type II in native osteoclasts and hClcn7 gene-expressing cells. Pflugers Arch. 458, 1049–1059 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Kittl M., et al. , Acid- and volume-sensitive chloride currents in human chondrocytes. Front. Cell Dev. Biol. 8, 583131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J., et al. , PAC, an evolutionarily conserved membrane protein, is a proton-activated chloride channel. Science 364, 395–399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ullrich F., et al. , Identification of TMEM206 proteins as pore of PAORAC/ASOR acid-sensitive chloride channels. eLife 8, e49187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osei-Owusu J., Yang J., Del Carmen Vitery M., Tian M., Qiu Z., PAC proton-activated chloride channel contributes to acid-induced cell death in primary rat cortical neurons. Channels (Austin) 14, 53–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H. Y., Shimizu T., Numata T., Okada Y., Role of acid-sensitive outwardly rectifying anion channels in acidosis-induced cell death in human epithelial cells. Pflugers Arch. 454, 223–233 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Osei-Owusu J., et al. , Proton-activated chloride channel PAC regulates endosomal acidification and transferrin receptor-mediated endocytosis. Cell Rep. 34, 108683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stauber T., Jentsch T. J., Chloride in vesicular trafficking and function. Annu. Rev. Physiol. 75, 453–477 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Ruan Z., Osei-Owusu J., Du J., Qiu Z., Lü W., Structures and pH-sensing mechanism of the proton-activated chloride channel. Nature 588, 350–354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jasti J., Furukawa H., Gonzales E. B., Gouaux E., Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449, 316–323 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Noreng S., Bharadwaj A., Posert R., Yoshioka C., Baconguis I., Structure of the human epithelial sodium channel by cryo-electron microscopy. eLife 7, e39340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vullo S., et al. , Conformational dynamics and role of the acidic pocket in ASIC pH-dependent gating. Proc. Natl. Acad. Sci. U.S.A. 114, 3768–3773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baconguis I., Bohlen C. J., Goehring A., Julius D., Gouaux E., X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel. Cell 156, 717–729 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng Z., et al. , Cryo-EM structure of a proton-activated chloride channel TMEM206. Sci. Adv. 7, eabe5983 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kots E., Shore D. M., Weinstein H., An equilibrium constant pH molecular dynamics method for accurate prediction of pH-dependence in protein systems: Theory and application. bioRxiv [Preprint] (2020). 10.1101/2020.11.23.394015 (Accessed 7 July 2022). [DOI]
- 22.Kots E., Shore D. M., Weinstein H., Simulation of pH-dependent conformational transitions in membrane proteins: The CLC-ec1 Cl-/H+ antiporter. Molecules 26, 6956 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeVine M. V., Weinstein H., NbIT—A new information theory-based analysis of allosteric mechanisms reveals residues that underlie function in the leucine transporter LeuT. PLoS Comput. Biol. 10, e1003603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khelashvili G., et al. , Spontaneous inward opening of the dopamine transporter is triggered by PIP2-regulated dynamics of the N-terminus. ACS Chem. Neurosci. 6, 1825–1837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C., Polovitskaya M. M., Delgado B. D., Jentsch T. J., Long S. B., Gating choreography and mechanism of the human proton-activated chloride channel ASOR. Sci. Adv. 8, eabm3942 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai R., Tang J., Chen X. Z., Ion permeation controlled by hydrophobic residues and proton binding in the proton-activated chloride channel. iScience 24, 103395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rook M. L., Musgaard M., MacLean D. M., Coupling structure with function in acid-sensing ion channels: Challenges in pursuit of proton sensors. J. Physiol. 599, 417–430 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gatenby R. A., Gillies R. J., A microenvironmental model of carcinogenesis. Nat. Rev. Cancer 8, 56–61 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Lardner A., The effects of extracellular pH on immune function. J. Leukoc. Biol. 69, 522–530 (2001). [PubMed] [Google Scholar]
- 30.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu E. L., et al. , CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Lee J., et al. , CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eastman P., et al. , OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 13, e1005659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J., MacKerell A. D. Jr., CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsson M. H., Søndergaard C. R., Rostkowski M., Jensen J. H., PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Lange O. F., Grubmüller H., Generalized correlation for biomolecular dynamics. Proteins 62, 1053–1061 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.