Abstract
Transcription of the Azospirillum brasilense ipdC gene, encoding an indole-3-pyruvate decarboxylase involved in the biosynthesis of indole-3-acetic acid (IAA), is induced by IAA as determined by ipdC-gusA expression studies and Northern analysis. Besides IAA, exogenously added synthetic auxins such as 1-naphthaleneacetic acid, 2,4-dichlorophenoxypropionic acid, and p-chlorophenoxyacetic acid were also found to upregulate ipdC expression. No upregulation was observed with tryptophan, acetic acid, or propionic acid or with the IAA conjugates IAA ethyl ester and IAA-l-phenylalanine, indicating structural specificity is required for ipdC induction. This is the first report describing the induction of a bacterial gene by auxin.
Auxins constitute a class of phytohormones that play important roles in the coordination of plant growth and development. Indole-3-acetic acid (IAA), the most abundant naturally occurring auxin, has been implicated in regulating a variety of developmental and cellular processes such as cell extension, cell division, vascular differentiation, root formation, apical dominance, and tropisms (18). Regulation of these processes by auxin is believed to involve auxin-induced changes in gene expression (1, 31). Over the past 10 years, a number of plant genes that are transcriptionally induced by auxin, and that may play roles in one or more of these processes, have been cloned and further characterized (28, 31). The signal transduction pathways leading to the auxin-mediated gene induction in plants, however, are still not well understood (13, 26).
Besides plants, many soil and rhizosphere bacteria, including phytopathogenic, epiphytic, and plant growth-stimulating bacteria, also produce IAA. IAA biosynthesis in these bacteria has been shown to occur through different biosynthetic pathways (6, 20). By means of feeding experiments with tritiated putative IAA precursors, we have previously demonstrated the existence of multiple routes for IAA biosynthesis in the plant growth-promoting rhizobacterium Azospirillum brasilense (24). One of the pathways was identified as the indole-3-pyruvic acid (IPyA) pathway (l-tryptophan [Trp]→IPyA→indole-3-acetaldehyde→IAA) by cloning of the A. brasilense ipdC gene encoding an IPyA decarboxylase (5). In addition, two amino acid aminotransferases that catalyze the transamination of Trp, the first step of this pathway, were also purified (29). An A. brasilense ipdC knockout mutant was found to synthesize less than 10% of the level of wild-type IAA production, indicating that the IPyA decarboxylase is a key enzyme for IAA biosynthesis in this bacterium (24). In this study, we show that the expression of the A. brasilense ipdC gene is upregulated by IAA and other auxins.
Construction of the translational ipdC-gusA fusion pFAJ64.
To study the expression of the A. brasilense ipdC gene, we constructed a translational ipdC-gusA fusion. First, the promoter region of the A. brasilense ipdC gene was amplified by means of PCR with primers annealing approximately 350 bp upstream of the ipdC start codon (avbgltx1, 5′-CGCCGGATCCAAAGACGCCCATCAGGCGTC-3′) and at codons 1 to 7 of the ipdC gene (avbipdC1, 5′-AGGGAGATCTCACCCGGAATCCCGAACATG-3′). The proofreading Vent DNA polymerase (New England Biolabs, Beverly, Mass.) was used to increase the fidelity of DNA synthesis. The amplified fragment was flanked by BamHI (avbgltx1) and BglII (avbipdC1) recognition sites (underlined in the sequences of the primers). Following digestion with BamHI and BglII, the fragment was then cloned into the BamHI site of pFAJ1171, a pUC18 derivative containing the promoterless gusA gene from pBI101.3 (15) on a 2-kb BamHI-EcoRI fragment. The orientation of the insert in the transformants was determined by PCR with the primer avbgltx1 and an internal gusA primer (5′-GATTTCACGGGTTGGGGTTTCT-3′). The DNA sequence of the 350-bp promoter fragment was verified by DNA sequence analysis using the primers avbgltx1 and avbipdC1. Nucleotide sequence analysis using the internal gusA primer confirmed that the fusion was in frame. Finally, the BamHI-EcoRI restriction fragment of approximately 2.4 kb, carrying the entire ipdC-gusA fusion, was inserted in the corresponding sites of the broad-host-range plasmid pLAFR3 (30), yielding pFAJ64. pFAJ64 was then transferred to Escherichia coli S17.1, from which it was mobilized to A. brasilense Sp245 and to the Sp245 IpdC− derivative FAJ009.
Expression of ipdC is cell density dependent.
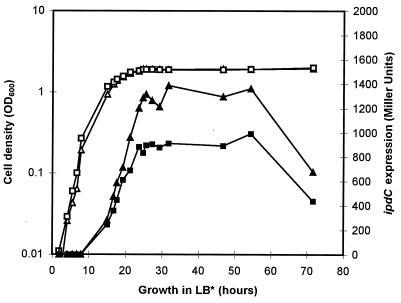
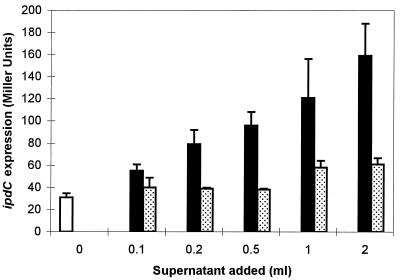
It was observed previously that the level of IAA biosynthesis in A. brasilense varies during bacterial growth and that the highest IAA production level is obtained during the stationary-growth phase (24). In a first experiment, expression of the ipdC-gusA fusion, pFAJ64, in the wild-type strain Sp245 and in the IpdC− mutant FAJ009 was measured quantitatively during growth in complex medium (Luria-Bertani medium supplemented with 2.5 mM MgSO4 and 2.5 mM of CaCl2 [LB* broth]). As shown in Fig. 1, expression of the ipdC-gusA fusion in both strains clearly increases with cell density and reaches its maximum when the cells enter the stationary-growth phase. Strikingly, the maximum level of gus expression in FAJ009 was found to be reduced to approximately 60% of that of the wild-type level. Growth phase and cell density-dependent induction of bacterial genes have often been shown to be mediated by small diffusible signal molecules. In E. coli, for instance, accumulation of weak acids in stationary-phase cultures serves as a signal to activate the expression of the sigma factor, RpoS, which in turn is required for the activation of other growth phase-specific sets of genes (27). Additionally, a variety of cellular processes in bacteria, including bioluminescence in Vibrio fischeri (4, 7, 9), production of exoenzymes and antibiotics in Erwinia carotovora (2, 16), and plasmid conjugal transfer in Agrobacterium tumefaciens (22, 34), are switched on by diffusible compounds, termed autoinducers and identified as N-acyl homoserine lactones (AHLs). Activation of the specific target genes occurs when a required threshold of AHLs is attained and in this way is only achieved at high cell densities. Therefore, the observed growth phase-dependent expression of the A. brasilense ipdC gene prompted us to investigate whether an extracellular product accumulating in spent medium is involved in ipdC induction. To test this possibility, different amounts of filter-sterilized culture supernatant (0.22-μm-pore size) obtained from a stationary-growth-phase culture of A. brasilense Sp245 were added to exponentially growing cells of Sp245(pFAJ64), and β-glucuronidase activity was determined after 2.5 h of additional growth. As can be concluded from Fig. 2, ipdC expression increases with increasing amounts of the stationary-phase culture supernatant. A twofold dilution of the reporter strain (2 ml of culture plus 2 ml of supernatant) resulted in a fivefold increase in β-glucuronidase activity compared to the activity detected in control cultures to which no supernatant was added.
FIG. 1.
Growth and expression of ipdC in A. brasilense Sp245 and in the IpdC− mutant FAJ009. Cultures of Sp245(pFAJ64) and FAJ009(pFAJ64) were grown in LB* broth. Periodically, samples were taken to determine the expression of the ipdC-gusA fusion, pFAJ64. Quantitative analysis of β-glucuronidase activity was carried out in microtiter plates with p-nitrophenyl-β-d-glucuronide as substrate (14). Growth was monitored by measuring the OD600. The data presented are from one representative experiment, which was confirmed twice in additional independent experiments. Triangles, Sp245(pFAJ64); squares, FAJ009(pFAJ64); open symbols, cell density (OD600); solid symbols, β-glucuronidase activity in Miller units (17).
FIG. 2.
Induction of the ipdC-gusA fusion by spent culture supernatants. An exponential-phase culture of Sp245(pFAJ64) (OD600 = 0.6) grown in LB* broth was centrifuged and resuspended in fresh LB* broth. Two-milliliter samples of this culture were then transferred to sterile test tubes, supplemented with different amounts of spent culture supernatant (ranging from 0 to 2 ml), and diluted up to a total volume of 4 ml with fresh LB* broth. After 2.5 h of growth at 30°C, the cultures were assayed for β-glucuronidase activity as detailed in the legend of Fig. 1. The spent culture supernatants were obtained from late-stationary-phase cultures (OD600 = 2.0) of Sp245 (solid bars) or FAJ009 (stippled bars) grown in LB* broth. Open bar, control culture to which no supernatant was added. All data are the means from three replicates. Error bars denote the standard deviations.
When this experiment was repeated with spent medium of a stationary-phase culture of the IpdC− strain FAJ009, only a slight effect on ipdC expression was observed (Fig. 2). Only when large amounts of supernatant of a FAJ009 culture were applied was pFAJ64 induction significantly greater than in the control cultures.
Expression of ipdC is upregulated by IAA.
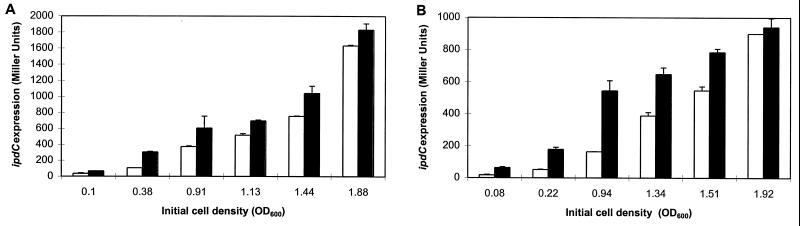
The difference in ipdC expression levels between the wild type and the IpdC− mutant and the observation that spent culture supernatant of the IpdC− mutant does not efficiently induce ipdC expression led us to speculate that the end product of the biosynthetic pathway, IAA, could be the inducing compound. It has been shown previously that IAA accumulates in the supernatant of wild-type culture (up to 200 μM in a stationary-growth-phase culture in LB* broth) but is reduced to 10% of the wild-type level in the supernatant of the IpdC− mutant (24). The idea of IAA as inducing compound was further tested by examining the effect of exogenously added IAA on ipdC expression. Sp245(pFAJ64) and FAJ009(pFAJ64) cultures at different growth phases (starter cultures) were supplemented with 1 mM IAA and, after 3.5 h of additional growth, compared for ipdC-gusA induction with cultures to which no IAA was added. As can be concluded from Fig. 3, in all growth phases, both for Sp245 and FAJ009, a higher expression level of pFAJ64 was found in cells exposed to exogenously added IAA compared to the level in untreated cells. As expected, upregulation by exogenously added IAA (i.e., the difference in the expression levels between a culture supplemented with IAA and an untreated culture) was most obvious in the IpdC− mutant background and was growth phase dependent. The difference in the extent of upregulation at the various growth phases and between the mutant and the wild type may be due to differences in endogenous IAA levels in the starter cultures. For Sp245, a threefold upregulation was detected in early-exponential-phase cells (optical densities at 600 nm [OD600] of 0.1 and 0.4). Upregulation then gradually decreased in the mid and late-exponential phases (1.4-fold increase at OD600 = 1.1 and 1.4) and stationary-phase cultures (1.1 fold increase at OD600 = 1.9) because of the higher endogenous IAA production in the corresponding starter cultures. In contrast to Sp245 and concomitant with the lower IAA production capacity of the IpdC− mutant, a fourfold upregulation could still be detected in late-exponential-phase FAJ009 culture (OD600 = 0.9). The lowest concentration of exogenously added IAA for which significant ipdC induction was observed in an exponential-phase culture of FAJ009 was 10 μM (Table 1).
FIG. 3.
Effect of exogenously added IAA on ipdC expression in Sp245 and the IpdC− mutant, FAJ009. To obtain cultures at various cell densities of Sp245(pFAJ64) (A) and FAJ009(pFAJ64) (B) different flasks containing 100 ml of LB* medium were inoculated with different volumes of preculture and grown overnight at 30°C. From each culture, six samples of 3 ml were transferred to sterile test tubes. Three test tubes of each series were supplemented with IAA to a final concentration of 1 mM (solid bars); to the remaining three test tubes no IAA was added (open bars). After 3.5 h of additional growth at 30°C, β-glucuronidase activity was assayed as detailed in the legend of Fig. 1. Data are the means from the three replicates. Error bars denote the standard deviations.
TABLE 1.
Effect of auxins on expression of translational gusA fusions with either ipdC (pFAJ64) or a constitutive A. brasilense promoter (pFAJ31.13)
| Substrate added (final concn)a | Mean β-glucuronidase activityb in:
|
|
|---|---|---|
| FAJ009(pFAJ64) | FAJ009(pFAJ31.13) | |
| None | 152 (5) | 296 (31) |
| IAA (1 mM) | 466 (18) | 304 (26) |
| IAA (0.1 mM) | 353 (15) | 261 (24) |
| IAA (0.01 mM) | 222 (30) | NDc |
| NAA (1 mM) | 439 (14) | 252 (17) |
| NAA (0.1 mM) | 367 (8) | 254 (7) |
| 2,4-DP (1 mM) | 473 (28) | ND |
| 4-CPA (1 mM) | 509 (39) | ND |
| IBA (1 mM) | 137 (10) | ND |
| Trp (1 mM) | 103 (6) | ND |
| Acetic acid (1 mM) | 154 (8) | ND |
| Propionic acid (1 mM) | 77 (7) | ND |
| IAA ethyl ester (1 mM) | 113 (23) | ND |
| IAA-L-phenylalanine (1 mM) | 175 (26) | ND |
Samples (3 ml) of exponential-phase cultures of FAJ009(pFAJ64) and FAJ009(pFAJ31.13) (OD600 = 0.4) grown in LB* broth were transferred to sterile test tubes and supplemented with the various substrates to the final concentrations indicated in parentheses. After 4 h of additional growth at 30°C, the cultures were assayed for β-glucuronidase activity as detailed in the legend of Fig. 1.
Standard deviations of three replicates are given in parentheses.
ND, not determined.
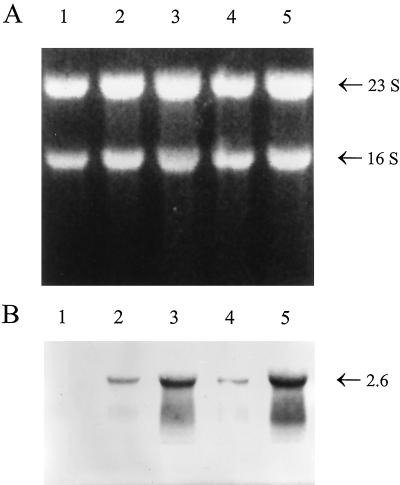
The observed induction of the ipdC-gusA fusion in cells growing in the presence of IAA was confirmed by quantitative Northern analysis. RNA of Azospirillum cells was isolated essentially as described by Eggermont et al. (8). RNA concentrations were determined fluorimetrically after staining with SYBR Green II in accordance with the manufacturer’s protocols (Molecular Probes). A total of 10 μg of each RNA sample was separated on a formaldehyde-agarose gel, transferred to a positively charged nylon membrane (Boehringer Mannheim), and hybridized as described previously (8). To verify equal loading and transfer of RNA, the loading buffer was supplemented with 50 μg of ethidium bromide/ml, allowing visualization of RNA in the gel and on the blot when illuminated with UV light (8) (Fig. 4A). To prepare the ipdC riboprobe, a 1.8-kb SmaI fragment containing approximately 200 bp of the ipdC promoter and almost the entire coding region was cloned into pBlueScriptIISK+ vector (Stratagene). The digoxigenin-labeled antisense transcript was then prepared by in vitro runoff transcription with the Dig RNA labeling kit from Boehringer Mannheim. Figure 4B shows the presence of a single transcript of approximately 2.6 kb hybridizing with the ipdC probe in RNA isolated from wild-type cells, which corresponds to the predicted size of a putative dicistronic mRNA product of the ipdC gene (1,635 bp) and a downstream located open reading frame (ORF) of 669 bp. The deduced amino acid sequence of this ORF is homologous to the E. coli anti-sigma cross-reacting protein (SCRP-27A [32]) (57% identity, unpublished results). The message was absent in the IpdC− mutant, proving the specificity of the ipdC probe (Fig. 4B, lane 1). In agreement with the results of the ipdC-gusA expression analysis, the level of ipdC mRNA in wild-type cells increased during growth (Fig. 4B, lanes 2 and 3) and was clearly higher in cells grown in the presence of exogenously added IAA (Fig. 4B, lanes 4 and 5).
FIG. 4.
Northern blot analysis of RNA extracted from cells of A. brasilense wild-type and IpdC− mutant strains. (A) Blot of total RNA (10 μg per lane) isolated from A. brasilense IpdC− mutant (lane 1) and wild-type strains (lanes 2 to 5). RNA was visualized by epi-illumination with UV light. Cells were harvested at an OD600 of 0.9 (lanes 1 and 3), 0.5 (lane 2), or 0.4 (lanes 4 and 5). To measure the effect of IAA on the transcription of the ipdC gene, the wild-type culture at OD600 of 0.4 was divided into two and further incubated for 3 h at 30°C in the absence (lane 4) or presence of exogenously added IAA at 1 mM (lane 5). The positions of the 16S and 23S ribosomal RNAs are indicated at the right. (B) RNA blot analysis of the same samples as in panel A after hybridization with a digoxygenin-labeled ipdC-derived riboprobe. The size of the ipdC transcript (in kilobases) is indicated at the right and was determined relative to RNA standards that were electrophoresed in the same gel.
Different auxins induce ipdC expression.
IAA-inducible genes in higher plants have been shown to respond to, besides IAA, synthetic auxins such as naphthaleneacetic acid (NAA) and chlorophenoxy acids (28). To evaluate whether the A. brasilense ipdC gene could also be induced by other auxins, exponential-growth-phase cultures of FAJ009(pFAJ64) (OD600 = 0.4) were supplemented with the naturally occurring auxin indole-3-butyric acid (IBA) or with the synthetic auxins NAA, 2,4-dichlorophenoxypropionic acid (2,4-DP), or p-chlorophenoxyacetic acid (4-CPA) and compared for ipdC induction. As indicated by the data in Table 1, the three tested synthetic auxins, NAA, 2,4-DP, and 4-CPA, were found to upregulate ipdC expression. Under the conditions tested, induction of the ipdC-gusA fusion by all three synthetic auxins was as strong as the induction observed with IAA. In contrast, addition of IBA to the cultures did not affect ipdC expression. In plants, the conversion of IBA to IAA has been reported (10, 11), and it is not yet clear whether IBA is itself an auxin or whether it exerts its auxin activity through its conversion to IAA (3). The observation that the A. brasilense ipdC gene is not upregulated by IBA might therefore be attributed to the absence of enzymes catalyzing the conversion of IBA into IAA in Azospirillum. The addition of the two IAA conjugates, IAA ethyl ester and IAA-l-phenylalanine, to the FAJ009(pFAJ64) culture had no effect on ipdC expression (Table 1). This might be due to the lack of intracellular transformation of these compounds into active auxins, although it cannot be excluded that these conjugates fail to induce ipdC gene expression because they are not taken up by A. brasilense cells. Treatment of the FAJ009(pFAJ64) culture with similar concentrations of Trp, acetic acid, and propionic acid also did not enhance ipdC induction (Table 1), indicating that the upregulation of ipdC expression by IAA and the synthetic auxins is auxin specific and not only caused by the acid group of the auxin molecules. In control experiments, incubation of FAJ009 containing pFAJ31.13, a pLAFR1 derivative carrying a fusion between a constitutive A. brasilense promoter and gusA (33), with IAA or NAA did not affect β-glucuronidase activity (Table 1).
General conclusion.
Evidence is presented here for the upregulation of expression of the A. brasilense IAA biosynthetic gene, ipdC, by IAA and synthetic auxins. This positive feedback regulation by IAA is responsible for the increasing ipdC transcription levels during growth of an A. brasilense culture, with the highest expression level of the ipdC gene observed in the stationary-growth phase. Reduced IAA accumulation in stationary-phase cells of the IpdC− mutant strain results in a lower ipdC expression maximum in this strain compared to that of the wild type. In contrast to negative transcription control of biosynthetic genes by the end product formed (19, 23), autoinduction of a biosynthetic pathway is rather exceptional. Other genes, besides the A. brasilense ipdC gene, that have been demonstrated to be autoinduced are the AHL biosynthetic genes found in various gram-negative bacteria (12) and the genes involved in the production of the siderophores yersiniabactin in Yersinia enterocolitica (21) and pyochelin in Pseudomonas aeruginosa (25). Until now auxin-responsive genes have only been identified in plants (1, 28, 31). The role of many of these auxin-inducible genes in plant developmental processes is not yet fully understood. This study presents data indicating for the first time the induction of a bacterial gene by auxin. This finding suggests that proteins involved in IAA perception and signal transduction are present not only in higher plants but also in bacteria.
Acknowledgments
A.V.B. and M.L. are recipients of post- and predoctoral fellowships of the Fund of Scientific Research-Flanders, respectively. We acknowledge financial support from K. U. Leuven (GOA [J.V.]), the Fund of Scientific Research-Flanders, and the Ministry of Agriculture of Belgium.
We thank B. Thomma and I. Nagy for advice concerning the Northern analysis and D. Haas for helpful discussions.
REFERENCES
- 1.Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bainton N J, Bycroft B W, Chhabra S R, Stead P, Gledhill L, Hill P J, Rees C E D, Winson M K, Salmond G P C, Stewart G S A B, Williams P A. A general role for the lux autoinducer in bacterial cell signalling: control of antibiotic biosynthesis in Erwinia. Gene. 1992;116:87–91. doi: 10.1016/0378-1119(92)90633-z. [DOI] [PubMed] [Google Scholar]
- 3.Bartel B. Auxin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:51–66. doi: 10.1146/annurev.arplant.48.1.51. [DOI] [PubMed] [Google Scholar]
- 4.Choi S H, Greenberg E P. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J Bacteriol. 1992;174:4064–4069. doi: 10.1128/jb.174.12.4064-4069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costacurta A, Keijers V, Vanderleyden J. Molecular cloning and sequence analysis of an Azospirillum brasilense indole-3-pyruvate decarboxylase gene. Mol Gen Genet. 1994;243:463–472. doi: 10.1007/BF00280477. [DOI] [PubMed] [Google Scholar]
- 6.Costacurta A, Vanderleyden J. Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol. 1995;21:1–18. doi: 10.3109/10408419509113531. [DOI] [PubMed] [Google Scholar]
- 7.Eberhard A, Burlingame A L, Eberhard C, Kenyon G L, Nealson K H, Oppenheimer N J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 8.Eggermont K, Goderis I J, Broekaert W F. High-throughput RNA extraction from plant samples based on homogenisation by reciprocal shaking in the presence of a mixture of sand and glass beads. Plant Mol Biol Rep. 1996;14:273–279. [Google Scholar]
- 9.Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 10.Epstein E, Lavee S. Conversion of indole-3-butyric acid to indole-3-acetic acid in cuttings of grapevine (Vitis vinifera) and olive (Olea europea) Plant Cell Physiol. 1984;25:697–703. [Google Scholar]
- 11.Epstein E, Ludwig-Müller J. Indole-3-butyric acid in plants: occurrence, synthesis, metabolism and transport. Physiol Plant. 1993;88:382–389. [Google Scholar]
- 12.Fuqua W C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 13.Guilfoyle T J. Aux/IAA proteins and auxin signal transduction. Trends Plant Sci. 1998;6:205–207. [Google Scholar]
- 14.Jefferson R A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 15.Jefferson R A. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones S, Yu B, Bainton N J, Birdsall M, Bycroft B W, Chhabra S R, Cox A J R, Golby P, Reeves P J, Stephens S, Winson M K, Salmond G P C, Stewart G S A B, Williams P. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 354–358. [Google Scholar]
- 18.Napier R M, Venis M A. Auxin action and auxin-binding proteins. New Phytol. 1995;129:167–201. doi: 10.1111/j.1469-8137.1995.tb04291.x. [DOI] [PubMed] [Google Scholar]
- 19.Neuhard J, Kelln R A. Biosynthesis and conversion of pyrimidines. In: Curtiss III R, Ingraham J L, Lin E C C, Brooks Low K, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella, cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 580–599. [Google Scholar]
- 20.Patten C L, Glick B R. Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol. 1996;42:207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- 21.Pelludat C, Rakin A, Jacobi C A, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piper K R, Beck von Bodman S, Farrand S K. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature. 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 23.Pittard A J. Biosynthesis of aromatic amino acids. In: Curtiss III R, Ingraham J L, Lin E C C, Brooks Low K, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella, cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 458–484. [Google Scholar]
- 24.Prinsen E, Costacurta A, Michiels K, Vanderleyden J, Van Onckelen H. Azospirillum brasilense indole-3-acetic acid biosynthesis: evidence for a non-tryptophan dependent pathway. Mol Plant-Microbe Interact. 1993;6:609–615. [Google Scholar]
- 25.Reimmann C, Serino L, Beyeler M, Haas D. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology. 1998;144:3135–3148. doi: 10.1099/00221287-144-11-3135. [DOI] [PubMed] [Google Scholar]
- 26.Rouse D, Mackay P, Stirnber P, Estelle M, Leyser O. Changes in auxin response from mutations in an AUX/IAA gene. Science. 1998;279:1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- 27.Schellhorn H E, Stones V L. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J Bacteriol. 1992;174:4769–4776. doi: 10.1128/jb.174.14.4769-4776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sitbon F, Perrot-Rechenmann C. Expression of auxin-regulated genes. Physiol Plant. 1997;100:443–455. [Google Scholar]
- 29.Soto-Urzua L, Xochinua-Corona Y G, Flores-Encarnacion M, Baca B E. Purification and properties of aromatic amino acid aminotransferases from Azospirillum brasilense UAP 14 strain. Can J Microbiol. 1996;42:294–298. doi: 10.1139/m96-043. [DOI] [PubMed] [Google Scholar]
- 30.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi Y, Ishida S, Nagata T. Auxin-regulated genes. Plant Cell Physiol. 1995;36:383–390. doi: 10.1093/oxfordjournals.pcp.a028965. [DOI] [PubMed] [Google Scholar]
- 32.Ueshima R, Fujita N, Ishihama A. Identification of Escherichia coli proteins cross-reacting with antibodies against region 2.2. peptide of RNA polymerase sigma subunit. Biochem Biophys Res Commun. 1992;184:634–639. doi: 10.1016/0006-291x(92)90636-y. [DOI] [PubMed] [Google Scholar]
- 33.Vande Broek A, Michiels J, Van Gool A, Vanderleyden J. Spatial-temporal colonization patterns of Azospirillum brasilense on the wheat root surface and expression of the bacterial nifH gene during association. Mol Plant-Microbe Interact. 1993;6:592–600. [Google Scholar]
- 34.Zhang L, Murphy P J, Kerr A, Tate M E. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]