Significance
Understanding the immunobiology of multiple sclerosis remains an unresolved challenge. Distinctive features of therapeutic response to effective treatments are one important source of insight into the underlying pathomechanisms. Here, we studied dimethyl fumarate, a routinely administered but mechanistically poorly understood compound. We developed an approach using multipanel immunophenotyping by mass cytometry in conjunction with a weakly supervised machine-learning algorithm to facilitate unprejudiced identification of rare but specific immune cell populations accounting for beneficial treatment response. This approach allowed identification of a subset of antigen-experienced T helper cells that are preferentially depleted in responding individuals. As a biomarker, this subset may guide clinical stratification.
Keywords: multiple sclerosis, mass cytometry, dimethyl fumarate, response marker, biomarker
Abstract
Dimethyl fumarate (DMF) is an immunomodulatory treatment for multiple sclerosis (MS). Despite its wide clinical use, the mechanisms underlying clinical response are not understood. This study aimed to reveal immune markers of therapeutic response to DMF treatment in MS. For this purpose, we prospectively collected peripheral blood mononuclear cells (PBMCs) from a highly characterized cohort of 44 individuals with MS before and at 12 and 48 wk of DMF treatment. Single cells were profiled using high-dimensional mass cytometry. To capture the heterogeneity of different immune subsets, we adopted a bioinformatic multipanel approach that allowed cell population–cluster assignment of more than 50 different parameters, including lineage and activation markers as well as chemokine receptors and cytokines. Data were further analyzed in a semiunbiased fashion implementing a supervised representation learning approach to capture subtle longitudinal immune changes characteristic for therapy response. With this approach, we identified a population of memory T helper cells expressing high levels of neuroinflammatory cytokines (granulocyte–macrophage colony-stimulating factor [GM-CSF], interferon γ [IFNγ]) as well as CXCR3, whose abundance correlated with treatment response. Using spectral flow cytometry, we confirmed these findings in a second cohort of patients. Serum neurofilament light-chain levels confirmed the correlation of this immune cell signature with axonal damage. The identified cell population is expanded in peripheral blood under natalizumab treatment, substantiating a specific role in treatment response. We propose that depletion of GM-CSF–, IFNγ-, and CXCR3-expressing T helper cells is the main mechanism of action of DMF and allows monitoring of treatment response.
Dimethyl fumarate (DMF) exerts beneficial immunomodulatory effects in several autoimmune conditions and is frequently used for treatment in relapsing–remitting multiple sclerosis (RRMS) (1–3). Various, partially conflicting (4) studies reported alterations in almost all leukocyte populations under therapy, including T cells (5–9), B cells (10–13), and antigen-presenting myeloid cells (6, 14). In contrast to recently developed therapies with designated immune targets, the mode of action of DMF in MS remains poorly understood. To elucidate which aspects of the drug’s complex effects on the immune system are most relevant to its therapeutic effect in MS, we made a detailed comparison between patients with good and poor response to DMF treatment. We developed a deep immunophenotyping approach based on mass cytometry in conjunction with a weakly supervised machine-learning algorithm (15) to characterize the longitudinal effect of DMF treatment on the immune system of MS patients and to elucidate the immune substrate of treatment response.
Results
DMF Modifies the Immune System of MS Patients.
To study the immunological effects of DMF in MS, we prospectively collected clinical information from 31 DMF-treated patients (hereafter “mass cytometry cohort”) and peripheral blood mononuclear cells (PBMCs) at three time points (T1, baseline; T2, 12 wk of treatment; T3, 48 wk of treatment). Among these patients, we distinguished responders without radiological or clinical signs of inflammatory disease activity from nonresponders, whose MRI revealed new or gadolinium-enhancing lesions at T3 and/or who experienced clinical relapses (for the study design, see Fig. 1A). Characteristics of this cohort are listed in SI Appendix, Table S1.
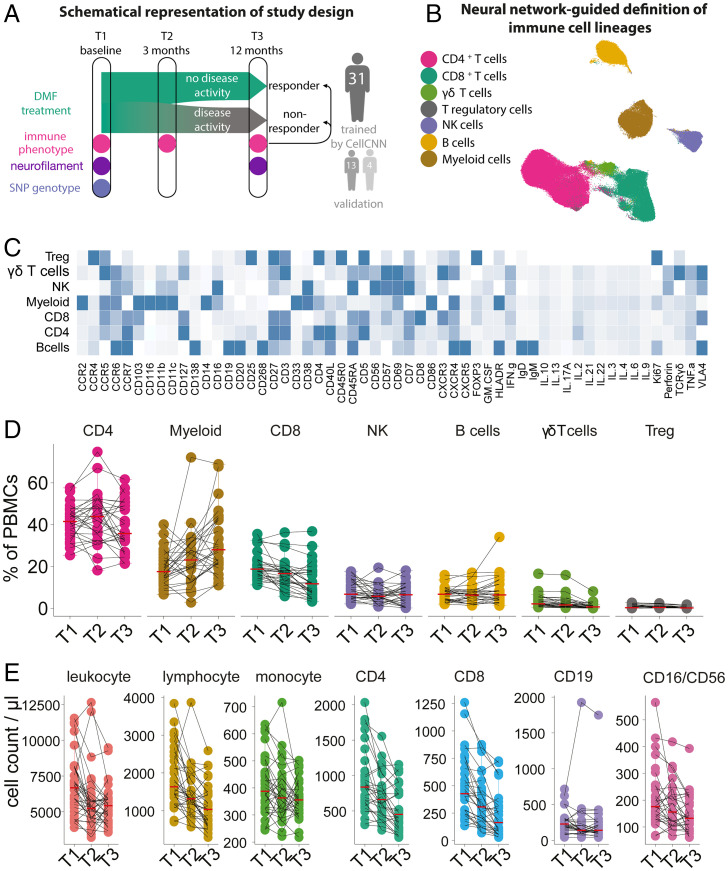
Fig. 1.
DMF treatment changes the peripheral immune profile of MS. (A) Blood samples and clinical data from a cohort of MS patients (n = 31, as well as two validation cohorts) before the initiation of DMF therapy and at 3- and 12-mo follow-up were longitudinally collected and analyzed for immune profile by mass cytometry and SNPs by DNA microarray. (B) UMAP representation of analyzed peripheral blood immune cells with overlaid color code as categorized by FlowSOM-defined clusters. (C) Immune compartments as defined by FlowSOM-defined clusters. Mean expression profile of all analyzed parameters in each defined population. (D) Longitudinal frequencies of the immune populations in blood in relative numbers. (E) Longitudinal cell counts of the main immune populations in blood in absolute numbers.
The first analysis aimed to characterize the effect of DMF therapy on the phenotype of PBMCs. Using two complementary, partially overlapping antibody panels (“stimulation panel” with phorbol 12-myristate 13-acetate [PMA]/ionomycin stimulation, and “conventional panel” without stimulation) for mass cytometry, we measured 54 characteristic features, including markers of lineage, activation, and maturation, transcription factors, chemokine receptors, and cytokines (SI Appendix, Tables S2 and S3). To capture the intercluster phenotypic identity, we exploited the partially overlapping design of our cytometry by time of flight (CyTOF) panels and the neural network–based algorithm of FlowSOM (16) to delineate the major immune populations (Fig. 1 B and C). This multipanel approach was further used to build a uniform manifold approximation and projection (UMAP) representation of the identified cell populations (Fig. 1B and all markers in SI Appendix, Fig. S1). We then analyzed the effect of DMF treatment and found a strong effect on the T cell compartment, with a sustained reduction in the frequency of CD8 (P = 0.0001) and γδ-T cells (P = 0.0001) already 3 mo after treatment initiation. A significant reduction of CD4 T cells and T regulatory cells (Tregs) was evident only after 1 y of treatment (P = 0.039 and P < 0.0001, respectively). Conversely, we observed an increased frequency of myeloid cells at each time point after the initiation of DMF. Natural killer (NK) and B cell frequencies were only minimally affected (Fig. 1D).
In addition to relative frequencies, we also examined the absolute numbers of the various cell types over time (Fig. 1E). As expected, DMF led to a decrease in total leukocyte and lymphocyte counts, with a strong reduction in CD4 and CD8 T cells (T1 vs. T2, P = 0.0004 and P = 0.00005, respectively). B cells also decreased (T1 vs. T2, P = 0.011), while NK cells and monocytes were minimally affected (T1 vs. T2, P = 0.050 and P = 0.067, respectively), indicating that the increase in their relative frequencies can largely be ascribed to a general contraction of the lymphocyte compartment.
Complex Rearrangement of Mature Lymphocytes Marks DMF Treatment.
Multiple cell types have been implicated in MS pathophysiology, especially T and B lymphocytes (17, 18). To determine the specific modulation pattern exerted by DMF on lymphocyte subsets, we mapped the longitudinal changes of cell density in a UMAP representation of T and B cells (for marker expression, see SI Appendix, Fig. S2A). In this map, the most drastically reduced cell sets clustered with the expression of CD45RO, while increases were seen in areas with CD45RA, CCR7, and CD27 expression (Fig. 2 A–C). The degree of change within these populations, however, was heterogeneous, suggesting a more complex rearrangement of lymphocyte populations (for longitudinal comparisons of T1 vs. T2 and T2 vs. T3, see SI Appendix, Fig. S2 B and C). We further characterized these alterations in T and B cell composition by canonical maturation patterns. After unsupervised clustering of T cells based on FlowSOM nodes, we defined subsets for naïve (Tn), effector memory (Tem), central memory (Tcm), and terminal effector (Teff) cells based on their expression profile and compared their frequencies. DMF profoundly affected the memory compartment, inducing a strong reduction in the frequencies of both Tem (P = 0.01) and Tcm (P = 0.02) already after 3 mo of treatment as compared with baseline. Conversely, Tn showed a significant relative increase (P = 0.002) (Fig. 2D and SI Appendix, Fig. S3 A–E). A similar pattern was observed in cytotoxic T (Tc) cells (SI Appendix, Fig. S3F). Analogously, we selected subsets of B cells based on their expression of maturation markers: naïve, immunoglobulin M (IgM) memory, class-switched memory, transitional, and plasma cells. We observed a significant decrease in both IgM (P = 0.009) and class-switched (P = 0.00004) memory B cells, with a concomitant increase in the naïve compartment (P = 0.0006). Interestingly, neither transitional nor plasma cells were significantly affected by the therapy (Fig. 2D and SI Appendix, Fig. S3 G and H). In conclusion, both B cell and T cell compartments undergo a drastic decrease of mature populations under DMF treatment.
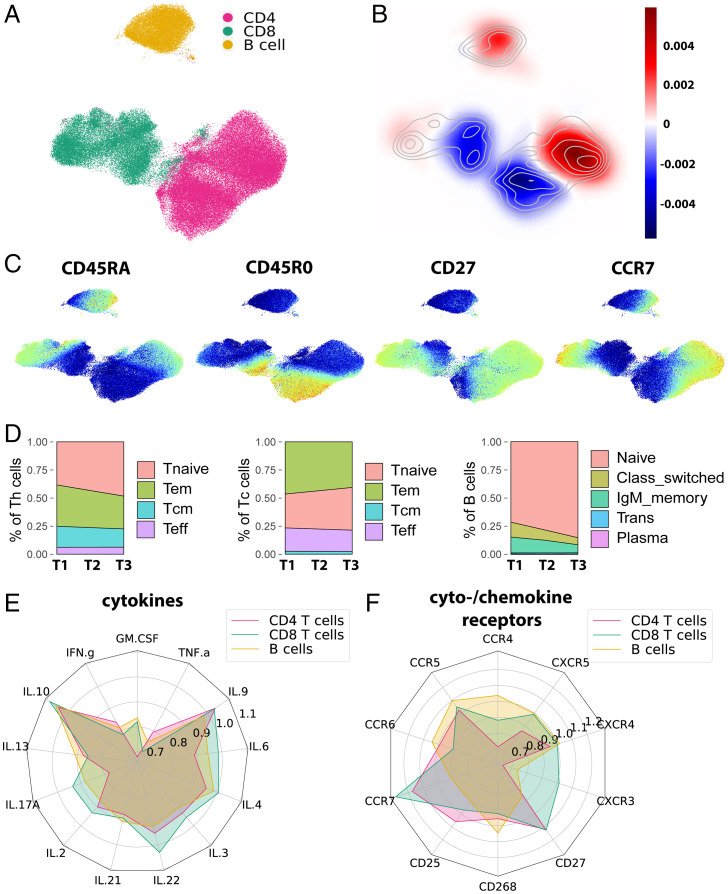
Fig. 2.
DMF treatment primarily affects mature T and B cells. (A) UMAP representation of B cell and Th and Tc cell subsets with overlaid color code as categorized by FlowSOM-defined clusters. (B) The change of density (blue indicating decrease and red indicating increase during follow-up) within the UMAP representation was computed via a Gaussian kernel density estimation for change between T1 and T3. (C) Expression maps of representative markers of lymphocyte maturation in the UMAP representation. (D) Frequencies of established maturation cell types of Th cells, Tc cells, and B cells at time points T1 to T3 visualized in area plots. (E) Radar chart depicting the change of mean cytokine expression (T3/T1) in each cell type for CD4 T cells, CD8 T cells, and B cells. (F) Radar chart depicting the change of mean cytokine and chemokine receptor expression (T3/T1) in each cell type for CD4 T cells, CD8 T cells, and B cells.
We then deepened the analysis of the most affected cell populations to include all measured markers. When assessing the longitudinal profile of T cells by distinct cytokines, granulocyte–macrophage colony-stimulating factor (GM-CSF), tumor necrosis factor α (TNFα), and interferon γ (IFNγ) were identified as the most strongly decreased cytokines (Fig. 2E and SI Appendix, Fig. S4). The expression of the chemokine receptors CXCR3 and CCR4 specifically decreased in Th cells under DMF treatment, contrasting with a rather slight decrease in CXCR4 and CXCR5 and a minor effect in Tc cells (Fig. 2F and SI Appendix, Fig. S4 D and E). Analogously, we assessed the influence of DMF on B cell composition. TNFα was the most frequently produced cytokine and most strongly decreased at 12 mo (Fig. 2E). The CXCR3 expression in B cells was significantly reduced after 3 and 12 mo of DMF therapy, while CXCR4 and CXCR5 were not significantly affected (Fig. 2F and SI Appendix, Fig. S4F). In summary, superimposed on the global reduction of lymphocytes with a memory phenotype, DMF selectively reshapes the cytokine polarization of T and B cells.
CellCnn Identifies a TNFα/GM-CSF/CXCR3 Effector-Memory T Cell Signature of DMF Treatment.
Given the complex impact of DMF on the immune system, we employed a weakly supervised machine-learning approach in order to identify the immune features most strongly associated with the effect of DMF. We utilized the CellCnn algorithm (15) to capture the longitudinal changes in our dataset, comparing T1 and T3. This approach was independently applied to the data obtained from both panels (Fig. 3 A–C). Both analyses identified an effector-memory population of mainly T helper cells (with minor contributions of Tc and γδ-T cells) (Fig. 3D). This treatment-associated phenotype featured high levels of GM-CSF and TNFα (stimulation panel, Fig. 3E) as well as CXCR3 (conventional panel, Fig. 3F) (for a complete panel of all markers, see SI Appendix, Fig. S5 A and B). When comparing the frequencies of the CellCnn-selected cells, we observed a dramatic reduction (P < 0.0001, stimulation panel; P = 0.0002, conventional panel) upon DMF therapy. This decrease developed over T1 to T2 and T2 to T3. To confirm that both panels identified a concordant cell population, we compared them regarding a set of 17 common markers by silhouette analysis (Fig. 3C). The calculated silhouette score of 0.008 identifies no separation between the two populations, as also indicated in the UMAP projection (Fig. 3A). To validate the specificity of the identified phenotype, we independently compared T2 and T3 in both panels, which resulted in analogous results (silhouette score: T1 vs. T3 and T2 vs. T3 for the conventional panel, 0.008; for the stimulation panel, 0.02; SI Appendix, Fig. S5 C and D). In conclusion, the analysis of both surface and intracellular markers of immune cells consistently identifies a circumscribed Tem population as the central target of DMF treatment.
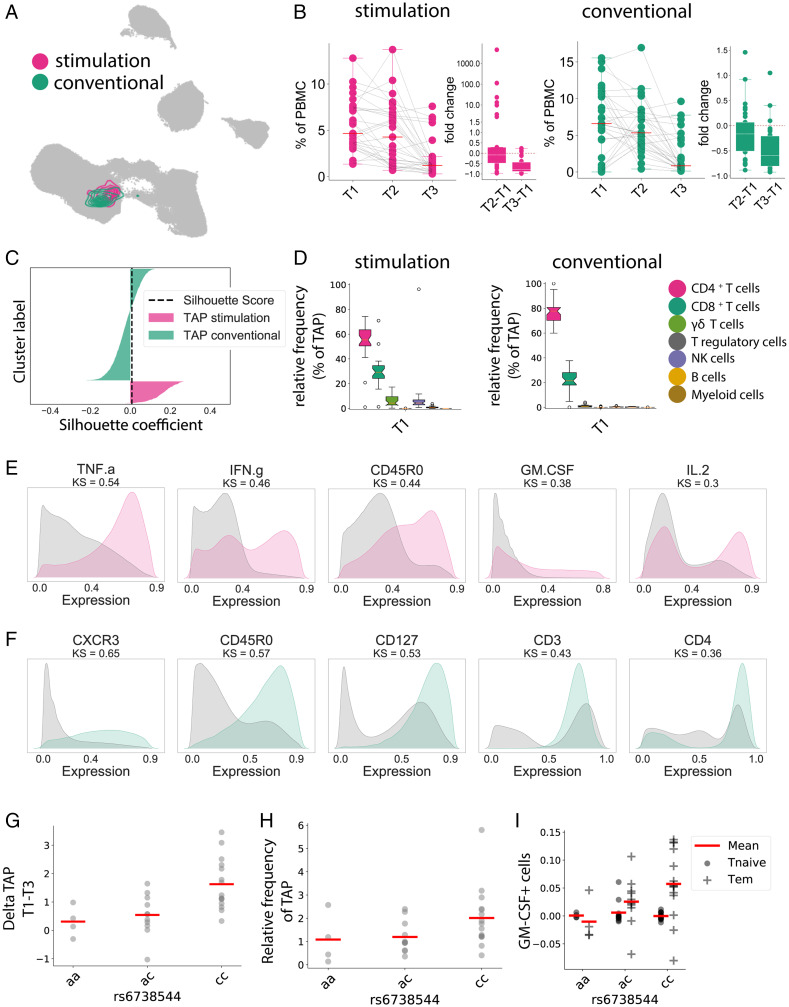
Fig. 3.
Automated multipanel analysis with CellCnn identifies an inflammatory Tem phenotype associated with DMF therapy. CellCnn analysis determined the core immune features between T1 and T3 from our patient cohort, termed the “treatment-associated phenotype” (TAP). (A) The UMAP algorithm was used to depict the identified cell filter in the conventional (green) and stimulated (red) panels. (B) Frequency of selected cells within the Th compartment at T1, T2, and T3, and the relative fold changes compared with baseline. (C) The overlap between both cell filters is estimated using silhouette analysis. The black dashed line represents the silhouette score. (D) Frequency of selected cell types within the treatment-associated phenotype at T1 in the stimulated panel and in the conventional panel. (E) Expression patterns of the five key discriminant markers between the treatment-associated phenotype and the reference cell population for the stimulated panel. Distance between patterns for each marker quantified by the Kolmogorov–Smirnov (KS) test. (F) Expression patterns of the five key discriminant markers between the treatment-associated phenotype and the reference cell population for the conventional panel. (G) Frequency change of the treatment-associated population between T1 and T3 in correlation with alleles of rs6738544. (H) Frequency of the treatment-associated population at T1 in correlation with alleles of rs6738544. (I) Frequency change of GM-CSF expression in Tem and Tn between T1 and T3 in correlation with alleles of rs6738544.
Since this immune modulation by DMF varied between patients, we aimed to identify potentially underlying genetic predispositions. For this purpose, we characterized our cohort regarding 198 single-nucleotide polymorphisms (SNPs) associated with MS (19). When polymorphisms were assessed for their correlation with change in the identified treatment-associated population, rs6738544 showed the highest association (P = 0.0009; Bonferroni correction 0.099; false discovery rate 0.089) (Fig. 3G). The disease-associated c allele of this intronic locus in the STAT4 gene was associated with both a higher baseline frequency and a more pronounced decrease in the specific Tem subpopulation (Fig. 3 G and H). This SNP correlated significantly with the expression of GM-CSF within the CD4 Tem cell pool (P = 0.05) (Fig. 3I).
CXCR3 Effector-Memory T Cells Constitute a Biomarker of Treatment Response.
Some patients experience disease activity despite DMF therapy. By comparing the deep immune phenotype of these patients with patients without evidence of clinical or radiological activity, we sought to identify possible predictors of therapy response. An adapted CellCnn algorithm was developed to identify common cell populations over all time points and assess their longitudinal profile change to predict clinical outcomes like disease activity. By this approach, the highest predictive accuracy of 79.14% was achieved on the conventional panel. It identified a response-associated phenotype of mainly effector-memory Th cells (with minor contributions of Tc and γδ-T cells) (Fig. 4 A–D) expressing CXCR3 as the best distinctive marker (Fig. 4E, all markers in SI Appendix, Fig. S6A, and relative frequency in SI Appendix, Fig. S6B). This cell population showed a constant and strong decrease (median decrease 77.5%) over the course of 12 mo in responders (Fig. 4B). At 12 mo, treatment nonresponders had a 2.9-fold higher proportion of this specific population (P = 0.0004). Using a cutoff of 50% of the maximal frequency of this population, 87.5% of nonresponders could be identified at a false positive rate of 26.1% (see the receiver operating characteristic [ROC] curve with an area under the curve [AUC] of 0.85 in Fig. 4F). Interestingly, the response-associated phenotype overlapped strongly with the above-described treatment-associated phenotype (Fig. 4A), illustrating the therapeutic relevance of this specific CXCR3+ T cell population in DMF treatment. A silhouette score of −0.0425 further affirmed the similarity of the identified populations (Fig. 4C). The prevalence of the response-associated population showed no significant association with age (Pearson correlation coefficient (PCC) −0.0550, P = 0.7765 at T1). Furthermore, therapy response did not significantly correlate with DMF-induced lymphopenia but rather depended on the recomposition within the Th cell pool—as illustrated by the distribution of absolute lymphocyte counts and the frequency of this phenotype in responders and nonresponders (Fig. 4G) and the respective ROC curves (Fig. 4F). A divergent cytokine profile of Tem between responding and nonresponding individuals further highlighted the central role of GM-CSF, IFNγ, and TNFα in the therapeutic effect (Fig. 4H). In conclusion, these data identify this specific Tem cell subpopulation as a marker of response to DMF in line with its proposed role in MS pathophysiology.
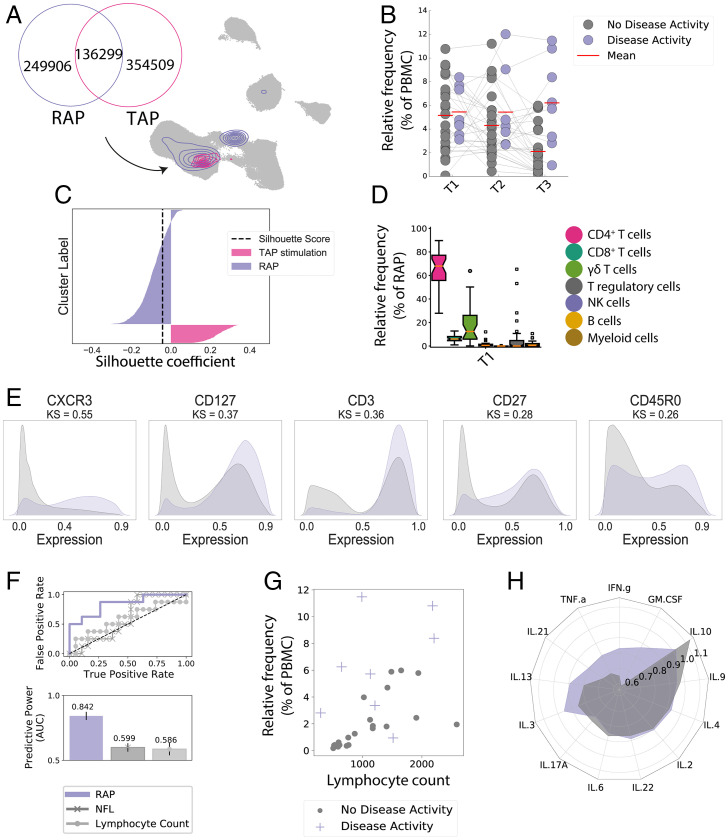
Fig. 4.
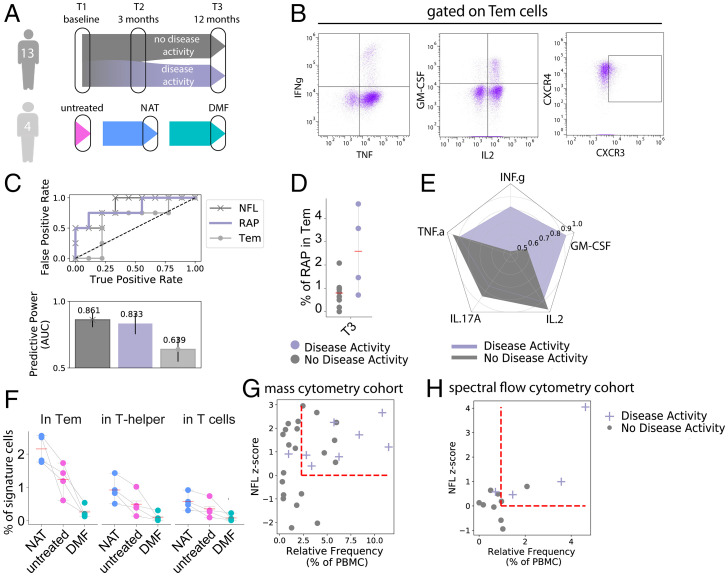
CXCR3 effector-memory T cell population correlates with DMF treatment response. Longitudinal CellCnn analysis determined the characterizing immune features of the therapy response from our patients’ cohort, as defined by relapses and/or new/active MRI lesions, termed the “response-associated phenotype” (RAP). (A) The UMAP algorithm was used to depict the identified response-associated (RAP, purple) and DMF treatment–associated cell filters (TAP, pink). (B) Frequency of the response-associated cell filter in patients with and without signs of disease activity, arranged for all three time points. (C) The silhouette analysis suggests the overlap of response-associated and DMF treatment–associated cell populations. The black dashed line represents the silhouette score. (D) Frequency of selected cell types within the response-associated phenotype at T1 in the stimulated panel. (E) Expression patterns of key discriminant markers between the response-associated population (purple) and the reference cell population (gray). (F) ROC curve depicting the accuracy of predicting disease activity at T3 for absolute lymphocyte counts at T3, NfL at T3, and response-associated population frequency at T3. Bars illustrate the predictive accuracy and SD at intragroup cross-validation. (G) Scatter plot of lymphocyte counts and frequency of the response-associated population at T3 with overlaid color code for disease activity. Each dot represents one patient. (H) Mean relative change between T1 and T3 of cytokine expression of analyzed cytokines in Tem. Each radius arm represents one cytokine and purple color indicates individuals with disease activity.
Independent Cohorts and Serum Neurofilament Confirm a DMF-Associated Immune Phenotype.
To confirm this predictive signature by an independent method in an independent patient population, we tested PBMCs from a second cohort (from here on termed “flow cytometry cohort”; for a scheme, see Fig. 5 A, Top) by spectral flow cytometry in a comparable analytical protocol (Fig. 5B and SI Appendix, Fig. S7A). In this cohort, 13 individuals with RRMS were assessed with clinical and laboratory follow-up matching the initial cohort. It included four nonresponders with relapse and MRI activity (of which one nonresponder experienced disease activity only within the first month of DMF treatment) and nine age-matching responders without radiological or clinical signs of disease activity. Baseline characteristics were similar for responders and nonresponders (for patient characteristics, see SI Appendix, Table S4). Both responders and nonresponders experienced a relative increase in CD4 cells within the 12 mo follow-up (12.7 vs. 6.1%, P = 0.26). As predicted from findings in the mass cytometry cohort, we saw a decrease of the CXCR3+ TNFα+ IFNγ+ GM-CSF+ Tem subpopulation by 46.5% in responders and an increase by 22.3% in nonresponders (P = 0.0427) within Tem cells (SI Appendix, Fig. S7A and Table S5), with GM-CSF (P = 0.0020) and IFNγ (P = 0.0047) as the main distinctive cytokines at T3 (Fig. 5 C–E). The high accuracy of its association with therapeutic response was consistent between the mass cytometry (AUC 0.85) and flow cytometry cohorts (AUC 0.84) (Fig. 5C). The analysis of specific cytokines revealed an analogous divergence for IFNγ and GM-CSF with response/nonresponse (Fig. 5E) in line with findings from the mass cytometry cohort.
Fig. 5.
Two independent cohorts and serum neurofilament levels confirm the algorithm-identified DMF-associated immune phenotype. (A) One cohort of 13 patients was analyzed in an analogous setup to the mass cytometry cohort by spectral flow cytometry. Another cohort of 4 patients with samples from untreated, DMF-treated, and natalizumab (NAT)-treated time points was assessed by conventional flow cytometry. (B) Representative illustration of cytokine and chemokine receptor staining on Tem subpopulations. (C) ROC curve depicting the accuracy of predicting disease activity at T3 using Tem frequency at T3, RAP frequency (within Tem) at T3, and NfL levels at T3. Bars illustrate the predictive accuracy and SD at intragroup cross-validation. (D) Frequency of the response/treatment–associated phenotype (as identified by CXCR3+ TNF+ IFNγ+ GM-CSF+ Tem cells) in patients with and without signs of disease activity at time point T3 in the flow cytometry cohort. (E) Mean relative change between T1 and T3 of cytokine expression in Tem. Each radius arm represents one cytokine and purple color indicates individuals with disease activity. (F) Frequency of the response/treatment–associated phenotype (as identified by CXCR3+ TNF+ IFNγ+ GM-CSF+ Tem cells) in the same individuals at untreated, DMF-treated, and natalizumab-treated time points in Tem, Th, and all T cells. (G and H) Scatter plots of the frequency of the response-associated population at T3 and NfL z scores at T3 in the mass cytometry cohort (G) and flow cytometry cohort (H) with overlaid color code for disease activity (purple) or no disease activity (gray). Each dot represents one patient. A z score of 0 and the mean value of the immune population are indicated by dashed red lines.
A possible alternative to our conclusion that the altered Th profile correlates with the DMF treatment response was the hypothesis that it might reflect decreased disease activity. To test this hypothesis, we took advantage of the fact that the CD49d-blocking antibody natalizumab also has a strong suppressive effect on disease activity but works by excluding pathogenic lymphocytes from the central nervous system (CNS) and other tissues (thereby increasing them in blood), while DMF depletes these cells (thereby decreasing them in blood). We therefore tested four patients with samples from three consecutive time points: without immunomodulatory medication, on natalizumab therapy (with good treatment response), and on DMF therapy (with good treatment response) (Fig. 5 A, Bottom; for baseline and patient characteristics, see SI Appendix, Table S6). As expected, DMF treatment led to drastically decreased abundance of the signature population in peripheral blood compared with the untreated time point (P = 0.026), while under natalizumab treatment the signature population was significantly increased (P = 0.0368) (Fig. 5F).
Serum neurofilament light chain (NfL) is a blood biomarker in MS specific for neuronal damage or loss caused by inflammatory activity or chronic neurodegeneration (20). To assess the association of the cell signature with neuroaxonal damage, NfL levels were determined at T1 and T3 in the mass and flow cytometry cohorts. Under DMF treatment, serum NfL age-adjusted z scores showed a moderate correlation with the frequency of the predictive population under DMF treatment (mass cytometry cohort, PCC 0.346; flow cytometry cohort, PCC 0.671; Fig. 5 G and H). Clinically and radiologically active patients were identified by both high neurofilament levels and the predictive population fraction as indicated in Fig. 5 G and H and by a stringent correlation in active patients (mass cytometry cohort: PCC 0.46, P < 0.0001; flow cytometry cohort: PCC 0.94, P = 0.05). As anticipated, NfL at T3 was reflective of disease activity during the observation period (mass cytometry cohort, P = 0.12; flow cytometry cohort, P = 0.05). In a combined approach, we added the z scores of both NfL and the response-associated phenotype to predict disease activity. This strategy increased the predictive accuracy compared with NfL alone in the mass cytometry cohort (AUC 0.73) and the flow cytometry cohort (AUC 0.94) (SI Appendix, Fig. S7B).
Discussion
This study approaches the challenging immunobiology of MS through examining the effects of treatment. Assuming that some type of immune cell is particularly important in causing the disease, such cells ought to be more abundant in patients than in the general population and influenced by effective treatments. This study focuses on the latter prediction. Some treatments, including DMF, are highly effective in suppressing relapses in some patients but less effective in others. This offers the opportunity to compare the impact of the same drug, when it works, and when it does not, and thereby to delimit therapeutically critical components of its effect.
In agreement with existing literature (6, 7, 21–23), our cohort shows alterations in several cell types under DMF treatment. The overall decrease of Tc cells was more pronounced than the global impact on Th cells or B cells (7, 9, 13), and mass cytometry confirmed the fundamental rearrangement of the most affected mature subsets of both T cells (5–9) and B cells (10–13). Among B cells, we observed a primary effect of DMF on IgM and class-switched memory B cells but not on transitional B cells or plasma cells, consistent with the target profile of the highly efficacious B cell–depleting therapies in MS (24, 25). More specifically, our study suggests that mature memory B cells expressing CXCR3 and TNFα are the main B cell target of DMF treatment. B cells expressing CXCR3 are reported to migrate into the CNS in MS and play roles in both disease activity (24, 26, 27) and as a therapeutic target (24, 28).
In the T cell compartment, a reduction in antigen-experienced cells (6, 7, 29) was paralleled by relative expansion of naïve T cells. Cytokine secretion and cytokine receptor expression patterns indicate a shift from Th1 to Th2 and Tfh polarization, and a strong decrease in proinflammatory cytokine-producing T cells, pathophysiologically plausible mediators (22, 30) of the therapeutic effect. Specifically, we identified a population of GM-CSF/IFNγ/CXCR3–expressing Tem cells as the main affected population upon DMF treatment. When postulating a pathophysiological role of GM-CSF–expressing Tem cells in the CNS, a genetic predisposition in the STAT4 gene is of specific interest (31). We here report a potential association of therapeutic response with the MS-associated allele at locus rs6738544 in the STAT4 gene, suggesting that DMF rebalances the aberration in this signaling cascade (32). We note, however, that the size of our cohort is insufficient for confident inference about genetic influences. It is instructive to compare the identified population of GM-CSF/IFNγ/CXCR3 Tem cells with the related cell type recently identified as being associated with MS pathogenesis (29). As expected for an empirically developed therapeutic like DMF, the signatures of disease and treatment effect diverge slightly. They are mainly discriminated by the predominant chemokine receptor, either CXCR4 [MS-associated phenotype (29)] or CXCR3 (DMF-impacted phenotype), a distinction well-explained by the known class-specific, presumably metabolic suppression of CXCR3 Th cells by fumaric acid esters (21, 22, 33). Mechanistically, the specific T cell polarization and proliferation patterns as well as the effector cell reduction are consistent with a postulated immunometabolic mode of action. The succination of GAPDH by DMF was described to inhibit glycolysis, thereby directly influencing the crucial energetic substrate required by the active Warburg-like metabolism of activated Th1 cells (6, 21).
Beyond characterizing the global effect of DMF treatment, the main goal of this study was to compare the phenotypes of responders versus nonresponders. While the cross-sectional analysis of baseline samples did not allow the identification of predictive biomarkers of response, thanks to the unique feature of our approach, we were able to screen for response-associated changes in the entire range of longitudinally collected PBMCs. This automated analysis identified the signature of Tem cells as the main marker associated with disease activity, with CXCR3 as the top descriptive feature. This circumscribed, response-associated immune signature accords with previously reported observations of suggestive Tem alterations (14, 34) but contrasts with other studies (35, 36). Specifically, in comparison with the previously suggested alterations in IL17, CD8 (35), and Th17 cells (36), the response-associated population was characterized by similarly elevated CXCR3 but not CCR6 or IL17a expression. An independent check on the significance of the response-associated cell population is offered by examination of the very different effects of natalizumab. By blocking α4-integrin, natalizumab prevents migration of lymphocytes from the blood into the CNS, preventing their pathological effect and leading to peripheral overrepresentation of brain-homing cells (37, 38). This mechanism of action yields the prediction that the pathologically critical cell type depleted by DMF ought to be increased by natalizumab. Indeed, this is the case—as examined in a cohort of patients with beneficial treatment response to both treatments—providing additional evidence that this cell type is a causal player in disease pathogenesis. Given the multiple disease-modifying therapies currently available for MS (18, 39, 40), it would be interesting to define immune biomarkers of response based on patients’ baseline immune characteristics. Future studies with broader cohorts will be required to further address this point, aiming to generalize our findings and expand them to other treatments.
We also examined the relationship between the blood cellular composition and the neuronal damage marker NfL. Since increased serum NfL reflects acute disease activity (i.e., relapse-associated worsening) and also progression independent of relapse activity (20, 41), the higher specificity of a response-associated signature for the former offers additional value in a clinical context. If validated by independent groups, we envisage that combined measurement of NfL in blood as a biomarker of axonal damage and cellular composition as a marker of inflammatory activity will allow discrimination of the different disease aspects and guide therapeutic strategies.
In summary, our findings indicate that GM-CSF/IFNγ/CXCR3–expressing T helper cells are a key target of DMF and that reduced depletion of this population is associated with relapse of MS. These findings suggest that immune monitoring of DMF-treated patients might be relevant to determine the therapeutic response and hence guide clinicians toward an alternative disease-modifying treatment. Further studies addressing the implication of CXCR3 and its ligands in MS might pave the way for the development of new therapeutic targets. Overall, we believe that our findings not only provide insights into disease pathophysiology but also substantially contribute to improved personalized treatment strategies in MS.
Materials and Methods
Study Design and Sample Processing.
Patients with evident diagnosis of RRMS according to revised McDonald criteria (42) planning to start DMF treatment were prospectively enrolled at the Multiple Sclerosis Center of University Hospital Basel. Inclusion criteria included no previous treatment with fumaric acid esters, a negative pregnancy test and effective contraception, and a signed informed consent. Follow-up regarding clinical parameters and biobanking was conducted before the start of treatment (T1) as well as at 3 (T2) and 12 mo (T3) under treatment (±45 d). Cerebral MRI with a standardized study protocol was conducted at T1 and T3 and assessed according to a standardized protocol by experienced board-certified neuroradiologists. Prospectively enrolled individuals were chronologically assigned to two cohorts: the mass cytometry cohort [SI Appendix, Table S1; referred to in Diebold et al. (43)] and flow cytometry cohort (SI Appendix, Table S4). All patients were instructed about the study design and signed an informed consent. The study was approved by the Ethics Committee for Northwest and Central Switzerland, EKNZ 48/12 (for DMF treatment samples: mass cytometry and flow cytometry cohorts) and EKNZ 49/06 (for the natalizumab cohort).
Blood samples obtained at each time point were freshly characterized by automated flow cytometry for main leukocyte and lymphocyte populations. PBMCs were collected from each donor’s blood (ethylenediaminetetraacetate [EDTA] Monovette, Sarstedt) by density gradient centrifugation (Lymphoprep, AXIS-Shield) and cryopreserved in liquid N2 in a medium of 10% dimethyl sulfoxide, 30% RPMI-1640, and 60% fetal calf serum (FCS). Serum samples (serum Monovette, Sarstedt) were centrifuged at 2,000 × g and stored at −80 °C until analysis.
Neurofilament.
We measured serum NfL levels in duplicates using the high-sensitivity single-molecule array (Simoa) NF-Light Advantage Kit (Quanterix) according to the manufacturer’s instructions on an HDX platform (Quanterix). We repeated measurements for a few samples with an intraassay coefficient of variation (CV) above 20%. Interassay CVs for three control samples were below 10%. Analyses were performed blinded to clinical data.
Identification of SNPs.
DNA was acquired from additionally sampled blood cells (1 to 3 × 106 cells per sample) with the DNeasy Blood & Tissue Kit (Qiagen). DNA was then analyzed for 654,027 SNPs by Global Screening Array Multidisease 2.0 (Illumina). In total, 198 SNPs were extracted for each patient, selected based on their correlation with the disease MS mentioned in recent literature (19). We encoded the genotypes using an additive method (44) with respect to disease-associated bases, extracted from the literature. ANOVA was used to test for differences in mean expression of immune cell populations of each patient (only SNPs with at least three available allele constellations were used for analysis). For each group of tests, we corrected the P values for multiple testing using Bonferroni, considering a cutoff of 0.1 as significant.
Ex Vivo Reactivation of PBMCs.
Short-term reactivation of cryopreserved PBMCs and subsequent mass cytometry analysis were performed as described recently (29). In short, PBMCs were kept in liquid N2 before thawing in a 37 °C water bath. Cells were resuspended in cell-culture medium (RPMI-1640, 10% FCS [Biochrom], 1× l-glutamine, 1× penicillin/streptomycin [both Life Technologies]) supplemented with 1:10,000 benzonase (Sigma) and centrifuged (300 relative centrifugal force, 7 min, 24 °C). Samples were then rested overnight at 37 °C before restimulation with 50 ng⋅mL−1 PMA (Sigma) and 500 ng⋅mL−1 ionomycin (Sigma) in the presence of 1× brefeldin A (BD Biosciences) for 4 h at 37 °C.
Antibodies.
For mass cytometry, monoclonal anti-human antibodies (SI Appendix, Tables S2 and S3) were purchased either preconjugated to heavy-metal isotopes (Fluidigm) or conjugated using the Maxpar X8 Chelating Polymer Kit (Fluidigm). Flow cytometry antibodies were purchased preconjugated (SI Appendix, Table S5).
Live-Cell Barcoding for Mass Cytometry.
To minimize intersample staining variability, sample handling time, and antibody consumption, we made use of a live-cell barcoding approach. In short, isothiocyanobenzyl-EDTA (Dojindo) was loaded with palladium isotopes (104Pd, 105Pd, 106Pd, 108Pd, and 110Pd; all from Trace Sciences International; 113In or 115In also from Trace Sciences International) as described previously, or niobium (93Nb) and tantalum (181Ta), and conjugated to anti-human CD45 (BioLegend). 89Y-CD45 (Fluidigm) was used as an additional barcoding reagent. Samples were stained with anti-CD45 antibodies following reactivation at 37 °C for 25 min in cell-staining medium (CSM; RPMI-1640 and 4% FCS) on a rotating shaker (500 rpm), washed twice with CSM, and combined into a single reaction vessel for further staining steps. Using this approach, up to 84 samples (all longitudinal samples were acquired within the same barcoded panel) were combined and processed together. In total, multiple barcoding reactions were performed on three independent occasions.
Surface Staining.
As previously described (29), the barcoded and combined sample for mass cytometry was resuspended in 400 μL of surface-antibody mixture [SI Appendix, Tables S2 and S3, for the conventional and stimulation panels, respectively; the stimulation panel was identical to Diebold et al. (43)] in CSM. Surface staining was performed for 20 min at 37 °C on a rotating shaker (500 rpm). After Fc blocking (BioLegend) and an additional wash, 2.5 μM cisplatin (Sigma-Aldrich) was added for 2 min on ice to identify dead cells. For flow cytometry, 50 μL surface-antibody mixture (SI Appendix, Tables S2 and S3) was added to the samples after Fc blocking, followed by incubation for 30 min at room temperature.
Intracellular Cytokine Staining.
For mass cytometry, the combined surface and live/dead–stained sample was fixed with 1.6% paraformaldehyde (Electron Microscopy Sciences) in phosphate-buffered saline (PBS) for 30 min at 4 °C on a rotary shaker (500 rpm) or in intranuclear fixative (Invitrogen, FoxP3/Transcription) for 1 h, washed, and resuspended in permeabilization buffer mix with intranuclear antibodies for staining. Fixed cells were washed twice with permeabilization buffer (PBS, 0.5% saponin, 2% bovine serum albumin, and 0.01% sodium azide [all Sigma-Aldrich]). Cells were resuspended in 400 μL intracellular antibody mixture (SI Appendix, Tables S2 and S3) in permeabilization buffer for 1 h at 4 °C on a rotary shaker (500 rpm). The sample was washed, the supernatant was removed, and the cells were resuspended in 1× iridium intercalator solution (Fluidigm) overnight. Lastly, the sample was washed twice with PBS/bovine serum albumin and once with double-distilled water before acquisition. For flow cytometry, similar fixation and permeabilization were performed. Cells were resuspended in 50 μL intracellular antibody mixture (SI Appendix, Table S2) in permeabilization buffer for 1 h at 4 °C on a rotatory shaker. Samples were then washed with CSM.
Data Acquisition and Preprocessing.
For acquisition with a CyTOF2 mass cytometer (Fluidigm), quality control and tuning were performed daily. Acquisitions and data normalization were achieved using five-element beads (Fluidigm). A shared reference sample across multiple runs was used as an independent control. For downstream analysis, live, single cells were identified based on event length, DNA (191Ir and 193Ir), and live cell (195Pt and 198Pt) channels using FlowJo (TreeStar). The combined sample was debarcoded using Boolean-gating or MATLAB-based software. Cytometry data were transformed with an inverse hyperbolic sine (arcsinh) function with a cofactor of 5 (45) using the R environment (R Development Core Team 2008, https://www.r-project.org/). To balance the influence of markers with different dynamic ranges, we performed background subtraction and channel-based percentile normalization using the 99th percentile of each marker across the whole barcoded dataset. Individual cytokine positivity thresholds were determined based on the 99th percentile of the residual staining in an unstimulated control sample.
Algorithm-Based High-Dimensional Analysis.
Preprocessed data were randomly down-sampled to a maximum of 300,000 cells per donor. Unless otherwise indicated, only samples with more than 5,000 cells were considered for the analysis. FlowSOM clustering was performed on cohort-combined samples after interrun normalization. Metaclustering of nodes was performed with the indicated k values (elbow criteria–based), and annotated according to their protein-expression pattern. UMAP visualization was performed on a reduced dataset of equal numbers of cells randomly selected (100,000 cells per condition) utilizing the UMAP package (n_component = 2, n_neighbors = 15/50, n_epochs = 400, min_dist = 0.1/0.8). All plots were drawn using ggplot2.
Statistical Analysis.
The change of density was computed via a Gaussian kernel density estimation using the two-dimensional UMAP representation as input for the three time points T1, T2, and T3, respectively. We defined a grid along both UMAP axes and computed the density based on the fitted kernels of the respective time points. Afterward, we subtracted the density grids T3–T1 and T3–T2.
CellCnn Longitudinal Application.
CellCnn (15) is a weakly supervised machine-learning model which can be applied to detect and define rare disease-associated cell subpopulations. The model predicts disease phenotypes based on relative cell subpopulation frequencies, which are encoded in a model intern CellCnn score defined via a convolutional filter. For more details on CellCnn, we refer to ref. 15. We adapted the method into a longitudinal setup, which allows taking all of our three time points (T1, T2, T3) in a single model into account, which was not possible before. Therefore, instead of predicting directly on the cell subpopulation frequencies, we computed the time residuals T3–T1 and T3–T2. This allows us to define disease-associated cell subpopulations which change over time and explain the disease phenotype.
We used a threefold cross-validation and trained multiple models with randomly selected hyperparameters (cell bag size ∈ [500, 1,000, 2,000, 3,000, 4,000, 5,000]; max pooling percentages ∈ [1, 5, 10, 15, 20]; number of filters ∈ [3, 4, …, 9, 10]; same patient in bags cell bags T1, T2, T3 ∈ [true, false] [otherwise from only the same phenotype]; L1 penalty coefficient on filters ∈ [0, 0.0001]; L2 penalty coefficient on filters ∈ [0, 0.0001]) which results in multiple above-mentioned filters per run, which all define a separate cell subpopulation. We then applied t tests to all filters at each time point between the disease phenotypes to select those filters which show a significant difference and therefore define phenotype-relevant cell subpopulations.
Silhouette Analysis.
A silhouette analysis (46) was performed to measure the separation and clustering properties between defined populations. This visual testing approach shows for each cell whether it lies well within its cluster or if it is rather located between clusters. The silhouette coefficient is defined as (b − a)/max(a,b), where a is the mean intracluster distance and b stands for the mean nearest-cluster distance. The average of all cells’ silhouette coefficients can be used as the silhouette score and is per definition between 1, which indicates a perfect separation of the clusters, and −1, which indicates incorrect clustering of cells. A silhouette score of 0 indicates overlapping clusters. All silhouette coefficients were computed on the maximum overlapping original marker space and only visualized in reduced dimensions.
Spectral and Conventional Flow Cytometry.
In-depth analysis of the flow cytometry cohort was performed in analogy to the mass cytometry approach on an Aurora spectral flow cytometer (Cytek) after stimulation with PMA/ionomycin (for antibodies, see SI Appendix, Table S5). Blood samples from the mass cytometry and flow cytometry cohorts were measured by automated flow cytometry for main leukocyte and lymphocyte populations on a FACSCanto II (BD Biosciences) and a FACSymphony (BD Biosciences) for cross-treatment comparison.
Supplementary Material
Acknowledgments
We thank Heidi Bodmer, Manuela Diederich, Annette Orleth, the team of the Multiple Sclerosis Center, Basel, as well as all participants for their continuous support. This study was supported as a driver project in the Swiss Personalized Health Initiative by the Swiss Academy of Medical Sciences (PHRT-103). Funding for this study was received from the Swiss National Science Foundation (733 310030_170320, 310030_188450, 199310 and CRSII5_183478) and from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme grant agreement No 882424. Ad-personam funding for this project was received from the Research Fund of the University of Basel (MD, EG), the German Research Foundation (MD) and Studienstiftung des deutschen Volkes (FI). Parts of this study were funded as an investigator-initiated trial with funding from Biogen Switzerland.
Footnotes
Competing interest statement: M.D. received speaker honoraria from Biogen Switzerland, which were used exclusively for research purposes. L.K. discloses research support to his institution (University Hospital Basel): steering committee, advisory board, and consultancy fees (Actelion, Bayer HealthCare, Biogen, BMS, Genzyme, Janssen, Merck, Novartis, Roche, Sanofi, Santhera, and TG Therapeutics); speaker fees (Bayer HealthCare, Biogen, Merck, Novartis, Roche, and Sanofi); support of educational activities (Allergan, Bayer HealthCare, Biogen, CSL Behring, Desitin, Genzyme, Merck, Novartis, Pfizer, Roche, Sanofi, Shire, and Teva); license fees for Neurostatus products; and grants (Bayer HealthCare, Biogen, European Union, InnoSwiss, Merck, Novartis, Roche, Swiss MS Society, and Swiss National Research Foundation). J.K. received speaker fees, research support, and/or travel support and/or served on advisory boards of ECTRIMS, Swiss MS Society, Swiss National Research Foundation (320030_160221), University of Basel, Bayer, Biogen, Genzyme, Merck, Novartis, Protagen AG, Roche, and Teva. T.D. received financial compensation for participation in advisory boards, steering committees, and data safety monitoring boards, and for consultation for Novartis Pharmaceuticals, Merck, Biogen, Celgene, GeNeuro, MedDay, Mitsubishi Tanabe Pharma, Roche, and Sanofi Genzyme. T.D. also received research support from Novartis, Biogen, National Swiss Science Foundation, European Union, and Swiss MS Society.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205042119/-/DCSupplemental.
Data Availability
Anonymized mass cytometry data can be accessed at: http://doi.org/10.17632/425ppfhjnv.1. The R-based custom workflow is available at https://github.com/GalliES/DMF_repository. been deposited in GitHub (https://github.com/GalliES/DMF_PNAS) (47).
References
- 1.Gold R., et al. ; DEFINE Study Investigators, Placebo-controlled phase 3 study of oral BG-12 for relapsing multiple sclerosis. N. Engl. J. Med. 367, 1098–1107 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Fox R. J., et al. ; CONFIRM Study Investigators, Placebo-controlled phase 3 study of oral BG-12 or glatiramer in multiple sclerosis. N. Engl. J. Med. 367, 1087–1097 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Blair H. A., Dimethyl fumarate: A review in relapsing-remitting MS. Drugs 79, 1965–1976 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Montes Diaz G., Hupperts R., Fraussen J., Somers V., Dimethyl fumarate treatment in multiple sclerosis: Recent advances in clinical and immunological studies. Autoimmun. Rev. 17, 1240–1250 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Spencer C. M., Crabtree-Hartman E. C., Lehmann-Horn K., Cree B. A., Zamvil S. S., Reduction of CD8(+) T lymphocytes in multiple sclerosis patients treated with dimethyl fumarate. Neurol. Neuroimmunol. Neuroinflamm. 2, e76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diebold M., et al. , Dimethyl fumarate influences innate and adaptive immunity in multiple sclerosis. J. Autoimmun. 86, 39–50 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Gross C. C., et al. , Dimethyl fumarate treatment alters circulating T helper cell subsets in multiple sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 3, e183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkovich R., Weiner L. P., Effects of dimethyl fumarate on lymphocyte subsets. Mult. Scler. Relat. Disord. 4, 339–341 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Montes Diaz G., Fraussen J., Van Wijmeersch B., Hupperts R., Somers V., Dimethyl fumarate induces a persistent change in the composition of the innate and adaptive immune system in multiple sclerosis patients. Sci. Rep. 8, 8194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R., et al. , Dimethyl fumarate treatment mediates an anti-inflammatory shift in B cell subsets of patients with multiple sclerosis. J. Immunol. 198, 691–698 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Smith M. D., Martin K. A., Calabresi P. A., Bhargava P., Dimethyl fumarate alters B-cell memory and cytokine production in MS patients. Ann. Clin. Transl. Neurol. 4, 351–355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lundy S. K., et al. , Dimethyl fumarate treatment of relapsing-remitting multiple sclerosis influences B-cell subsets. Neurol. Neuroimmunol. Neuroinflamm 3, e211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Longbrake E. E., et al. , Dimethyl fumarate selectively reduces memory T cells in multiple sclerosis patients. Mult. Scler. 22, 1061–1070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlström K. E., et al. , Therapeutic efficacy of dimethyl fumarate in relapsing-remitting multiple sclerosis associates with ROS pathway in monocytes. Nat. Commun. 10, 3081 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arvaniti E., Claassen M., Sensitive detection of rare disease-associated cell subsets via representation learning. Nat. Commun. 8, 14825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Gassen S., et al. , FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Pröbstel A. K., Sanderson N. S. R., Derfuss T., B cells and autoantibodies in multiple sclerosis. Int. J. Mol. Sci. 16, 16576–16592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reich D. S., Lucchinetti C. F., Calabresi P. A., Multiple sclerosis. N. Engl. J. Med. 378, 169–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Multiple Sclerosis Genetics Consortium, Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eaav7188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Disanto G., et al. ; Swiss Multiple Sclerosis Cohort Study Group, Serum neurofilament light: A biomarker of neuronal damage in multiple sclerosis. Ann. Neurol. 81, 857–870 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornberg M. D., et al. , Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Q., et al. , Dimethyl fumarate selectively reduces memory T cells and shifts the balance between Th1/Th17 and Th2 in multiple sclerosis patients. J. Immunol. 198, 3069–3080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm Hansen R., Højsgaard Chow H., Sellebjerg F., Rode von Essen M., Dimethyl fumarate therapy suppresses B cell responses and follicular helper T cells in relapsing-remitting multiple sclerosis. Mult. Scler. 25, 1289–1297 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Johansson D., et al. , Mass cytometry of CSF identifies a multiple sclerosis associated B cell population. Neurol. Neuroimmunol. Neuroinflamm. 8, e943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gingele S., et al. , Ocrelizumab depletes CD20+ T cells in multiple sclerosis patients. Cells 8, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sørensen T. L., Roed H., Sellebjerg F., Chemokine receptor expression on B cells and effect of interferon-β in multiple sclerosis. J. Neuroimmunol. 122, 125–131 (2002). [DOI] [PubMed] [Google Scholar]
- 27.van Langelaar J., et al. , Induction of brain-infiltrating T-bet-expressing B cells in multiple sclerosis. Ann. Neurol. 86, 264–278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saraste M., Penttilä T. L., Airas L., Natalizumab treatment leads to an increase in circulating CXCR3-expressing B cells. Neurol. Neuroimmunol. Neuroinflamm. 3, e292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galli E., et al. , GM-CSF and CXCR4 define a T helper cell signature in multiple sclerosis. Nat. Med. 25, 1290–1300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner C. A., Roqué P. J., Goverman J. M., Pathogenic T cell cytokines in multiple sclerosis. J. Exp. Med. 217, e20190460 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McWilliams I. L., Rajbhandari R., Nozell S., Benveniste E., Harrington L. E., STAT4 controls GM-CSF production by both Th1 and Th17 cells during EAE. J. Neuroinflammation 12, 128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canto E., Isobe N., Didonna A., Hauser S. L., Oksenberg J. R.; MS-EPIC Study Group, Aberrant STAT phosphorylation signaling in peripheral blood mononuclear cells from multiple sclerosis patients. J. Neuroinflammation 15, 72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm Hansen R., Højsgaard Chow H., Christensen J. R., Sellebjerg F., von Essen M. R., Dimethyl fumarate therapy reduces memory T cells and the CNS migration potential in patients with multiple sclerosis. Mult. Scler. Relat. Disord. 37, 101451 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Medina S., et al. , Optimal response to dimethyl fumarate associates in MS with a shift from an inflammatory to a tolerogenic blood cell profile. Mult. Scler. 24, 1317–1327 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Lückel C., et al. , IL-17+ CD8+ T cell suppression by dimethyl fumarate associates with clinical response in multiple sclerosis. Nat. Commun. 10, 5722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Langelaar J., et al. , T helper 17.1 cells associate with multiple sclerosis disease activity: Perspectives for early intervention. Brain 141, 1334–1349 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Traub J. W., et al. , Natalizumab promotes activation and pro-inflammatory differentiation of peripheral B cells in multiple sclerosis patients. J. Neuroinflammation 16, 228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Börnsen L., et al. , Effect of natalizumab on circulating CD4+ T-cells in multiple sclerosis. PLoS One 7, e47578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callegari I., Derfuss T., Galli E., Update on treatment in multiple sclerosis. Presse Med. 50, 104068 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Diebold M., Derfuss T., Immunological treatment of multiple sclerosis. Semin. Hematol. 53 (suppl. 1), S54–S57 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Kappos L., et al. , Contribution of relapse-independent progression vs relapse-associated worsening to overall confirmed disability accumulation in typical relapsing multiple sclerosis in a pooled analysis of 2 randomized clinical trials. JAMA Neurol. 77, 1132–1140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson A. J., et al. , Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 17, 162–173 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Diebold M., et al. , Immunological predictors of dimethyl fumarate-induced lymphopenia. Ann. Neurol. 91, 676–681 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gumpinger A. C., Roqueiro D., Grimm D. G., Borgwardt K. M., “Methods and tools in genome-wide association studies” in Computational Cell Biology, von Stechow L., Santos Delgado A., Eds. (Methods in Molecular Biology, Humana Press, 2018), pp. 93–136. [DOI] [PubMed] [Google Scholar]
- 45.Bendall S. C., et al. , Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rousseeuw P. J., Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987). [Google Scholar]
- 47.Diebold M., et al., High-dimensional immune profiling identifies a biomarker to monitor dimethyl fumarate response in multiple sclerosis. GitHub. https://github.com/GalliES/DMF_repository. Deposited 15 July 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized mass cytometry data can be accessed at: http://doi.org/10.17632/425ppfhjnv.1. The R-based custom workflow is available at https://github.com/GalliES/DMF_repository. been deposited in GitHub (https://github.com/GalliES/DMF_PNAS) (47).