Significance
Interaction between peptide–major histocompatibility complexes (pMHCs) and T cell receptors (TCRs) is the key mediator for tumor cell recognition by cytotoxic T cells. We performed a multimodal analysis on a prostate tissue antigen, PAP, to reveal peptides restricted by MHC I. Cognate TCRs were then isolated against those candidate targets. Information regarding both the epitopes and TCRs can be beneficial in designing new treatments for prostate cancer. Our platform can also be generalizable to other cancer antigens and different HLA types.
Keywords: T cell receptor (TCR), prostate cancer, immunopeptidome, prostatic acid phosphatase, major histocompatibility complexes (MHC)
Abstract
Tissue-specific antigens can serve as targets for adoptive T cell transfer-based cancer immunotherapy. Recognition of tumor by T cells is mediated by interaction between peptide–major histocompatibility complexes (pMHCs) and T cell receptors (TCRs). Revealing the identity of peptides bound to MHC is critical in discovering cognate TCRs and predicting potential toxicity. We performed multimodal immunopeptidomic analyses for human prostatic acid phosphatase (PAP), a well-recognized tissue antigen. Three physical methods, including mild acid elution, coimmunoprecipitation, and secreted MHC precipitation, were used to capture a thorough signature of PAP on HLA-A*02:01. Eleven PAP peptides that are potentially A*02:01-restricted were identified, including five predicted strong binders by NetMHCpan 4.0. Peripheral blood mononuclear cells (PBMCs) from more than 20 healthy donors were screened with the PAP peptides. Seven cognate TCRs were isolated which can recognize three distinct epitopes when expressed in PBMCs. One TCR shows reactivity toward cell lines expressing both full-length PAP and HLA-A*02:01. Our results show that a combined multimodal immunopeptidomic approach is productive in revealing target peptides and defining the cloned TCR sequences reactive with prostatic acid phosphatase epitopes.
Tissue antigens are encoded by nonmutated genes but can serve as potential targets for cancer therapies (1–3). Adoptive T cell therapies reactive with tissue antigens have enabled clinical trials targeting MART1, NY-ESO-1, and WT1 (4). T cell receptor (TCR) immunotherapy has demonstrated its potential in treating different cancers.
A major concern for TCR immunotherapy is toxicity on benign tissues. This “on-target off-tumor toxicity” is linked to expression of the target or cross-reactive antigens in normal organs. Several clinical trials have been discontinued because of observed off-tumor toxicities (4, 5).
One way to minimize “on-target off-tumor” toxicity is to select tissue antigens expressed on nonessential organs. Patients with late-stage prostate cancers have often received a radical prostatectomy to remove the prostate gland (6). We chose prostatic acid phosphatase (PAP) among many previously defined prostate tissue antigens because 1) the expression of PAP is highly restricted to the prostate and prostate cancer (7), 2) PAP expression can be found in >95% of prostate cancers (8), 3) serum PAP elevation is found in >60% patients with relapsed prostate cancers (9), 4) an elevated level of PAP is a strong predictor for high-risk recurrence (10), and 5) the secreted form of PAP will not compete with TCR–PAP recognition, because the interaction is restricted to peptides bound to MHC I.
Previous efforts to target PAP led to the first Food and Drug Administration–approved cancer vaccine, sipuleucel-T (Provenge) (11). Clinical trials showed a median improvement in overall survival of 4.1 mo in men with metastatic castration-resistant prostate cancer (11). Recent studies have also provided video evidence that T cells from sipuleucel-T-treated patients are capable of lysing PAP+ target cells (12). Neither the PAP epitopes presented nor the cognate TCR sequences have been defined at the molecular level. Recovery of TCRs that specifically recognize PAP epitopes could have potential therapeutic value.
In Silico Prediction of Epitopes on MHC I Is an Important Strategy but Not Sufficient
Multiple computational methods have been developed to predict peptide–MHC (pMHC) binding affinities with knowledge based on experimentally defined epitopes (13–15). In silico prediction can rapidly generate results and has been widely used (14). Previous efforts to identify PAP epitopes mainly relied on motif-based predictions (16, 17). In a recent study, Wells et al. (18) assembled a consortium (Tumor Neoantigen Selection Alliance or TESLA) including 25 different prediction platforms for comparison. Only 6% of predicted peptides were found to be immunogenic by pMHC multimer staining (18).
Using Physical Assays to Define the Immunopeptidome
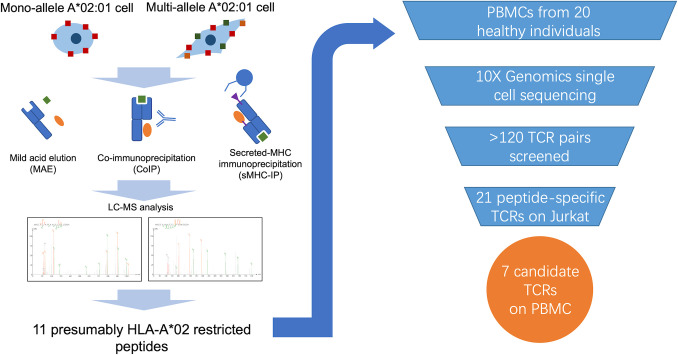
An alternative way to define the immunopeptidome is to directly isolate peptides bound to MHC I and identify them by liquid chromatography and mass spectrometry (LC-MS). Multiple physical methods using MS to define the immunopeptidome have been previously developed, including mild acid elution (MAE), MHC coimmunoprecipitation (CoIP), and secreted MHC immunoprecipitation (sMHC-IP). MAE was one of the earliest approaches to isolate peptides from MHC I by using an isotonic acid buffer to destabilize pMHC complexes (19). Although fast and convenient, this method can yield non-MHC-bound peptides from other extracellular proteins. CoIP purifies pMHC I complexes with monoclonal antibodies to generate results with better specificity (20, 21). This requires large quantities of antibody as well as expression of both the antigens of interest and the desired HLA types in target cells. The sMHC-IP technique requires the engineering and expression of soluble single-chain MHC in cell lines for affinity capture (22, 23). This protocol requires creation of modified cell lines and might generate peptides only presentable on artificial constructs. There is no consensus for the single best approach. To capture a more comprehensive immunopeptidomic signature of PAP, we performed all three approaches on HLA-A*02:01, one of the most common subtypes (24).
Identification of TCRs from Antigen-Reactive Single T Cells
To date, no single-cell MHC I–restricted TCR sequence has been defined and disclosed against PAP epitopes on the publicly available Immune Epitope Database (IEDB) (25). One of the major challenges has been to enrich and to identify cognate T cells for single-cell sequencing. We recently developed a technique, CLInt-seq, to enable single–T cell isolation of activated cells. Cells were fixed with a disulfide bond-based reversible cross-linker (dithiobis[succinimidyl propionate] or DSP) and sorted based on intracellular activation markers by fluorescence-activated cell sorting (FACS) (26). Utilization of a reversible cross-linker allows the cells’ messenger RNA (mRNA) to be released from mRNA–protein cross-linked complexes. These mRNAs can then be efficiently reverse transcribed and are compatible with the 10× Genomics single-cell TCR sequencing platform (26, 27). T cells stimulated by cognate peptides can produce cytokines such as IFNγ and TNFα, which can be trapped and intracellularly stained. Using the physically determined PAP epitopes, peptide-reactive TCRs were successfully isolated with CLInt-seq from multiple healthy donor peripheral blood mononuclear cell (PBMC) samples.
Results
Multimodal Immunopeptidomic Profiling of PAP on HLA-A*02:01.
Both physical and in silico approaches were used to define a thorough HLA-A*02:01 immunopeptidomic signature of PAP. A commonly used algorithm, NetMHCpan 4.0, was applied to profile PAP epitopes on HLA-A*02:01 (14). Forty PAP peptides were selected as potential good binders on HLA-A*02:01 using the top two percentile as a cutoff (Table 1 and SI Appendix, Tables S1 and S2).
Table 1.
Summary of recovered PAP peptides that presumably restricted to HLA-A*02:01 according to NetMHC 4.0 prediction (<1000 nM), T2 assays, or sSCT assays (>0.2)
| Peptide | Length | Name | By MAE M202-PAP | By MAE K562-A2-PAP | By CoIP M202-PAP | By CoIP K562-A2-PAP | By sMHC-IP | NetMHC predicted affinity (nM) | NetMHCpan percentile (%) | T2 assay | sSCT assay | Cognate TCRs in Jurkat | Cognate TCRs in PBMC | Full-length PAP recognition |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ILLWQPIPV | 9 | PAP_A2_14 | √ | 5.2 | 0.1557 | Positive | 0.81 | 5 | 1 | |||||
| TLMSAMTNL | 9 | PAP_A2_22 | √ | √ | 9.4 | 0.0794 | Positive | 0.73 | 1 | 1 | Yes | |||
| VLAKELKFV | 9 | PAP_A2_27 | √ | 51.2 | 0.1858 | Positive | 0.11 | |||||||
| SVHNFTLPSW | 10 | PAP_A2_24 | √ | 21204.2 | 24.2338 | 1.40 | ||||||||
| IMYSAHDTTV | 10 | PAP_A2_25 | √ | 82.3 | 0.8042 | Positive | 0.50 | 2 | ||||||
| KVYDPLYCESV | 11 | PAP_A2_20 | √ | 615.5 | 0.401 | Positive | 0.61 | 2 | ||||||
| LLLARAASLSL | 11 | PAP_A2_21 | √ | 977.9 | 5.1611 | Positive | 0.32 | 9 | 5 | |||||
| WQPIPVHTVPLS | 12 | PAP_A2_15 | √ | 22400.3 | 44 | 0.68 | ||||||||
| LLFFWLDRSVLA | 12 | PAP_A2_23 | √ | 647.2 | 2.2948 | 0.13 | 1 | |||||||
| YSAHDTTVSGLQM | 13 | PAP_A2_2 | √ | 27417.1 | 33.2593 | 0.21 | ||||||||
| YSAHDTTVSGLQMA | 14 | PAP_A2_1 | √ | 19437.6 | 19.7418 | 0.74 |
Information includes methods used, predicted affinity and rank of HLA-A*02:01 binding, in vitro MHC I binding results, and number of recovered cognate TCRs. Green: positive results or predicted binders on HLA-A*02:01; Yellow: predicted non-binders on HLA-A*02:01 but are near cutoff; Red: negative results or predicted non-binders on HLA-A*02:01.
To determine the presence of these predicted peptides and others, three previously published physical methods were performed, including MAE, CoIP, and sMHC-IP (Fig. 1). The MAE protocol uses an acidic buffer (pH 3.3) to dissociate pMHC I complexes. It was applied on both monoallelic (K562-A2-PAP) and multiallelic (M202-PAP) HLA-A*02:01+ cell lines. K562-A2-PAP is considered a mono-HLA-allele cell line because wild-type K562 cells are deficient in surface MHC I (28). K562-PAP (without HLA-A2) and wild-type M202 (without PAP) were treated by MAE and used as controls for background signals. This strategy identified 11 PAP peptides in total (SI Appendix, Fig S1A and Table S1 and Table 1). It is worth noting that none of the peptides from the MAE results have high predictive affinity by either NetMHC4.0 or NetMHCpan4.0. This is possibly due to the nonspecific release of surface peptides/proteins, which has been reported in other publications (29).
Fig. 1.
Diagram of overall project flowchart and summary of TCR screening process.
An alternative approach, CoIP, was performed on the same two cell lines. This method uses a monoclonal antibody (clone W6/32) to enrich for MHC I released from cell surfaces after mild detergent lysis (20, 21, 29). K562-PAP and M202 were also treated by the same protocol as negative controls. Peptides bound to MHC I are then dissociated from purified products and analyzed by LC-MS/MS. Twelve PAP peptides were recovered by CoIP (SI Appendix, Fig S1A and Table S1 and Table 1). Two peptides overlapped with those found by MAE (SI Appendix, Fig. S1A and Table S1). Both peptides from K562-A2-PAP CoIP results have moderate predicted affinities for binding to HLA-A*02:01 (Table 1). Peptides from M202-PAP CoIP have relatively lower predicted ranking (SI Appendix, Table S1), possibly due to other HLA alleles expressed on M202.
The sMHC-IP was previously developed to enforce higher expression of an engineered soluble form of MHC I as a single-chain dimer (22, 23). A recently published sMHC-IP platform, ARTEMIS, achieves robust expression and secretion of soluble HLA-A*02:01 molecules (23). This engineered form contains a hexa-histidine-tag (6× His tag) to increase enrichment efficacy by Ni-NTA agarose. Eight peptides were recovered in sMHC-IP, including six not found in the other two physical methods (SI Appendix, Fig S1A and Table S1 and Table 1). Six out of eight peptides from sMHC-IP have high predicted affinity (<1,000 nM) by netMHC 4.0 (Table 1 and SI Appendix, Table S1).
Combined, these three physical methods yielded 27 PAP peptides in total. Basic Local Alignment Search Tool (BLAST) analysis was then performed on all the physically recovered PAP epitopes against the human protein library to test their specificity for PAP (30). All 27 PAP peptides are unique to PAP sequences. Peptides with similar sequences mostly came from other members of the acid phosphatase family such as lysosomal acid phosphatase and testicular acid phosphatase (SI Appendix, Table S3).
Evaluating HLA-A*02:01 Specificity of Recovered PAP Peptides.
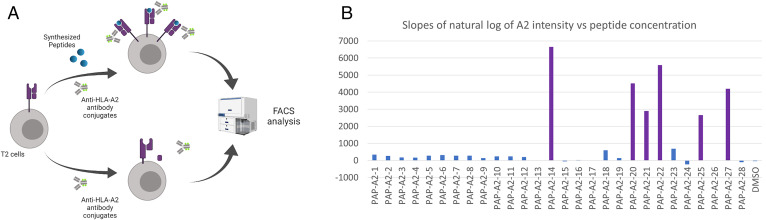
A subset of the 27 peptides might not be HLA-A2 restricted: They could originate from either contaminating peptides or non-A2 HLA alleles. To assess the HLA-A*02:01 specificity of recovered PAP epitopes, T2 cell binding assays were performed. The T2 cell line is deficient in the MHC I assembly machinery. As a result, stable MHC I can only form when exogenous peptides are added (31). The level of HLA-A*02:01 can then be quantified by anti-A2 antibodies (clone BB7.2) conjugated with Fluorescein isothiocyanate(FITC) using a flow cytometer (Fig. 2A).
Fig. 2.
Using T2 stabilization assay to assess stability of pMHC I. (A) Schematic for the overall process of T2 assays. (B) Slopes of natural log of A2 fluorescent intensity vs. diluted peptide concentration. Purple, positive candidates in T2 assays; blue, negative candidates in T2 assays.
Six PAP peptides show high HLA-A2 signal when exogenously pulsed on T2 cells (Table 1, SI Appendix, Table S1, and Fig. 2B). All six peptides can be detected by sMHC-IP, including one epitope found by both sMHC-IP and CoIP. Five out of these six peptides passed the 2% selection cutoff of NetMHCpan 4.0 as strong HLA-A*02:01 binders (Table 1 and SI Appendix, Table S1).
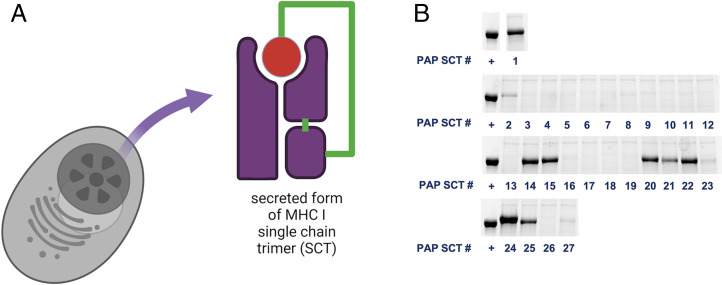
A recently developed technique, sMHC single-chain trimer (SCT), was also used to evaluate relative stability of pMHC of interest in addition to T2 assays (32). SCT assays can overcome limitations of exogenously added peptides including 1) potential posttranslational modifications and 2) peptide solubility in aqueous buffers. In the secreted SCT construct, MHC I heavy chain (HLA-A*02:01 alpha chain with H74L and Y84C mutations), light chain (beta-microglobulin), and corresponding peptide were tethered by linkers as one single-chain molecule (Fig. 3A). Constructs were expressed in cells and released into the culture supernatant. Only peptides favored by HLA-A*02:01 are expected to generate a stable SCT and have higher yields. The quantity of SCTs was measured by band intensity on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) and normalized against a well-known WT1 cancer epitope that is restricted on HLA-A*02:01 (RMFPNAPYL) (Fig. 3B) (33, 34).
Fig. 3.
Using the secreted form of MHC I single-chain trimer to assess the stability of pMHC. (A) Diagram of the SCT constructs. (B) SDS/PAGE gel results of the relative yield of each PAP SCTs comparing to positive control (+) WT1 peptide RMFPNAPYL.
Nine peptides were found to have relatively high stability on HLA-A*02:01 when using a cutoff of 0.2 normalized to the control WT1 epitope (Fig. 3B and SI Appendix, Table S1). Five out of these nine PAP peptides were not among the selected candidates by NetMHCpan 4.0 (top 2%) (Table 1). Only one peptide that scored positive in T2 assays did not form a stable SCT (SI Appendix, Table S1). This shows good overlap between the two assays. Notably, PAP_A2_24 shows a higher yield (1.40) than the positive control, but it has a poor predicted score by NetMHCpan 4.0 (24.22%) (Table 1 and Fig. 3B).
UV-labile MHC monomers were then used to test whether candidate peptides can be made into stable MHC tetramers for future use as immunostaining reagents (35). Enzyme-linked immunosorbent assay (ELISA) was used to quantify the intact MHC I monomers after UV treatment. A previously published PAP peptide, PAP-A2-14 (ILLWQPIPV), was used as positive control (17). All readouts were normalized against PAP-A2-14 (SI Appendix, Fig. S2). In general, the UV-exchange efficiency correlates well with NetMHCpan 4.0 prediction results (SI Appendix, Table S1). Combined in vitro and in silico analyses show that 11 candidate PAP peptides are restricted to HLA-A*02:01 (Table 1).
A Posttranslationally Modified PAP Peptide Shows Increased Binding Affinity to HLA-A*02:01.
PAP-A2-24 shows contradictory results of HLA-A*02:01 binding in different stability assays. One possible explanation is that PAP-A2-24 has been posttranslationally modified. Previous literature reports N-glycosylation on the asparagine of PAP-A2-24 (N220 of PAP) (36). To investigate whether the N-glycosylated form of PAP-A2-24 is presented, SCT products of both PAP-A2-24 (SVHNFTLPSW) and PAP-A2-25 (IMYSAHDTTV) were treated with PNGase F, which can specifically remove N-glycan (37). SDS/PAGE analysis of PAP-A2-24 SCT showed a band of apparent higher molecular weight than PAP-A2-25 prior to PNGase F treatment. Both SCTs migrate similar distances in the gel after deglycosylation (SI Appendix, Fig S3A). This result indicates that an additional N-glycan exists on PAP-A2-24 in addition to glycosylation sites on the MHC I heavy and light chains. The only sequence difference between PAP-A2-24 SCT and PAP-A2-25 SCT resides in their peptide fragments. It is very likely that the additional N-glycan was within the peptide (SVHNFTLPSW).
Spectrums were also reanalyzed to confirm whether the deglycosylated form of PAP-A2-24 also exists. Previous reports suggests that N-glycosylated asparagine (N) can undergo enzymatic deamidation to aspartate (D) (38). Both forms were detected by LC-MS in CoIP results: SVHNFTLPSW and SVHDFTLPSW (SI Appendix, Fig. S3B).
Isolation of PAP Peptide-Specific TCRs from Healthy Individuals’ PBMCs.
PBMC cells collected from multiple commercially available normal donors (n > 20) were screened to find TCRs reactive to PAP peptides. Twenty-seven chemically synthesized peptides were added to total PBMCs, which contain a mixture of antigen presenting cells (e.g., monocytes and B cells) that are able to prime T cells. The T cells were then allowed to expand for 10 d in culture. The CLInt-seq protocol was then applied on those stimulated cells to isolate reactive candidate T cells (26). The TNFα+/IFNγ+ positive CD8 T cell population was sorted by FACS to enrich for the reactive population. TCR pairs appearing more than once in 10× Genomics sequencing results were selected as potential PAP-reactive clones. One hundred twenty-four candidate α/β pairs were recovered from eight healthy individuals, including three females, four males, and one unknown (SI Appendix, Table S4).
TCR variable regions of both alpha and beta chains from all selected candidates were then synthesized into DNA fragments for cloning. Constant regions of both alpha chain and beta chain (TRAC and TRBC) were replaced by mouse constant regions to decrease mispairing with endogenous human TCRs. Paired TCR alpha chain and beta chain were linked with a mutated self-cleaving 2A peptide linker (F2Aopt) to ensure equal expression (39).
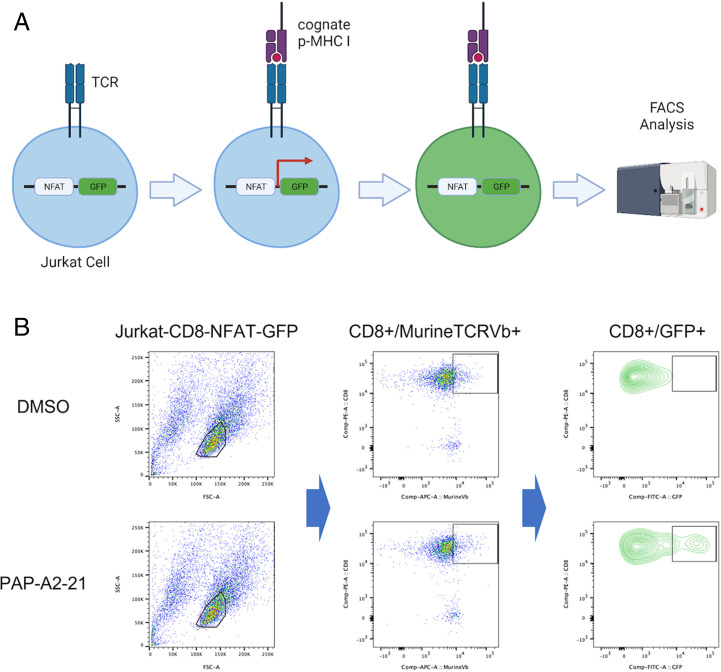
Engineered TCR sequences were then cloned into the pMAX-Cloning vector for rapid functional screening using electroporation. The pMAX constructs containing a TCR of interest were electroporated into the Jurkat-CD8-NFAT-GFP cell line, which is used as a reporter system. In Jurkat-CD8-NFAT-GFP cells, GFP expression is induced by the binding and activation of NFAT promoter repeats after TCR activation (Fig. 4A). GFP expression can then be quantified by flow cytometry to determine whether a TCR recognized cognate pMHC I. Murine TCR beta chain was measured by FACS to estimate transfection efficiency. K562 cells were transduced with HLA-A*02:01-IRES-GFP (K562-A2) by lentivirus and used as target cells during the test. Individual chemically synthesized PAP peptides were added into and presented by K562-A2 cells. Effector cells (Jurkat) and target cells (K562) were mixed at a ratio of 2:1. From 124 candidate clones, 21 TCRs (17%) were found to recognize seven distinct PAP peptides defined previously by LC-MS (Table 1 and SI Appendix, Tables S4 and S5).
Fig. 4.
Testing candidate TCRs in Jurkat-NFAT-GFP for rapid screening. (A) Schematic illustration of the Jurkat-NFAT-GFP system for TCR screening. (B) Example (TCR-218) FACS results of Jurkat-NFAT-GFP screening. (Top) Dimethyl sulfoxide (DMSO) as negative control with TCR-218. (Bottom) PAP-A2-21 as positive hit with TCR-218.
Functional Validation of Candidate TCRs in Human PBMCs.
Candidate TCRs that showed reactivity in the Jurkat-CD8-NFAT-GFP system were then tested in human PBMC cells. The selected TCR constructs with mouse constant regions were coexpressed with truncated low-affinity nerve growth factor receptor (delta LNGFR) as a transduction marker. Candidate TCRs were transduced into human PBMCs with the pMSGV retroviral system (5) (see Materials and Methods). Surface dLNGFR level was measured by FACS to ensure similar levels of TCR expression. Murine TCR beta chain was also quantified by FACS to assess whether TCRs traffic to the cell surface.
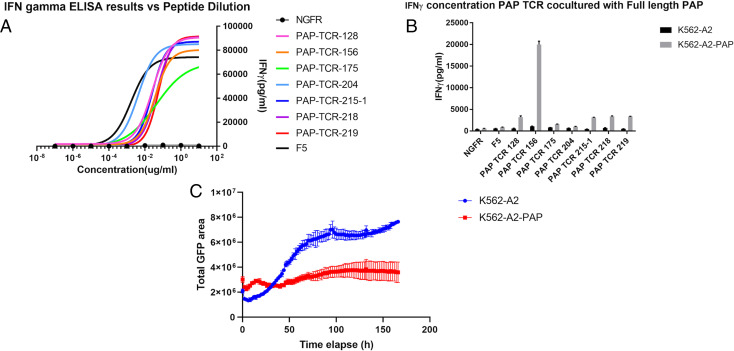
Stimulated T cells that recognize cognate peptide bound to MHC I can release cytokines such as IFNγ. ELISA was performed to quantify released IFNγ by using recombinant IFNγ as a standard (see Materials and Methods). Individual PAP peptides were added exogenously to K562-A2 cells. Engineered PBMCs and target K562-A2 cells were mixed at a ratio of 2:1 (effector:target). The supernatants of the coculture experiments were then collected after 48 h. Seven TCRs showed significant IFNγ signal against three distinct PAP peptides when expressed in human PBMCs (Fig. 5A and Table 1). MHC tetramers made from cognate peptide (PAP-A2-14,21 or 22) showed specific staining with their matched TCRs (SI Appendix, Figs. S2 and S4 and Table 1).
Fig. 5.
Functional test of candidate TCRs with various methods. (A) IFNγ results of different TCR constructs on PBMCs with peptide dilution; F5, positive control against MART1 peptide (EAAGIGILTV); NGFR, negative control with DMSO. (B) IFNγ results of candidate TCR constructs on PBMCs with cell lines with or without full-length PAP; black bars, TCR-engineered PBMCs with K562-A2; gray bars, TCR-engineered PBMCs with K562-A2-PAP. (C) Cytotoxicity curve of TCR-156 by Incucyte using total GFP signals of target cells to quantify target cell number; blue, K562-A2 target cells with TCR-156 engineered PBMCs; red, K562-A2-PAP target cells with TCR-156 engineered PBMCs.
TCRs generating significant IFNγ signal in PBMCs were tested with a serial dilution of cognate peptides to compare their relative potency with a clinically tested TCR, F5. This TCR was previously isolated from a melanoma patient reactive with a MART1 epitope (EAAGIGILTV) (40). Chemically synthesized peptides were tested at various concentrations on K562-A2. PBMCs expressing candidate PAP TCRs were mixed at a ratio of 2:1 (effector:target). IFNγ ELISA was performed on the collected supernatant after 48 h. One PAP TCR (PAP-TCR-204) shows a similar level of activation compared to F5 by peptide dilutions, while the remaining six TCRs showed weaker activation (Fig. 5A).
PBMCs expressing these seven TCRs were then cocultured with target cells expressing full-length PAP to test their ability for recognizing processed PAP epitopes. Full-length PAP isoform 2 (TM-PAP) was transduced into the K562-A2 cell line by lentiviral vector. TCR-engineered PBMCs were mixed with target K562-A2-PAP cells at a ratio of 16:1 (effector:target). The F5 TCR and dLNGFR only (without TCR) empty vector transduced PBMCs were used as negative controls. ELISA was performed on coculture supernatant after 48 h. One TCR (PAP-TCR-156) showed specific full-length PAP recognition with IFNγ produced at 20,000 pg/mL (Fig. 5B), compared to 80,000 pg/mL when cocultured with exogenously added peptides (Fig. 5A). Four TCRs (128, 215-1, 218, and 219) produced a low IFNγ signal (around 3,000 pg/mL) (Fig. 5B). PAP-TCR-204, which shows the highest reactivity by peptide dilution assay, surprisingly did not produce a significant level of IFNγ (Fig. 5B). The uncorrelated IFNγ release between peptide pulsing and full-length PAP is possibly the result of different efficiencies of peptide presentation following cellular processing.
Cytotoxicity of the candidate PAP TCRs was assessed by recording total live target cells using the IncuCyte platform. Target K562-A2-PAP cells coexpress GFP and can be distinguished from GFP− PBMC cells by real-time imaging and analysis. Live-cell imaging was taken every 2 h to record the number of target cells over a time course of 120 h. GFP signals were then processed by the IncuCyte analysis tool to estimate the area of target cells. PAP-TCR-156, which shows moderate IFNγ signal by ELISA, can inhibit growth of cells expressing full-length PAP (Fig. 5C). The total GFP area of K562-A2-PAP is maintained at a similar level during the 150-h coculture with PBMCs expressing PAP-TCR-156 (Fig. 5C). The total GFP area for K562-A2 cells showed a threefold increase compared to the experimental group (Fig. 5C). The target of PAP-TCR-156, PAP-A2-22, is the only peptide that is recovered by both CoIP and sMHC-IP while meeting all the A2-restriction verification steps (SCT assay, T2 assay, and in silico prediction) (Table 1).
Discussion
Cell-mediated therapy of cancer has been proven quite effective in treating lymphoma (CD19 target) and myeloma (BCMA target) with Chimeric Antigen Receptor(CAR) modified autologous T cell populations, with several recently approved drugs now available (41–44). Dozens of additional cell surface expressed proteins are being developed as targets on a wide range of blood cell–derived and solid tissue cancers for new CARs (45). All of these programs must consider the expression of the target antigen on nontumor tissues and the possible toxicities which may result.
Strategies to target patient-specific or shared neoantigens generated from mutated cellular proteins with TCRs can also provide selective killing of cancer cells with lower toxicity, as the targeted mutation sequence is only found within the tumor (46). The restriction of the target antigen(s) to an individual or low mutational burdens in certain tumor types can limit this approach (46).
Tissue antigens which are commonly expressed among specific groups of cancers can present epitopes with specific MHC restriction and be targeted by TCRs. Excellent examples include NY-ESO-1 in certain sarcomas and MART1 in melanoma (1, 4, 47, 48). Of the over 162,868 reported TCR alpha/beta paired sequences available in the IEDB public database to date, less than 1% (1,620) are categorized as cancer related (25). Our evaluation of PAP as a potential TCR target was motivated by its abundant expression in a very high percentage of prostate cancers, its highly restricted tissue expression pattern, and T cell activation data available from patients treated with Sipuleucel-T (8–12, 49).
We anticipated that specific TCRs reactive with PAP epitopes would be rare and of modest affinity, since previous efforts to isolate melanoma antigen-specific TCRs reported an 87% false positive rate (48). We used a combination of physical and computational approaches to define 11 peptide epitopes of PAP that could be presented by the most common HLA-A*02:01 allele (24). Although the number of peptides defined from any one approach was fairly small, the most informative approach seemed to be the sMHC technology by the ARTEMIS platform, which is easily adaptable to multiple alleles of HLA and any protein antigen (23). We used these PAP peptides to stimulate large numbers of PBMCs from normal individuals. From these stimulated T cell populations, we were able to molecularly clone a set of TCRs with specificity for PAP peptides. Some can respond to processed PAP antigen in cell culture models.
The affinity of selected TCRs will have to be improved for further evaluation in preclinical models. We are currently employing a number of strategies, including targeted mutagenesis of the complementarity-determining regions and the introduction of residues with “catch bond” function for affinity maturation of selected PAP-specific TCRs (47, 50).
Alternative sources of T cells with reactivity for PAP peptides are also being evaluated. Recent improvements in the in vitro culture of T cells derived from stem cell sources in artificial thymic organoids can provide cells which have not undergone negative selection and may produce cells of higher affinity and alternative HLA restriction (51–53).
Materials and Methods
MAE.
MAE protocol to elute MHC I–associated peptides was mainly based on previously published protocol with a few changes (54); 1 × 108 to 2 × 108 cells were used. M202-PAP cells were dissociated with 1× Phosphate Buffered Saline(PBS) + 1mM (ethylenedinitrilo)tetraacetic acid (EDTA), while K562-A2-PAP cells were collected by spinning down at 1,500 RPM for 5 mins. Target cells were then washed three times with 1× Hanks’ balanced salt solution buffer (Thermo Fisher); 25 mL of MAE buffer (0.131M citric acid, 0.066M Na2HPO4, 150 mM NaCl, 0.3 μM Aprotinin, 5 mM Iodoacetamide, pH = 3.3) was applied to target cells and gently rocked for 2 min at room temperature. Samples were then spin at 4,000 × g for 5 mins at 4 °C, and supernatant was harvested. Formic acid was added to the samples to reach a final concentration of 0.1% (vol/vol); 3 mL of C18 solid phase extraction cartridge (3M) was prerinsed by 99.9% acetonitrile (ACN) + 0.1% formic acid three times. MAE samples were then added to the C18 column followed by washing three times with 0.1% formic acid in water. The C18 column was then eluted with 200 μL of 40% ACN + 5% formic acid + 55% H2O three times. Samples were then passed through 3-kDa centrifugal filters (Millipore) for 90 mins at 4 000 × g at 4 °C. Flow-through was then dried by vacuum centrifugation and stored in −20 °C until MS analysis.
MHC I CoIP.
CoIP protocol was modified based on previous published procedures (55, 56); 1 × 108 to 2 × 108 M202-PAP or K562-A2 PAP cells were collected either by nonenzymatic dissociation reagents (1XPBS+1mM EDTA) or by spinning down with 1,500 rpm for 5 mins. Cells were first washed three times with 1XPBS. Cells were then lysed with CoIP lysis buffer (20 mM Tris [pH8.0], 1 mM EDTA, 100 mM NaCl, 1% Triton X-100, 60 mM n-octyl glucoside, 1 mM phenylmethylsulfonyl fluoride[PMSF] [Sigma-Aldrich], protease inhibitor [Roche Life Science] and 1mg/mL DNase I [Roche Life Science]) with 1 mL of lysis buffer per 107 cells. Samples were then rocked for 1 h at 4 °C. Lysates were then centrifuge at 10,000 × g for 20 min to pellet debris. Supernatant was then combined with GammaBind Plus Sepharose beads (GE Lifesciences) that have been conjugated with W6/32 antibodies (BioXCell) at the ratio of 1 mL of beads per 108 cells. Mixture of beads and lysates were rocked at 4 °C for 180 min. Mixture was then loaded on to Poly-Prep Chromotography Column (Bio-Rad). The column was then washed four times with 10 mL of wash buffer I (CoIP wash buffer I: 20mM Tris [pH8.0], 1 mM EDTA, 100 mM NaCl, 60 mM n-octyl glucoside, and 1mg/mL DNase I), four times with 10 mL of wash buffer II (CoIP wash buffer II: 10mM Tris [pH8.0]), and one time with 10 mL of Ultrapure H2O (Thermo Fisher). Peptides were released from beads by adding 10% Acetic Acid (Sigma) for 2 min and cleaned up by spinning for 30 s at 3,000 × g with 0.45-μm Costar Spin-X centrifuge tube filters (Corning). Samples were then snap frozen and stored at −70 °C until further processing.
sMHC-IP with ARTEMIS Protocol.
The ARTEMIS protocol was based on a previously published protocol (23). Expression of both the secreted form of HLA-A2 and PAP was achieved by using lentiviral transduction system in free-style 293-F cells (Thermo Fisher); 400 mL of supernatant containing sMHC I was purified by Ni-NTA agarose (1 μL of slurry per 1 mL of supernatant). Slurry was loaded and washed in Poly-Prep Chromatography column. Samples after denaturation were stored in −70 °C until further processing.
LC-MS Analysis.
Eluted samples were loaded to HyperSep C18 Column (Thermo Scientific 60108–390) and washed three times with 0.1% formic acid, then eluted with elution buffer (40% Acetonitrile, 0.1% formic acid). Desalted samples were lyophilized by speed vacuum and then reconstituted in the water. Next, samples were processed with a detergent removal kit to remove residual detergent from the lysis buffer. Finally, the samples were acidified to contain 5% formic acid before being loaded to LC-MS. Samples were delivered to Orbitrap Fusion Lumos hybrid mass spectrometer by a 140-min gradient (0 min to 5 min, 1 to 5.5% B; 5 min to 128 min, 5.5 to 27.5% B; 128 min to 135 min, 27.5 to 35% B; 135 min to 136 min, 35 to 80% B; 136 min to 138 min, 80% B; 138 min to 138.5 min, 80 to 1% B; 138.5 min to 140 min, 1% B; B: 80% CAN+0.1% formic acid). The acquisition was conducted under data-dependent acquisition mode: The full MS scan was acquired under 120K resolution in the Orbitrap mass analyzer, and singly charged ions with >800 m/z and multicharged ions were selected to be fragmented with high-energy collision dissociation at 32% collision energy, and then we performed an MS/MS scan under 15K resolution in Orbitrap. Dynamic exclusion was enabled to not repeat selecting ions with same m/z in 60 s. A database search was performed using Crux pipeline (v3.2) against European Molecular Biology Laboratory(EMBL) human reference proteome (UP000005640human_9606), with nonspecific digestion. Peptide-Spectrum Match(PSM) and peptide False Discovery Rate(FDR) is set to 1% threshold.
T2 Peptide Binding Assay.
T2 cells (ATCC) were cultured in Iscove's Modified Dulbecco's Medium(IMDM) (Thermo Fisher) with 20% fetal bovine serum (FBS) (Omega Scientific). Before peptide loading, 2 × 105 cells were resuspended in 100 μL of serum-free Roswell Park Memorial Institute(RPMI) media(Thermo Fisher) and added into each well of 96 U-bottom tissue culture plates (Corning). Chemically synthesized peptides were diluted into multiple concentrations with serum-free RPMI and added into designated wells with T2 cells. Cells with peptides were cocultured overnight in an incubator at 37 °C. Cells were then washed two times with 1XPBS and stained with 2 μL per well anti-HLA-A2 FITC antibodies (clone BB7.2, Biolegend). The quantity of HLA-A2 molecules was quantified by FACS.
SCT Quantification Assay.
SCT constructs (mutant H74L/Y84C) with individual PAP peptides were synthesized according to the previously published protocol (32).
Cell Culture.
K562 (ATCC), M202 (gift from A. Ribas at University of California, Los Angeles, CA [UCLA]), and Jurkat-NFAT-ZsGreen (gift from D. Baltimore at California Institute of Technology, Pasadena, CA) were cultured in RPMI 1640 (Thermo Fisher) with 10% FBS (Omega Scientific) and Glutamine (Fisher Scientific); 293T (ATCC) was cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher) with 10% FBS and Glutamine. Naïve PBMCs for stimulation were cultured in TCRPMI with 50 U/mL IL-2 (Peprotech) and chemically synthesized PAP peptides of interest (>80% purity, Elim Biopharm) as previously described (57). TCRPMI media includes RPMI 1640 (Thermo Fisher), 10% FBS (Omega Scientific), Glutamax (Thermo Fisher), 10 mM Hepes (Thermo Fisher), nonessential amino acids (Thermo Fisher), sodium pyruvate (Thermo Fisher), and 50 μM β-mercaptoethanol (Sigma). PBMCs for retroviral transduction were first activated by CD3/CD28 dynabeads (Thermo Fisher) and cultured in T cell media: AIM V media (Thermo Fisher), 5% Human AB serum (Omega Scientific), 50 U/mL IL-2 (Peprotech), 0.5 ng/mL IL-15 (Peprotech), Glutamax (Thermo Fisher), and 50 μM β-mercaptoethanol (Sigma).
Generation of Cell Lines Expressing Full-Length PAP.
The pCCL-MNDU3-PAP2-IRES-mSTR is a gift from G.M.C.’s laboratory and C.S.S.’s laboratory at UCLA, Los Angeles, CA. Lentivirus was generated as described previously and infected K562-A*02:01 and M202 cells (51). Transduced cells were then single-cell cloned by FACS deposition based on high A*02:01 and PAP expression. Stable A*02:01 expression is confirmed by FACS staining using anti-HLA-A2 antibodies (clone BB7.2), and PAP expression is confirmed by Western blot (clone 15840-1-AP).
CLInt-seq.
Isolation of reactive T cells by CLInt-seq was performed on stimulated PBMCs according to a previously published protocol (26). After 7- to 10-d coculture with the PAP peptide pool, PBMCs were transferred into a 96-well U plate and rested overnight. Cells were then cultured with 10 μg/mL peptide pool and 1 μg/mL CD28/49d antibodies (BD Biosciences) for 1 h before adding Brefeldin A (Biolegend). After about 8 h incubation at 37 °C, cells were treated as previously described and stained for CD3+/CD4−/CD8+/TNFα+/IFNγ+ population by FACS (57).
Single-Cell TCR Sequencing.
CD8+ T cells that produce both TNFα and IFNγ were sorted into ∼30 μL of 0.04% bovine serum albumin solution. If fewer than 1,000 cells were isolated, 5,000 to 10,000 K562 cells would be sorted into the same tube as carrier population. The 10× Genomics’ single-cell TCR V(D)J library was then constructed by the UCLA Technology Center for Genomics & Bioinformatics. TCR pairs were then sequenced on MiSeq (Illumina).
Jurkat-NFAT-GFP Assay.
Candidate TCRs were rapidly screened in Jurkat-NFAT-GFP cells as described previously (57).
Transduction of TCRs in PBMC.
Engineering of candidate TCRs in PBMC was performed according to previous publication (57).
Preparation of MHC Tetramers.
MHC tetramers used to stain candidate PAP TCRs were synthesized and prepared according to a previously published protocol (35).
T Cell Activation Analysis.
For peptide pulsing coculture experiments, target cells were mixed with TCR-engineered PBMCs at a ratio of 1:2 (T:E) in the media desired by target cells and supplemented with 1 µg/mL of anti-CD28/CD49d antibodies (BD Biosciences). For cell lines expressing full-length PAP, target cells were first treated with 2 ng/mL IFNγ and 3 ng/mL TNFα for 8 h to 10 h. Target cells and PBMCs were then mixed at a ratio of 1:16 (T:E) for coculture analysis. Supernatants were collected after 48 h and analyzed by ELISA (BD Biosciences) to estimate IFNγ concentration.
Cytotoxicity Analysis by IncuCyte.
Target cells were plated onto 96-well tissue culture plates coated with 0.001% poly-L-lysine (Sigma) and kept in 37 °C for ∼2 h. TCR-engineered PBMCs were then added to desired wells with effector: target ratio of 2:1 (peptide pulsed target cells) or 16:1 (full length PAP target cells). Plates with cell mixtures were analyzed by the IncuCyte system for 120 h using GFP surface area to estimate killing of T cells.
Supplementary Material
Acknowledgments
We thank Dr. Antoni Ribas for providing M202 cell lines. The Jurkat-NFAT-GFP reporter system is a gift from C.S.S.’s laboratory (UCLA) and Dr. David Baltimore’s laboratory (California Institute of Technology). TCR libraries and sequencing were constructed and performed by Technology Center for Genomics and Bioinformatics at UCLA (National Cancer Institute Grant P30CA016042). This project is supported by NIH R01 Grant AI121242 (R.K.S.), Fred Hutchinson Human Biology Division Pilot Project Award (R.K.S. and J.K.L.), NIH R01 Grant CA264090-01 (J.R.H.), National Cancer Institute U01 Grant CA233074 (O.N.W. and G.M.C.), Parker Institute for Cancer Immunotherapy (O.N.W. and J.R.H.), and UCLA Broad Stem Cell Research Center (O.N.W.). D.C.D. is supported by Department of Defense Prostate Cancer Research Program Award W81XWH2010119. P.A.N. was supported by the UCLA Tumor Immunology Training Grant T32 CA009056. Z.M. is supported by the UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Training Program predoctoral fellowship. Figs. 2–4 were created with BioRenders.com.
Footnotes
Reviewers: V.E., University of Virginia; and P.W.K., Convergent Therapeutics.
Competing interest statement: Z.M., P.A.N., J.M., J.K.L., and O.N.W. are inventors on a provisional patent application titled “Human T cell receptor pairs reactive with HLA-A*02:01 restricted human prostatic acid phosphatase (PAP) epitopes.” O.N.W. currently has consulting, equity and/or board relationships with Trethera Corporation, Kronos Biosciences, Sofie Biosciences, Breakthrough Properties, Vida Ventures, Nammi Therapeutics, Two River, Iconovir, Appia BioSciences, Neogene Therapeutics, 76Bio, and Allogene Therapeutics. J.R.H. is a board member of PACT Pharma and Isoplexis. J.R.H. also received research funding from Merck and Gilead Sciences for COVID-19 research. None of these companies contributed to or directed any of the research reported in this article.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2203410119/-/DCSupplemental.
Data Availability
TCR sequences will be deposited into the IEDB online. All other study data are included in the article and/or SI Appendix. All data are included in the article and/or SI Appendix. TCR and epitope information will also be uploaded and disclosed by the publicly available IEDB online database (https://www.iedb.org) after publication.
References
- 1.Rosenberg S. A., A new era for cancer immunotherapy based on the genes that encode cancer antigens. Immunity 10, 281–287 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Srinivasan R., Wolchok J. D., Tumor antigens for cancer immunotherapy: Therapeutic potential of xenogeneic DNA vaccines. J. Transl. Med. 2, 12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinrichs C. S., Restifo N. P., Reassessing target antigens for adoptive T-cell therapy. Nat. Biotechnol. 31, 999–1008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson L. A., June C. H., Driving gene-engineered T cell immunotherapy of cancer. Cell Res. 27, 38–58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson L. A., et al. , Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 114, 535–546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebello R. J., et al. , Prostate cancer. Nat. Rev. Dis. Primers 7, 9 (2021). [DOI] [PubMed] [Google Scholar]
- 7.GTEx Consortium, The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jöbsis A. C., De Vries G. P., Meijer A. E., Ploem J. S., The immunohistochemical detection of prostatic acid phosphatase: Its possibilities and limitations in tumour histochemistry. Histochem. J. 13, 961–973 (1981). [DOI] [PubMed] [Google Scholar]
- 9.Seamonds B., et al. , Evaluation of prostate-specific antigen and prostatic acid phosphatase as prostate cancer markers. Urology 28, 472–479 (1986). [DOI] [PubMed] [Google Scholar]
- 10.Moul J. W., Connelly R. R., Perahia B., McLeod D. G., The contemporary value of pretreatment prostatic acid phosphatase to predict pathological stage and recurrence in radical prostatectomy cases. J. Urol. 159, 935–940 (1998). [PubMed] [Google Scholar]
- 11.Kantoff P. W., et al. ; IMPACT Study Investigators, Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 363, 411–422 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Kibel A. S., et al. , Videos of sipuleucel-T programmed T cells lysing cells that express prostate cancer target antigens. J. Natl. Cancer Inst. 114, 310–313 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoof I., et al. , NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurtz V., et al. , NetMHCpan-4.0: Improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 199, 3360–3368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Donnell T. J., et al. , MHCflurry: Open-source class I MHC binding affinity prediction. Cell Syst. 7, 129–132.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Fong L., et al. , Dendritic cell-based xenoantigen vaccination for prostate cancer immunotherapy. J. Immunol. 167, 7150–7156 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Becker J. T., et al. , DNA vaccine encoding prostatic acid phosphatase (PAP) elicits long-term T-cell responses in patients with recurrent prostate cancer. J. Immunother. 33, 639–647 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells D. K., et al. ; Tumor Neoantigen Selection Alliance, Key parameters of tumor epitope immunogenicity revealed through a consortium approach improve neoantigen prediction. Cell 183, 818–834.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storkus W. J., Salter R. D., Cresswell P., Dawson J. R., Peptide-induced modulation of target cell sensitivity to natural killing. J. Immunol. 149, 1185–1190 (1992). [PubMed] [Google Scholar]
- 20.Hunt D. F., et al. , Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Engelhard V. H., Structure of peptides associated with MHC class I molecules. Curr. Opin. Immunol. 6, 13–23 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Bassani-Sternberg M., et al. , Soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proc. Natl. Acad. Sci. U.S.A. 107, 18769–18776 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finton K. A. K., et al. , ARTEMIS: A novel mass-spec platform for HLA-restricted self and disease-associated peptide discovery. Front. Immunol. 12, 658372 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao K., et al. , Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 62, 1009–1030 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Vita R., et al. , The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 47, D339–D343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nesterenko P. A., et al. , Droplet-based mRNA sequencing of fixed and permeabilized cells by CLInt-seq allows for antigen-specific TCR cloning. Proc. Natl. Acad. Sci. U.S.A. 118, e2021190118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng G. X., et al. , Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Britten C. M., et al. , The use of HLA-A*0201-transfected K562 as standard antigen-presenting cells for CD8(+) T lymphocytes in IFN-gamma ELISPOT assays. J. Immunol. Methods 259, 95–110 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Purcell A. W., Ramarathinam S. H., Ternette N., Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc. 14, 1687–1707 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Madden T. L., Tatusov R. L., Zhang J., Applications of network BLAST server. Methods Enzymol. 266, 131–141 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Henderson R. A., et al. , HLA-A2.1-associated peptides from a mutant cell line: A second pathway of antigen presentation. Science 255, 1264–1266 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Chour W., et al. , Shared antigen-specific CD8+ T cell responses against the SARS-COV-2 spike protein in HLA-A*02:01 COVID-19 participants. medRxiv [Preprint] (2020). 10.1101/2020.05.04.20085779. Posted May 08, 2020 [DOI]
- 33.Maslak P. G., et al. , Vaccination with synthetic analog peptides derived from WT1 oncoprotein induces T-cell responses in patients with complete remission from acute myeloid leukemia. Blood 116, 171–179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dao T., et al. , Targeting the intracellular WT1 oncogene product with a therapeutic human antibody. Sci. Transl. Med. 5, 176ra33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodenko B., et al. , Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 1, 1120–1132 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Jakob C. G., Lewinski K., Kuciel R., Ostrowski W., Lebioda L., Crystal structure of human prostatic acid phosphatase. Prostate 42, 211–218 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Maley F., Trimble R. B., Tarentino A. L., Plummer T. H. Jr., Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180, 195–204 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Skipper J. C., et al. , An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med. 183, 527–534 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu K. K., et al. , Use of mutated self-cleaving 2A peptides as a molecular rheostat to direct simultaneous formation of membrane and secreted anti-HIV immunoglobulins. PLoS One 7, e50438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chodon T., et al. , Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma. Clin. Cancer Res. 20, 2457–2465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davila M. L., Brentjens R. J., CD19-Targeted CAR T cells as novel cancer immunotherapy for relapsed or refractory B-cell acute lymphoblastic leukemia. Clin. Adv. Hematol. Oncol. 14, 802–808 (2016). [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson H. J., Rafiq S., Brentjens R. J., Driving CAR T-cells forward. Nat. Rev. Clin. Oncol. 13, 370–383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park J. H., Geyer M. B., Brentjens R. J., CD19-targeted CAR T-cell therapeutics for hematologic malignancies: Interpreting clinical outcomes to date. Blood 127, 3312–3320 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roex G., et al. , Chimeric antigen receptor-T-cell therapy for B-cell hematological malignancies: An update of the pivotal clinical trial data. Pharmaceutics 12, 194 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albinger N., Hartmann J., Ullrich E., Current status and perspective of CAR-T and CAR-NK cell therapy trials in Germany. Gene Ther. 28, 513–527 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poorebrahim M., et al. , TCR-like CARs and TCR-CARs targeting neoepitopes: An emerging potential. Cancer Gene Ther. 28, 581–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robbins P. F., et al. , Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J. Immunol. 180, 6116–6131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson L. A., et al. , Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. J. Immunol. 177, 6548–6559 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamey T. A., et al. , Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N. Engl. J. Med. 317, 909–916 (1987). [DOI] [PubMed] [Google Scholar]
- 50.Zhao X., et al. , Tuning T cell receptor sensitivity through catch bond engineering. Science 376, eabl5282 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seet C. S., et al. , Generation of mature T cells from human hematopoietic stem and progenitor cells in artificial thymic organoids. Nat. Methods 14, 521–530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shukla S., et al. , Progenitor T-cell differentiation from hematopoietic stem cells using Delta-like-4 and VCAM-1. Nat. Methods 14, 531–538 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Klein L., Kyewski B., Allen P. M., Hogquist K. A., Positive and negative selection of the T cell repertoire: What thymocytes see (and don’t see). Nat. Rev. Immunol. 14, 377–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lanoix J., et al. , Comparison of the MHC I immunopeptidome repertoire of B-cell lymphoblasts using two isolation methods. Proteomics 18, e1700251 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Bassani-Sternberg M., et al. , Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 7, 13404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abelin J. G., et al. , Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nesterenko P. A., et al. , HLA-A∗02:01 restricted T cell receptors against the highly conserved SARS-CoV-2 polymerase cross-react with human coronaviruses. Cell Rep. 37, 110167 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TCR sequences will be deposited into the IEDB online. All other study data are included in the article and/or SI Appendix. All data are included in the article and/or SI Appendix. TCR and epitope information will also be uploaded and disclosed by the publicly available IEDB online database (https://www.iedb.org) after publication.