Significance
Posttranslational processing enzymes that generate mature bioactive peptides from prepropeptides have been conserved throughout eukaryotic evolution. Often acting through G protein-coupled receptors, peptides play a central role in signaling between cells, tissues, and organisms. An amidated chemotactic peptide produced by Chlamydomonas reinhardtii, a ciliated green alga, was identified in ectosomes from cilia of mating gametes. This bioactive peptide is contained within a heavily glycosylated 91-kDa peptide precursor, proGATI, which contains stably folded domains. Trafficking and endoproteolytic processing of this peptide precursor are coupled to its release in ciliary ectosomes and retention of the bioactive product on the ectosomal surface. This represents an ancient mode of regulated secretion; metazoans may employ similar mechanisms to control ciliary release of bioactive products.
Keywords: amidation, Chlamydomonas, chemotaxis, ectosomes, peptidergic signaling
Abstract
Cilia are sensory and secretory organelles that both receive information from the environment and transmit signals. Cilia-derived vesicles (ectosomes), formed by outward budding of the ciliary membrane, carry enzymes and other bioactive products; this process represents an ancient mode of regulated secretion. Peptidergic intercellular communication controls a wide range of physiological and behavioral responses and occurs throughout eukaryotes. The Chlamydomonas reinhardtii genome encodes what appear to be numerous prepropeptides and enzymes homologous to those used to convert metazoan prepropeptides into bioactive peptide products. Since C. reinhardtii, a green alga, lack the dense core vesicles in which metazoan peptides are processed and stored, we explored the hypothesis that propeptide processing and secretion occur through the regulated release of ciliary ectosomes. A synthetic peptide (GATI-amide) that could be generated from a 91-kDa peptide precursor (proGATI) serves as a chemotactic modulator, attracting minus gametes while repelling plus gametes. Here we dissect the processing pathway that leads to formation of an amidated peptidergic sexual signal specifically on the ciliary ectosomes of plus gametes. Unlike metazoan propeptides, modeling studies identified stable domains in proGATI. Mass spectrometric analysis of a potential prohormone convertase and the amidated proGATI-derived products found in cilia and mating ectosomes link endoproteolytic cleavage to ectosome entry. Extensive posttranslational modification of proGATI confers stability to its amidated product. Analysis of this pathway affords insight into the evolution of peptidergic signaling; this will facilitate studies of the secretory functions of metazoan cilia.
Cilia are membrane-delimited, microtubule-based cell extensions that protrude into the extracellular space and function as key motile, sensory, and secretory organelles in many eukaryotes (1). These complex organelles, which were present in the last eukaryotic common ancestor, both receive and transmit signals (2, 3). Proteins encoded by ∼5% of the human genome contribute to their assembly, structure, and function (4). Mutations in many of these genes cause ciliopathies, with phenotypes ranging from neurological malformations, skeletal abnormalities and kidney disease to infertility, obesity, and insulin resistance (5). The ciliary localization of receptors for peptides such as Wnt, Hedgehog, insulin, somatostatin, and α-melanocyte stimulating hormone (αMSH) plays an essential role in their signaling ability (6–8). Chlamydomonas reinhardtii, a chlorophyte green alga, has served as a key model organism for dissecting ciliary assembly, function, and signaling (9, 10).
Secreted peptides are an ancient means of intercellular communication. Genes encoding potential peptide precursors can be identified based on the presence of an N-terminal presequence, which directs the nascent chain into the lumen of the endoplasmic reticulum (ER), multiple sites subject to cleavage by subtilisin-like prohormone convertases, and multiple sites at which C-terminal amidation could occur (11). Posttranslational modification—including disulfide bond formation, N- and O-glycosylation, lipidation, endo- and exo-proteolysis, and peptide amidation—occur as the newly synthesized peptide precursors travel from the ER to the Golgi complex and are packaged for secretion (12–16). Peptidylglycine α-amidating monooxygenase (PAM), an ancient copper-dependent monooxygenase, catalyzes a late step (amidation) in the biosynthesis of a broad array of peptides, such as mammalian αMSH and vasopressin, the sea urchin sperm attractant resact, as well as numerous invertebrate venom peptide toxins. Amidation occurs via a two-step reaction catalyzed by the sequential actions of the monooxygenase (peptidylglycine α-hydroxylating monooxygenase; PHM) and lyase (peptidyl-α-hydroxyglycine α-amidating lyase; PAL) catalytic cores of PAM (17).
Active PAM localizes to Golgi membranes and to the ciliary membrane, and plays an essential role in ciliogenesis in C. reinhardtii; its ciliary localization is conserved in metazoans (18, 19). Although their phenotypes differ substantially, a role for PAM in ciliary formation and maintenance is also observed in mice, zebrafish, and planaria (18–20). How an enzyme essential for a final step in the biosynthesis of secreted peptides contributes to ciliogenesis remains unclear. In metazoans, the processing of peptide precursors and packaging of product peptides for release in response to an appropriate stimulus is regulated in part by luminal pH. Lacking dense core vesicles, C. reinhardtii rely on the secretion of soluble cargo from Golgi-derived vesicles and the regulated release of extracellular vesicles (ectosomes) by outward budding from the ciliary membrane (21–25).
Bioactive ectosomes released from the cilia of vegetative cells contain a subtilisin-like endoprotease (VLE1) that degrades the mother cell wall (22, 24, 26), but do not contain PAM (27). Under plentiful nutrient conditions, haploid C. reinhardtii cells divide by mitosis. Starvation triggers a genetically encoded developmental process resulting in the formation of sexual gametes of opposite mating type (termed minus and plus). Ectosome release increases rapidly when minus and plus gametes are mixed (21). Mating ectosomes contain PAM and a variety of amidated products, including one that could be derived from preproGATI, one of the many putative prepropeptides encoded by the C. reinhardtii genome (27). The interaction of mating cell cilia triggers a complex intraciliary signaling pathway that leads to loss of gametic cell walls, formation of mating structures, and cell fusion, yielding a quadriciliate cell that ultimately develops into a diploid zygote. When nutrient conditions improve, the zygote hatches, releasing haploid meiotic progeny (28).
The sequence of the peptidylglycine precursor to the 23-residue GATI-amide peptide, which attracts minus gametes and repels plus gametes, is contained at the C-terminus of proGATI (27). Amidated peptides from echinoderms, Hydra, vespids, and humans have also been reported to induce chemotaxis (29–32). By developing the tools needed to evaluate proGATI processing and secretion during mating and taking advantage of structural predictions made for proGATI using RoseTTAFold (33) and AlphaFOLD 2 (34), a role for extensive posttranslational processing and the regulated entry of proGATI and its cleavage products into mating ectosomes were identified. As PAM, peptide processing enzymes, and cilia are broadly conserved in eukaryotes, this study provides a paradigm for understanding how peptides that transmit chemotactic sexual and other signals through cilia can be generated and released in a controlled manner.
Results
Processing of proGATI Is Mating-Type Specific.
Transcriptomic studies identified mating as a developmental stage during which expression of both PAM and preproGATI is increased (35). PAM protein and activity have been identified in mating ciliary ectosomes, but not in vegetative ectosomes (27). Most importantly, several amidated peptides that could be derived from putative C. reinhardtii prepropeptides were identified in tryptic digests of mating ectosomes (27). Based on its increased expression in resting and activated gametes, we focused on one of these, preproGATI. In-gel tryptic digestion of high molecular mass proteins (>100 kDa) found in mating ectosomes yielded an amidated 23-amino acid peptide that could be derived from proGATI and was identical to the synthetic GATI-amide peptide (27) (Fig. 1A). Like the original 23-residue GATI-amide peptide, a shorter 11-residue amidated peptide also produced a mating-type–specific chemotactic response by gametes; amidation of these peptides was essential for this biological activity.
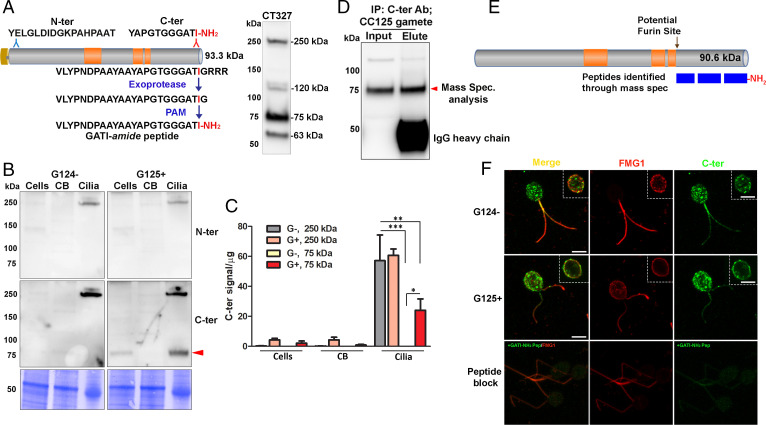
Fig. 1.
Processing and localization of proGATI in minus and plus gametes. (A) Diagram shows Cre03.g204500 (preproGATI) with Prorich regions in orange. The N-terminal and C-terminal peptides used as antigens are shown above the diagram. The pathway leading to C-terminal amidation is illustrated below the diagram. The immunoblot to the right of the diagram shows the bands detected by serum from one of the rabbits (CT327) inoculated with a mixture of N-ter and C-ter peptide conjugates (SI Appendix, Fig. S1). (B) Immunoblot of cells, deciliated cell bodies (CB) and cilia of minus (G124−) and plus (G125+) resting gametes using affinity-purified proGATI N-ter and C-ter antibodies. Equal amounts of protein (20 µg) were loaded. (C) Quantification of C-ter signal revealed significant enrichment of both 250-kDa and 75-kDa bands in cilia. Results are the average of duplicates. Means were compared with ± range. Asterisks indicate significant differences between groups: *P < 0.05, **P < 0.01, ***P < 0.001. (D) Affinity-purified C-ter antibody was used to immunoprecipitate cross-reactive material from mating type plus (CC125) gamete cell lysates. The 75-kDa fragment (red arrowhead) was excised and analyzed by mass spectrometry. (E) Only peptides between Met697 and Ile904-amide were identified, covering 76.4% of this region; all of the spectra identified from the C terminus were amidated. (F) Maximal projection confocal images of minus and plus resting gametes stained with the C-ter proGATI antibody (green) and antibody to FMG1 (red). Insets show a single Z-plane. Plus gametes probed with antibody preincubated with the GATI-amide peptide (Peptide block) exhibit reduced staining (green) in cell bodies and cilia (Bottom). Similar localization of proGATI in gametes was obtained in three independent experiments. (Scale bars, 5 µm apply to all panels.)
The amidation of proGATI requires removal of three Arg residues by a carboxypeptidase E-like exoprotease (Fig. 1A), generating a Gly-extended protein that serves as the PAM substrate; following α-hydroxylation of this Gly residue by PHM, PAL-mediated cleavage produces a protein terminating with a C-terminal Ile-amide. To explore the posttranslational processing, trafficking, and secretion of amidated products generated from proGATI, we produced and validated affinity-purified rabbit polyclonal antibodies to a synthetic peptide located near its N terminus (N-ter peptide) and to the chemomodulatory amidated 11-residue peptide (C-ter peptide); together, these antibodies detect four specific bands in ciliary ectosomes (Fig. 1A; see SI Appendix, Fig. S1 for antibody validation). Preincubation of the antiserum with C-ter antigenic peptide eliminated the 250- and 75-kDa bands while preincubation with N-ter antigenic peptide eliminated the 250-, 120-, and 63-kDa bands. When used for immunoblots or immunofluorescence, addition of the amidated peptide blocked the signal produced by the affinity-purified C-ter antibody; the signal was not blocked by the same peptide terminating with a free carboxyl group or by its glycine-extended precursor, demonstrating its specificity for the presence of Ile-amide; signals produced by affinity purified N-ter antibody were also blocked by the N-ter antigenic peptide (SI Appendix, Fig. S1).
Gametic cells, their deciliated cell bodies, and isolated cilia were subjected to immunoblot analysis. Use of the N-ter and C-ter antibodies revealed enrichment of a 250-kDa band recognized by both antibodies in the cilia of minus and plus gametes (Fig. 1B). Although the mass predicted for proGATI is 91 kDa, extensive N- and O-glycosylation of proteins can result in apparent molecular mass estimates that far exceed those of unmodified proteins, leading us to hypothesize that the 250-kDa band, the only product detected by both the N-ter and C-ter antibodies, represented proGATI. The C-ter antibody also detected a 75-kDa band in the cilia of plus gametes; the N-ter antibody detected only the 250-kDa protein. The 75-kDa C-ter band was detectable in whole cells of plus gametes, was greatly enriched in isolated cilia, and was essentially undetectable in deciliated cell bodies (Fig. 1C).
The specificity of the affinity-purified C-ter antibody suggested that the 75-kDa product was an amidated C-terminal fragment of proGATI. To determine its identity and confirm its amidation, the affinity-purified C-ter antibody was used to isolate the 75-kDa proGATI fragment from plus gamete cell lysates (Fig. 1D). Mass spectroscopy after in-gel tryptic digestion confirmed complete amidation of its C terminus. The peptides identified provided 76.4% coverage of the region from Met697 to Ile904-amide, yielding a predicted molecular mass of 23 kDa. The underlined sequence immediately preceding the experimentally determined N terminus of 75-kDa GATI (-SVRFSR↓M697EA-) matches the criteria for cleavage by a furin-like endoprotease ([R/K]-Xn-[R/K]↓X; n = 0, 2, or 4) (36). This proGATI region contains two potential N-glycosylation sites (N814TT, N833QT) and two potential hydroxyproline O-glycosylation sites (SP723L and AP896G), raising the possibility that these modifications are responsible for its greatly increased apparent molecular mass.
Following removal of its N-terminal signal peptide in the ER, newly synthesized proGATI would be expected to travel through the lumen of the secretory pathway, undergoing posttranslational modifications, ranging from disulfide bond formation and N- and O-glycosylation to endoproteolytic cleavage by subtilisin-like endoproteases, trimming by exoproteases and C-terminal amidation. We used immunofluorescence microscopy to determine the subcellular localization of amidated proGATI-derived proteins in resting gametes. Maximal Z-projection confocal images of minus and plus gametes showed that the C-ter GATI signal was localized to discrete puncta throughout the cytoplasm (Fig. 1F); this signal could represent intact 250-kDa proGATI or its 75-kDa C-ter product. In contrast, simultaneous visualization of flagellar membrane glycoprotein 1 (FMG1), a type I ciliary membrane glycoprotein, revealed more signal in the cilia and around the margins of the cell body. Single Z-stack images showed the accumulation of C-ter GATI signal at the cell surface, colocalized with FMG1 (Fig. 1 F, Insets). Diffuse C-ter GATI signal along the length of the cilia was also observed in both mating types (Fig. 1F). To confirm staining specificity, the C-ter antibody was preincubated with antigenic peptide; signal intensity (green) was greatly reduced (Fig. 1 F, Bottom). The punctate staining in the cell body may represent Golgi-derived vesicles; puncta are distributed over a larger cytoplasmic region than the perinuclear Golgi stacks (SI Appendix, Fig. S2) (see for example ref. 18). Fractionation of gametic cytoplasm revealed that proGATI was mainly present in the microsomal fraction as was FMG1; in contrast, most ARF1 remained in the cytosol (SI Appendix, Fig. S3). Although proGATI lacks a transmembrane domain, the C-ter GATI antibody signal highlights the ciliary membrane, suggesting an interaction of proGATI with other ciliary membrane proteins.
Mating Selectively Triggers the Ectosomal Trafficking and Processing of proGATI.
The initiation of mating triggers the formation and rapid release of ciliary ectosomes (21). This process involves the outward budding of the ciliary membrane (24), placing the catalytic domains of PAM on the outer surface of ectosomes within a few minutes; strikingly, neither PAM activity nor PAM protein has been identified in the soluble secretome (18, 27). In contrast, amidated GATI products were identified in both mating ectosomes and occasionally in the soluble secretome (11). The presence of GATI proteins in ectosomes suggests that they interact with the ectosomal surface, which contains two lectin receptor-related proteins that might recognize sugar moieties.
To explore this possibility, ectosomes prepared from mating gametes were embedded in agarose and imaged by thin section transmission electron microscopy (EM); the vesicles observed ranged from ∼80 nm to ∼260 nm in diameter, with minimal contamination from other cell debris (Fig. 2A). Exposure of membranous organelles to high pH buffers removes many proteins whose interactions with membranes are noncovalent; the surface of ectosomes washed in pH 11.5 Na2CO3 appeared smoother than before washing, and had a selective effect on the ectosomal association of proGATI and its products (SI Appendix, Fig. S4).
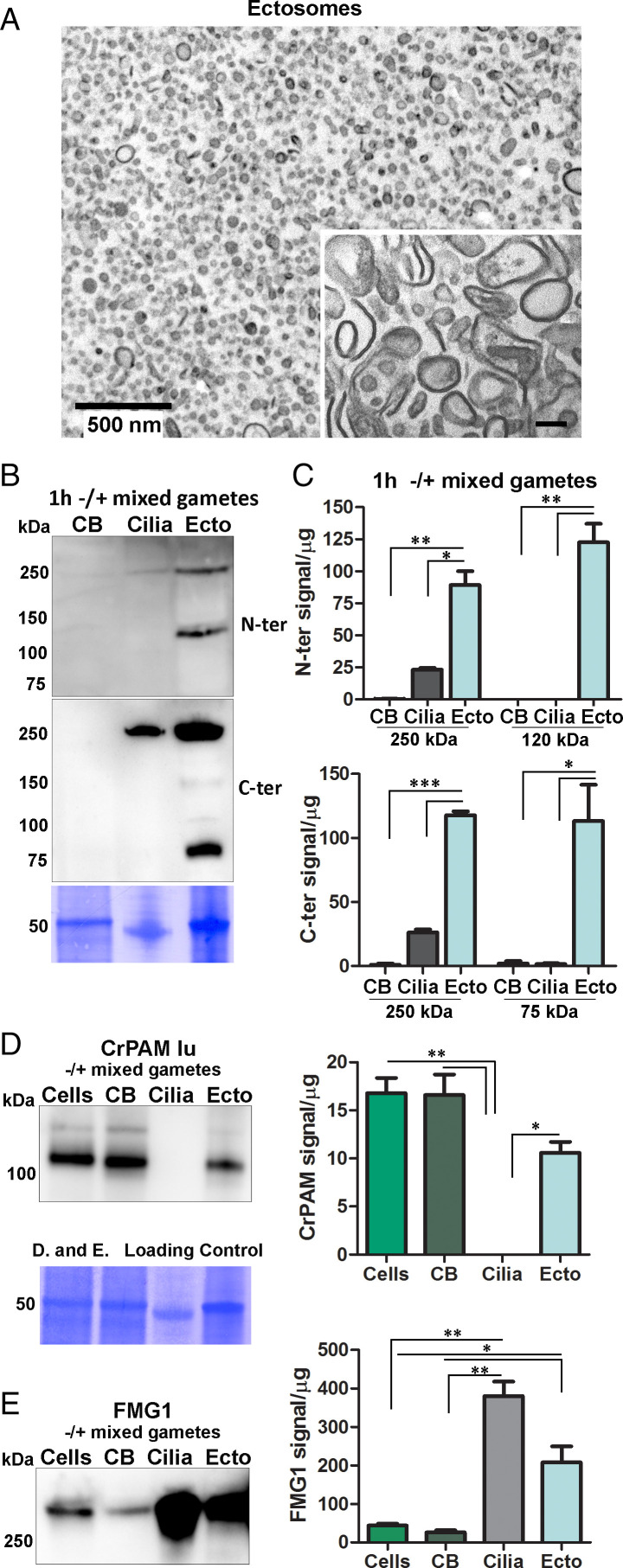
Fig. 2.
Processing of proGATI in mating ectosomes. (A) Cross-section transmission electron micrograph of an agarose-embedded ectosome pellet isolated from 1 h -/+ mixed gametes. Inset shows a higher magnification image of ectosomes treated with Na2CO3 to remove peripheral membrane proteins. (Scale bar, 100 nm.) Images are representative of three independent experiments. (B) Deciliated cell bodies (CB), cilia, and ectosomes (Ecto) isolated from mixed gametes were fractionated by SDS/PAGE, blotted, and probed with the affinity purified N-ter and C-ter proGATI antibodies. (C) Graphs showing enrichment of N-ter and C-ter signals for the 250-kDa band and smaller fragments. Results are average of two independent experiments; mean ± range. Asterisks indicate a statistically significant difference between two groups: *P < 0.05, **P < 0.001, ***P < 0.0001. Immunoblots from the same membrane showing CrPAM (D) and FMG1 (E) levels in cells, cell bodies (CB), cilia and ectosomes (Ecto) isolated from mixed gametes; quantification was as in C. The loading control applies to both panels D and E.
Deciliated mixed gamete cell bodies, cilia, and ectosomes prepared 1 h after the initiation of mating were subjected to immunoblot analysis (Fig. 2B). Based on use of both the N-ter and C-ter antibodies, proGATI products were undetectable in the cell bodies, and cilia contained only the 250-kDa proGATI protein. In contrast, mating ectosomes contained 250-kDa proGATI along with a 120-kDa N-ter fragment and the 75-kDa C-ter fragment. (Fig. 2B). Quantification revealed enrichment of 250-kDa proGATI in mating ectosomes and an even greater enrichment of the 120 kDa N-ter and 75 kDa C-ter fragments (Fig. 2C). These observations suggest that the cleavage creating the N-ter and C-ter fragments of proGATI occurs during ectosome formation or on the ectosomal surface.
For comparison, we evaluated the specificity and selectivity with which two type I integral membrane proteins, PAM and FMG1, move from cilia into ectosomes during mating (Fig. 2 D and E). PAM was previously identified in mating ectosomes, but was not found in vegetative ectosomes (27), while FMG1 is present in both (22, 27). Immunoblot analysis confirmed the presence of PAM and FMG1 in mating ectosomes. After an hour of mating, negligible PAM remained in the cilia; although mating ectosomes contained PAM, its ectosomal levels did not exceed those in the cell body (Fig. 2D). FMG1 levels in cilia and mating ectosomes greatly exceeded those in cell bodies, but FMG1 levels in mating ectosomes did not exceed those in cilia (Fig. 2E). One hour after the initiation of mating, most of the proGATI had moved into ectosomes, with little remaining in the cell bodies or cilia (Fig. 2B).
Mating Ectosomes Contain an Extensively Modified 75-kDa Fragment that Includes the Amidated Chemomodulatory Peptide.
As described for cell lysates (Fig. 1D), the 75-kDa C-ter fragment found in mating ectosomes was enriched by immunoprecipitation using the C-ter antibody (Fig. 3A). Analysis of in-gel tryptic digests revealed its complete C-terminal amidation. The other proGATI tryptic peptides identified in this sample provided almost complete coverage (79.7%) of the region from M697 to Ile904-amide, the amidated C terminus of the chemomodulatory peptide (Fig. 3B). Both the presence of extended Prorich regions and extensive glycosylation of proGATI could contribute to the aberrant molecular mass estimates provided by SDS/PAGE. As in all eukaryotes, N-glycosylation in C. reinhardtii involves the assembly of a lipid-linked oligosaccharide that is transferred to target Asn residues in the lumen of the ER followed by oligosaccharide maturation in the ER and the Golgi complex (37). However, lacking several of the enzymes required for the synthesis of a canonical lipid-linked oligosaccharide, C. reinhardtii N-glycans have a unique core structure (37). Using the NetNGlyc tool, a total of five N-glycosylation sites are predicted in proGATI, with two in the 23-kDa C-ter proGATI region (Fig. 3B). While most of the O-glycans identified in mammals are attached to Ser or Thr residues, in C. reinhardtii O-glycans are more often attached to hydroxyproline (HyP) residues (37–39); two predicted sites of HyP O-glycosylation (Pro728, Pro896) occur in the 23-kDa C-ter proGATI region (Fig. 3B).
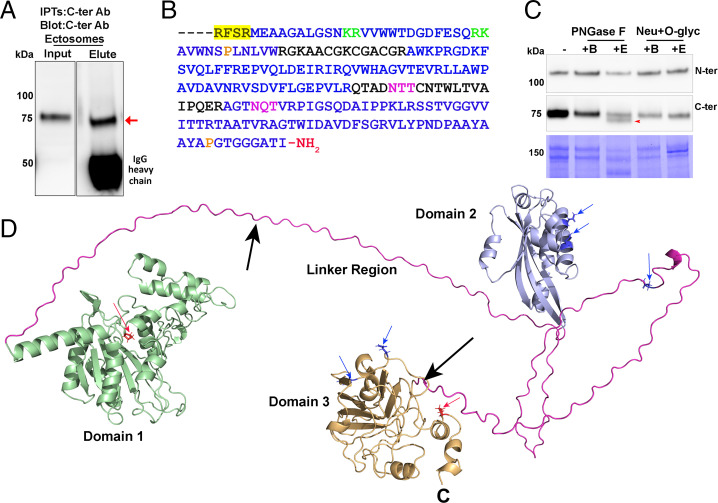
Fig. 3.
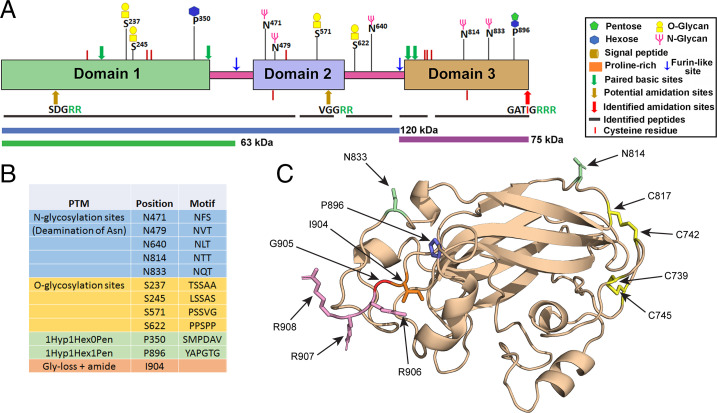
Structural modeling predicts the presence of stably folded domains in proGATI. (A) The 75-kDa fragment (red arrow) was immunoprecipitated from mating ectosomes with affinity-purified C-ter antibody and analyzed by mass spectrometry. (B) Sequence of the C-terminal region identified by mass spectrometry. The residues identified are shown in blue; the location of the furin-like cleavage site (yellow highlight), dibasic sites (green), potential N-glycosylation (purple), and proline hydroxylation (orange) sites are indicated. The terminal peptide is amidated. (C) Mating ectosomes (15 µg protein) were analyzed without treatment (−) or after digestion with PNGase F (buffer alone, +B; with enzyme, +E), revealing a reduction in molecular mass (red arrow) of the 75-kDa C-ter product but not the 120-kDa N-ter proGATI product. Digestion with neuraminidase and O-glycosidase (Neu+O-glyc) (buffer alone, +B; with enzyme, +E) had no effect. The results were replicated in independent experiments. (D) The structural model of proGATI generated using RoseTTAFold (33) contains three well-folded domains (domain 1, residues 51 to 370, green; domain 2, residues 446 to 593, blue; domain 3, residues 696 to 908, orange) connected by long, highly flexible, Prorich linkers (pink). Although individual domains are well structured, their relative orientation with respect to each other is variable. Predicted Asn N-glycosylation (blue residues and arrows) and HyP O-glycosylation (red residues and arrows) sites are shown, along with two potential endoproteolytic cleavage sites (black arrows) and the C terminus (C). Structural overlays between RosseTTAFold and AlphaFOLD 2 domain predictions using DALI (49) gave excellent alignment scores of z = 11.4, 9.4, and 18.7 with RMSDs of 6.0, 3.2, and 3.0 Å for domains 1, 2 and 3, respectively.
Although the unique features of N- and O-glycosylation in C. reinhardtii made it unlikely that enzymes optimized for the removal of mammalian sugars would be very effective, mating ectosomes were treated with PNGase F, which removes many mammalian N-linked sugars, and with a mixture of neuraminidase and O-glycosidase. PNGase F treatment slightly reduced the apparent molecular mass of about half of the 75-kDa product detected by the C-ter antibody (Fig. 3C); treatment with the O-glycosidase/neuraminidase mixture was without effect on the 75-kDa product. The mobility of the 120-kDa N-ter proGATI fragment was unaltered by either treatment (Fig. 3C). The noncanonical lipid-linked oligosaccharide identified in C. reinhardtii, along with its lack of N-acetylglucosaminyltransferase I, which is required for the canonical maturation of N-linked oligosaccharides, likely compromise the efficacy of PNGase F; the unique composition of C. reinhardtii O-glycans would limit the efficacy of the O-glycosidase mixture used (37, 39).
Structural Modeling Suggests the Presence of Folded Domains in proGATI.
Like preproGATI, many of the putative prepropeptides identified in the C. reinhardtii genome are substantially larger than typical metazoan prepropeptides. The algorithms used to identify prepropeptides usually exclude proteins larger than about 30 kDa or require that putative endoproteolytic processing sites produce products less than 100 amino acids long (40). While precursors like proinsulin and proopiomelanocortin must fold in a manner that facilitates formation of the correct disulfide bonds, their endoproteolytic cleavage, not their stability, is critical (41).
To explore the possibility that proGATI might fold into stable domains, structural models were generated using RoseTTAFold (33) and AlphaFOLD 2 (34). Both programs predict the formation of three well-folded domains connected by highly extended, Prorich flexible linkers (Fig. 3D). The signal sequence was not included in these structural models. N-terminal domain 1 consists of 323 residues (green), terminating just before a Prorich region. Domain 2 includes 153 residues and domain 3 has 213 residues, ending at the C terminus. The 70-residue linker between domains 1 and 2 contains 37 Pro residues and a motif (K407PRK) that fits the selectivity pattern for several vertebrate prohormone convertases (42, 43), while 43 of the 99 residues in the second linker are Pro residues. Domain 3 forms an antiparallel β-sandwich and has a nominal molecular mass of 23 kDa, with a pI of 10 (Fig. 3D). This domain corresponds precisely to the C-terminal region identified by mass spectrometry of the 75-kDa amidated products from cell bodies (Fig. 1E) and mating ectosomes (Fig. 3B). The cleavage that generates the 75-kDa amidated product would release domains 1 and 2 (predicted to represent the 120-kDa N-terminal fragment), while further proteolysis at K407 might generate the 63-kDa N-terminal product.
Using proGATI Secreted by a Human Cell Line to Understand C. reinhardtii proGATI.
Our identification of a stable 75-kDa amidated proGATI product is consistent with the presence of stably folded domains in C. reinhardtii proGATI. To take advantage of the similar manner in which N- and O-glycosylation sites are recognized in all eukaryotes, and facilitate our understanding of proGATI, a cDNA encoding preproGATI was stably expressed in a human embryonic kidney cell line (HEK-293). In C. reinhardtii, as in other species, maturation of newly synthesized glycoproteins and their ability to move from the ER into the Golgi complex are monitored by their interactions with calnexin and calreticulin (37). We reasoned that the efficient secretion of proGATI by HEK-293 cells would indicate proper folding and allow usage of tools available to study vertebrate N- and O-glycosylation. Affinity-purified proGATI antibodies were used to evaluate cell extracts and spent medium (Fig. 4A and SI Appendix, Fig. S5 A and B). Specific bands of 120- and 170-kDa were detected in cell extracts by both N-ter and C-ter antibodies; a minor ∼37-kDa doublet detected by the C-ter antibody was present in cell extracts, but not spent medium (Fig. 4A). That spent medium contains a single 170-kDa protein recognized by both N-ter and C-ter antibodies identified it as HEK-proGATI; changes in the oligosaccharides attached to proGATI in HEK cells and C. reinhardtii likely account for the difference in their apparent molecular masses. To further test whether HEK-293 cells amidate proGATI, bathocuproine disulfonate (BCS) was used to deplete cellular copper, inhibiting the activity of the amidating enzyme, PAM (44). The 170-kDa C-ter signal, but not the N-ter signal, fell dramatically following BCS treatment, indicating a reduction in amidation (Fig. 4 B and C). HEK-proGATI is bioactive as CC124 gametes bound to HEK-GATI cells plated in a microfluidic chamber, but showed little interaction with control (untransfected HEK-293) cells (SI Appendix, Fig. S6).
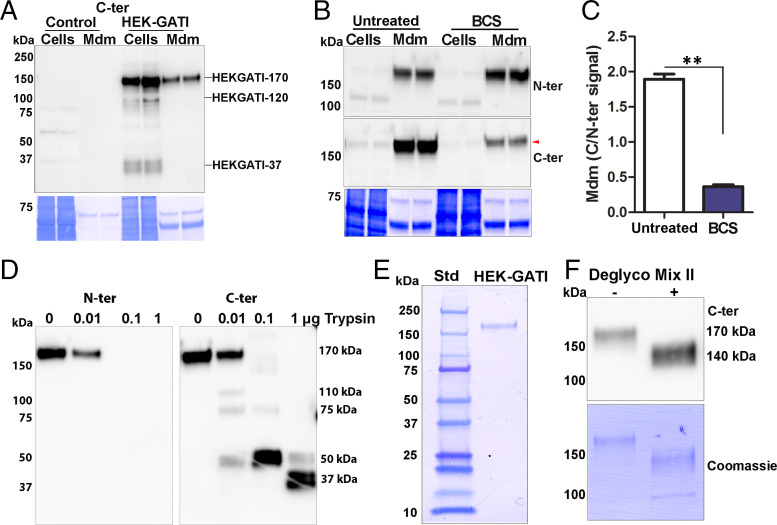
Fig. 4.
HEK cell proGATI contains a stable, amidated C-ter domain. (A) Immunoblot of cell extracts (Cells) and medium (Mdm) prepared from nontransfected cells (Control) and HEK-GATI cells; 20 μg cell extract protein (∼10% of total) and ∼1% of the medium collected over an 18 h time period were analyzed using affinity-purified C-ter antibody. A 170-kDa protein (HEK-proGATI) was detected in both cells and spent medium while 120-kDa and 37-kDa bands containing the amidated C terminus were detected only in cells. (B) Analysis of C-terminal amidation of full-length 170-kDa HEK-proGATI. Medium (Mdm; 5% of total) and cell lysates (Cells; 15 µg, ∼20% of total) from Untreated and BCS-treated cells grown in serum-free media were analyzed. The C-ter signal for HEK-proGATI in the medium was reduced following BCS treatment (red arrowhead), whereas the N-ter signal was unaffected. (C) The C-ter/N-ter signal ratio for HEK-GATI-170 was reduced following BCS treatment. Results are the average of duplicates; **P < 0.001. (D) Medium collected from HEK-proGATI cells was digested by addition of trypsin (10 µL medium plus indicated amount of trypsin); samples were fractionated by SDS/PAGE and probed with C-ter antibody, which detected the indicated cleavage products. The N-ter antibody epitope contains a Lys residue, and is destroyed by trypsin treatment. The results were replicated in independent experiments. (E) SDS/PAGE of purified HEK-proGATI (SI Appendix, Fig. S5C); Coomassie-stained PVDF membrane is shown. (F) Digestion of purified HEK-proGATI with protein deglycosylation Mix II reduced its apparent molecular mass by ∼30 kDa.
Spent medium containing HEK-proGATI was used to determine whether a stable C-ter domain could be detected. Exposure to increasing amounts of trypsin eliminated the N-ter signal and generated a series of smaller products recognized by the C-ter antibody (Fig. 4D). Cleavage at the single Lys residue in the N-ter antigenic peptide is consistent with this result (Fig. 1A). Trypsin produced a sequence of smaller products detected by the C-ter antibody. C-ter signal intensity was not diminished, with essentially complete conversion of 170-kDa HEK-proGATI into a 50-kDa and then a 37-kDa product, which may resemble the amidated C-ter fragment found in mating ectosomes. Successful ectosome-mediated delivery of a chemomodulatory peptide such as GATI-amide may require it to be resistant to proteolysis.
Mass Spectroscopic Identification of Posttranslational Modifications of HEK-proGATI.
HEK-proGATI was purified from medium collected daily from HEK-GATI cells maintained in complete serum-free medium (Fig. 4E and SI Appendix, Fig. S5C). Enzymatic deglycosylation indicated the presence of multiple sugar chains on the HEK-GATI product (Fig. 4F). Five N-glycosylation sites were identified following tryptic digestion of deglycosylated HEK-proGATI (Fig. 5 A and B); two are located in domain 2, two in domain 3, and one in the linker between them. Four Ser O-glycosylation sites were identified following tryptic digestion of purified native HEK-proGATI; two in domain 1, one in domain 2, and one in the linker between domains 2 and 3. Two sites of HyP O-glycosylation were identified, one in domain 1 and one in domain 3; the domain 3 site (HyP896) is separated from Ile904-amide by only seven residues. In the structural models, all identified glycosylation sites (Fig. 5B) are predicted to be solvent exposed and accessible (Figs. 3D and 5C). Importantly for domain 3, in addition to the Pro residue that is O-glycosylated (P896), and both Asn residues that are N-glycosylated (Asn814ThrThr and Asn833GlnThr), the experimentally confirmed C-terminal amidation site that requires exoproteolytic removal of three Arg residues and conversion of the exposed Gly to an amide are also readily accessible for modification (Fig. 5C). Domain 3 also includes four Cys residues; AlphaFOLD 2 predicts that these form two intramolecular disulfide bonds (C739/C745 and C742/C817) (Fig. 5C).
Fig. 5.
Posttranslational modification of HEK-proGATI. (A) Peptides identified by mass spectrometry (black bars) cover most of the three stable domains (domains 1, 2, and 3 are shown) and the linker regions of proGATI; the regions predicted to correspond to the 120-, 75-, and 63-kDa products are indicated by colored bars. The symbols used to indicate the various modifications are shown. All C-terminal peptides had been proteolytically processed and amidated; C-terminal peptides ending in –Gly, –Gly-Arg, –Gly-Arg-Arg, or –Gly-Arg-Arg-Arg were not found. (B) Table indicating the posttranslational modifications identified. (C) Detailed AlphaFOLD 2 structural prediction for domain 3. The two disulfide bonds, glycosylated Asn and HyP residues are shown. The C-terminus has three Arg residues (R906-R908) that are proteolytically removed. The exposed Gly residue (Gly905) is then α-hydroxylated by the PHM domain of PAM and cleavage by its lyase domain releases glyoxylate and generates a C-terminal Ile904-amide.
Distribution, Processing, and Activation of Prohormone Convertases in Cilia and Ectosomes.
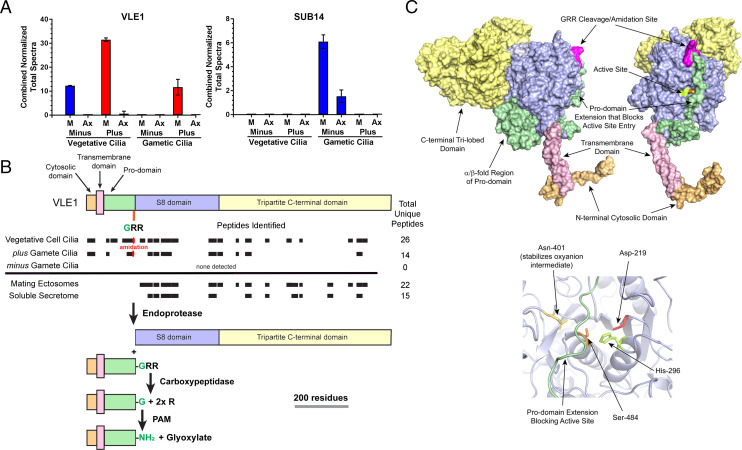
The accumulation of the 75-kDa amidated proGATI product on plus (but not minus) gametic cilia, and of both the N-ter 120-kDa and C-ter 75-kDa proGATI products on mating ectosomes, suggested that proteolytic processing occurs on the ciliary and ectosomal surface or during the sorting and transit of the precursor from cilia into nascent ectosomes. Mating ectosomes contain two subtilisin-like proteases, SUB14 and VLE1; they are the closest C. reinhardtii homologs of mammalian prohormone convertases PC2 and PCSK7, respectively (27). To address the ciliary distribution of these putative prohormone convertases, we performed comparative proteomics of cilia from vegetative and gametic cells of both mating types (45). For this analysis, cilia were separated into membrane and axonemal fractions (Fig. 6A and Dataset S1). We confirmed the presence of VLE1 in vegetative cilia of both mating types (26); SUB14 was not detectable in vegetative cilia. Strikingly, VLE1 was identified in the cilia of plus gametes, but was not detected in minus gamete cilia. SUB14 expression was also mating type-specific, but it was present in the cilia of minus, but not plus, gametes. In consequence, VLE1 is the only putative prohormone convertase present in ciliary samples that contain proteolytically processed proGATI products. Mutant cells lacking VLE1 cannot escape the mother cell wall and grow in large clumps, making their cilia inaccessible. To test whether VLE1 acts as a convertase for proGATI, we isolated ectosomes from vegetative cell cilia that contain active VLE1 but lack other subtilisin-like enzymes (22, 24, 26), and incubated them with purified HEK-proGATI. Ectosome addition lead to formation of a C-terminal proGATI fragment of ∼37 kDa detected by the C-ter antibody, consistent with cleavage at the furin-like cleavage site immediately preceding the N-terminus of domain 3 (SI Appendix, Fig. S7).
Fig. 6.
Ciliary distribution and processing of subtilisin-like proteases. (A) Normalized total spectral counts for VLE1 and SUB14 in the ciliary membrane/matrix (M) and axonemal (Ax) fractions from minus and plus vegetative and gametic cilia (mean ± SEM; n = 3) (45). (B) Diagram showing the cytosolic (light orange), transmembrane (pink), pro- (green), S8 (purple) and C-terminal (yellow) domains of VLE1. The VLE1 peptides identified in the cilia of vegetative cells, plus and minus gametes and in mating ectosome and the soluble secretome are indicated by black boxes. The cleavage/amidation site (GRR) that immediately precedes the S8 catalytic domain and the processing pathway proposed for VLE1 are shown. The α-amidated peptide (K)APTDITDPTAASSSS-amide was found in both vegetative and plus gamete cilia. (C) Two views of the molecular surface of a model for VLE1 calculated using RoseTTAFold are shown. The protein consists of a short N-terminal cytosolic domain (light orange), a single transmembrane region (pink), an unusual prodomain (green), the catalytic S8 domain (blue), and a large C-terminal domain (yellow). The cleavage/amidation site is indicated in magenta. The Right panel shows a ribbon diagram of the active site. Side chains of the catalytic triad residues and the Asn that stabilizes the transition state are shown. The prodomain strand that arches across the active site is shown in green. Comparison of RosseTTAFold and AlphaFOLD 2 predictions for VLE1 using DALI (49) gave alignment scores of z = 51.2 and 13.1 with RMSDs of 5.8 and 4.5 Å for the N-terminal to S8 and C-terminal regions, respectively.
Subtilisin-like prohormone convertases are synthesized as inactive proenzymes (42); their activation generally requires both an autoproteolytic cleavage immediately preceding the S8 catalytic domain and dissociation of the S8 domain from the prodomain (42). Unlike PCSK7, which is a type I integral membrane protein, VLE1 is type II integral membrane protein (Fig. 6B); thus, activation of VLE1 would be expected to release it from its transmembrane domain. The VLE1 present in the cilia of vegetative and plus gamete cells contained peptides from its Cytosolic, Pro, S8, and C-terminal domains (Fig. 6B). In contrast, the VLE1 present in mating ectosomes and in the soluble secretome contained only peptides from its S8 and C-terminal domains (Fig. 6B), suggesting that activation had occurred. Intriguingly, the C-terminal domain is predicted to have a tripartite organization, with each lobe consisting of two antiparallel β-sheets (Fig. 6C); each lobe exhibits considerable structural similarity to the CEA1 N-acetylglucosamine-binding adhesin from the methylotrophic yeast Komagataella pastoris (z score = 11.8, RMSD = 4.0 Å; 5A3L).
Clustal analysis identified the –Gly-Arg-Arg site that immediately precedes the catalytic S8 domain as the likely site for autoactivation (Fig. 6B). Autoproteolytic cleavage at this site, followed by exoproteolytic removal of the two Arg residues, would produce a potential amidation site. Mass spectrometry revealed that all of the ciliary VLE1 had been proteolytically processed at this site and was amidated (Fig. 6B); partially processed peptides derived from this region of VLE1 and ending in –Gly, –Gly-Arg, or –Gly-Arg-Arg were not observed. Cleavage at this site is a prerequisite for activation, but dissociation of the S8 catalytic domain from the proregion is also usually required. Detailed analysis of the VLE1 structure predicted by RosETTAFold (Fig. 6C) and the VLE1 peptides identified in cilia suggests that the prodomain may remain associated with the S8 domain, tethering it to the ciliary membrane in an inactive form even after autoproteolytic cleavage and amidation.
Differential Release of proGATI Products from Mating Ectosomes.
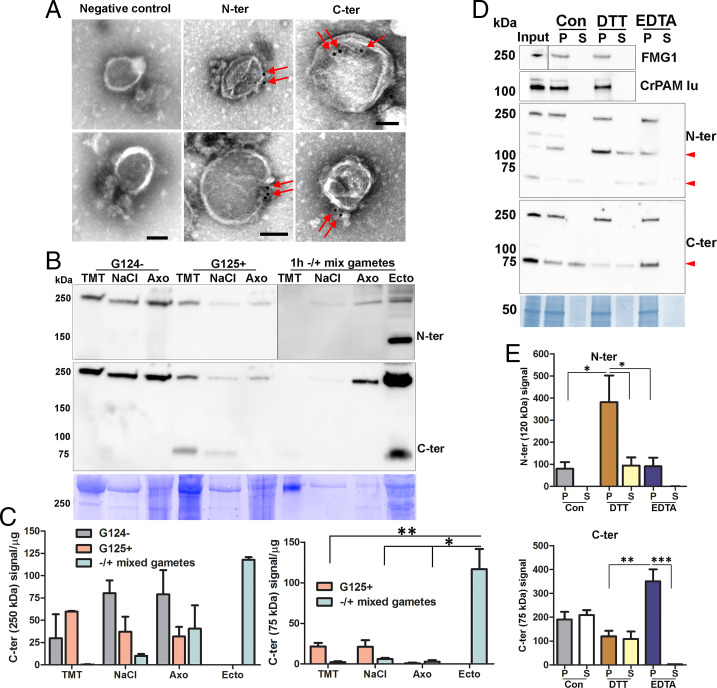
The proGATI associated with the ciliary membrane and with ectosomes is predicted to be on the surface. We utilized affinity-purified proGATI antibodies and negative-stain immunogold-EM to examine ectosomal localization. Following incubation with intact ectosomes, affinity-purified proGATI antibodies were visualized using a gold-tagged anti-rabbit secondary antibody and whole-mount negative-stain EM; signals obtained with both antibodies were localized on the ectosome surface (Fig. 7A). Ectosomes incubated only with gold-conjugated secondary antibody served as a negative control.
Fig. 7.
Ciliary localization and association of proGATI and its fragments with ectosomes. (A) Whole-mount negative-stain immunogold EM images of intact ectosomes incubated with affinity-purified N-ter or C-ter antibodies and a gold-tagged secondary antibody; both epitopes localized to the ectosomal surface. Ectosomes incubated with gold-tagged secondary anti-rabbit antibody alone served as a negative control. (Scale bars, 100 nm.) (B) Cilia were sequentially treated with buffers containing 1% Triton X-100 (TMT) and 0.6 M NaCl (NaCl); the resulting axonemal pellet (Axo) was solubilized in 1% SDS-buffer. The subciliary fractions from resting minus (G124−) and plus (G125+) gametes, mixed gametes, and mating ectosomes (Ecto) were fractionated by SDS/PAGE, blotted and probed with affinity-purified N-ter and C-ter antibodies. Equal amounts of protein (20 µg) were loaded for each sample. (C) Immunoblot quantification of the 250- and 75-kDa C-ter products. Means are average of duplicates and error bars indicate ± range, where *P < 0.05, **P < 0.01. (D) Freshly isolated mating ectosomes (Input) were washed with buffer alone (10 mM Hepes, control) or with buffer containing 10 mM DTT or 10 mM EDTA; after centrifugation, the resulting supernatants (S) and pellets (P) were analyzed for the presence of proGATI (using N-ter and C-ter antibodies), PAM, and FMG1. Red arrowheads mark the 120-, 75-, and 63-kDa bands. Samples loaded represent the pellets and corresponding supernatants derived from an initial 15 µg of ectosomes. (E) Quantification of 120-kDa N-ter signal (n = 4) and 75-kDa C-ter signal (n = 3); means ± SEM are shown. Asterisks indicate significant differences between the groups: *P < 0.05, **P < 0.01, ***P < 0.001.
We previously found that both CrPAM protein and enzyme activity associate with the ciliary axoneme; this interaction is disrupted by treatment with 0.6 M NaCl following detergent extraction (18). To explore the ciliary distribution of proGATI and its products, we isolated cilia from resting gametes of both mating types and from 1-h mixed gametes. Isolated cilia were first treated with Triton X-100 to release membrane proteins and soluble matrix components. This was followed by treatment with 0.6 M NaCl to extract proteins that were tightly bound to the axoneme; the resulting extracted axoneme pellet was solubilized in 1× SDS buffer.
The amidated 75-kDa GATI product was detected in the cilia of plus but not minus gametes (Fig. 7 B and C). This fragment was largely recovered in the detergent-soluble fraction, with a smaller amount released by 0.6 M NaCl; it was not present in the axonemal fraction of plus gametes. The 250-kDa proGATI protein was in the detergent-soluble, 0.6 M NaCl, and axonemal fractions from both plus and minus gametes (Fig. 7 B and C). During gamete mating, the amidated 75-kDa GATI product and the 250-kDa proGATI protein were released into ectosomes (Fig. 7 B and C). The presence of an N-terminal fragment of proGATI in ectosomes but not in the ciliary fractions suggests that cleavage of 250-kDa proGATI occurred on the ectosomal surface.
To evaluate how 250-kDa proGATI (which lacks a transmembrane domain) associates with the ectosome surface, freshly isolated ectosomes were washed with 10 mM Hepes buffer (control) or with buffer containing 10 mM dithiothreitol (DTT) or 10 mM EDTA, and the resulting supernatants examined for the release of GATI products, CrPAM, and FMG1 (Fig. 7 D and E). Neither 250-kDa proGATI, CrPAM, nor FMG1 was solubilized, with signal detected only in the ectosomal pellets. In contrast, the amidated 75-kDa C-terminal product and N-terminal 63-kDa segment were both released by washing with low ionic strength Hepes buffer; release did not occur following chelation of divalent cations with EDTA. Although not released by buffer alone or by EDTA treatment, the N-terminal 120-kDa GATI fragment was partially displaced from ectosomes by 10 mM DTT. This effect was DTT-specific; treatment with 10 mM β-mercaptoethanol had no effect (SI Appendix, Fig. S8). These results suggest that all three domains individually mediate associations with the ectosomal surface. This tripartite attachment mechanism likely explains why release of 250-kDa amidated proGATI was not observed under any of the conditions tested including carbonate washing (SI Appendix, Fig. S4).
Discussion
Identification of an amidated peptide that has a mating-type–specific effect on C. reinhardtii mobility led us to explore the properties of its putative precursor, the manner in which this precursor might be converted into smaller products, and the regulated secretion of its product peptides.
ProGATI Undergoes Extensive Posttranslational Modification and Contains Multiple Domains.
The C. reinhardtii genome encodes hundreds of proteins with the general characteristics of prepropeptides (27). As observed in the ER of metazoans, preproGATI undergoes signal peptide removal, along with the first steps of N- and O-glycosylation (Fig. 8A). ProGATI, like many other putative C. reinhardtii propeptides, is quite large, with a predicted molecular mass of 90.6 kDa, and multiple domains connected by Prorich linker regions. In addition to the Asn and Ser/Thr sites subject to N- and O-glycosylation in metazoan propeptides, hydroxy-Pro residues in domains 1 and 3 of proGATI are O-glycosylated. In plants and algae, hydroxy-Pro residues are major O-glycosylation sites for addition of pentose (arabinogalactan) sugars (38, 46). With a unique core structure to their N-glycans and unique O-glycosyl transferases, peptide precursors synthesized by C. reinhardtii differ in important ways from metazoan propeptides (37, 39, 47, 48).
Fig. 8.
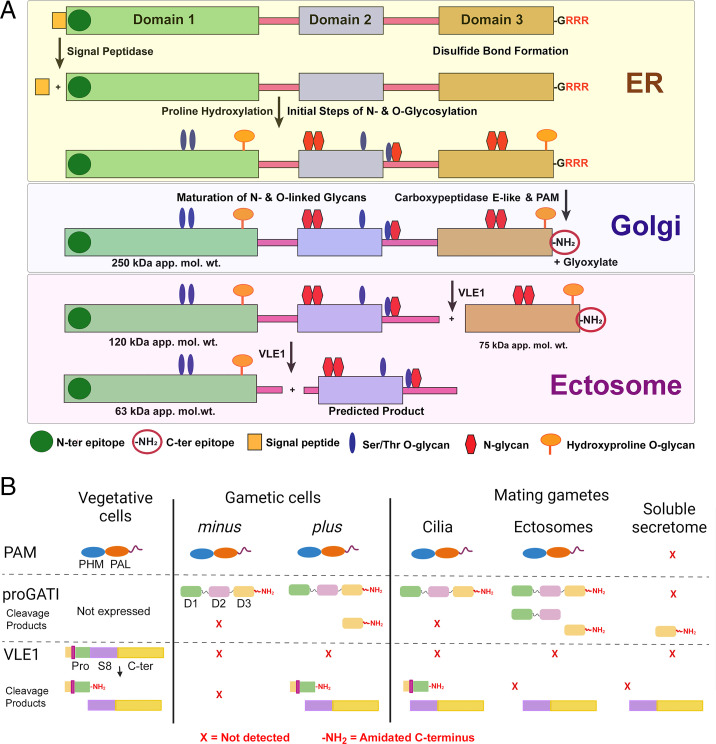
ProGATI processing pathway. (A) Diagram illustrating the processing pathway of preproGATI that occurs as it traffics through the ER and Golgi and subsequently enters cilia and ectosomes. The signal peptide (orange box) is removed in the ER by signal peptidase. Addition of N-linked sugars (red) begins in the ER, as does modification of Pro to HyP by prolyl hydroxylases. As proGATI moves into the Golgi complex, more complex sugars and O-linked sugars on HyP (orange) and Ser/Thr (blue) residues are added, leading to the higher apparent molecular mass (∼250 kDa) of proGATI. A carboxypeptidase sequentially removes the C-terminal Arg residues and generates a Gly-extended substrate for PAM. PAM converts this into the amidated product proGATI-amide (indicated by -NH2) in a two-step reaction, and releases glyoxylate as a byproduct. This 250-kDa amidated proGATI then moves to the ciliary membrane. Once on cilia, or as it moves from cilia into nascent ectosomes, 250-kDa proGATI is cleaved by VLE1 producing the 120-kDa N-terminal and amidated 75-kDa C-terminal fragments. Cleavage of the 120-kDa product at a second putative prohormone convertase site located in the linker between domains 1 and 2, or at a dibasic site at the C-terminal end of domain 1, might produce the 63-kDa N-terminal fragment and a second product containing domain 2 for which no probe currently exists. (B) Diagram illustrating the presence and absence of PAM, amidated full-length proGATI (indicated by -NH2) and its various fragments, and the cleaved/amidated subtilisin-like endoprotease VLE1 in cilia of minus and plus vegetative and gametic cells, and in ectosomes and the soluble secretome obtained from mating gametes. PAM is present in vegetative and gametic cell cilia and is released into ectosomes but not into the secretome. In contrast, amidated proGATI is undetectable in vegetative cilia and only appears following gametogenesis. The amidated C-terminal fragment is generated in plus gamete cilia, and released into ectosomes and the secretome during mating; other proGATI products are also variably present in these samples. VLE1 is found in vegetative and plus gamete cilia, but not minus gamete cilia. All ciliary VLE1 is proteolytically processed within the prodomain and amidated. As VLE1 moves to ectosomes and is released into the soluble secretome, it undergoes a change in domain architecture with the catalytic S8 and C-terminal domains dissociating from the amidated N-terminal segment.
With the endoproteolytic cleavage of proGATI limited to the surface of mating ectosomes (Fig. 2B), we considered the possibility that its structural domains might play a role in its localization. Despite sharing little sequence similarity, a DALI search (49) revealed structural relatives for each proGATI domain: domain 1 is distantly similar to halohydrin dehalogenase from Ilumobacter coccineus (z score = 6.2, RMSD = 13.6 Å; 6I9W); domain 2 is related to EPR3 (a carbohydrate receptor) from Lotus japonicus (z score = 4.2, RMSD = 2.4 Å; 6QUP); and domain 3 resembles a chitosanase from Paenibacillus sp. (z score = 9.3, RMSD = 2.5 Å; 4ZXE). The ability of EPR3 and chitosanase to interact with carbohydrates (50–52) suggests that domains 2 and 3 might play a role in the tripartite interaction of proGATI with the ectosomal surface, with subsequent endoproteolytic cleavages facilitating release of specific fragments.
For signaling peptides released on ectosomes, protease resistance may be especially important. Domain 3 corresponds precisely to the 75-kDa C-ter fragment (Fig. 3A). N-glycosylation of the two potential sites in domain 3, along with O-glycosylation of a hydroxy-Pro located close to the amidated C terminus, likely accounts for the ∼50-kDa discrepancy between its apparent molecular mass and the mass of its polypeptide chain (23 kDa) (Fig. 8A). Although the C-terminus of proGATI is accessible to PAM, which converts its C-terminus from -GATI-Gly into -GATI-amide, the amidated C-terminus is trypsin resistant and stable when exposed on the ciliary membrane and the surface of mating ectosomes.
Examination of the first C. reinhardtii protein known to serve as a peptide precursor indicates that it shares many similarities with vertebrate peptide precursors. However, its larger size, more complex domain organization, and extensive modifications suggest that this precursor carries additional information needed to ensure that its signaling task can be accomplished.
Controlling the Endoproteolytic Cleavage of proGATI.
ProGATI cleavage is linked to both mating type and subcellular location (Fig. 8B). The cell bodies of plus and minus gametes contain intact proGATI, but the 75-kDa C-ter fragment is found only in the cilia of plus gametes. Both N- and C-ter proGATI fragments accumulate in mating ectosomes. In metazoans, the cell-type–specific cleavage of propeptides, such as proopiomelanocortin (53) and proglucagon (54), reflects the cell-type–specific expression of subtilisin-like prohormone convertases. Mating ectosomes contain only two subtilisin-like proteases, VLE1 and SUB14 (11). The presence of VLE1 but not SUB14 in the cilia of plus gametes, where proGATI cleavage occurs, suggests that VLE1 serves as a proGATI convertase. VLE1 is also localized to the ciliary membrane in vegetative cells (24, 26); its release into vegetative ectosomes provides access to the mother cell wall, which it degrades, allowing release of mitotic progeny. A matrix metalloproteinase (gametolysin), not VLE1, cleaves the gametic cell wall prior to fusion (55), suggesting that VLE1 has additional targets on plus gamete cilia and in the extracellular milieu. VLE1 cleaves to the C-terminal side of basic residues, although the required sequence context is poorly understood (56). Endoproteolytic cleavage of proGATI after a basic residue within a furin-like cleavage site (R693FSR↓) produces the amidated 75-kDa C-ter fragment (Figs. 3B and 8A). That amidation precedes endoproteolytic cleavage is a distinctive feature of the proGATI processing pathway, but has also been observed for chromogranin A (57). Treatment of HEK-proGATI with VLE1-containing ectosomes from vegetative cells yielded an amidated C-terminal domain 3 fragment, supporting identification of VLE1 as a proGATI convertase (SI Appendix, Fig. S7).
In metazoans, luminal pH plays a central role in controlling prohormone convertase activation and the storage of product peptides in secretory granules (58). With proGATI cleavage products accumulating on the surface of mating ectosomes, luminal pH cannot serve as a regulatory factor. The prodomains of subtilisin-like endoproteases facilitate catalytic domain folding and inhibit activity. Protease activation requires autoproteolytic cleavage, separating the prodomain from the catalytic domain (42). Additional cleavages within the prodomain may also be required for prodomain release and S8 domain activation. Consistent with this, active VLE1 purified from culture medium following hatching lacked its prodomain (26). Our analysis of the soluble mating secretome identified the intact VLE1 S8/C-terminal domain, but not the N-terminal/prodomain (Fig. 6B).
Sequence analysis revealed an unusual prodomain in VLE1, with homologous sequences found only in other members of the volvocine algae [e.g., the protease VheA, required for release of juvenile Volvox from the parental spheroid (59)]. To understand how VLE1 activation might occur, a structural model was built using RoseTTAFold (33) (Fig. 6C and SI Appendix, Fig. S9). The active site contains a classic Ser-His-Asp catalytic triad and an Asn residue that stabilizes the transition state in the oxyanion hole (Fig. 6C) (42). The VLE1 prodomain consists of an α/β fold that makes extensive contact with one face of the S8 domain. Emanating from this α/β region is an extended strand that arches over the active site, occluding it; the –Gly-Arg-Arg cleavage/amidation site is exposed on the surface. Given the large surface area buried by the prodomain, cleavage at the –Gly-Arg-Arg site seems unlikely to result in prodomain release from the catalytic core.
For amidation to occur, the extended strand must swing away from the catalytic site, enabling carboxypeptidases to remove remaining Arg residues and allowing PAM to access the exposed Gly residue. The functional consequences of amidation at this site remain to be determined. Binding of the amidated prodomain C-terminus to a target protein might facilitate retention of the N-terminal/prodomain of this type II membrane protein in the ciliary membrane, allowing the enzymatically active S8/C-terminal domain to enter mating ectosomes.
Ciliary Ectosomes as an Ancient Mode of Rapid, Regulated Secretion.
Changes in protein expression allow unicellular organisms like C. reinhardtii to regulate secretion of the enzymes needed to acquire specific nutrients, but this type of response requires time. In metazoans, peptides stored in secretory granules can be released within milliseconds of signal receipt. Our data indicate that ciliary ectosomes serve as an ancient mode of rapid, regulated secretion. Like the assembly of secretory granules, the assembly of ciliary ectosomes is a highly regulated process. The cilia of both vegetative cells and mating gametes release bioactive ectosomes; their compositions are unique and developmentally regulated (21, 22, 24, 27). Since ectosomes are formed from the ciliary membrane, proteins targeted to ectosomes must first gain access to the cilium. The transition zone plays an essential role in establishing and maintaining the unique lipid and protein composition of the ciliary membrane (60–62).
The entry of ciliary proteins into ectosomes is also regulated. Differences in the ectosomal trafficking of PAM, VLE1, and proGATI illustrate key features of this regulatory step (Fig. 8B). CrPAM is found in mating ectosomes but not in vegetative ectosomes (27); cleavage of CrPAM does not occur and active enzyme does not appear in the soluble secretome. While VLE1 is found in the cilia of plus gametes, the presence of the N-terminal/prodomains, along with the intact S8/C-terminal domains, suggests that ciliary VLE1 is not active. VLE1 recovered from mating ectosomes and the secretome lacks the N-terminal/prodomains, indicating that it has been activated. While proGATI is present in the cilia of plus and minus gametes, cleavage occurs only in the cilia of plus gametes; further cleavage of proGATI is linked to its release in mating ectosomes, where both N-ter and C-ter fragments accumulate. Although metazoan secretory granules generally store mature product peptides, the cleavage of proatrial natriuretic factor by corin, a type II plasma membrane enzyme like VLE1, is tied to the exocytosis of atrial granules (63).
Metazoan peptide-containing secretory granules can be stored for long periods of time, with release responding rapidly to receptor-mediated secretagogue stimulation. In C. reinhardtii, ciliary adhesion of mating gametes causes ectosomes to appear on the ciliary surface in a process that requires receptor-mediated signaling; strikingly, activating gametes directly with dibutyryl-cAMP does not lead to ectosome release (21). Similarly, the signals that control food intake in mammals require localization of the melanocortin-4 receptor to the primary cilia of hypothalamic neurons (8). The ciliary localization of free fatty acid receptor-4 and prostaglandin-E receptor-4 in α- and β-cells plays an essential role in hormone secretion (64), and mice lacking primary cilia on their β-cells exhibit impaired glucose homeostasis and develop diabetes (65).
As observed in mammals, multiple receptors have been identified in C. reinhardtii cilia (27, 66, 67). By taking advantage of the ease with which cilia can be isolated from C. reinhardtii, its precisely delineated sexual reproductive cycle, and the identification of a bioactive amidated peptide in mating ectosomes, it is now clear that cilia provide a means of controlling endoproteolytic processing of propeptides and the release of mature bioactive peptide products. Although the stimulus-dependent secretion of neuropeptides from dense core vesicles stored at the presynaptic endings of axons or exported to dendrites has been well studied (68), whether bioactive peptides are released from the primary cilia of neurons and endocrine cells remains to be determined.
In summary, this study provides support for a mechanism through which amidated peptide products are synthesized, posttranslationally modified, trafficked into cilia, and then processed and released into ciliary ectosomes by a unicellular organism, C. reinhardtii. As both cilia and the peptidergic signaling machinery are conserved throughout eukaryotes, this study should shed light on the mechanisms through which cilia-based secretion is regulated in health and dysregulated in various ciliopathies.
Materials and Methods
Extended descriptions of the methods are provided in SI Appendix.
Chlamydomonas Cell Culture and Gametogenesis Induction.
Wild-type C. reinhardtii mating type minus (CC124) and plus (CC125) strains were cultured in R-medium (28). To induce gametogenesis, vegetative cells of both mating types were washed and resuspended in nitrogen-deficient minimal medium (M-N medium) for 24 to 36 h under aeration and a 12-h light/12-h dark cycle.
Preparation of Cell Lysates, Cilia, and Ectosomes from Mating Gametes.
Cell lysates were prepared by resuspending gametes in TMT buffer (20 mM 2-[Tris(hydroxymethyl)-methylamino]-ethanesulfonic acid [TES], pH 7.4, 10 mM mannitol, 1% Triton X-100) containing 0.2 M NaCl and protease inhibitors. Gametes were resuspended in fresh nitrogen-free M-N medium, mixed, and ectosomes isolated by differential centrifugation (27). Gametes were deciliated using dibucaine and cilia isolated by standard methods and resuspended in HMS buffer (10 mM Hepes, pH 7.4, 5 mM MgSO4, and 4% sucrose) (69, 70).
Antibody Generation.
Synthetic peptides from the N-terminal (YELGLDIDGKPAHPAAT-NH2, 1.5 mg) and C-terminal (YAPGTGGGATI-NH2, 1.5 mg) regions of proGATI were individually conjugated to keyhole limpet hemocyanin. Three rabbits (CT327, CT330, and CT332) were immunized with a mixture of KLH-conjugated N-ter and C-ter peptides by Covance Immunology Services (procedure approved by the UConn Health Institutional Animal Care and Use Committee). N-ter and C-ter antibodies were purified by peptide affinity chromatography.
Microscopy.
Resting gametes were treated with methanol for 10 min at −20 °C. Primary antibodies used were affinity-purified rabbit N-ter and C-ter proGATI antibodies (from CT327; 1:500) and mouse FMG1 (1:1,000). Alexa 488 anti-rabbit (1:500) and Cy3 anti-mouse (1:2,000) conjugates were used as secondary antibodies. Images were obtained using a Zeiss 880 confocal microscope with a 63× oil objective. Electron microscopy was performed as described previously (27); glutaraldehyde-fixed ectosomes were embedded in low-melting point agarose prior to processing.
PreproGATI from HEK-293 Cells.
HEK-293 cells were maintained in DMEM/F12 medium containing 10% fetal calf serum (HyClone), 100 units/mL penicillin-streptomycin and 25 mM Hepes, pH 7.4 at 37 °C in a 5% CO2 incubator. A cDNA encoding preproGATI in pCI-neo was transfected and stable lines expressing preproGATI selected in DMEM/F12 medium containing 0.5 mg/mL G418 disulfate. Following transfer to serum-free medium and 16- to 18-h growth, spent medium was collected, pooled and HEK-proGATI purified using ion exchange and gel-filtration chromatography. To assess the effect of reduced copper, HEK-GATI cells in serum-free media were treated with 50 µM bathocuproine disulfonic acid.
HEK-GATI Bioassay.
Control and HEK-GATI cells were plated in microfluidic channel slides for 16 to 18 h at 37 °C. After washing, the channel and reservoirs were filled with 50 μL gametic medium and an aliquot (10 μL, 1.2 × 106 gametes/mL) of minus CC124 gametes added to one reservoir. Following incubation, phase-contrast images of HEK cells were taken. Gametes were identified based on their auto-fluorescence. HEK cell areas were quantified using ImageJ and the number of minus gametes located in these areas was counted manually.
Deglycosylation Assays.
The presence of N-linked oligosaccharides was examined using PNGase F (New England Biolabs # P0708S) and the presence of O-linked sugars was assessed by combined treatment with O-glycosidase (New England Biolabs #P0733S) and α2-3,6,8 neuraminidase (New England Biolabs #P0720S). Mating C. reinhardtii ectosomes (20 µg) and spent medium (9 µL) from HEK-293 cells expressing proGATI were denatured by heating at 100 °C for 10 min with 1× denaturing buffer and enzymatically deglycosylated.
Immunoprecipitation.
Immunoprecipitation was performed using slight modifications of previous protocols (71). Cross-reactive proteins were immunoprecipitated from plus gametic cell lysates and from mating ectosomes using affinity-purified C-ter antibody and Protein A agarose beads.
Mass Spectrometry.
Peptides from excised electrophoretic gel bands and purified soluble HEK-proGATI were analyzed using nanoflow liquid chromatography coupled to tandem mass spectrometry. Raw data were searched against the C. reinhardtii proteome using variable modifications to identify sites of glycosylation, amidation, proline hydroxylation, and phosphorylation. Comparative proteomics of VLE1 and SUB14 was performed as described previously (45).
Supplementary Material
Acknowledgments
We thank Dr. Jeremy L. Balsbaugh, Director of the Proteomics & Metabolomics Facility (a component of the Center for Open Research Resources and Equipment at the University of Connecticut) for quantitative proteomics analysis; Maya Yankova for assistance with electron microscopy; Dr. Miho Sakato-Antoku for assistance with chromatography; Dr. Xin-Ming Ma for access to his cell culture facility; and Dr. R. Bloodgood (University of Virginia) for the gift of FMG-1 antibody. This study was supported by NIH Grants R01-DK032949 (to B.A.E. and R.E.M.), R01-GM125606 (to S.M.K. and B.A.E.), and R35-GM140631 (to S.M.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206098119/-/DCSupplemental.
Data Availability
All underlying data are included within the main text and the supporting information. Mass spectrometry data have been deposited in Dryad at https://doi.org/10.5601/dryad.fn2z34txn (72) .
References
- 1.Marshall W., Basto R., Cilia (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2017). [Google Scholar]
- 2.Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J. B., Bettencourt-Dias M., Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165–175 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malicki J. J., Johnson C. A., The cilium: Cellular antenna and central processing unit. Trends Cell Biol. 27, 126–140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Dam T. J. P., et al. , CiliaCarta: An integrated and validated compendium of ciliary genes. PLoS One 14, e0216705 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reiter J. F., Leroux M. R., Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18, 533–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anvarian Z., Mykytyn K., Mukhopadhyay S., Pedersen L. B., Christensen S. T., Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol. 15, 199–219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green J. A., et al. , Recruitment of β-arrestin into neuronal cilia modulates somatostatin receptor subtype 3 ciliary localization. Mol. Cell. Biol. 36, 223–235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., et al. , Melanocortin 4 receptor signals at the neuronal primary cilium to control food intake and body weight. J. Clin. Invest. 131, e142064 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar D., Mains R. E., Eipper B. A., King S. M., Ciliary and cytoskeletal functions of an ancient monooxygenase essential for bioactive amidated peptide synthesis. Cell. Mol. Life Sci. 76, 2329–2348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasso S., Stibor H., Mittag M., Grossman A. R., From molecular manipulation of domesticated Chlamydomonas reinhardtii to survival in nature. eLife 7, e39233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luxmi R., et al. , Proteases shape the Chlamydomonas secretome: Comparison to classical neuropeptide processing machinery. Proteomes 6, 36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsubayashi Y., Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol. 52, 5–13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuda A., Jones L. S., Shigeri Y., The multiplicity of post-translational modifications in pro-opiomelanocortin-derived peptides. Front. Endocrinol. (Lausanne) 4, 186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madsen T. D., et al. , An atlas of O-linked glycosylation on peptide hormones reveals diverse biological roles. Nat. Commun. 11, 4033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindberg I., Fricker L. D., Obesity, POMC, and POMC-processing enzymes: Surprising results from animal models. Endocrinology 162, bqab155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chrétien M., Mbikay M., 60 years of POMC: From the prohormone theory to pro-opiomelanocortin and to proprotein convertases (PCSK1 to PCSK9). J. Mol. Endocrinol. 56, T49–T62 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Luxmi R., Mains R. E., King S. M., Eipper B. A., “Peptidylglycine α-amidating monooxygenase (PAM)” in Encyclopedia of Biological Chemistry, Jez J., Ed. (Elsevier, Oxford, 2021, 3rd ed.). pp. 88–104. [Google Scholar]
- 18.Kumar D., et al. , Early eukaryotic origins for cilia-associated bioactive peptide-amidating activity. J. Cell Sci. 129, 943–956 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar D., et al. , A bioactive peptide amidating enzyme is required for ciliogenesis. eLife 6, e25728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar D., et al. , Microvillar and ciliary defects in zebrafish lacking an actin-binding bioactive peptide amidating enzyme. Sci. Rep. 8, 4547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao M., et al. , Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. elife 4, e05242 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long H., et al. , Comparative analysis of ciliary membranes and ectosomes. Curr. Biol. 26, 3327–3335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., et al. , C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr. Biol. 24, 519–525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood C. R., Huang K., Diener D. R., Rosenbaum J. L., The cilium secretes bioactive ectosomes. Curr. Biol. 23, 906–911 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luxmi R., King S. M., Cilia-derived vesicles: An ancient route for intercellular communication. Semin. Cell Dev. Biol., 10.1016/j.semcdb.2022.03.014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubo T., et al. , The Chlamydomonas hatching enzyme, sporangin, is expressed in specific phases of the cell cycle and is localized to the flagella of daughter cells within the sporangial cell wall. Plant Cell Physiol. 50, 572–583 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Luxmi R., Kumar D., Mains R. E., King S. M., Eipper B. A., Cilia-based peptidergic signaling. PLoS Biol. 17, e3000566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris E., The Chlamydomonas Sourcebook. 1: Introduction to Chlamydomonas and Its Laboratory Use (Elsevier, San Diego, 2009, 2nd ed.). [Google Scholar]
- 29.Palma M. S., “Insect venom peptides” in Handbook of Biologically Active Peptides, Kastin A. J., Ed. (Academic Press, Burlington, 2006), pp. 389–396. [Google Scholar]
- 30.Rowe M. L., Elphick M. R., The neuropeptide transcriptome of a model echinoderm, the sea urchin Strongylocentrotus purpuratus. Gen. Comp. Endocrinol. 179, 331–344 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Szabó R., et al. , Effect of SXWS/WSXWS peptides on chemotaxis and adhesion of the macrophage-like cell line J774. J. Mol. Recognit. 28, 253–260 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Takahashi T., et al. , Systematic isolation of peptide signal molecules regulating development in hydra: LWamide and PW families. Proc. Natl. Acad. Sci. U.S.A. 94, 1241–1246 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baek M., et al. , Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ning J., et al. , Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev. 27, 1198–1215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amare A., et al. , Bridging neuropeptidomics and genomics with bioinformatics: Prediction of mammalian neuropeptide prohormone processing. J. Proteome Res. 5, 1162–1167 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathieu-Rivet E., Mati-Baouche N., Walet-Balieu M.-L., Lerouge P., Bardor M., N- and O-glycosylation pathways in the microalgae polyphyletic group. Front Plant Sci 11, 609993 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bollig K., et al. , Structural analysis of linear hydroxyproline-bound O-glycans of Chlamydomonas reinhardtii—Conservation of the inner core in Chlamydomonas and land plants. Carbohydr. Res. 342, 2557–2566 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Joshi H. J., et al. , SnapShot: O-glycosylation pathways across kingdoms. Cell 172, 632–632.e2 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Mirabeau O., et al. , Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res. 17, 320–327 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M., et al. , Deciphering the hidden informational content of protein sequences: Foldability of proinsulin hinges on a flexible arm that is dispensable in the mature hormone. J. Biol. Chem. 285, 30989–31001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shakya M., Lindberg I., Mouse models of human proprotein convertase insufficiency. Endocr. Rev. 42, 259–294 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein-Szanto A. J., Bassi D. E., Proprotein convertase inhibition: Paralyzing the cell’s master switches. Biochem. Pharmacol. 140, 8–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonnemaison M. L., et al. , Adaptor protein-1 complex affects the endocytic trafficking and function of peptidylglycine α-amidating monooxygenase, a luminal cuproenzyme. J. Biol. Chem. 290, 21264–21279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakato-Antoku M., King S. M., Developmental changes in ciliary composition during gametogenesis in Chlamydomonas. Mol. Biol. Cell 33, br10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan L., Leykam J. F., Kieliszewski M. J., Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiol. 132, 1362–1369 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu N., et al. , Altered N-glycan composition impacts flagella-mediated adhesion in Chlamydomonas reinhardtii. elife 9, e58805 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulze S., et al. , N-glycoproteomic characterization of mannosidase and xylosyltransferase mutant strains of Chlamydomonas reinhardtii. Plant Physiol. 176, 1952–1964 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holm L., Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29–42 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Lopez-Moya F., Suarez-Fernandez M., Lopez-Llorca L. V., Molecular mechanisms of chitosan interactions with fungi and plants. Int. J. Mol. Sci. 20, 332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawaharada Y., et al. , Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523, 308–312 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Wong J. E. M. M., et al. , Structural signatures in EPR3 define a unique class of plant carbohydrate receptors. Nat. Commun. 11, 3797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar D., Mains R. E., Eipper B. A., 60 years of POMC: From POMC and α-MSH to PAM, molecular oxygen, copper, and vitamin C. J. Mol. Endocrinol. 56, T63–T76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drucker D. J., Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metab. 27, 740–756 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Kinoshita T., Fukuzawa H., Shimada T., Saito T., Matsuda Y., Primary structure and expression of a gamete lytic enzyme in Chlamydomonas reinhardtii: Similarity of functional domains to matrix metalloproteases. Proc. Natl. Acad. Sci. U.S.A. 89, 4693–4697 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuda Y., Koseki M., Shimada T., Saito T., Purification and characterization of a vegetative lytic enzyme responsible for liberation of daughter cells during the proliferation of Chlamydomonas reinhardtii. Plant Cell Physiol. 36, 681–689 (1995). [PubMed] [Google Scholar]
- 57.Simpson P. D., et al. , Striking oxygen sensitivity of the peptidylglycine α-amidating monooxygenase (pam) in neuroendocrine cells. J. Biol. Chem. 290, 24891–24901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Halban P. A., Structural domains and molecular lifestyles of insulin and its precursors in the pancreatic beta cell. Diabetologia 34, 767–778 (1991). [DOI] [PubMed] [Google Scholar]
- 59.Fukada K., Inoue T., Shiraishi H., A posttranslationally regulated protease, VheA, is involved in the liberation of juveniles from parental spheroids in Volvox carteri. Plant Cell 18, 2554–2566 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takao D., Verhey K. J., Gated entry into the ciliary compartment. Cell. Mol. Life Sci. 73, 119–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nachury M. V., Mick D. U., Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Biol. 20, 389–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Long H., Huang K., Transport of ciliary membrane proteins. Front. Cell Dev. Biol. 7, 381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glembotski C. C., Dixon J. E., Gibson T. R., Secretion of atrial natriuretic factor-(1-98) by primary cardiac myocytes. J. Biol. Chem. 263, 16073–16081 (1988). [PubMed] [Google Scholar]
- 64.Wu C. T., et al. , Discovery of ciliary G protein-coupled receptors regulating pancreatic islet insulin and glucagon secretion. Genes Dev. 35, 1243–1255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hughes J. W., et al. , Primary cilia control glucose homeostasis via islet paracrine interactions. Proc. Natl. Acad. Sci. U.S.A. 117, 8912–8923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang K., Kunkel T., Beck C. F., Localization of the blue-light receptor phototropin to the flagella of the green alga Chlamydomonas reinhardtii. Mol. Biol. Cell 15, 3605–3614 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ranjan P., Awasthi M., Snell W. J., Transient internalization and microtubule-dependent trafficking of a ciliary signaling receptor from the plasma membrane to the cilium. Curr. Biol. 29, 2942–2947.e2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding K., et al. , Imaging neuropeptide release at synapses with a genetically engineered reporter. eLife 8, e46421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.King S. M., Large-scale isolation of Chlamydomonas flagella. Methods Cell Biol. 47, 9–12 (1995). [DOI] [PubMed] [Google Scholar]
- 70.Witman G. B., Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280–290 (1986). [DOI] [PubMed] [Google Scholar]
- 71.Miller M. B., et al. , Brain region and isoform-specific phosphorylation alters kalirin SH2 domain interaction sites and calpain sensitivity. ACS Chem. Neurosci. 8, 1554–1569 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R. Luxmi, R.E. Mains, B.A. Eipper, S.M. King, Regulated processing and secretion of a peptide precursor in cilia. Dryad. 10.5601/dryad.fn2z34txn. Deposited 8 July 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All underlying data are included within the main text and the supporting information. Mass spectrometry data have been deposited in Dryad at https://doi.org/10.5601/dryad.fn2z34txn (72) .