Our thinking about protein interactions is still largely constrained by the structure–function dogma. We tend to associate specific protein complexes with a well-defined, structured interface, implying a unique mode of optimizing the polypeptide chain for function. Under cellular conditions, however, protein interactions must be tightly regulated, which generates a wide variety of energetic conflicts. This leads to multiple, suboptimal states which encode different biological activities enabling adaptation. Increasing experimental evidence shows that, similarly to unbound proteins, protein assemblies also sample a wide range of states from ordered to disordered conformations (1, 2). It has been recently demonstrated that frustration in protein folding and fuzziness in protein interactions are analogous concepts, which emerge from the ruggedness of the energy landscape (3) (Fig. 1). It still remains to be explored, however, how the affinity of fuzzy complexes is tuned to the cellular context. The key questions address the biophysical forces which govern formation of fuzzy complexes and determine their specificity (4).
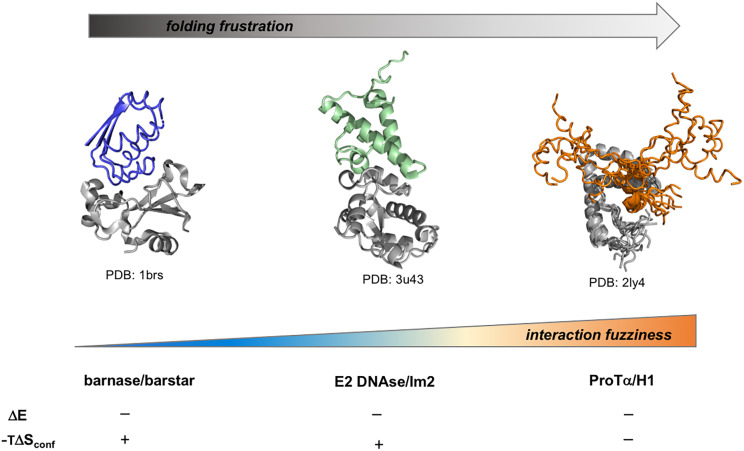
Fig. 1.
Entropy modulates the affinity of ordered and disordered complexes. High-affinity binding can be achieved through both structured and disordered interfaces. An ordered complex is represented by the ribonuclease barnase and its inhibitor barstar [blue, Protein Data Bank (PDB) code 1brs (25)]. A partially ordered complex is represented by the endonuclease colicin E2 with its cognate immunity protein [Im2, green, PDB code 3u43 (26)]. A disordered complex is represented by the complex between HMGB1 and p53 [orange, PDB code 2ly4 (27)], as the high-resolution structure of the ProTα/H1 complex is not available. Increasing folding frustration decreases structural order from barnase/barstar to HMGB1/p53 complex (from left to right, top arrow). Following a similar trend, frustration of the interface contacts also increases from ordered (blue) to disordered (orange) complexes. A high degree of frustration results in a conformationally heterogeneous complex and positive change in configurational entropy (TΔSconf). Electrostatic interactions provide major contributions to binding affinity in all these scenarios. While charged residues in the ordered complex are engaged in specific contacts, in disordered complexes they modulate the degree of disorder and binding entropy (4, 27).
Structure–function studies revealed four decades ago that within the transcriptional machinery components are assembled without structural precision and their binding characteristics are distinct from those of ordered complexes (5). The binding affinity, in particular, is strongly dependent on the length of the protein construct, while not substantially influenced by sequence variations (6). The Tau protein, for example, binds to microtubules via a flexible array of weak interaction sites which are embedded in nonconserved regions (7). Similarly, tandem repeats or redundant interaction motifs in low-complexity sequences tolerate sequence scrambling without major impact on function (8). These sequences often retain conformational flexibility upon interacting with their specific partners, leading to the formation of disordered complexes (9). Strikingly, reducing conformational heterogeneity in these assemblies may compromise their biological activities. Stabilization of the GCN4 transcription factor in complex with the Med15 transcriptional coactivator, for example, reduces transcriptional activity (10). In contrast, synthetic activators recapitulating structural mobility in the bound state exhibit improved transcription (11). Increasing experimental evidence demonstrates that a wide range of biological functions are associated with conformational and interaction heterogeneity (12). Such fuzziness confers robustness and flexibility for versatile cellular conditions (13).
A pertinent example of fuzzy interactions is the picomolar affinity complex of the linker histone H1 with the nuclear protein prothymosin-α (ProTα), which plays a role in chromatin remodeling. The ProTα–H1 assembly has low secondary structure content and low dispersion of the 1H–15N heteronuclear single quantum coherence spectra similar to the free proteins, indicating structural disorder in the bound complex (14). The high-affinity interactions are mediated by the opposite charges, making the association strongly dependent on the ionic strength. Hazra and Levy have studied binding thermodynamics in 52 ProTα–H1 variants by swapping charges between the two proteins and shuffling the charged residues within each protein, while keeping the net charge of the complex constant (4). They have found that the interaction free energy profile strongly depends on the actual charge of each protein and to a lesser extent on their charge patterns. Charge swapping significantly impacts complex affinity by 15 to 20 orders of magnitude, while shuffling of the charges alters the dissociation constant by 10 orders of magnitude. In addition, they show that formation of fuzzy complexes, in contrast to formation of ordered complexes, is coupled to the increase of configurational entropy (TΔSconf ∼15 kcal/mol) (4). In addition, counterion release further increases the binding entropy in a sequence-specific manner (TΔScounterions ∼40 to 80 kcal/mol). In the ProTα–H1 complex the binding entropy (-TΔS) and enthalpy (ΔH) are linearly correlated, in contrast to ordered complexes, where these terms are compensatory. More uniform charge distributions and increased disorder facilitate such entropy–enthalpy reinforcement. On the contrary, increasing net charge and separation of charged patches promotes compactness of the structure and decreases binding entropy. These results illustrate how binding entropy can be fine-tuned in a conformationally heterogeneous complex in a sequence-specific manner.
Previously it was found that scrambling the H1 C-terminal domain (CTD) sequence does not affect chromatin condensing functions (15). In contrast, specific mutations which shift the amino acid composition in different CTD regions influence chromatin folding. These results suggest that the local composition of the sequence is critical for biological activity (16). Analysis of about 2,000 protein complexes showed that local composition determines the degree of disorder in the assembly and modulates the entropic contribution to binding (17). In such a scenario, mutations that optimize protein interactions lower the degree of folding to avoid energetically unfavorable structured bound configurations (18). In addition, mutations can affect hydration and counterion release, which can serve as “fingerprints” to modulate entropic contributions in a sequence-specific manner. Thus, changing local sequence complexity fine-tunes binding affinity and specificity through modulating binding entropy. Application of these principles was demonstrated in the case of small-molecule interactions with disordered proteins (19, 20).
In the cell, a wide range of proteins form liquid–liquid phase separated condensates through multiple interaction sites (21, 22). Multivalent interactions inevitably generate a wide variety of energetic conflicts leading to the formation of frustrated, fuzzy complexes (23) (Fig. 1). Conformational ensembles of these higher-order assemblies are sensitive to the cellular conditions. It was shown that altering the populations of different substates leads to distinct biological activities (24). We may reason that assembly of protein droplets is also under entropic control, which can modulate both affinity and specificity as seen in case of the ProTα–H1 complex (4). It is yet to be explored how such mechanisms operate in protein condensates and how to exploit these principles for modulating their functions. Understanding the physical principles of fuzzy interactions will contribute to these efforts by providing a framework for cellular context dependence of protein interactions.
Footnotes
The author declares no competing interest.
See companion article, “Affinity of disordered protein complexes is modulated by entropy–energy reinforcement,” 10.1073/pnas.2120456119.
References
- 1.Fuxreiter M., Fuzziness in protein interactions-A historical perspective. J. Mol. Biol. 430, 2278–2287 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Fuxreiter M., Fold or not to fold upon binding - Does it really matter? Curr. Opin. Struct. Biol. 54, 19–25 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Gianni S., et al. , Fuzziness and frustration in the energy landscape of protein folding, function, and assembly. Acc. Chem. Res. 54, 1251–1259 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazra M. K., Levy Y., Affinity of disordered protein complexes is modulated by entropy–energy reinforcement. Proc. Natl. Acad. Sci. U.S.A. 119, 10.1073/pnas.2120456119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigler P. B., Transcriptional activation. Acid blobs and negative noodles. Nature 333, 210–212 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Hope I. A., Mahadevan S., Struhl K., Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature 333, 635–640 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Butner K. A., Kirschner M. W., Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 115, 717–730 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross E. D., Baxa U., Wickner R. B., Scrambled prion domains form prions and amyloid. Mol. Cell. Biol. 24, 7206–7213 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tompa P., Fuxreiter M., Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 33, 2–8 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Brzovic P. S., et al. , The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol. Cell 44, 942–953 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warfield L., Tuttle L. M., Pacheco D., Klevit R. E., Hahn S., A sequence-specific transcription activator motif and powerful synthetic variants that bind Mediator using a fuzzy protein interface. Proc. Natl. Acad. Sci. U.S.A. 111, E3506–E3513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatos A., Monzon A. M., Tosatto S. C. E., Piovesan D., Fuxreiter M., FuzDB: A new phase in understanding fuzzy interactions. Nucleic Acids Res. 50, D509–D517 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschner M., Gerhart J., Evolvability. Proc. Natl. Acad. Sci. U.S.A. 95, 8420–8427 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borgia A., et al. , Extreme disorder in an ultrahigh-affinity protein complex. Nature 555, 61–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu X., Hamkalo B., Parseghian M. H., Hansen J. C., Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry 48, 164–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen J. C., Lu X., Ross E. D., Woody R. W., Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 281, 1853–1856 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Miskei M., Horvath A., Vendruscolo M., Fuxreiter M., Sequence-based prediction of fuzzy protein interactions. J. Mol. Biol. 432, 2289–2303 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Hadži S., Mernik A., Podlipnik Č., Loris R., Lah J., The thermodynamic basis of the fuzzy interaction of an intrinsically disordered protein. Angew. Chem. Int. Ed. Engl. 56, 14494–14497 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Heller G. T., et al. , Sequence specificity in the entropy-driven binding of a small molecule and a disordered peptide. J. Mol. Biol. 429, 2772–2779 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Heller G. T., Sormanni P., Vendruscolo M., Targeting disordered proteins with small molecules using entropy. Trends Biochem. Sci. 40, 491–496 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Li P., et al. , Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuxreiter M., Vendruscolo M., Generic nature of the condensed states of proteins. Nat. Cell Biol. 23, 587–594 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Freiberger M. I., Wolynes P. G., Ferreiro D. U., Fuxreiter M., Frustration in fuzzy protein complexes leads to interaction versatility. J. Phys. Chem. B 125, 2513–2520 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H., Fuxreiter M., The structure and dynamics of higher-order assemblies: Amyloids, signalosomes, and granules. Cell 165, 1055–1066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckle A. M., Schreiber G., Fersht A. R., Protein-protein recognition: Crystal structural analysis of a barnase-barstar complex at 2.0-A resolution. Biochemistry 33, 8878–8889 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Wojdyla J. A., Fleishman S. J., Baker D., Kleanthous C., Structure of the ultra-high-affinity colicin E2 DNase–Im2 complex. J. Mol. Biol. 417, 79–94 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Rowell J. P., Simpson K. L., Stott K., Watson M., Thomas J. O., HMGB1-facilitated p53 DNA binding occurs via HMG-Box/p53 transactivation domain interaction, regulated by the acidic tail. Structure 20, 2014–2024 (2012). [DOI] [PubMed] [Google Scholar]