Significance
Rationalizing how the abiological Kemp eliminase is optimized in laboratory evolution remains a great challenge. Previous mechanistic studies only cover very few designs, leading to a partial understanding of the optimization process. Here, we demonstrate that the evolutionary information distilled from the homologs of natural protein scaffold correlates with the Kemp elimination activity of various mutants introduced in laboratory evolution. Therefore, even if an active site is replaced to catalyze a new reaction, the underlying evolutionary pressures that shaped the natural protein scaffold are still relevant. The present study sheds light on enzyme architecture, enzyme evolution, and the power of interpolating the catalytic landscape in enzyme design.
Keywords: designer enzyme, natural evolution, directed evolution, enzyme design, enzyme architecture
Abstract
Laboratory evolution combined with computational enzyme design provides the opportunity to generate novel biocatalysts. Nevertheless, it has been challenging to understand how laboratory evolution optimizes designer enzymes by introducing seemingly random mutations. A typical enzyme optimized with laboratory evolution is the abiological Kemp eliminase, initially designed by grafting active site residues into a natural protein scaffold. Here, we relate the catalytic power of laboratory-evolved Kemp eliminases to the statistical energy () inferred from their natural homologous sequences using the maximum entropy model. The of designs generated by directed evolution is correlated with enhanced activity and reduced stability, thus displaying a stability-activity trade-off. In contrast, the for mutants in catalytic-active remote regions (in which remote residues are important for catalysis) is strongly anticorrelated with the activity. These findings provide an insight into the role of protein scaffolds in the adaption to new enzymatic functions. It also indicates that the valley in the landscape can guide enzyme design for abiological catalysis. Overall, the connection between laboratory and natural evolution contributes to understanding what is optimized in the laboratory and how new enzymatic function emerges in nature, and provides guidance for computational enzyme design.
Enzymes exhibit remarkable catalytic power, reflecting life’s struggle for adaptation during billions of years of evolution (1). It is challenging to generate efficient biocatalysts for novel chemical reactions (2, 3). Early strategies for computational enzyme design started from calculating the relevant transition state in the gas phase, which was then engineered into a known protein scaffold so that the active site residues may stabilize the transition state (4). Such designs exhibit low catalytic efficiencies compared to natural enzymes (5, 6). Although more consistent computational strategies that model the mutation effects in the actual enzyme-substrate complex led to important insight, they have not yet provided very effective designs (7–9). The absence of a robust computational method for creating efficient enzymes reflects the challenge in understanding the protein sequence-function relationship, which hinders the creation of new biocatalysts for industrial and biomedical purposes. Meanwhile, laboratory-directed evolution (commonly known as directed evolution) with iterative rounds of mutation and selection process has dramatically improved the computational design’s catalytic power (4, 10). Thus, exploring the optimization pathway of the de novo enzymes may provide clues for producing efficient enzymes and shed light on how new enzymatic function emerges in nature (11).
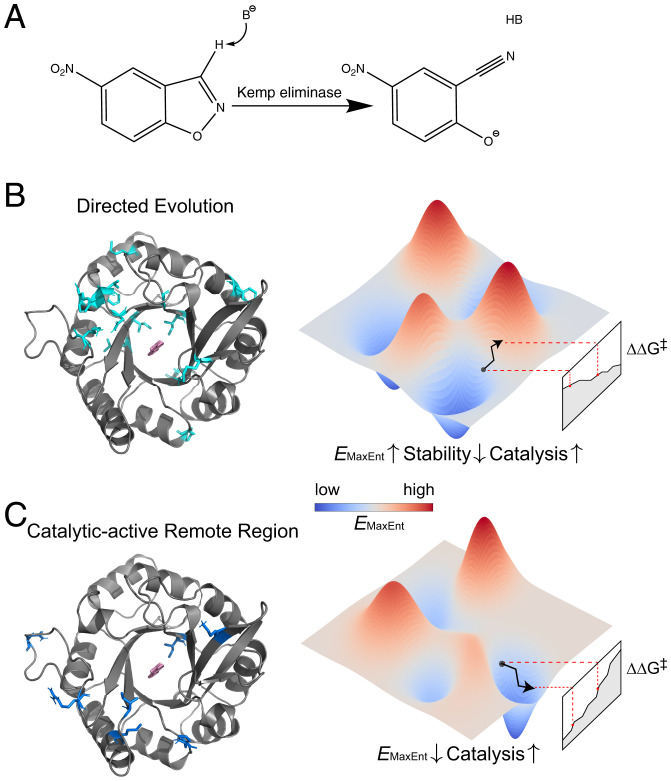
As the most studied de novo enzyme (4, 6, 12–23), Kemp eliminase was designed to catalyze Kemp elimination that has not been known to have a natural biocatalyst (Fig. 1A) (4). The best Kemp eliminase evolved in directed evolution can accelerate the deprotonation of 5-nitrobenzisoxazole with a very high turnover number (12). Nevertheless, the efficiency is still three to four orders of magnitude below the diffusion limit, and it is unclear how to advance with rational strategies for further improvement. It is also unclear how the catalytic power of Kemp eliminase is achieved during directed evolution, partially because previous mechanistic studies only consider very few designs or mutants (14–17, 19, 21, 23). A study covering all the Kemp eliminases is still missing and clearly needed.
Fig. 1.
The maximum entropy model for natural sequences underlies the laboratory evolution of Kemp eliminase. (A) Scheme of the Kemp elimination with 5-nitrobenzisoxazole as substrate. The elimination reaction involves a catalytic base that deprotonates the carbon. In the KE07 series, the active site contains a glutamic acid residue as the catalytic base and a lysine residue to stabilize the negative charge of the oxygen. (B and C) The connection between the MaxEnt model for naturally evolved sequences and laboratory evolution of the abiological Kemp eliminase. The schematic pictures of the landscape from the model are plotted for laboratory evolution for the KE07 series. is the reaction barrier for different Kemp eliminases. PDB entry used in rendering the structure is 2RKX with substrate docked. (B) Directed evolution: (Left) Residues that are mutated in the KE07 series of directed evolution are highlighted in cyan. (Right) The increases as a result of sacrificing stability in directed evolution; the catalytic power is enhanced due to the stability-activity trade-off. (C) Catalytic-active remote regions: (Left) Residues with experimentally generated single-mutation data are highlighted in marine. (Right) The anticorrelates with catalytic power for such regions, and optimizing the statistical energy could improve the activity of Kemp elimination.
Here, we investigate reported designs (or mutants) for Kemp eliminase in a comprehensive way and show that their catalytic power is strongly correlated with the statistical energy () obtained from the maximum entropy (MaxEnt) model for their natural homologs. In directed evolution, it is found the evolved mutants tend to decrease the enzyme stability (with increasing ) and achieve stronger catalytic power (Fig. 1B). The finding can be rationalized by the trade-off between enzyme catalysis and protein stability, and the MaxEnt model appears to capture protein stability even for the laboratory-evolved sequences. On the other hand, single-mutants in catalytic-active remote regions can directly enhance Kemp elimination activity with decreasing (Fig. 1C). The results connect the emergence of new enzymatic function to the natural evolution of the old enzyme used as the scaffold in the design.
Results
The maximum entropy model for enzyme.
To explore the possible connection between laboratory and natural evolution, we leveraged the maximum entropy model (24) and tried to quantify the probability of homologs that share the same evolutionary origin during natural evolution. The MaxEnt model has enjoyed great success in predicting protein residue-residue contact and protein fitness (25–29). For each homologous sequence (), the MaxEnt model considering pair-wise epistasis specifies a statistical energy , where and are site energy and pair-wise coupling between amino acids at two different residue sites and , respectively. More details of the derivation and parameterization of the model can be found in the SI Appendix, SI Text. Shifting the statistical energy by a constant will not affect any physics due to the gauge invariance of the model. The probability of having a given protein sequence follows the Boltzmann distribution , where is the partition function or normalization constant. A lower value for a sequence suggests a higher probability of survival in nature and might reflect a particular evolutionary advantage and vice versa. Thus, the landscape described by the MaxEnt model can also be viewed as a fitness landscape (10) that relates protein fitness to its sequence, and sequences having lower frequently have higher fitness measured in high-throughput screening experiments (28, 29).
Our recent study (30) of natural enzymes found that the statistical energies for mutants in and far away from the catalytic center show strong negative correlations with enzyme activity and stability, respectively. Such results demonstrate that different physical properties of enzymes can be predicted from a collection of natural homologous sequences, which go beyond fitness predictions. In principle, other generative models in machine learning should give consistent results. Obviously, it is of great interest whether the above considerations can help improve the design of artificial enzymes. Thus, we focus in this work on Kemp eliminase and use the MaxEnt model to investigate the connection between sequences produced in the laboratory and their natural homologs.
As the starting point for building the MaxEnt model, we collected the homologs of the studied enzyme by searching sequence databases (SI Appendix, SI Text). The initial computational designs for Kemp eliminase involve a large number of mutations to the template (4). In particular, the KE07 series start with 13 mutations compared to the imidazole glycerol phosphate synthase template before performing directed evolution (SI Appendix, Fig. S1). As shown in the Protein Data Bank (PDB) entry 1THF, the template has a triose-phosphate isomerase (TIM) barrel fold. Interestingly, we were able to obtain 15,685 natural homologs (4,178 after down-weighting similar sequences), using the KE07 initial design as the target sequence in a multiple sequence alignment (MSA) search (SI Appendix, Table S1).
The number of homologs is large enough to parameterize the MaxEnt model, and the reproduction of constraints calculated from MSA and prediction of higher-order MSA statistics validate the parameterization (SI Appendix, Fig. S2). The statistical energy was then calculated for each Kemp eliminase in the KE07 series of directed evolution (4) and each mutant in the catalytic-active remote region (31).
We also used the TIM barrel template (PDB entry: 1THF) as the target sequence to build MSA. The statistics calculated from the two sets of MSAs are nearly identical (SI Appendix, Fig. S3), showing that the natural scaffold used in the KE07 initial design is critical in extracting the natural evolutionary information here. Nearly all the mutations studied here occur at the natural scaffold (at least in the second shell around the substrate) (Fig. 1B and Fig. 1C) (4, 31), indicating that the difference in is due to the change of properties in the natural scaffold region.
Positive correlation between and Kemp elimination activity in directed evolution.
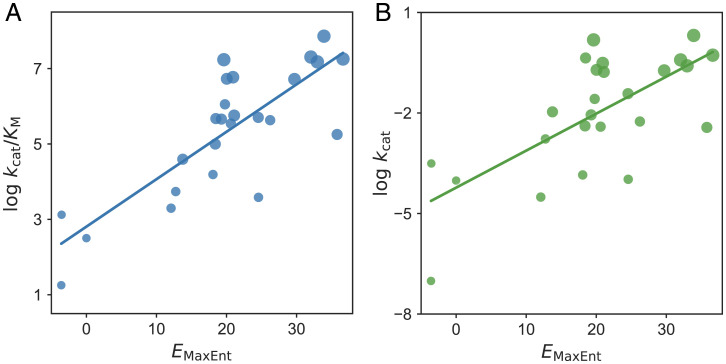
We explored whether Kemp eliminases evolved in directed evolution have any connection with its natural homologs by correlating catalytic efficiency for Kemp elimination and . These Kemp eliminases share the same active sites and differ in the undesigned natural scaffold region. Somewhat surprisingly, the has a significant positive correlation with the catalytic power (expressed both by and ) of Kemp elimination, and the correlation values with and are 0.81 (P value < 0.001, Fig. 2A) and 0.69 (P value < 0.001, Fig. 2B), respectively. [The correlation values reported in this work are all Pearson correlation with P value from the two-tailed test calculated using SciPy 1.0 (32).] The strong correlation is unexpected because no known natural enzymes catalyze Kemp elimination. Thus, it is not clear how the statistical energy that measures sequence fitness in nature could be correlated with the abiological Kemp elimination at first glance.
Fig. 2.
The maximum entropy model for natural sequences is correlated with the catalytic power of designs in directed evolution. Strong positive correlations between and (A) and (B) for the KE07-directed evolution are shown with the least-squares regression lines included. Each dot represents a design with a larger dot size indicating later rounds in directed evolution. We shifted by a constant so that the initial design has a zero value.
Note that the catalytic power of the KE07 designs is highly correlated with the number of mutations relative to the initial computational design, which is used as the target sequence to construct MSA. Such a fact reflects how directed evolution works, accumulating mutations with improved activity through increasing rounds of evolution (33). One may suspect the results shown in Fig. 2 are indirectly caused by the number of mutations in reported designs. Nine of the 25 sequences have five mutations compared to the initial design. Using the subdataset of these nine sequences, the correlation between and (or ) remains (SI Appendix, Fig. S4). Such a control study suggests that the strong correlations are not due to the number of accumulated mutations in directed evolution.
Stability-activity trade-off in directed evolution of Kemp eliminase.
The strong positive correlations between and catalytic power of Kemp eliminases connect the laboratory optimization pathway for the abiological Kemp elimination and the natural evolution of its scaffold. Given the above finding, it is essential to ask how the connection is established. It seems that only a negative correlation with indicates an evolutionary advantage of enzyme property, such as higher protein stability or enhanced catalytic power when decreasing (30). Such a logic is quite intuitive, since it is reasonable to assume that the lower that follows Boltzmann distribution means a higher probability of survival during evolution. However, the correlations we have found here in Fig. 2 are all positive.
We reason that these positive correlations reflect a trade-off between the Kemp elimination activity and the scaffold’s property. The catalytic mechanisms of Kemp elimination and template synthase function are unrelated, suggesting these two functions may not directly counteract each other. For natural enzymes studied in ref (30), we have found a negative correlation between and protein stability in the scaffold region. Therefore, we hypothesize that the stability of KE07 designs shows a negative correlation with , and there is a stability-activity trade-off for the newly evolved Kemp elimination function.
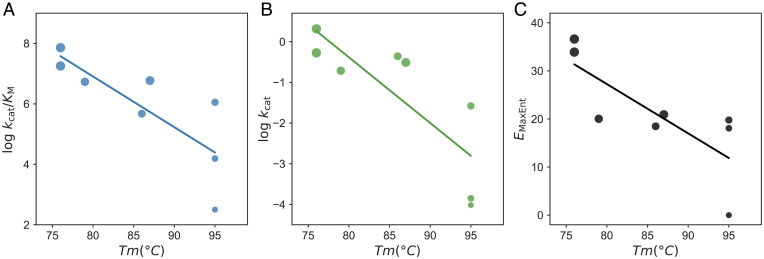
Fortunately, the melting temperature () for several KE07 designs has been measured (34). The quantifies protein stability; higher indicates a more stable protein. In agreement with the above hypothesis, we indeed observed a strong anticorrelation between protein stability and catalytic power for the laboratory-evolved designs. The correlation values between and or are −0.80 (P value = 0.017, Fig. 3A) and −0.81 (P value = 0.014, Fig. 3B), respectively. Consistently, has a strong negative correlation with a correlation value of −0.77 (P value = 0.025, Fig. 3C), supporting our reasoning.
Fig. 3.
Enzyme catalytic power and stability trade-off in directed evolution of designer enzyme. (A and B) Strong negative correlations between protein stability quantified by melting temperature () and enzyme catalytic power expressed by (A) and (B) for the KE07-directed evolution. (C) A strong negative correlation between and . The is the same as that in Fig. 2 for the same mutant.
In addition to the KE07 series, several other directed evolution experiments exist for Kemp elimination. For the KE59 and HG series, we were able to get at least seven designs with biochemical data to check the correlation between and catalytic power. The KE59 series starts from imidazole glycerol phosphate synthase as template (4), while the template of HG-directed evolution is a xylanase (35). Similarly, the correlations are positive and strong for both the KE59 and the HG series. For the KE59 series, the correlation values between and and are, respectively, 0.85 (P value = 0.0072) and 0.80 (P value = 0.058) (SI Appendix, Fig. S5). The corresponding correlation values for the HG series are 0.96 (P value < 0.001) and 0.95 (P value = 0.003) (SI Appendix, Fig. S6), respectively. For the HG series, we also found values for five designs, and the stability-activity trade-off also applies (SI Appendix, Fig. S7).
Although the contribution of some second-shell mutations around the substrate may be rationalized, the role of the majority of mutations introduced in the KE07 series remains elusive (34). The stability-activity trade-off (36–38) emerges in the directed evolution path of this designer enzyme. That is, individual mutation may not contribute significantly to the stability; their combination shows a clear destabilizing trend with increasing activity, as revealed here. Recent studies of other enzymes also highlighted the importance of destabilization in enhancing biocatalysis in directed evolution (39).
At present, the underlying molecular mechanism is still not clear. It is obviously unlikely that any destabilization mutation will increase the activity. There may be insufficient constraints (or evolution time) in directed evolution to maintain stability. Perhaps the fastest way to increase a new activity involves destabilization of the scaffold evolved for the old function, and increases preorganization of the active site region to accommodate the new reaction. More information on unseen intermediates that connect reported designs could help to understand why a particular mutation is picked up in directed evolution. Previous studies also demonstrated the emergence of new active site configurations in evolving the KE07 series (18). Connecting our findings to the conformational plasticity of protein at the molecular level would also be exciting.
It is important to note that theoretical studies found that the folding energy pays for the preorganization of the active site residues which is used for transition state stabilization in natural enzymes, which explains the trade-off (40). In the case of the KE07 series, it was found that the catalysis in these artificial enzymes is due to ground state destabilization by excluding internal water molecules (6, 14). This theoretical finding is consistent with the observed correlation here.
Negative correlation between and Kemp elimination activity in catalytic-active remote regions.
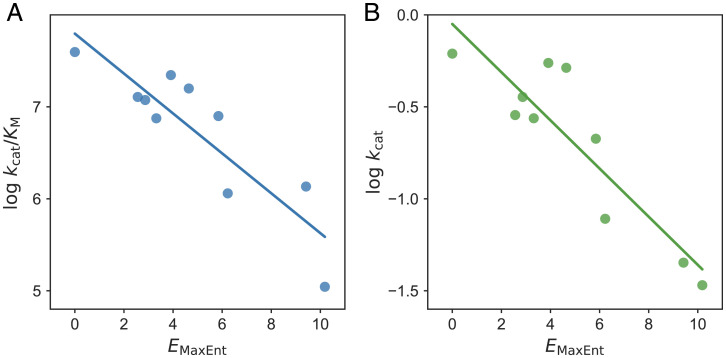
Besides directed evolution, Kemp eliminase has been recently studied, focusing on the remote region that shows coupled conformational dynamics with active site residues (31). The residues were identified based on the mutual information of residue dynamics in molecular simulation. For the best-evolved KE07 design, such residues were shown to be important for enzyme catalysis by single-site mutagenesis (31). Therefore, we term them “catalytic-active remote regions.” The majority of such residues are in the protein loop region distal from the active site (Fig. 1C). Surprisingly, strongly anticorrelates with and with correlation values of −0.88 (P value < 0.001) and −0.89 (P value < 0.001), respectively (Fig. 4). Using the correlation to improve enzyme catalysis can be done in a similar way to what we have proposed for natural enzymes by optimizing in the landscape (Fig. 1C) (supporting information in ref (30)).
Fig. 4.
The maximum entropy model for natural sequences is anticorrelated with the catalytic power of mutants in catalytic-active remote regions. Strong negative correlations between and (A) and (B) for the catalytic-active remote region are shown with the least-squares regression lines included. Each dot represents one single mutation. We shifted by a constant so that the best-evolved design has a zero value.
The strong negative correlation discussed above argues for the importance of remote loop regions in enzyme catalysis from an evolutionary perspective, along with increasingly experimental evidence (41, 42). In the case of the KE07 template, TIM barrel, the loops can close to exclude water molecules and thus tune the electrostatics within the barrel where the chemical reaction happens (41). Such a mechanism seems insensitive to the reaction type and thus can be inherited by the abiological Kemp elimination. Interestingly, previous studies emphasized the shifting of a substate population when evolving new enzyme functions (43, 44). In contrast, our finding suggests that some conformational flexibility of protein scaffold can be shared among various biocatalytic reactions.
In addition, the Kemp elimination activity of active site mutants does not correlate with the (SI Appendix, Fig. S8), which is expected considering that the natural evolution of active sites is for the template function instead of the Kemp elimination. Note that the correlation between catalytic power and for the whole enzyme scaffold far away from the substrate is not strong in general for natural enzymes (30). The strong negative correlation shown in Fig. 4 suggests that the enzyme scaffold can be classified into more refined categories in terms of the evolution-biocatalysis relationship. In particular, the flexible loop regions with conformational dynamics coupled with active sites may stand out compared to other scaffold regions.
Discussion
Current laboratory evolution procedures only access a tiny fraction of sequence space to achieve an impressive performance. It has been a major conceptual challenge in directed evolution to relate or predict enzyme evolution behavior in the laboratory with natural evolution (45, 46). The strong correlations found in this work indicate that there are indeed such connections between the laboratory and natural evolution, which can be used to prioritize sequence space and design better mutation libraries to reduce the time and effort in laboratory evolution or experimental validation. The positive correlations between and activity we found for directed evolution datasets suggest a stability-activity trade-off. Here, the MaxEnt model can be used to predict the stability of designs when mutations occur at the natural scaffold region, which helps to suggest mutations to tune the stability to enhance evolvability (47). The negative -activity correlation for the catalytic-active remote regions can be straightforwardly used to improve activity, similar to what we have suggested for improving natural enzymes (30). The suggested mutations may be further screened by other approaches, e.g., QM/MM-EVB (quantum mechanics/molecular mechanics-empirical valance bond) calculation (6, 40).
Although the current study is closely related to our previous work on natural enzymes (30), their scopes are quite different. For natural enzymes we have studied, the evolutionary information of the enzyme catalytic center and the enzyme scaffold (or surface) is correlated with enzyme activity and stability, respectively (30). It is unclear whether the sequence entropy of the natural enzyme scaffold is informative for a new function when assembled with designed active sites. To address this question, we examined designs from laboratory evolution for the abiological Kemp eliminase. The different Kemp eliminases share identical active sites while mutating the natural enzyme scaffold. It turns out that the stability of such de novo designs can still be captured by natural evolution, consistent with our findings for natural enzymes (30). Meanwhile, the evolutionary information for catalytic-active remote regions is correlated with new enzyme activity, and we hypothesize that the negative correlation is not very sensitive to the type of enzymatic reaction. This can be tested experimentally by checking the same mutations for the imidazole glycerol phosphate synthase from which the natural scaffold is obtained. In contrast, the natural evolution of active sites only reflects the natural synthase activity instead of the designed Kemp elimination, which is indicated in our previous work (30) and confirmed here (SI Appendix, Fig. S8).
By distilling information from natural sequences, we hope to discover new biophysical and biochemical rules of enzymes. Our results suggest that enzymes can be dissected into several parts mainly shaped by different evolutionary pressures. As for now, we may have identified three modules, including the active center and catalytic-active remote regions, which are mainly shaped by enzyme efficiency, and other enzyme scaffolds, which are responsible for a stable enzyme. We anticipate that residues interacting with another domain or protein will form a different module influenced by the strength of the protein-protein interaction. Further studies will be needed to examine the generalization of these findings across all known enzymes. Moreover, our work echoes Tawfik's efforts to apprehend natural evolution from protein engineering (48, 49).
Adopting machine learning to laboratory evolution is promising. Supervised learning on a few measured mutants can provide useful models in improving protein properties (50, 51). In particular, evolutionary information can assist this process for natural functions, including enzyme activity (51). Besides, the consensus design using MSA has been employed to prepare mutation libraries in directed evolution to increase protein stability (47). Here, our work establishes a quantitative connection between the laboratory evolution of a designer enzyme and the natural evolution of its homologs with the MaxEnt model.
In summary, our findings connect the laboratory evolution of an abiological enzyme function to natural evolution, contribute to the understanding of the emergence of new enzyme functions, and provide fresh perspectives for utilizing the enormous natural sequences for guiding enzyme design. Besides, the correlations between the MaxEnt model and enzyme properties show evolutionary evidence of how the protein scaffold plays a role in biocatalysis. One implication might be that enzyme scaffold is still broadly adopted when evolved to a new function, supporting the hypothesis (52) that modularity promotes functional innovation of enzyme. It is also likely that the valley in the landscape (Fig. 1) provides a direction that can guide further mutations. Overall, the perspective from the landscape quantified by the MaxEnt model (or other generative machine learning models) could fill the considerable gaps in the protein sequence-function relationship, which is an essential step toward truly rational protein engineering.
Supplementary Material
Acknowledgments
This work was supported by the NIH R35 GM122472 and the NSF Grant MCB 1707167. We thank the University of Southern California High Performance Computing and Communication Center for computational resources. We thank Tianmin Fu and Aoxuan Zhang for insightful discussions.
Footnotes
Reviewers: J.-K.H., National Yang Ming Chiao Tung University; and S.K., Uppsala Universitet.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207904119/-/DCSupplemental.
Data Availability
The code for parameterizing the Maximum Entropy Model for Enzyme is available at https://github.com/Wenjun-Xie/MEME. The data and other code are available at https://github.com/Wenjun-Xie/Kemp_eliminase, including collected experimental data, multiple sequence alignments, and a jupyter-notebook for a step-by-step guide to reproduce the figures in the main text.
References
- 1.Davidi D., Longo L. M., Jabłońska J., Milo R., Tawfik D. S., A bird’s-eye view of enzyme evolution: Chemical, physicochemical, and physiological considerations. Chem. Rev. 118, 8786–8797 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Bolon D. N., Mayo S. L., Enzyme-like proteins by computational design. Proc. Natl. Acad. Sci. U.S.A. 98, 14274–14279 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaplan J., DeGrado W. F., De novo design of catalytic proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 11566–11570 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Röthlisberger D., et al. , Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Baker D., An exciting but challenging road ahead for computational enzyme design. Protein Sci. 19, 1817–1819 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frushicheva M. P., Cao J., Chu Z. T., Warshel A., Exploring challenges in rational enzyme design by simulating the catalysis in artificial Kemp eliminase. Proc. Natl. Acad. Sci. U.S.A. 107, 16869–16874 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuxreiter M., Mones L., The role of reorganization energy in rational enzyme design. Curr. Opin. Chem. Biol. 21, 34–41 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Risso V. A., et al. , Enhancing a de novo enzyme activity by computationally-focused ultra-low-throughput screening. Chem. Sci. (Camb.) 11, 6134–6148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaissier Welborn V., Head-Gordon T., Computational design of synthetic enzymes. Chem. Rev. 119, 6613–6630 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Romero P. A., Arnold F. H., Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10, 866–876 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bloom J. D., Arnold F. H., In the light of directed evolution: Pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. U.S.A. 106 (suppl. 1), 9995–10000 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blomberg R., et al. , Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature 503, 418–421 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Osuna S., Jiménez-Osés G., Noey E. L., Houk K. N., Molecular dynamics explorations of active site structure in designed and evolved enzymes. Acc. Chem. Res. 48, 1080–1089 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Jindal G., Ramachandran B., Bora R. P., Warshel A., Exploring the development of ground-state destabilization and transition-state stabilization in two directed evolution paths of Kemp eliminases. ACS Catal. 7, 3301–3305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otten R., et al. , How directed evolution reshapes the energy landscape in an enzyme to boost catalysis. Science 370, 1442–1446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunzel H. A., et al. , Evolution of dynamical networks enhances catalysis in a designer enzyme. Nat. Chem. 13, 1017–1022 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Bhowmick A., Sharma S. C., Head-Gordon T., The importance of the scaffold for de novo enzymes: A case study with Kemp eliminase. J. Am. Chem. Soc. 139, 5793–5800 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Hong N. S., et al. , The evolution of multiple active site configurations in a designed enzyme. Nat. Commun. 9, 3900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaissier V., Sharma S. C., Schaettle K., Zhang T., Head-Gordon T., Computational optimization of electric fields for improving catalysis of a designed Kemp eliminase. ACS Catal. 8, 219–227 (2018). [Google Scholar]
- 20.Broom A., et al. , Ensemble-based enzyme design can recapitulate the effects of laboratory directed evolution in silico. Nat. Commun. 11, 4808 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bunzel H. A., et al. , Emergence of a negative activation heat capacity during evolution of a designed enzyme. J. Am. Chem. Soc. 141, 11745–11748 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Khersonsky O., et al. , Optimization of the in-silico-designed Kemp eliminase KE70 by computational design and directed evolution. J. Mol. Biol. 407, 391–412 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Åqvist J., Computer simulations reveal an entirely entropic activation barrier for the chemical step in a designer enzyme. ACS Catal. 12, 1452–1460 (2022). [Google Scholar]
- 24.Jaynes E. T., Information theory and statistical mechanics. Phys. Rev. 106, 620–630 (1957). [Google Scholar]
- 25.Marks D. S., et al. , Protein 3D structure computed from evolutionary sequence variation. PLoS One 6, e28766 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morcos F., et al. , Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. U.S.A. 108, E1293–E1301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamisetty H., Ovchinnikov S., Baker D., Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc. Natl. Acad. Sci. U.S.A. 110, 15674–15679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopf T. A., et al. , Mutation effects predicted from sequence co-variation. Nat. Biotechnol. 35, 128–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Figliuzzi M., Jacquier H., Schug A., Tenaillon O., Weigt M., Coevolutionary landscape inference and the context-dependence of mutations in beta-lactamase tem-1. Mol. Biol. Evol. 33, 268–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie W. J., Asadi M., Warshel A., Enhancing computational enzyme design by a maximum entropy strategy. Proc. Natl. Acad. Sci. U.S.A. 119, e2122355119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhowmick A., Sharma S. C., Honma H., Head-Gordon T., The role of side chain entropy and mutual information for improving the de novo design of Kemp eliminases KE07 and KE70. Phys. Chem. Chem. Phys. 18, 19386–19396 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Virtanen P., et al. ; SciPy 1.0 Contributors, SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., et al. , Directed evolution: Methodologies and applications. Chem. Rev. 121, 12384–12444 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Khersonsky O., et al. , Evolutionary optimization of computationally designed enzymes: Kemp eliminases of the KE07 series. J. Mol. Biol. 396, 1025–1042 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Privett H. K., et al. , Iterative approach to computational enzyme design. Proc. Natl. Acad. Sci. U.S.A. 109, 3790–3795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoichet B. K., Baase W. A., Kuroki R., Matthews B. W., A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. U.S.A. 92, 452–456 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Minasov G., Shoichet B. K., Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 320, 85–95 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Tokuriki N., Tawfik D. S., Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 19, 596–604 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Stimple S. D., Smith M. D., Tessier P. M., Directed evolution methods for overcoming trade-offs between protein activity and stability. AIChE J. 66, e16814 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roca M., Liu H., Messer B., Warshel A., On the relationship between thermal stability and catalytic power of enzymes. Biochemistry 46, 15076–15088 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malabanan M. M., Amyes T. L., Richard J. P., A role for flexible loops in enzyme catalysis. Curr. Opin. Struct. Biol. 20, 702–710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nestl B. M., Hauer B., Engineering of flexible loops in enzymes. ACS Catal. 4, 3201–3211 (2014). [Google Scholar]
- 43.Campbell E. C., et al. , Laboratory evolution of protein conformational dynamics. Curr. Opin. Struct. Biol. 50, 49–57 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Tokuriki N., Tawfik D. S., Protein dynamism and evolvability. Science 324, 203–207 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Arnold F. H., Wintrode P. L., Miyazaki K., Gershenson A., How enzymes adapt: Lessons from directed evolution. Trends Biochem. Sci. 26, 100–106 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Martínez R., Schwaneberg U., A roadmap to directed enzyme evolution and screening systems for biotechnological applications. Biol. Res. 46, 395–405 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Khersonsky O., et al. , Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc. Natl. Acad. Sci. U.S.A. 109, 10358–10363 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peisajovich S. G., Tawfik D. S., Protein engineers turned evolutionists. Nat. Methods 4, 991–994 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Trudeau D. L., Tawfik D. S., Protein engineers turned evolutionists-the quest for the optimal starting point. Curr. Opin. Biotechnol. 60, 46–52 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Yang K. K., Wu Z., Arnold F. H., Machine-learning-guided directed evolution for protein engineering. Nat. Methods 16, 687–694 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Biswas S., Khimulya G., Alley E. C., Esvelt K. M., Church G. M., Low-N protein engineering with data-efficient deep learning. Nat. Methods 18, 389–396 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Dellus-Gur E., Toth-Petroczy A., Elias M., Tawfik D. S., What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J. Mol. Biol. 425, 2609–2621 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code for parameterizing the Maximum Entropy Model for Enzyme is available at https://github.com/Wenjun-Xie/MEME. The data and other code are available at https://github.com/Wenjun-Xie/Kemp_eliminase, including collected experimental data, multiple sequence alignments, and a jupyter-notebook for a step-by-step guide to reproduce the figures in the main text.