Significance
N-degron pathways target proteins for degradation by recognizing their N-terminal residues. A destabilizing N-terminal Arg residue can be generated by a proteolytic cleavage of a protein either directly or after N-terminal arginylation of the resulting C-terminal fragment by the Ate1 arginyl-tRNA-protein transferase (R-transferase). A three-dimensional structure of Ate1 is unknown. We describe here the crystal structure of the Ate1 R-transferase from the yeast Kluyveromyces lactis. We also describe results of enzymatic and functional assays with wild-type Ate1 and its mutants to address specific structural findings. These and related results advance the understanding of R-transferase and the Arg/N-degron pathway.
Keywords: arginine, Ate1, hemin, degron, ubiquitin
Abstract
N-degron pathways are proteolytic systems that target proteins bearing N-terminal (Nt) degradation signals (degrons) called N-degrons. Nt-Arg of a protein is among Nt-residues that can be recognized as destabilizing ones by the Arg/N-degron pathway. A proteolytic cleavage of a protein can generate Arg at the N terminus of a resulting C-terminal (Ct) fragment either directly or after Nt-arginylation of that Ct-fragment by the Ate1 arginyl-tRNA-protein transferase (R-transferase), which uses Arg-tRNAArg as a cosubstrate. Ate1 can Nt-arginylate Nt-Asp, Nt-Glu, and oxidized Nt-Cys* (Cys-sulfinate or Cys-sulfonate) of proteins or short peptides. Ate1 genes of fungi, animals, and plants have been cloned decades ago, but a three-dimensional structure of Ate1 remained unknown. A detailed mechanism of arginylation is unknown as well. We describe here the crystal structure of the Ate1 R-transferase from the budding yeast Kluyveromyces lactis. The 58-kDa R-transferase comprises two domains that recognize, together, an acidic Nt-residue of an acceptor substrate, the Arg residue of Arg-tRNAArg, and a 3′-proximal segment of the tRNAArg moiety. The enzyme’s active site is located, at least in part, between the two domains. In vitro and in vivo arginylation assays with site-directed Ate1 mutants that were suggested by structural results yielded inferences about specific binding sites of Ate1. We also analyzed the inhibition of Nt-arginylation activity of Ate1 by hemin (Fe3+-heme), and found that hemin induced the previously undescribed disulfide-mediated oligomerization of Ate1. Together, these results advance the understanding of R-transferase and the Arg/N-degron pathway.
The ubiquitin (Ub)-proteasome system (UPS) covalently conjugates Ub, a 76-residue protein, to other intracellular proteins and thereby mediates, in particular, the processive degradation of ubiquitylated proteins by the 26S proteasome (1–11). In eukaryotes, proteolytic systems called N-degron pathways are a part of the UPS (12). Prior to 2019, N-degron pathways were called “N-end rule pathways” (12).
Different N-degron pathways have in common their ability to recognize proteins that contain N-terminal (Nt) degradation signals (degrons) called N-degrons. This recognition causes degradation of the targeted proteins by the 26S proteasome and autophagy in eukaryotes and by the proteasome-like ClpAP protease in bacteria (SI Appendix, Fig. S1). A eukaryotic N-degron comprises, in particular, a destabilizing Nt-residue of a protein and its internal Lys residue(s) that act as a site of polyubiquitylation. All 20 amino acids of the genetic code can function, in cognate sequence contexts, as destabilizing Nt-residues targeted by distinct N-degron pathways (SI Appendix, Fig. S1) (12–16).
Eukaryotes contain the Arg/N-degron pathway (it recognizes, in particular, specific unacetylated Nt-residues); the Ac/N-degron pathway (it recognizes, in particular, the Nα-terminally acetylated [Nt-acetylated] Nt-residues); the Pro/N-degron pathway (it recognizes, in particular, the Nt-Pro residue); the Gly/N-degron pathway (it recognizes unmodified Nt-Gly); and the fMet/N-degron pathway (it recognizes Nt-formylated proteins) (SI Appendix, Fig. S1) (12–67).
Initially, most N-degrons are cryptic (preN-degrons). Nearly all Nt-residues that can be recognized by the Arg/N-degron pathway (Fig. 1A and SI Appendix, Fig. S1G) cannot be exposed at the N-termini of nascent proteins by Met-aminopeptidases (MetAPs), since the initially present Nt-Met would not be cleaved off by MetAPs if the second residue, to become N-terminal after the cleavage, is larger than Val (68). However, a multitude of nonprocessive intracellular proteases—including caspases, calpains, separases, and non-MetAP aminopeptidases—can function as components of N-degron pathways by mediating cleavages of specific proteins that produce C-terminal (Ct) fragments bearing destabilizing Nt-residues (20, 34, 36, 62, 69, 70). Active N-degrons can also be formed through the enzymatic Nt-deamidation, Nt-oxidation, Nt-arginylation, Nt-acetylation, Nt-leucylation, and Nt-formylation of specific proteins or their natural Ct-fragments (Fig. 1A and SI Appendix, Fig. S1).
Fig. 1.
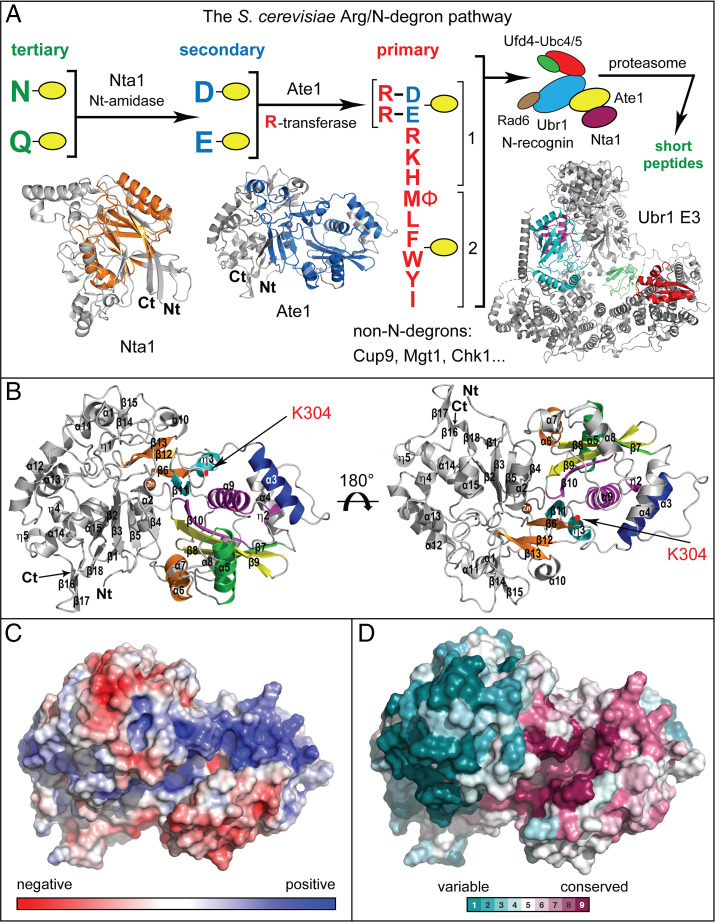
The S. cerevisiae Arg/N-degron pathway and the 3D structure of K. lactis klAte1 R-transferase. (A) The Arg/N-degron pathway (12, 49). Nt-residues are denoted by single-letter abbreviations. Yellow ovals denote the rest of a protein substrate. “Primary,” “secondary,” and “tertiary” refer to distinct classes of destabilizing Nt-residues. “1” and “2” on the right denote two sets of Nt-residues that are recognized by two distinct binding sites of the Ubr1 E3. The latter contains additional binding sites that can target not only N-degrons but also other degrons in proteins that lack N-degrons, including Cup9, Mgt1, and Chk1 (19, 48, 77, 107). Three-dimensional structures of the 52-kDa Nta1 Nt-amidase (32), the 58-kDa Ate1 R-transferase (its 3D structure was determined in the present study), and the 225-kDa Ubr1 E3 N-recognin (18) are shown as well (not to scale, because of a much larger size of Ubr1). Orange (Nta1) and blue (Ate1) colors denote strongly conserved parts of these enzymes, whose N and C termini are also marked. A multiprotein diagram on the upper right depicts the previously characterized multienzyme targeting complex of the yeast Arg/N-degron pathway (49). See first section of the article for additional references. (B) Ribbon diagrams of the 3D structure of klAte1. The conserved motifs A to D of the GNAT-fold are in green, yellow, magenta, and cyan, respectively. Yet another region of GNAT, in orange, is less conserved than the A to D motifs. The positively charged α3 α-helix, conserved among aminoacyltransferases, is in blue. Lys304, a highly conserved residue of the inferred klAte1 active site, is indicated by an arrow in both 3D orientations. (C) Electrostatic potential surface of klAte1, with negatively and positively charged regions in red and blue, respectively. (D) A surface map of sequence conservation in klAte1 vis-à-vis its sequelogs (81), produced using the OrthoDB database (108). Note a much higher evolutionary conservation of residues near the inferred active site of klAte1.
The remarkably broad functions of N-degron pathways include a selective degradation of misfolded and retrotranslocated proteins; the sensing of small compounds, such as oxygen, nitric oxide (NO), heme, and short peptides; the regulation of DNA transcription, replication, repair, and chromosome cohesion/segregation; the regulation of peptide transport, meiosis, chaperones, cytoskeletal proteins, gluconeogenesis, autophagy, apoptosis, immunity and inflammation, cardiovascular development, neurogenesis, spermatogenesis, and circadian rhythms; diverse involvements in diseases, such as cancer, neurodegeneration, and defects of immunity; a variety of roles in bacteria; and specific functions in plants, including seed germination and oxygen/NO sensing (refs. 12–67 and references therein).
The recognition components of N-degron pathways, called N-recognins, are E3 Ub ligases or other proteins that can recognize N-degrons, such as mammalian SQSTM1/p62 (a regulator of autophagy) and bacterial ClpS (a ligand of the ClpAP protease) (12, 18, 56–59, 63, 71). The extensively characterized Arg/N-degron pathway, the subject of the present study, was the first N-degron system and the first specific pathway of UPS to be discovered (Fig. 1A and SI Appendix, Fig. S1G) (12, 13, 18).
In the yeast Saccharomyces cerevisiae, Ubr1, an E3 Ub ligase, is the N-recognin of the Arg/N-degron pathway. Unmodified N-terminal Arg, Lys, His, Leu, Phe, Tyr, Trp, Ile, and Met (if Met is followed by a bulky hydrophobic residue) are “primary” destabilizing residues in that they can be directly bound by Ubr1 (Fig. 1A) (12, 18, 30, 31). In contrast, Nt-Asp and Nt-Glu are “secondary” destabilizing residues, in that they can be Nt-arginylated by the Ate1 Arg-tRNA-protein transferase (arginyltransferase or R-transferase), a nuclear/cytosolic enzyme that is present in all examined eukaryotes but is absent in bacteria. The resulting (conjugated) Nt-Arg can be bound by Ubr1. R-transferase can also Nt-arginylate an oxidized Nt-Cys* residue (Nt-Cys-sulfinate or Nt-Cys-sulfonate) of proteins or short peptides. Oxidized Nt-Cys* can form at least in some Nt-Cys–bearing proteins of multicellular eukaryotes (but apparently not in unstressed S. cerevisiae) through enzymatic reactions that involve oxygen/NO (22, 26, 52, 64, 67, 72).
Nt-Asn and Nt-Gln are “tertiary” destabilizing residues in that they can be deamidated by the S. cerevisiae Nta1 Nt-amidase, yielding Nt-arginylatable Nt-Asp and Nt-Glu (Fig. 1A) (32, 73). Multicellular eukaryotes lack the dual-specificity yeast Nta1 but contain the Nt-Asn–specific Ntan1 Nt-amidase and the Nt-Gln–specific NtNt-amidase (12, 74–76). The Ubr1 E3 N-recognin contains binding sites that can target not only N-degrons but also other degrons in proteins that lack N-degrons (12, 19, 48, 77). In S. cerevisiae, Ubr1 is the sole N-recognin of the Arg/N-degron pathway. The 225-kDa Ubr1 E3 binds to its cognate Ub-conjugating (E2) enzyme Rad6, and functions as a part of a double-E3 complex that also contains the Nta1 Nt-amidase, the Ate1 R-transferase, and the 168-kDa Ufd4 E3 bound to its E2 enzyme (Ubc4 or Ubc5). Ufd4 E3 is not an N-recognin (Fig. 1A) (49, 78, 79).
The 58-kDa Ate1 R-transferase uses Arg-tRNAArg as a cosubstrate for Nt-arginylation of proteins or short peptides (Fig. 1A). A single gene encodes R-transferase in both S. cerevisiae and mammals (21, 80). In plants, Ate1 is encoded by two sequelogous (similar in sequence) (81) genes (82). A number of physiological Ate1 substrates were either identified or inferred over the last decades (12, 26). R-transferase-lacking ate1Δ S. cerevisiae mutants are viable and not obviously abnormal (80). In contrast, a deletion of mouse Ate1 results in a midgestation embryonic lethality (22). If mouse Ate1 is deleted conditionally, in adulthood, a significant fraction of adult ate1Δ mice die, while surviving ate1Δ mice exhibit a variety of abnormal phenotypes (83).
At least in mammals, some (possibly most) proteins that are Nt-arginylated by R-transferase can be recognized not only by N-recognin E3 Ub ligases of the mammalian Arg/N-degron pathway but also by the p62 N-recognin, a regulator of autophagy (63, 84, 85). Thus, physiological substrates of the Arg/N-degron pathway can be destroyed not only by the 26S proteasome but also through autophagy. The partitioning of Arg/N-degron substrates between these proteolytic routes is a function of specific substrates and physiological states of a cell.
An earlier study described the inhibition of Nt-arginylation activity of both S. cerevisiae and mouse Ate1 R-transferases by hemin (Fe+3-heme) (53). The in vivo levels of R-transferase in mouse cells were decreased upon the addition of hemin to a growth medium, apparently through an accelerated degradation of R-transferase (53).
Ate1 genes of fungi, animals and plants have been cloned decades ago (21, 80, 82), but a three-dimensional (3D) structure of Ate1 remained unknown. A detailed mechanism of Nt-arginylation is unknown as well. We describe here the crystal structure of klAte1, the Ate1 R-transferase of Kluyveromyces lactis, a thermostable budding yeast. R-transferase is shown to comprise two spatial domains that recognize, together, an acidic Nt-residue of an acceptor substrate, the basic Arg residue of the Arg-tRNAArg cosubstrate, and a 3′-proximal segment of tRNAArg in Arg-tRNAArg. The active site of klAte1 is located, at least in part, between the two domains. Arginylation assays with site-directed klAte1 mutants that were suggested by structural results yielded inferences about specific binding sites of Ate1. We also show that the inhibition of klAte1 arginylation activity by hemin (Fe3+-heme) induces the previously undescribed disulfide-mediated oligomerization of Ate1.
Results and Discussion
Crystal Structure of klAte1.
We used the LC3B-fusion technique (86) to construct klAte1 DNA plasmids that expressed EVAA-klAte1 and DVAA-klAte1. The Nt-EVAA and Nt-DVAA extensions contained the arginylatable Nt-residues Asp (D) or Glu (E). Purified EVAA-klAte1 and DVAA-klAte1 were successfully crystallized in the space groups P43212 and P212121, and the structures were determined at 1.8- and 2.2-Å resolution, respectively (Figs. 1B–D, 2A, B, E, and F, 3A and B, and SI Appendix, Figs. S2 and S3, and Table S1). In solution, all forms of analyzed klAte1 were monomeric proteins, as indicated by gel filtration profiles of purified klAte1 (Fig. 4A and B and SI Appendix, Fig. S8D). Contrary to the (hoped for) possibility that the four-residue Nt-EVAA or the Nt-DVAA extension might interact, in klAte1 crystals, with a substrate-binding site of an adjacent klAte1 molecule, these extensions were disordered (not observed) in any of the solved crystal structures of klAte1.
Fig. 2.
Arginylation activity of klAte1 mutants. (A) The pocket of klAte1 and its residues comprising the inferred site that recognizes an acceptor substrate. A ribbon diagram is on the left and the electrostatic potential surface of the same region is on the right, with negatively and positively charged regions in red and blue, respectively. (B) The same region as in A, but in a different orientation. (C) Results of arginylation assays with L-[14C]-Arg, with 13-residue test peptides bearing either Nt-Asp (D-p) or Nt-Glu (E-p), and with purified wild-type klAte1 versus its site-directed mutants at position 80 (Arg80). (D) Same as in C but with wild-type klAte1 versus its mutants at positions 25 and 87 (Tyr25, Tyr87). (E) The region and residues of klAte1 which comprise the inferred site that recognizes the Arg residue of the Arg-tRNAArg cosubstrate, with a ribbon diagram on the left and the electrostatic potential surface of the same region on the right. (F) Same region as in E but in a different orientation. (G) Same as in C but with wild-type klAte1 versus its mutants at position 277 (Glu277). (H) Same as in C but with wild-type klAte1 versus its mutants at position 304 (Lys304). As indicated on the y axis, measured 14C-dpm (disintigrations per minute) ranged from 0 to more than 150,000 dpm. See Results and Discussion and SI Appendix, Table S2 for the values of kcat and Km determined through the use of these assays.
Fig. 3.
Arginylation assays and a model of the ternary complex of klAte1 with a substrate and cosubstrate. (A) The conserved, positively charged α3 helix of klAte1 that binds to a 3′-proximal segment of tRNAArg in Arg-tRNAArg. (B) The conserved region near the one in A that also plays a role in the binding of klAte1 to a 3′-proximal segment of tRNAArg (Results and Discussion). (C) Arginylation assays (see also Fig. 2C, D, G, and H) with wild-type klAte1 versus its indicated mutants. (D) Same as in C but with wild-type klAte1 versus its H178A and Y303F mutants. (E) A working model of the ternary complex of klAte1 with an acidic Nt-residue–bearing protein substrate (in light blue) and the cosubstrate Arg-tRNAArg on the right (Results and Discussion).
Fig. 4.
Hemin-dependent oligomerization of klAte1. (A) Gel filtration of purified klAte1 as a function of hemin concentration. The levels of klAte1 were determined using Bradford assay. (B) Same as in A but with detection using absorbtion at 280 nm (blue and dark green curves) or at the hemin-specific Soret wavelength of 372 nm. Green curve: klAte1 in the absence of added hemin. Blue (A280) and red (A372) curves: klAte1 in the presence of hemin at the 20-fold molar excess over klAte1. (C) Nonreducing SDS/PAGE of klAte1 in the absence of hemin. (D) Same as in C but the 20-fold molar excess of hemin. (E) Same as in D, but reducing SDS/PAGE. See also SI Appendix, Fig. S9.
The 3D structure of the 503-residue untagged klAte1 comprises two (not clearly separated) domains, termed ATE_N and ATE_C (Fig. 1B–D and SI Appendix, Fig. S3). ATE_N comprises the evolutionarily conserved residues 1 to 109 plus a Ct-region of klAte1 (residues 286 to 503; the residue numbers are of untagged klAte1.) The ATE_C domain comprises the evolutionarily conserved residues 110 to 285 (Fig. 1B–D and SI Appendix, Figs. S3 and S4).
A distinct feature of the klAte1 ATE_N domain is a C4-type metal-binding zinc-finger motif. It comprises four Cys residues (Cys23, Cys26, Cys95, and Cys96) that coordinate with a Zn2+ ion (see below for details) (SI Appendix, Fig. S2B). The zinc-finger motif is located near the putative active-site cleft and is likely to contribute to its structural integrity (Fig. 1A and C and SI Appendix, Fig. S2B).
The ATE_C domain contains a GCN5-related N-acetyltransferase (GNAT)-fold, which comprises eight antiparallel β-sheets and four α-helices (Fig. 1B and SI Appendix, Figs. S2A, S5, and S6). GNAT-folds are present in enzymes that catalyze the acetyl group transfer to a primary amine of a target protein (87). GNAT-folds are also present in bacterial aminoacyl-tRNA transferases, such as FemX; in the alanyl-phosphatidylglycerol synthase (A-PGS), in the lysyl-phosphatidylglycerol synthase (L-PGS), and in the bacterial Leu/Phe‐tRNA‐protein transferase (L/F-transferase, a component of the Leu/N-degron pathway) (SI Appendix, Fig. S1D), all of which use aminoacyl-tRNAs to mediate nonribosomal peptide bond formation (88–92) (SI Appendix, Figs. S5 and S6).
GNAT-folds comprise four evolutionarily conserved motifs A, B, C, and D (92). In acetyltransferases, the A-motif functions in the binding of a GNAT-fold to acetyl-CoA, the cosubstrate. In the structure of klAte1, the motifs A and B of the ATE_C domain form (together with the ATE_N domain) a cleft containing two pockets (Fig. 3E and SI Appendix, Figs. S3–S6). The first pocket is located between the A-motif and the ATE_N domain. The second pocket is located at the interface between the motifs A and B in the ATE_C domain (Fig. 1B–D and SI Appendix, Fig. S3).
The first pocket contains positively charged residues (Figs. 1C and 2A and B), suggesting the binding of this pocket to acceptor substrates, which contain the negatively charged Nt-Asp, Nt-Glu, or oxidized Nt-Cys* residues (Fig. 1A). The second pocket is largely negatively charged (save for a positively charged region in the pocket’s upper part), suggesting the binding of the second pocket to the Arg moiety of Arg-tRNAArg, the cosubstrate of klAte1 (Figs. 1C and 2E and F). The sequences of both pockets as well as corresponding charge distributions are highly conserved in Ate1 R-transferases of different eukaryotes (Fig. 1D and SI Appendix, Fig. S4). Together, these findings strongly suggested that the active site of klAte1 is located, at least in part, in the cleft formed by the two pockets (Figs. 1C and D and 3E).
Structural Comparisons of Ate1 with Enzymes That Share an Ate1-Like Catalytic Mechanism.
Only the ATE_C domain of klAte1 exhibited significant 3D similarities [searches using the DALI server (93)] to other proteins that are a part of the GNAT superfamily (SI Appendix, Figs. S5 and S6). We focused on aminoacyltransferases, which use aminoacyl-tRNAs as their donor cosubstrates, specifically L-PGS (PDB ID code 4V36, Z = 9.8 [Z is a spatial similarity score]) (90); A-PGS (PDB ID code 4V34, Z = 9.6) (90); FemX (UDP-N-acetylmuramoylpentapeptide-lysine N(6)-alanyltransferase; PDB ID code 4II9, Z = 8.7) (88); and the Aat L/F-transferase (PDB ID code 2Z3N, Z = 6.8) (89) (SI Appendix, Fig. S6). GNAT-folds of these enzymes function as substrate-recognizing domains.
Our further analyses identified yet another feature in common between the above proteins and the 3D structure of the klAte1 R-transferase: the positively charged residues of the α3 helix of klAte1 are conserved in all of the above-cited enzymes (SI Appendix, Figs. S5 and S6), but are absent in other enzymes that also contain GNAT-folds (92). In A-PGS and L-PGS, the α3 helix is known to interact with the tRNA moiety of an aminoacyl-tRNA (90). This fact suggested the involvement of the α3 helix of klAte1 in the transfer of Arg from Arg-tRNAArg. Specifically, it suggested, by analogy with the cited enzymes (90), the binding of the positively charged α3 helix of klAte1 to the negatively charged tRNAArg moiety of Arg-tRNAArg through electrostatic interactions between the α3 helix and tRNA phosphates (Fig. 3E).
An earlier study (94) pointed out a similarity between the Ate1 R-transferase and enzymes of the FemABX family that mediate the biosynthesis of bacterial peptidoglycans and use an aminoacyl-tRNA as a donor cosubstrate for the transfer of specific amino acid residues to UDP-MurNAc-peptapeptide (95). In FemX of the bacterium Weissella viridescens, Lys305 is a key catalytic residue whose positively charged ε-amino group interacts with the negatively charged carbonyl oxygen of L-alanine (Ala) that is formed through the nucleophilic attack of UDP-MurNAc-pentapeptide (88).
This functionally essential lysine of W. viridescens FemX is strictly conserved in Ate1 R-transferases of different eukaryotes (SI Appendix, Figs. S5 and S6). Superpositions of the 3D structures of klAte1 and the above aminoacyl-tRNA transferases, including FemX, revealed that the placement and 3D geometry of Lys304 in klAte1 are similar to these parameters for the relevant Lys residues in other aminoacyl-tRNA transferases (Lys813 in L-PGS, Lys840 in A-PGS, and Lys305 in FemX) (SI Appendix, Fig. S6). Specifically, Lys304 of the klAte1 R-transferase forms a hydrogen bond with the main-chain carbonyl group of Leu291 and Tyr293 (SI Appendix, Fig. S6F). These facts and the strict evolutionary conservation of Lys304 among Ate1 R-transferases strongly suggested that klAte1 catalyzes the transfer of Arg from Arg-tRNAArg owing in part to stabilization of a critical reaction intermediate by the ε-amino group of Lys304.
Through surveys of sequelogies (sequence similarities) (81) among Ate1 R-transferases and through surveys of 3D structures of the above-mentioned aminoacyltransferases, we focused on three distinct 3D parts of the klAte1 R-transferase (Fig. 1B–D) as regions likely to mediate the following functions: the recognition of an acidic Nt-residue of an acceptor substrate; the transfer of the Arg residue from Arg-tRNAArg to the acceptor substrate; and the binding of klAte1 to the tRNAArg moiety of Arg-tRNAArg.
Recognition of an Acidic Residue of an Acceptor Substrate.
Ate1 arginylates Nt-Asp, Nt-Glu, and oxidized Nt-Cys* of proteins or short peptides (Fig. 1A) (12, 17, 21, 80). Key recognition determinants of a cognate Ate1 substrate are its α-amino group and an acidic side chain of Nt-residue. For reasons described above, it was likely that a specific pocket in the ATE_N domain of klAte1 recognizes an acidic Nt-residue of a substrate, given both the pocket’s positive charge and its high evolutionary conservation not only among Ate1 R-transferases but also in functional analogs (and sequelogs) of Ate1 such as the Bpt Leu-tRNALeu-protein transferase (L-transferase) (SI Appendix, Fig. S5). Bpt is a component of the bacterial Leu/N-degron pathway that recognizes, similarly to Ate1, an acidic Nt-residue, but (in contrast to Ate1) conjugates Leu, not Arg, to the N termini of acceptor substrates (SI Appendix, Figs. S1D and S5) (66). The conjecture about recognition, by a positively charged klAte1 pocket, of a substrate’s acidic Nt-residue was also consistent with a low sequence conservation of positive residues in the analogous regions of aminoacyltransferases that do not recognize acidic Nt-residues (SI Appendix, Fig. S5).
To address the significance of the positively charged pocket (Figs. 1C and 2A and B), we employed site-directed mutagenesis and in vitro arginylation assays with wild-type klAte1 and its mutants within the pocket, using otherwise identical Ct-biotinylated 13-residue peptides bearing Nt-Asp or Nt-Glu as Ate1 substrates (Fig. 2C and D). A charge-reversing R80E mutation within the pocket completely abolished the Nt-arginylation activity of klAte1. However, and tellingly, the R80K mutation, which partially retained the positive charge, yielded a mutant enzyme with a low but detectable Nt-arginylation activity (Fig. 2C). Thus, a positive charge of a residue at position 80 is essential for the activity of klAte1, most likely because of its role in the binding of substrates that bear an acidic (Asp or Glu) Nt-residue (Fig. 1A and C and Fig. 2A–D, and SI Appendix, Table S2).
Tyr25 and Tyr87 are highly conserved Ate1 residues (SI Appendix, Fig. S4). They are a part of the surface that abuts the positively charged pocket of klAte1 (Fig. 2A and B). To address the relevance of that surface to the activity of klAte1, we used arginylation assays to compare wild-type klAte1 with its Y25F and Y87F mutants (Fig. 2D and SI Appendix, Table S2). The Y25F conversion (i.e., the loss of the hydroxyl group of Tyr) led to the nearly complete abrogation of arginylation activity with the Nt-Glu-peptide substrate, and to a smaller (but still major) decrease of activity with the Nt-Asp-peptide (Fig. 2D). The Y87F mutation had a detectable but much weaker effect, in that the activity of klAte1-Y87F was essentially identical to that of wild-type klAte1 with the Nt-Asp-peptide but was significantly lower than wild-type activity with the Nt-Glu-peptide (Fig. 2D).
We also used fluorescence polarization assays to measure physical affinity of purified wild-type klAte1 and its single-residue mutants to fluorescently Ct-labeled six-residue peptides bearing Nt-Asp, Nt-Glu, or other Nt-residues. However, no significant binding could be detected, indicating a Kd larger than at least 0.1 mM and suggesting that a higher-affinity interaction between wild-type klAte1 and an acceptor substrate would require, in addition, the presence and binding of the Arg-tRNAArg cosubstrate.
In addition, we examined klAte1 and its mutants by expressing them in S. cerevisiae that lacked the endogenous scAte1, the endogenous scUra3 enzyme (required for the synthesis of uracil), and expressed, through the use of the Ub fusion technique (96), the previously described ha-epitope-tagged model Arg/N-degron substrates X-eK-ha-Ura3 (X = Asp or Glu) (SI Appendix, Fig. S7) (14, 27, 65, 96). The acronym eK (extension [“e”] containing lysine [“K”]) denotes the previously characterized 45-residue extension containing “ubiquitylatable” Lys residues (27, 65, 96). Arginylation of Asp-eK-ha-Ura3 (produced from Ub-Asp-eK-ha-Ura3) or Glu-eK-ha-Ura3 (produced from Ub-Glu-eK-ha-Ura3) converted these proteins into short-lived substrates of the Arg/N-degron pathway (Fig. 1A). Since the viability of cells in uracil-lacking media required Ura3, the extent of the Ate1-dependent degradation (in the ura3Δ background) of expressed Asp-eK-ha-Ura3 or Glu-eK-ha-Ura3 could be monitored using cell growth assays (27, 97).
Expression of either Asp-eK-ha-Ura3 or Glu-eK-ha-Ura3 in [ate1Δ ura3Δ] S. cerevisiae allowed cell growth, since an enzymatically active Ura3-based reporter (e.g., Asp-eK-ha-Ura3) was long-lived (and therefore more abundant) in the absence of endogenous scAte1 (SI Appendix, Fig. S7 A, B, and G, row 1) (27, 97). In contrast, little or no growth took place upon coexpression of wild-type klAte1, because of the rapid degradation of Asp-eK-ha-Ura3 under these conditions (SI Appendix, Fig. S7 A and B, row 2, and G, rows 1–3). Tellingly, however, and in agreement with in vitro arginylation data (Fig. 2C), cell growth was rescued upon coexpression of the enzymatically inactive klAte1-R80E mutant (SI Appendix, Fig. S7 A and B, row 3, compare with rows 1 and 2).
The Transfer of Arg Residue.
To address the conjugation of the Arg moiety of Arg-tRNAArg to Nt-Asp or Nt-Glu, we examined the negatively charged pocket in the motif A, located in the ATE_C domain (Figs. 1C and 2E and F, and SI Appendix, Fig. S3). This pocket comprises four residues (Ser255, Ser273, Glu277, and Tyr289), all of which are highly conserved among Ate1 R-transferases of different eukaryotes. Tellingly, the “equivalent” residues of the bacterial Bpt L-transferase (66) (which is sequelogous to eukaryotic Ate1) are, overall, significantly more hydrophobic (Ala168, Ala185, Gln189, and Leu201) than those in the Ate1 pocket (SI Appendix, Fig. S5). This difference between otherwise sequelogous Ate1 and Bpt is consistent with the fact that although the Bpt L-transferase recognizes (similarly to Ate1) Nt-Asp and Nt-Glu of proteins or short peptides, it conjugates Leu (not Arg) to these Nt-residues (66).
In the above klAte1 pocket, Glu277 is the sole charged (negatively charged) residue among the four conserved residues (Fig. 2E and F and SI Appendix, Fig. S4). In agreement with the conjecture about that pocket’s importance for the binding of klAte1 to the Arg moiety of Arg-tRNAArg, the klAte1-E277K mutant (Glu277 to Lys, a basic residue) and klAte1-E277A mutant (Glu277 to Ala, an uncharged weakly hydrophobic residue) exhibited, respectively, the undetectable (klAte1-E277K) or very low (klAte1-E277A) levels of arginylation activity, even with the more efficacious Nt-Asp-peptide substrate (Fig. 2G).
In contrast and tellingly, the klAte1-E277D mutant (Glu277 to the smaller but also acidic Asp residue) exhibited essentially wild-type levels of Nt-arginylation activity, indicating the critical importance of negative charge at that (conserved) position, most likely because the above pocket mediates the functionally essential binding of klAte1 to the positively charged Arg moiety of Arg-tRNAArg (Fig. 2G). While these data strongly supported the above conjecture, they did not prove it directly, as the latter would also require a “complementary” physical-affinity evidence. As described above, we could not detect (using in vitro fluorescence polarization binding assays in the absence of Arg-tRNAArg cosubstrate) the physical binding of enzymatically active klAte1 to the Nt-Asp-bearing or Nt-Glu-bearing peptides. It is likely that a higher-affinity binding would require three components together (klAte1, a substrate, and the Arg-tRNAArg cosubstrate), and also a still to be identified mutant of klAte1 that is inactive enzymatically but retains the bulk of its affinity to both a substrate and the cosubstrate.
The transfer of Arg from Arg-tRNAArg to an acidic N terminus of a substrate is expected to occur upon the formation of a tripartite substrate/enzyme/cosubstrate complex. As mentioned above, the conjugation reaction mediated by the bacterial FemX aminoacyltransferase involves its Lys305 residue, which stabilizes the reaction intermediate (89). Remarkably, this key Lys305 of FemX is also conserved in klAte1, in which it is Lys304 (SI Appendix, Figs. S4–S6). In agreement with that view, the K304A mutant of klAte1 was found to be completely inactive, strongly suggesting the correctness of this interpretation, through the analogy with FemX and its previously understood enzymatic mechanism (Fig. 2H and SI Appendix, Fig. S6F and Table S2).
Similar results were obtained using cell growth assays (SI Appendix, Fig. S7), save for a particularly high sensitivity of those “qualitative” in vivo tests (as distinguished from quantitative in vitro arginylation assays) to a very low level of arginylation activity of a klAte1 mutant. For example, the apparently inactive klAte1-E277K mutant was also classed as inactive in cell growth assays with either Asp-eK-ha-Ura3 or Glu-eK-ha-Ura3 as reporter substrates (Fig. 2G and SI Appendix, Fig. S7 C and D, row 3). In contrast, the klAte1-E277A mutant, which was also (nearly) inactive in in vitro arginylation assays, could still rescue cell growth with the Asp-eK-ha-Ura3 (but, significantly, not with the Glu-eK-ha-Ura3), indicating a very low but nonzero Nt-arginylation activity of the E277A allele of klAte1 (Fig. 2G and SI Appendix, Fig. S7 C and D, row 2).
The Binding of klAte1 to the tRNAArg Moiety of Arg-tRNAArg.
Studies of eukaryotic Ate1 R-transferases and bacterial Aat L/F-transferases indicated that these enzymes recognize, in particular, a 3′-proximal segment of a cognate tRNA in a corresponding aminoacyl-tRNA cosubstrate (98, 99). What segments of klAte1 may interact with tRNAArg of Arg-tRNAArg? In searching for such regions, we focused, initially, on electrostatic interactions through a negatively charged ribose-phosphate backbone, and also on π–π stacking interactions via bases of tRNAArg.
Sequence alignments of Ate1 R-transferases from different eukaryotes pinpointed a cluster of highly conserved basic residues (SI Appendix, Fig. S4). This cluster is a part of the α3 helix and forms a positively charged patch (Figs. 1C and 3A). Interestingly, this patch is located near a negatively charged pocket that is highly likely (as described above) to play a role in the interaction between klAte1 and the (positively charged) Arg moiety of Arg-tRNAArg. Together, these features of the positively charged patch suggested its binding to the tRNAArg moiety of Arg-tRNAArg, in part through electrostatic interactions (Fig. 3A).
To address this possibility, we mutated basic residues of the conserved basic patch (Lys112, Lys115, Lys119, Arg120, and Lys123) to the acidic Glu residues, and assayed the resulting klAte1 mutant for its arginylation activity. The pentaglutamate klAte1 mutant was completely inactive (Fig. 3C). However, the arginylation activity could be fully rescued when all five residues were converted to Arg (in wild-type klATe1, only one basic residue, at position 120, was Arg) (Fig. 3C). Thus, it is the positive charge of the patch in the α3 helix (but not whether the residues involved were Lys or Arg) that is essential for the arginylation activity of klAte1 (Fig. 3E).
In further searches for functionally significant sites of klAte1, we considered the mostly aromatic residues Tyr173, His178, Trp260, and Tyr303 near the putative active site of klAte1 that were highly conserved among Ate1 enzymes of different eukaryotes (SI Appendix, Fig. S4). Among these residues, His178 and Tyr303 of klAte1 are the equivalents of His206 and Phe304 in the above-mentioned FemX enzyme (SI Appendix, Figs. S5 and S6). In particular, His206 of FemX is required for the binding of FemX to the tRNAAla moiety of Ala-tRNAAla, owing to the formation of hydrogen bonds between His206 and phosphates of C75 and A76, near the 3′-end of tRNAAla (88).
Given the above, we replaced His178 of klAte1 with Ala. The arginylation activity of the resulting klAte1-H178A mutant was considerably lower than the activity of wild-type klAte1 (Fig. 3D), in agreement with the conjecture about its participation (by analogy with His206 of FemX) in the binding of klAte1 to the tRNAArg moiety of Arg-tRNAArg.
Tyr303 of klAte1 is strictly conserved among Ate1 R-transferases of different eukaryotes (SI Appendix, Fig. S4). The equivalent residue in the FemABX family is also conserved, but as Phe, not Tyr (SI Appendix, Fig. S5). To address the importance of the hydroxyl group (the sole difference between Tyr and Phe) in the Tyr303 of klAte1, we constructed the klAte1-Y303F mutant and found it to be completely inactive (Fig. 3D and SI Appendix, Table S2), indicating the critical importance of the hydroxyl group of Tyr303 in that (conserved) residue of klAte1.
Thus, it is highly likely (but remains to be verified directly) that, in contrast to the previously identified π-stacking interaction between C75-ribose of the tRNAAla moiety of Ala-tRNAAla and the equivalent (also conserved) Phe residue of FemX (88), the conserved hydroxyl-containing Tyr303 of klAte1 is involved in a different mode of binding of klAte1 to a different tRNA, the tRNAArg moiety of Arg-tRNAArg. These aspects of klAte1 would be illuminated through a still to be determined 3D structure of a complex between klAte1 and Arg-tRNAArg.
Tyr173 is conserved in Ate1 R-transferases of all examined eukaryotes (SI Appendix, Fig. S4). Trp260 Ate1 is also highly conserved, but is replaced by Tyr in mammalian Ate1. Significantly, these residues of Ate1 are not conserved in either FemX or other non-Ate1 aminoacyltransferases that do not bind to Arg-tRNAArg. Together, this evidence suggested that Tyr173 and Trp260 of Ate1, in addition to the above Tyr303 residue, play a role in the binding of klAte1 to the tRNAArg moiety of Arg-tRNAArg.
The results of arginylation assays with the klAte1-Y173F and klAte1-W260A mutants are shown in SI Appendix, Fig. S8A. The kcat and Km of klAte1-Y173F were, respectively, 1.9-fold and 6.3-fold larger with the Nt-Asp-peptide, and 1.1-fold and 6.6-fold larger with the otherwise identical Nt-Glu-peptide, in comparison to wild-type klAte1 (SI Appendix, Table S2). The klAte1-W260A mutant exhibited up to 2-fold increases in kcat (1.9-fold for the Nt-Asp-peptide, 1.5-fold for the Nt-Glu-peptide) and 18-fold increase in Km (in comparison to wild-type klAte1) with both the Nt-Asp-peptide and the Nt-Glu-peptide (SI Appendix, Table S2). Strong (6-fold and 18-fold) increases in Km indicated a significantly weaker binding of the two mutant enzymes either to a substrate or (more likely) to the cosubstrate Arg-tRNAArg. The results of cell growth assays (SI Appendix, Fig. S7 E and F, rows 5 and 6) were in agreement with those of in vitro arginylation assays.
Hemin-Induced Oligomerization of Ate1 R-Transferase.
In addition to structural and reverse-genetics studies of klAte1, we also examined the previously described finding that micromolar levels of hemin (Fe+3-heme) inhibited the arginylation activity of both S. cerevisiae and mouse Ate1 R-transferases (53). In its crystal structure, klAte1 coordinates an electron-rich metal ion (apparently Zn2+) with four Cys residues (Cys23, Cys26, Cys95, and Cys96) (SI Appendix, Fig. S2B). These cysteines are strictly conserved among Ate1 R-transferases of different eukaryotes (SI Appendix, Fig. S4), and were considered to be a part of a putative heme regulatory motif (HRM) (53).
To determine the identity of metal in the crystallized klAte1, we performed elemental analysis using inductively coupled plasma-mass spectrometry and the same purified klAte1 that was used for crystallization, in the same buffer as well. As expected, the bound metal atom was found to be zinc (Zn2+) (SI Appendix, Table S3). Two vicinal Cys residues, Cys95 and Cys96 in S. cerevisiae Ate1 (Cys71 and Cys72 in mouse Ate1), were shown to form, upon hemin binding, a disulfide bond. This transition was inferred to be the likely cause of the hemin-mediated inactivation of Ate1 (53). Besides the above HRM, Ate1 R-transferases, including klAte1, contain yet another evolutionarily conserved HRM-like motif (53), specifically Cys411, Pro412 in mouse Ate1 and Cys298, Pro299 in klAte1. Remarkably, in the crystal structure of klAte1, these two HRMs, termed HRM1 and HRM2, are located spatially close to each other. The first HRM1 is partially exposed to solvent, whereas the second HRM2 is fully exposed (Fig. 3E and SI Appendix, Fig. S3D).
Purified klAte1 is a monomer in solution (Fig. 4B, a dark green curve, and SI Appendix, Fig. S8D). Examining klAte1 in the presence of different concentrations of hemin, we found that hemin increased the apparent size of klAte1, determined by gel filtration (Fig. 4A and B). The absorbance was measured using both 280 nM (the absorbance of aromatic residues) and the hemin-specific Soret wavelength of 372 nm, with a shoulder at 423 nm that is characteristic of hemin-protein complexes (53, 100). In later gel-filtration analyses, the concentration of protein fractions was also determined using the Bradford assay. These results indicated a hemin-mediated oligomerization of klAte1, a previously undescribed finding (Fig. 4A and B).
Hemin-induced oligomerization of klAte1 was confirmed using nonreducing SDS/PAGE. The latter result indicated that klAte1 oligomers were resistant to a (nonreducing) SDS-containing buffer, presumably because of intermolecular disulfide bonds between different (denatured by SDS) klAte1 protein molecules (Fig. 4C and D and SI Appendix, Fig. S9). This interpretation was confirmed when the same samples were examined using reducing SDS/PAGE, as the klAte1 oligomer bands disappeared nearly completely in the presence of both SDS and a reducing agent (Fig. 4C–E and SI Appendix, Fig. S9). We also measured the activity of klAte1 in the presence of hemin, and found that hemin inhibited arginylation by klAte1 (SI Appendix, Fig. S8 B and C), in agreement with the earlier study (53).
Concluding Remarks.
The Nt-arginylation of proteins, which was known since the 1960s but not understood biologically, has “acquired” a physiological function with the 1986 discovery of the first N-degron pathway (prior to 2019, it was called the “N-end rule pathway” and is now called the Arg/N-degron pathway) (Fig. 1A and SI Appendix, Fig. S1G) (12–14, 21, 22, 67, 80, 101). The 58-kDa Ate1 R-transferase uses the Arg-tRNAArg cosubstrate to Nt-arginylate Nt-Asp, Nt-Glu and oxidized Nt-Cys* of proteins or short peptides (see the first section of the article and Fig. 1A). Residues immediately downstream of these “arginylatable” Nt-residues can modulate or even preclude Nt-arginylation (102, 103). An example of the influence of downstream residues is the recent demonstration that mammalian β-actin, despite bearing (after Nt-processing) the arginylatable Nt-Glu residue, is not arginylated (102), contrary to previous (technically circumstantial) claims about a significant Nt-arginylation of β-actin (104, 105).
We report here a crystal structure of an R-transferase, klAte1 of the yeast K. lactis (Figs. 1B–D and 3E and SI Appendix, Figs. S2 and S3). As described in detail above, the 3D structure of klAte1, determined at 1.8- to 2.2-Å resolution, has revealed telling spatial proximities between specific 3D regions of klAte1 that were unlinked at the level of amino acid sequences. In addition, our 3D results led, in a structurally informed way, to the construction and analyses of specific site-directed mutants of klAte1.
As was also described above, quantitative arginylation assays with these klAte1 mutants could be interpreted in the context of the klAte1 3D structure, and yielded strong inferences about 3D locations, within klAte1, of its specific binding sites for cognate substrates (which bear an acidic Nt-residue), for the Arg residue of the Arg-tRNAArg cosubstrate, and for a 3′-proximal segment of the tRNAArg moiety in Arg-tRNAArg (Figs. 2 and 3 and SI Appendix, Figs. S7 and S8A). The resulting advances led to the working model of the ternary complex of klAte1 with an acidic Nt-residue–bearing protein substrate and the cosubstrate Arg-tRNAArg, and also to a plausible (and mechanistically parsimonious) reaction diagram for the Ate1-catalyzed Nt-arginylation (Fig. 3E and SI Appendix, Fig. S10).
A direct and deeper understanding of these aspects of Ate1 would require a determination of a 3D structure of a ternary complex of klAte with its cognate substrate and the Arg-tRNAArg cosubstrate. Rendering such a complex enzymatically inactive (to preclude Nt-arginylation of a substrate within a crystal) would require a still to be discovered klAte1 mutant that is inactive enzymatically but retains its affinity to both a substrate and the cosubstrate.
In addition to the crystal structure and reverse-genetic analyses of klAte1, we also studied the previously described inhibition of arginylation activity of the S. cerevisiae and mouse Ate1 R-transferases by hemin (Fe3+-heme) (53). We found that hemin, besides inhibiting the enzymatic activity of klAte1, could also induce the previously undescribed disulfide-mediated oligomerization of klAte1 (Fig. 4 and SI Appendix, Fig. S9).
A distinct feature of the klAte1 ATE_N domain is a C4-type metal-binding zinc-finger motif that comprises four Cys residues (Cys23, Cys26, Cys95, and Cys96) that coordinate with a Zn2+ ion (SI Appendix, Fig. S2B). Very recently, Van et al. (106) deposited a preprint at BioRxiv that described analyses of S. cerevisiae Ate1 (scAte1) as a potential [Fe-S] cluster-binding enzyme. The authors’ data suggested that a [Fe-S] cluster binds to the above C4-type metal-binding domain of Ate1 (106). Our analyses of purified klAte1, described above and utilizing the inductively coupled plasma-mass spectrometry, identified Zn2+ (not iron) as the sole metal ligand of that C4-type domain (SI Appendix, Table S3). Future studies in this arena will determine whether physiologically relevant metal-containing ligands of Ate1 comprise Zn2+, [Fe-S] clusters, and hemin, or only some of these ligands.
A large part of the remarkably broad functions of the Arg/N-degron pathway, including its role as an oxygen/NO sensor in animals and plants, requires the Nt-arginylation branch of this pathway (see the first section of the article and Fig. 1A). Physiological regulation of the Ate1 R-transferase, as well as inhibition or activation of this enzyme in clinical settings, are likely to be one focus of future studies. The structural and mechanistic insights of the present work advanced the understanding of both Ate1 and the Arg/N-degron pathway.
Materials and Methods
Further information is in SI Appendix, SI Materials and Methods.
Cloning and Mutagenesis.
The klATE1 gene was amplified from genomic DNA of K. lactis. The LC3B-fusion technique (86) was used to construct specific DNA plasmids that expressed klAte1 and its derivatives.
Protein Expression and Purification.
The wild-type klAte1 protein and its site-directed mutants were expressed in Escherichia coli as LC3B-based fusions. Purification details are described in SI Appendix, SI Materials and Methods.
Crystallization and Structure Determination.
Purified klAte1 proteins were crystallized using hanging drop plates and 1:1 mixing of proteins (8 mg/mL) and mother liquors. Crystals were flash-frozen using liquid nitrogen with 25% (v/v) glycerol as a cryoprotectant in the original mother liquor. Other details are described in SI Appendix, SI Materials and Methods.
Nt-arginylation Activity Assay.
The in vitro Nt-arginylation of 13-residue peptides XAGAIISDWIPPK-biotin (X = Asp or Glu) was determined by quantifying the incorporation of L-[14C]-arginine. Other details are described in SI Appendix, SI Materials and Methods.
Yeast Strains and Yeast Genetic Methods.
Standard techniques were used for yeast strain construction and transformation. Details are described in SI Appendix, SI Materials and Methods.
Hemin-klAte1 Binding and Size-Exclusion Chromatography.
Before incubation with hemin, purified klAte1 was passed through a Superdex-200 column in 0.15 M NaCl, 20 mM Hepes (pH 7.5) to remove the reducing agent in the initial buffer. Other details are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank colleagues at the beamlines 5C and 11C, Pohang Accelerator Laboratory, South Korea, the beamline BL17A at the Photon Factory, Japan, and at the beamline BL44XU, Spring-8, Japan, for assistance with collecting X-ray data. This work was supported by National Research Foundation of Korea Grants 2020R1A2C3008285, 2020R1A5A1019023, 2021M3A9G8024747, and 2021M3A9I4030068 (to H.K.S.); Grants 2020R1A3B2078127 and 2017R1A5A1015366 (to C.-S.H); and NIH Grant GM031530 (to A.V.).
Footnotes
Reviewers: U.H., Max Planck Institute of Biochemistry; and W.T., Vanderbilt University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209597119/-/DCSupplemental.
Data Availability
All relevant data in the paper are available in the main text, in SI Appendix, or in the Protein Data Bank under the following accession codes: 7WFX (EV_Ortho) (109), 7WG1 (DV_Ortho) (110), 7WG2 (EV_Tetra) (111), and 7WG4 (DV_Tetra) (112).
References
- 1.Hershko A., Ciechanover A., Varshavsky A., The ubiquitin system. Nat. Med. 6, 1073–1081 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A., Discovery of cellular regulation by protein degradation. J. Biol. Chem. 283, 34469–34489 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finley D., Ulrich H. D., Sommer T., Kaiser P., The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192, 319–360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vittal V., Stewart M. D., Brzovic P. S., Klevit R. E., Regulating the regulators: Recent revelations in the control of E3 ubiquitin ligases. J. Biol. Chem. 290, 21244–21251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji C. H., Kwon Y. T., Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol. Cells 40, 441–449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng N., Shabek N., Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Yu H., Matouschek A., Recognition of client proteins by the proteasome. Annu. Rev. Biophys. 46, 149–173 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Rape M., Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 19, 59–70 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Sakata E., Eisele M. R., Baumeister W., Molecular and cellular dynamics of the 26S proteasome. Biochim. Biophys. Acta. Proteins Proteomics 1869, 140583 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Bard J. A. M., et al. , Structure and function of the 26S proteasome. Annu. Rev. Biochem. 87, 697–724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budenholzer L., Cheng C. L., Li Y., Hochstrasser M., Proteasome structure and assembly. J. Mol. Biol. 429, 3500–3524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varshavsky A., N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. U.S.A. 116, 358–366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachmair A., Finley D., Varshavsky A., In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 (1986). [DOI] [PubMed] [Google Scholar]
- 14.Bachmair A., Varshavsky A., The degradation signal in a short-lived protein. Cell 56, 1019–1032 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T., Varshavsky A., Degradation signals in the lysine-asparagine sequence space. EMBO J. 18, 6017–6026 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inobe T., Fishbain S., Prakash S., Matouschek A., Defining the geometry of the two-component proteasome degron. Nat. Chem. Biol. 7, 161–167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varshavsky A., The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan M., et al. , Structural insights into Ubr1-mediated N-degron polyubiquitination. Nature 600, 334–338 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner G. C., Du F., Varshavsky A., Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405, 579–583 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Rao H., Uhlmann F., Nasmyth K., Varshavsky A., Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410, 955–959 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Kwon Y. T., Kashina A. S., Varshavsky A., Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol. Cell. Biol. 19, 182–193 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon Y. T., et al. , An essential role of N-terminal arginylation in cardiovascular development. Science 297, 96–99 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Tasaki T., Sriram S. M., Park K. S., Kwon Y. T., The N-end rule pathway. Annu. Rev. Biochem. 81, 261–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winter N., Novatchkova M., Bachmair A., Cellular control of protein turnover via the modification of the amino-terminus. Int. J. Mol. Sci. 22, 3545 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Timms R. T., Koren I., Tying up loose ends: The N-degron and C-degron pathways of protein degradation. Biochem. Soc. Trans. 48, 1557–1567 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holdsworth M. J., Vicente J., Sharma G., Abbas M., Zubrycka A., The plant N-degron pathways of ubiquitin-mediated proteolysis. J. Integr. Plant Biol. 62, 70–89 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Hwang C. S., Shemorry A., Varshavsky A., N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shemorry A., Hwang C. S., Varshavsky A., Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell 50, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aksnes H., Ree R., Arnesen T., Cotranslational, posttranslational, and noncatalytic roles of N-terminal acetyltransferases. Mol. Cell 73, 1097–1114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi W. S., et al. , Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat. Struct. Mol. Biol. 17, 1175–1181 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Matta-Camacho E., Kozlov G., Li F. F., Gehring K., Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat. Struct. Mol. Biol. 17, 1182–1187 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Kim M. K., Oh S. J., Lee B. G., Song H. K., Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 113, 12438–12443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim L., et al. , Structural basis for the N-degron specificity of ClpS1 from Arabidopsis thaliana. Protein Sci. 30, 700–708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piatkov K. I., Brower C. S., Varshavsky A., The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc. Natl. Acad. Sci. U.S.A. 109, E1839–E1847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brower C. S., Piatkov K. I., Varshavsky A., Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol. Cell 50, 161–171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piatkov K. I., Oh J.-H., Liu Y., Varshavsky A., Calpain-generated natural protein fragments as short-lived substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 111, E817–E826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S. J., Wu X., Wadas B., Oh J.-H., Varshavsky A., An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 355, 366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hämmerle M., et al. , Proteins of isolated mutants and N-terminal proline are essential for degradation of yeast fructose-1,6-bisphosphatase. J. Biol. Chem. 273, 25000–25005 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Dong C., et al. , Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway. Nat. Chem. Biol. 14, 466–473 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Dougan D. A., Varshavsky A., Understanding the Pro/N-end rule pathway. Nat. Chem. Biol. 14, 415–416 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Melnykov A., Chen S. J., Varshavsky A., Gid10 as an alternative N-recognin of the Pro/N-degron pathway. Proc. Natl. Acad. Sci. U.S.A. 116, 15914–15923 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S. J., Melnykov A., Varshavsky A., Evolution of substrates and components of the Pro/N-degron pathway. Biochemistry 59, 582–593 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao S., et al. , Interconversion between anticipatory and active GID E3 ubiquitin ligase conformations via substrate receptor assembly. Mol. Cell 77, 150–163.e159 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Sherpa D., Chrustowicz J., Schulman B. A., How the ends signal the end: Regulation by E3 ubiquitin ligases recognizing protein termini. Mol. Cell 82, 1424–1438 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karayel O., Michaelis A. C., Mann M., Schulman B. A., Langlois C. R., DIA-based systems biology approach unveils E3 ubiquitin ligase-dependent responses to a metabolic shift. Proc. Natl. Acad. Sci. U.S.A. 117, 32806–32815 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong K. E., et al. , Timer-based proteomic profiling of the ubiquitin-proteasome system reveals a substrate receptor of the GID ubiquitin ligase. Mol. Cell 81, 2460–2476.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J. M., et al. , Formyl-methionine as an N-degron of a eukaryotic N-end rule pathway. Science 362, eaat0174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh J. H., Hyun J. Y., Varshavsky A., Control of Hsp90 chaperone and its clients by N-terminal acetylation and the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 114, E4370–E4379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oh J. H., Hyun J. Y., Chen S. J., Varshavsky A., Five enzymes of the Arg/N-degron pathway form a targeting complex: The concept of superchanneling. Proc. Natl. Acad. Sci. U.S.A. 117, 10778–10788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vu T. T. M., Varshavsky A., The ATF3 transcription factor is a short-lived substrate of the Arg/N-degron pathway. Biochemistry 59, 2796–2812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vu T. T. M., Mitchell D. C., Gygi S. P., Varshavsky A., The Arg/N-degron pathway targets transcription factors and regulates specific genes. Proc. Natl. Acad. Sci. U.S.A. 117, 31094–31104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu R.-G., et al. , The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Hu R.-G., Wang H., Xia Z., Varshavsky A., The N-end rule pathway is a sensor of heme. Proc. Natl. Acad. Sci. U.S.A. 105, 76–81 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hwang C. S., Varshavsky A., Regulation of peptide import through phosphorylation of Ubr1, the ubiquitin ligase of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 19188–19193 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao X., Yeom J., Groisman E. A., The expanded specificity and physiological role of a widespread N-degron recognin. Proc. Natl. Acad. Sci. U.S.A. 116, 18629–18637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeom J., Gao X., Groisman E. A., Reduction in adaptor amounts establishes degradation hierarchy among protease substrates. Proc. Natl. Acad. Sci. U.S.A. 115, E4483–E4492 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang K. H., Roman-Hernandez G., Grant R. A., Sauer R. T., Baker T. A., The molecular basis of N-end rule recognition. Mol. Cell 32, 406–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tobias J. W., Shrader T. E., Rocap G., Varshavsky A., The N-end rule in bacteria. Science 254, 1374–1377 (1991). [DOI] [PubMed] [Google Scholar]
- 59.Erbse A., et al. , ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439, 753–756 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Shrader T. E., Tobias J. W., Varshavsky A., The N-end rule in Escherichia coli: Cloning and analysis of the leucyl, phenylalanyl-tRNA-protein transferase gene aat. J. Bacteriol. 175, 4364–4374 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dougan D. A., Micevski D., Truscott K. N., The N-end rule pathway: From recognition by N-recognins, to destruction by AAA+ proteases. Biochim. Biophys. Acta 1823, 83–91 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Chen S. J., Kim L., Song H. K., Varshavsky A., Aminopeptidases trim Xaa-Pro proteins, initiating their degradation by the Pro/N-degron pathway. Proc. Natl. Acad. Sci. U.S.A. 118, e2115430118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cha-Molstad H., et al. , Regulation of autophagic proteolysis by the N-recognin SQSTM1/p62 of the N-end rule pathway. Autophagy 14, 359–361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Masson N., et al. , Conserved N-terminal cysteine dioxygenases transduce responses to hypoxia in animals and plants. Science 365, 65–69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim H. K., et al. , The N-terminal methionine of cellular proteins as a degradation signal. Cell 156, 158–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graciet E., et al. , Aminoacyl-transferases and the N-end rule pathway in a human pathogen. Proc. Natl. Acad. Sci. U.S.A. 103, 3078–3083 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferber S., Ciechanover A., Role of arginine-tRNA in protein degradation by the ubiquitin pathway. Nature 326, 808–811 (1987). [DOI] [PubMed] [Google Scholar]
- 68.Xiao Q., Zhang F., Nacev B. A., Liu J. O., Pei D., Protein N-terminal processing: Substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry 49, 5588–5599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Piatkov K. I., Colnaghi L., Békés M., Varshavsky A., Huang T. T., The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol. Cell 48, 926–933 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Justa-Schuch D., et al. , DPP9 is a novel component of the N-end rule pathway targeting the tyrosine kinase Syk. eLife 5, e16370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwon Y. T., et al. , The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 7898–7903 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee M. J., et al. , RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 15030–15035 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baker R. T., Varshavsky A., Yeast N-terminal amidase. A new enzyme and component of the N-end rule pathway. J. Biol. Chem. 270, 12065–12074 (1995). [DOI] [PubMed] [Google Scholar]
- 74.Grigoryev S., et al. , A mouse amidase specific for N-terminal asparagine. The gene, the enzyme, and their function in the N-end rule pathway. J. Biol. Chem. 271, 28521–28532 (1996). [DOI] [PubMed] [Google Scholar]
- 75.Kwon Y. T., et al. , Altered activity, social behavior, and spatial memory in mice lacking the NTAN1p amidase and the asparagine branch of the N-end rule pathway. Mol. Cell. Biol. 20, 4135–4148 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang H., Piatkov K. I., Brower C. S., Varshavsky A., Glutamine-specific N-terminal amidase, a component of the N-end rule pathway. Mol. Cell 34, 686–695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du F., Navarro-Garcia F., Xia Z., Tasaki T., Varshavsky A., Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. U.S.A. 99, 14110–14115 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hwang C. S., Shemorry A., Auerbach D., Varshavsky A., The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat. Cell Biol. 12, 1177–1185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dohmen R. J., Madura K., Bartel B., Varshavsky A., The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc. Natl. Acad. Sci. U.S.A. 88, 7351–7355 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balzi E., Choder M., Chen W. N., Varshavsky A., Goffeau A., Cloning and functional analysis of the arginyl-tRNA-protein transferase gene ATE1 of Saccharomyces cerevisiae. J. Biol. Chem. 265, 7464–7471 (1990). [PubMed] [Google Scholar]
- 81.Varshavsky A., ‘Spalog’ and ‘sequelog’: Neutral terms for spatial and sequence similarity. Curr. Biol. 14, R181–R183 (2004). [DOI] [PubMed] [Google Scholar]
- 82.Yoshida S., Ito M., Callis J., Nishida I., Watanabe A., A delayed leaf senescence mutant is defective in arginyl-tRNA:protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 32, 129–137 (2002). [DOI] [PubMed] [Google Scholar]
- 83.Brower C. S., Varshavsky A., Ablation of arginylation in the mouse N-end rule pathway: Loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS One 4, e7757 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cha-Molstad H., et al. , Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 17, 917–929 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kwon D. H., et al. , Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter. Nat. Commun. 9, 3291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim L., Kwon D. H., Heo J., Park M. R., Song H. K., Use of the LC3B-fusion technique for biochemical and structural studies of proteins involved in the N-degron pathway. J. Biol. Chem. 295, 2590–2600 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salah Ud-Din A. I., Tikhomirova A., Roujeinikova A., Structure and functional diversity of GCN5-related N-acetyltransferases (GNAT). Int. J. Mol. Sci. 17, 1018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fonvielle M., et al. , The structure of FemX(Wv) in complex with a peptidyl-RNA conjugate: Mechanism of aminoacyl transfer from Ala-tRNA(Ala) to peptidoglycan precursors. Angew. Chem. Int. Ed. Engl. 52, 7278–7281 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Watanabe K., et al. , Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature 449, 867–871 (2007). [DOI] [PubMed] [Google Scholar]
- 90.Hebecker S., et al. , Structures of two bacterial resistance factors mediating tRNA-dependent aminoacylation of phosphatidylglycerol with lysine or alanine. Proc. Natl. Acad. Sci. U.S.A. 112, 10691–10696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maruyama C., Hamano Y., tRNA-dependent amide bond-forming enzymes in peptide natural product biosynthesis. Curr. Opin. Chem. Biol. 59, 164–171 (2020). [DOI] [PubMed] [Google Scholar]
- 92.Dyda F., Klein D. C., Hickman A. B., GCN5-related N-acetyltransferases: A structural overview. Annu. Rev. Biophys. Biomol. Struct. 29, 81–103 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holm L., DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rai R., Mushegian A., Makarova K., Kashina A., Molecular dissection of arginyltransferases guided by similarity to bacterial peptidoglycan synthases. EMBO Rep. 7, 800–805 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dare K., Ibba M., Roles of tRNA in cell wall biosynthesis. Wiley Interdiscip. Rev. RNA 3, 247–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Varshavsky A., Ubiquitin fusion technique and related methods. Methods Enzymol. 399, 777–799 (2005). [DOI] [PubMed] [Google Scholar]
- 97.Ghislain M., Dohmen R. J., Levy F., Varshavsky A., Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15, 4884–4899 (1996). [PMC free article] [PubMed] [Google Scholar]
- 98.Avcilar-Kucukgoze I., et al. , tRNA(Arg)-derived fragments can serve as arginine donors for protein arginylation. Cell Chem. Biol. 27, 839–849.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Suto K., et al. , Crystal structures of leucyl/phenylalanyl-tRNA-protein transferase and its complex with an aminoacyl-tRNA analog. EMBO J. 25, 5942–5950 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ishikawa H., et al. , Involvement of heme regulatory motif in heme-mediated ubiquitination and degradation of IRP2. Mol. Cell 19, 171–181 (2005). [DOI] [PubMed] [Google Scholar]
- 101.Gonda D. K., et al. , Universality and structure of the N-end rule. J. Biol. Chem. 264, 16700–16712 (1989). [PubMed] [Google Scholar]
- 102.Drazic A., et al. , The final maturation state of β-actin Involves N-terminal acetylation by NAA80, not N-terminal arginylation by ATE1. J. Mol. Biol. 434, 167397 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wadas B., Piatkov K. I., Brower C. S., Varshavsky A., Analyzing N-terminal arginylation through the use of peptide arrays and degradation assays. J. Biol. Chem. 291, 20976–20992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karakozova M., et al. , Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science 313, 192–196 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Zhang F., Saha S., Shabalina S. A., Kashina A., Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science 329, 1534–1537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Van V., et al. , Iron-sulfur clusters are involved in posttranslational arginylation. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/2021.04.13.439645v1.full. Accessed 15 April 2022.
- 107.Hwang C.-S., Shemorry A., Varshavsky A., Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc. Natl. Acad. Sci. U.S.A. 106, 2142–2147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zdobnov E. M., et al. , OrthoDB in 2020: Evolutionary and functional annotations of orthologs. Nucleic Acids Res. 49 (D1), D389–D393 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.B.H. Kim et al., Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates. Protein Data Bank. https://www.rcsb.org/structure/unreleased/7WFX. Deposited 27 December 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.B.H. Kim et al., Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates. Protein Data Bank. https://www.rcsb.org/structure/unreleased/7WG1. Deposited 27 December 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.B.H. Kim et al., Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates. Protein Data Bank. https://www.rcsb.org/structure/unreleased/7WG2. Deposited 28 December 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.B.H. Kim et al., Crystal structure of the Ate1 arginyl-tRNA-protein transferase and arginylation of N-degron substrates. Protein Data Bank. https://www.rcsb.org/structure/unreleased/7WG4. Deposited 28 December 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data in the paper are available in the main text, in SI Appendix, or in the Protein Data Bank under the following accession codes: 7WFX (EV_Ortho) (109), 7WG1 (DV_Ortho) (110), 7WG2 (EV_Tetra) (111), and 7WG4 (DV_Tetra) (112).