SUMMARY
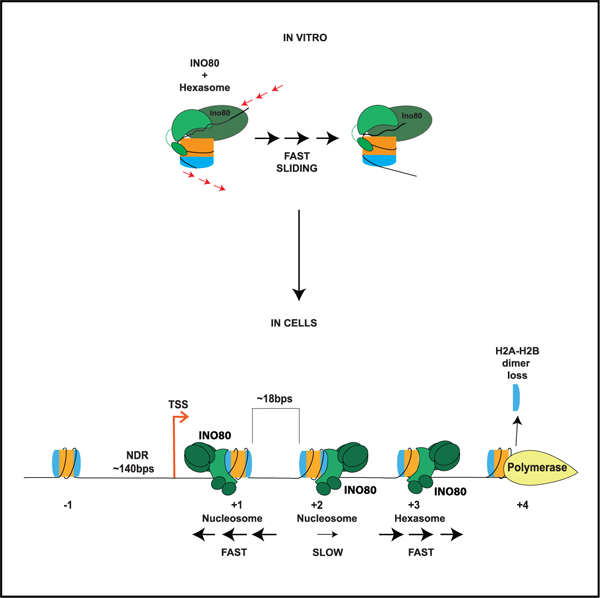
The critical role of the INO80 chromatin remodeling complex in transcription is commonly attributed to its nucleosome sliding activity. Here, we have found that INO80 prefers to mobilize hexasomes over nucleosomes. INO80’s preference for hexasomes reaches up to ~60 fold when flanking DNA overhangs approach ~18-bp linkers in yeast gene bodies. Correspondingly, deletion of INO80 significantly affects the positions of hexasome-sized particles within yeast genes in vivo. Our results raise the possibility that INO80 promotes nucleosome sliding by dislodging an H2A-H2B dimer, thereby making a nucleosome transiently resemble a hexasome. We propose that this mechanism allows INO80 to rapidly mobilize nucleosomes at promoters and hexasomes within gene bodies. Rapid repositioning of hexasomes that are generated in the wake of transcription may mitigate spurious transcription. More generally, such versatility may explain how INO80 regulates chromatin architecture during the diverse processes of transcription, replication, and repair.
Graphical abstract
In brief
Emerging evidence shows that hexasomes are formed in gene bodies during transcription. How these particles are regulated is poorly understood. Here, Hsieh et al. use mechanistic enzymology and genomics to show that the INO80 remodeler preferentially slides hexasomes compared with nucleosomes in vitro and affects hexasome positioning in vivo.
INTRODUCTION
In eukaryotes, most DNA-dependent processes have to contend with chromatin. The most prevalent building block of chromatin is a nucleosome, which is composed of ~147 bp of DNA wrapped around a histone octamer containing two H2A-H2B dimers and an H3-H4 tetramer. Several studies indicate that the octameric histone composition changes during processes such as transcription and replication, which require transient disruption of histone-DNA contacts (Henikoff, 2016). In particular, transcription results in accumulation of subnucleosomal particles, many of which are hexasomes that are missing a single H2A-H2B dimer (Ramachandran et al., 2017). Extensive work has addressed how nucleosome positions are regulated during transcription (Lowary and Widom, 1998; Mavrich et al., 2008; Radman-Livaja and Rando, 2010; Segal et al., 2006). In comparison, whether and how hexasome positions are regulated is poorly understood.
Nucleosome positions during transcription are regulated by ATP-dependent chromatin remodeling enzymes, which can evict octamers, exchange histones, and distort and slide the histone octamer (Zhou et al., 2016). These highly conserved enzymes often fall into four main classes, ISWI, SWI/SNF, CHD, and INO80. Together, these enzymes collaborate to maintain a nucleosome-depleted region (NDR) at promoters and specific nucleosome positions in the gene body (Figure 1A) (Klein-Brill et al., 2019; Krietenstein et al., 2016). In vitro, enzymes from these different classes, such as S. cerevisiae Chd1, INO80, and ISW2, superficially show a similar ability to slide nucleosomes (Zhou et al., 2016). Yet, in vivo, these enzymes play non-overlapping roles. In such comparisons, it is commonly assumed that nucleosomes are the preferred substrates. Indeed, specific features of nucleosomes are recognized by these enzymes, suchas the length of the DNA overhang flanking a nucleosome, an acidic patch found on the H2A-H2B dimer, and histone post-translational modifications (McGinty and Tan, 2015; Zhou et al., 2016). Among these features, interactions with the acidic patch play a significant role in nucleosome remodeling by the ISWI, SWI/SNF, CHD, and INO80 classes of enzymes (Dao et al., 2020; Eustermann et al., 2018; Gamarra et al., 2018; Levendosky and Bowman, 2019; Valencia et al., 2019). At the same time, however, the prevalence of subnucleosomal particles in vivo (Ramachandran et al., 2017; Rhee et al., 2014) provokes the question of whether the action of these remodeling enzymes differs on hexasomes, thereby contributing to some of their unique functions in vivo.
Figure 1. INO80 regulates positions of subnucleosomal particles in vivo.
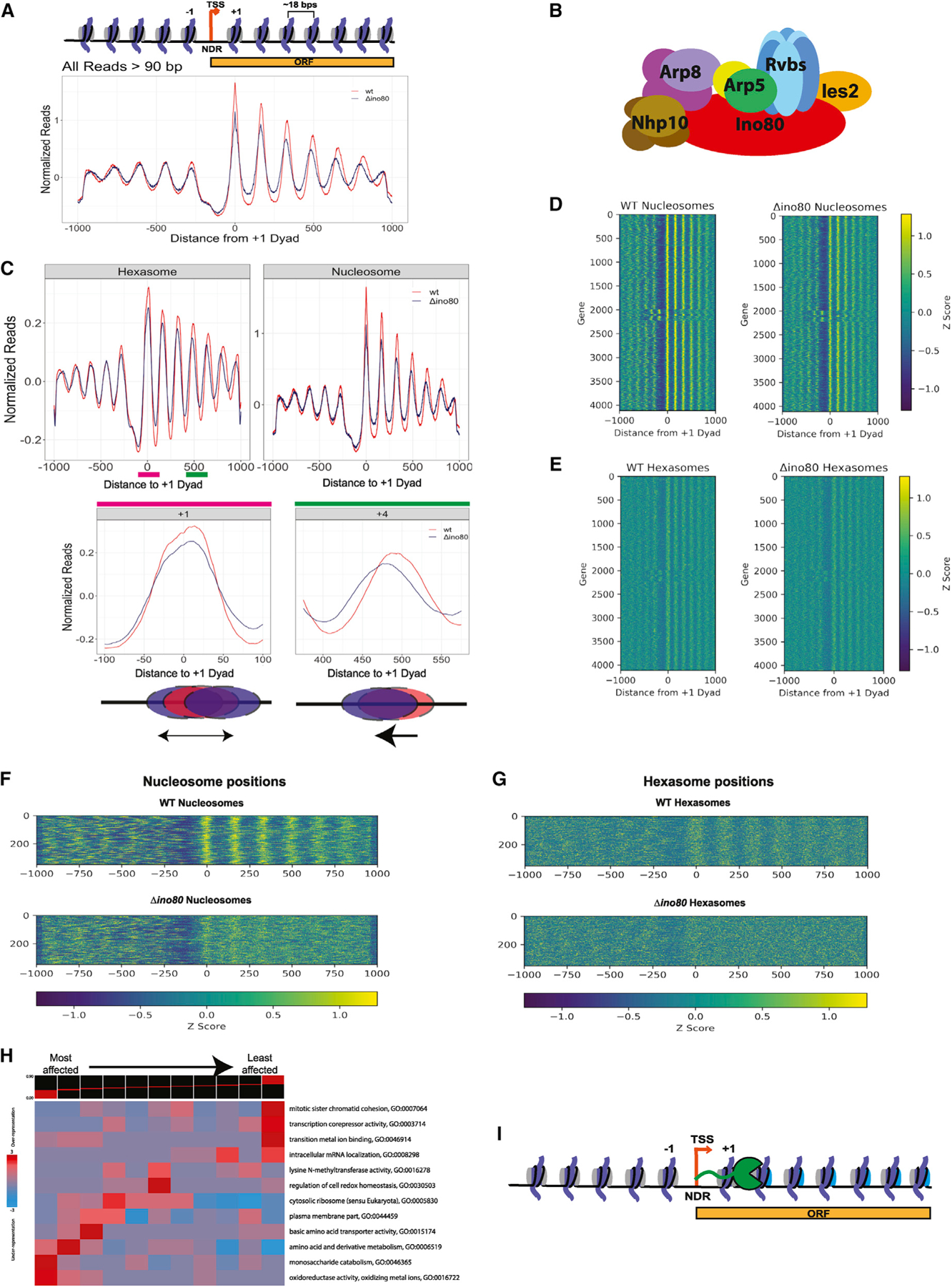
(A and B) (A) Normalized MNase signal at the TSSs of all genes for WT, in orange, and Δino80, in dark blue, for all fragment lengths greater than 90 bps. The x axis represents distance from +1 nucleosome dyad. Upper panel: schematic of the corresponding chromatin architecture in (B) illustration of the INO80 complex. Only one subunit per module is labeled.
(C) Upon deletion of Ino80, nucleosomes and hexasomes are less well positioned at +1 location and display shifted positions in gene body. Upper panel: fragments were binned by sizes representing either hexasomes (100 ± 10 bps) or nucleosomes (147 ± 10 bps), and the average signal at TSSs and in the gene body are plotted for WT and Δino80. Lower panel: pink and green lines in the hexasome plot from the upper panel are magnified to show hexasome footprints at +1 and +4 positions. Below the graphs are illustrations of the respective changes in positioning that occur from WT (red) to Δino80 (blue).
(D) Heatmap of nucleosome footprint signals across all genes for WT and Δino80 cells.
(E) Same as (D) but for the hexasome footprint signals.
(F) Data for genes where nucleosome positions are most affected by deletion of Ino80. A heatmap representing genes that had the lowest Spearman Rho correlation (i.e., most affected by deletion of Ino80) for nucleosome footprint signals ranging from the −100 to +1,000 bps of the +1 dyad between WT and Δino80.
(G) Data for the genes in (F) but focusing on hexasome positions.
(H) An iPAGE heatmap of the correlations of the nucleosome footprint signal between WT and Δino80 and their annotated GO terms. The genes have been binned into 11 groups, with the lowest correlation group (most affected in terms of nucleosome footprints) on the left, and the highest correlation group (least affected in terms of nucleosome footprints) on the right.
(I) Illustration of RNA Pol II traversing through the gene body. The promoter distal dimer that is lost during elongation is depicted in cyan.
Recent studies on S. cerevisiae Chd1 provide some insight. Chd1 can bi-directionally slide nucleosomes (Qiu et al., 2017). However, removal of one H2A-H2B dimer inhibits sliding in one direction, resulting in unidirectional sliding (Levendosky et al., 2016). Whether other remodeling enzymes are similarly regulated is unclear. Here, we addressed this question in the context of the multi-subunit S. cerevisiae INO80 complex, which plays central roles in DNA repair, replication, and transcription. Unlike CHD and ISWI enzymes, where the ATPase subunit is sufficient for remodeling, INO80 sliding is highly regulated by its additional subunits. S. cerevisiae INO80 has 14 subunits in addition to the Ino80 ATPase subunit (Shen et al., 2000). These additional subunits are organized in separable modules (Figure 1B). In particular, the Arp5-Ies6 module plays an activating role for sliding nucleosomes (Brahma et al., 2017; Eustermann et al., 2018; Shen et al., 2003; Yao et al., 2016). Furthermore, unlike the CHD and ISWI ATPases, which bind at the internal location of super helical location (SHL) + 2 on a nucleosome, the Ino80 ATPase binds near the entry-exit site of the nucleosome at SHL-5/-6 (Ayala et al., 2018; Eustermann et al., 2018) (Figure 2A). These significant biochemical and structural differences between INO80 versus CHD and ISWI enzymes led us to examine more carefully INO80’s substrate specificity in vivo and in vitro.
Figure 2. Hexasomes are better substrates for INO80 in vitro.
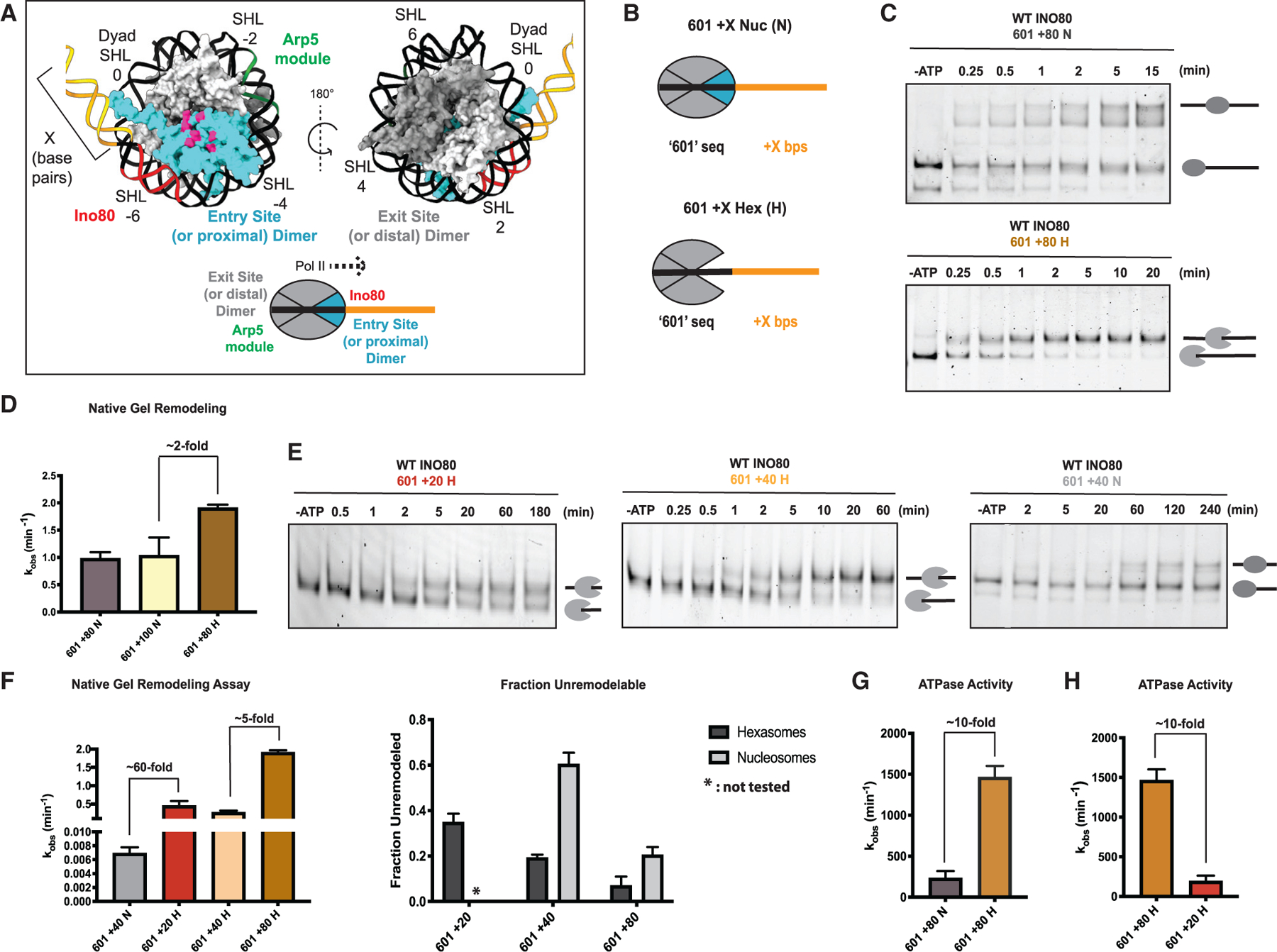
(A) Upper panel: two different rotations of nucleosome structure (PDB: 1KX5) highlighting the dimer (cyan) at the entry site (or proximal to flanking DNA) and its acidic patch (pink). Regions of DNA where Arp5-Ies6 binds (green), Ino80 ATPase binds (red), that are in flanking DNA (yellow), and are in super helical locations (SHLs) are indicated. Lower panel: illustration of nucleosome, indicating the exit site (or distal) and entry site (or proximal) H2A-H2B dimers, and regions where the Arp5-Ies6 and Ino80 ATPase binds. Direction of elongating RNA Pol II based on the loss of the distal dimer is shown with a black dotted line.
(B) Depiction of 601 nucleosome (N) and hexasome (H) constructs with flanking DNA used in this study. The H2A-H2B dimer missing in hexasomes is in cyan.
(C) Example native gels showing remodeling by INO80 of 601 + 80 nucleosomes (top) and 601 + 80 hexasomes (bottom). Substrates (end-positioned nucleosomes or end-positioned hexasomes) are labeled by illustrations next to the respective bands in gels.
(D) Quantification of rate constants from multiple repeats of data, such as in (C). Rate constant for remodeling of 601 + 100 N is also shown.
(E) Example gels of INO80 remodeling with 601 + 20 H (left), 601 + 40 H (middle), and 601 + 40 N (right). Substrates (end-positioned nucleosomes or end-positioned hexasomes) are labeled by illustrations next to the respective bands in gels.
(F) Left panel: quantification of rate constants for remodeling of 601 + 40 N (gray), 601 +20 H (red), 601 + 40 H (peach), and 601 + 80 H (tan) by INO80 as assayed using the native gel assay. Note that the data for 601 + 80 H are the same data shown in (D) and are shown again here for ease of comparison. Right panel: fraction of unremodeled substrate, assayed via the native gel assay and measured at the longest time point (where all reactions have mostly gone to completion). Hexasomes are in dark gray, and nucleosomes are in light gray.
(G) Observed rate constants of INO80 ATPase activity on 601 + 80 N in gray and 601 + 80 H in tan. ATPase assays were performed under the same conditions as native-gel-based remodeling.
(H) Rate constants of INO80 ATPase activity on 601 + 80 H (tan) (data from G) and 601 + 20 H (red). Note that the data for 601 + 80 H are the same data as shown in (G) and are shown again here for ease of comparison. Error bars represent SEM from n > 3.
Here, using a combination of in vivo and biochemical studies, we have uncovered a new biological activity of INO80. We find that, in addition to regulating nucleosome positioning, INO80 contributes to the steady-state positioning of subnucleosomal particles in gene bodies. Surprisingly, in vitro, INO80 is not just capable of sliding hexasomes but prefers hexasomes over nucleosomes. The preference for hexasomes was unexpected given INO80’s reliance on the acidic patch of H2A-H2B for nucleosome sliding. However, our results raise the possibility that during nucleosome sliding, the Arp5-Ies6 module of INO80 enables transient detachment of an H2A-H2B dimer through interactions with the acidic patch, allowing a hexasome-like intermediate. Overall, our work shows that INO80’s specific biochemical mechanism uniquely gives it the versatility to act on both nucleosome and hexasome substrates based on genomic context.
RESULTS
INO80 regulates both nucleosomal and subnucleosomal spacing in vivo
Most genes in S. cerevisiae have a stereotypical chromatin architecture near promoters, which includes an NDR at the transcription start site (TSS). The NDR is flanked by two well-positioned nucleosomes, a +1 nucleosome, the first nucleosome in the gene body, and a −1 nucleosome, the first upstream nucleosome (Mavrich et al., 2008) (Figure 1A). Nucleosomes further in the gene body (+2 to +9) show some degree of defined positioning with an average inter-nucleosomal linker DNA spacing of ~18 bp (Mavrich et al., 2008) (Figure 1A).
INO80 has been shown to position the +1 nucleosome at TSSs, specifically at metabolic genes in budding yeast (Klein-Brill et al., 2019; Krietenstein et al., 2016; Yao et al., 2016; Yen et al., 2013). However, although there is increasing evidence for subnucleosomal particles at promoters and gene bodies of active genes arising from the high rates of nucleosome turnover during transcription, little is known on how remodelers affect these particles (Ramachandran et al., 2017; Rhee et al., 2014). To identify potential roles of INO80 in regulating subnucleosomal particles, we performed MNase-seq in the context of wild-type (WT) S. cerevisiae cells and cells deleted for the ATPase subunit of INO80, Ino80 (Δino80 cells), and mapped nucleosomal and subnucleosomal particles as previously described (Ramachandran et al., 2017).
A prior study demonstrated that a prevalent set of subnucleosomal particles found near TSSs correspond to MNase-protected fragments of ~100 bp, which were identified as hexasomes, (i.e., nucleosomes missing one H2A-H2B) (Ramachandran et al., 2017). We, therefore, first mapped all fragments >90 bps to TSSs of the yeast genome using chemical cleavage mapping data (Brogaard et al., 2012; see materials and methods) in WT and Δino80 cells (Figure 1A). We then filtered the fragments by size to differentiate potential hexasomes from nucleosomes (Figure 1C). Consistent with prior data, we observed that in the Δino80 strain, nucleosomes at the +1 position are not as well positioned compared with WT cells (Klein-Brill et al., 2019; Yao et al., 2016) (Figure 1C, top right panel). Previous studies have focused on the role of INO80 in positioning the +1 nucleosome (Klein-Brill et al., 2019; Yao et al., 2016). Here, in addition to the changes in positioning of the +1 nucleosome, we observe that nucleosomes further into the gene body (up to +6) also show altered positions in Δino80 cells (Figure 1C, top right panel). Thus, our data suggest that INO80 is also important for maintaining spacing of nucleosomes within the gene body.
Intriguingly, we found that particles suggestive of hexasomes at the +1 position also show disrupted positioning in Δino80 cells compared with WT cells, similar to the effects seen on +1 nucleosomes. Furthermore, there is a clear directional change in the positioning of potential hexasomes within the gene bodies in Δino80 beyond the +1 position, analogous to results observed with nucleosomes (Figure 1C, top left panel, bottom right, and bottom left panels). The mispositioning of nucleosomes and hexasomes is also obvious in a heatmap representation showing all genes, where the +1 to +6 nucleosomes show fuzzier profiles in Δino80 cells compared with WT cells (Figures 1D and 1E). Overall, these results show that INO80 is important for both nucleosome and hexasome positioning at the TSS and in the gene body.
Next, we analyzed the genes that were most affected by deletion of Ino80 by calculating the Spearman Rho correlation coefficient for WT and Δino80 nucleosome signals (see STAR Methods). The genes with the lowest Spearman Rho value represent the genes that had most differences in nucleosome positioning between WT and Δino80 cells (Figure 1F). In other words, the nucleosome positions within these genes in the Δino80 cells are most disrupted relative to those in the WT cells (Figure 1F). These same genes also have substantially mispositioned hexasomes (Figure 1G), suggesting that INO80 contributes to positioning both nucleosomes and hexasomes at the same class of genes. An iPAGE heatmap of genes (Goodarzi et al., 2009) sorted by least to most correlated between WT and Δino80 shows the gene ontology (GO) enrichment terms of the different classes (Figure 1H). The GO enrichment terms are consistent with previously reported roles for INO80 at metabolism-related genes (Yao et al., 2016). Our data show that genes involved in metabolism not only have changes in nucleosome positions but also have changes in hexasome positions upon loss of INO80. Together, these results for the first time demonstrate that the locations of subnucleosomal particles at TSSs and within genes are regulated by INO80.
INO80 shows a large preference for remodeling hexasomes over nucleosomes
The effects of INO80 on hexasome positions in vivo described above could arise directly from the action of INO80 on hexasomes, or indirectly through INO80’s action on nucleosomes, which are then partially disassembled by other factors. To distinguish between these possibilities, we asked whether INO80 can act on a hexasome substrate in vitro.
Prior biochemical work has shown that RNA polymerase elongation through nucleosomes results in asymmetric loss of the H2A-H2B dimer in the direction of RNA polymerase II (RNA Pol II) transcription (Figure 1I) (Kulaeva et al., 2009). Consistent with these biochemical findings, in vivo MNase footprinting in Drosophila cells suggests that elongating RNA polymerase results in hexasome formation within the gene body with a bias for losing the promoter-distal H2A-H2B dimer (Ramachandran et al., 2017). Our data mentioned above suggest that INO80 is important for sliding hexasomes within the gene body away from the promoter (+2 onward, Figure 1C). Based on the prior biochemical and Drosophila studies, we interpret this to mean that INO80 plays a role in sliding hexasomes toward the direction that the dimer is lost from. Therefore, we focused on testing INO80’s activity on a hexasome lacking the dimer proximal to the flanking DNA, which would mimic a nucleosome with the dimer lost in the direction of RNA Pol II transcription in vivo (Figures 1I, 2A, and 2B). We define this dimer as the proximal (or entry side) dimer to reflect its proximity to the side of the DNA that enters into the nucleosome during nucleosome sliding. The other dimer is referred to as the distal or exit side dimer.
Previously, we have found that INO80 displays maximal remodeling activity on an end-positioned nucleosome assembled on the 601 Widom sequence followed by at least 80 bps of flanking DNA (Zhou et al., 2018). We, therefore, assembled hexasomes on the same 601 DNA template using recently described methods to obtain specifically oriented hexasomes (Levendosky et al., 2016) (Figure 2B). INO80 remodeled the hexasome, which we refer to as 601 + 80 hexasome (H), ~2 fold faster than the 601 + 80 nucleosome (N) (Figures 2C, 2D, and S3; Table 2). The products of hexasome remodeling are consistent with sliding of the hexamer toward the center of the DNA based on comparisons with 40-601-40 hexasome standards (Figure S7). The products of nucleosome remodeling also migrate at locations consistent with centered nucleosomes as seen previously (Figure S7). Additionally, INO80 remodeled 601 + 100 nucleosomes with comparable rates as 601 + 80 nucleosomes (Figure 2D; Table 2).
Table 2.
Rate constants with the SEM for all gel-remodeling-based assays
| Construct | WT INO80 kobs (min−1) | SEM | INO80(Δarp5) kobs (min−1) | SEM |
|---|---|---|---|---|
| +40 WT N | 0.007 | 0.0005 | – | – |
| +80 WT N | 0.977 | 0.0695 | 0.006 | 0.0009 |
| +100 WT N | 1.035 | 0.1908 | – | – |
| +80 APM N | 0.005 | 0.0007 | 0.010 | 0.0011 |
| +20 WT H | 0.450 | 0.0751 | – | – |
| +40 WT H | 0.273 | 0.0338 | – | – |
| +80 WT H | 1.908 | 0.0349 | *no remodeling detected | *no remodeling detected |
| +80 WT hex + WT dim | 1.639 | 0.0803 | – | – |
| +80 APM hex + APM dim | 0.015 | 0.0006 | – | – |
| +80 WT hex + APM dim | 0.007 | 0.0018 | – | – |
| +80 APM hex + WT dim | 0.998 | 0.1023 | – | – |
Although 80 bps of flanking DNA is required for maximal remodeling of nucleosomes by INO80 in vitro, within gene bodies in yeast, the average linker DNA length is ~18 bp. In vitro, the flanking DNA on a nucleosome provides a context to mimic the linker DNA found in vivo. Interestingly, in vitro, reducing nucleosomal flanking DNA to 40 bp or less reduces rates of remodeling by INO80 by 300-fold, such that remodeling occurs on the order of hours (Zhou et al., 2018). This observation raised the question of how INO80 acts on nucleosomes and subnucleosomal particles in gene bodies. Given that hexasomes are substrates for INO80, we asked if these substrates are more readily mobilized on shorter flanking DNAs resembling the linker DNAs found in gene bodies.
We found that a 601 + 40 hexasome is remodeled ~60-fold faster than a 601 + 40 nucleosome (Figures 2E and 2F; Table 2). A missing H2A-H2B dimer proximal to the linker DNA will release ~20 bp of DNA effectively increasing the length of the flanking DNA. To test if the faster hexasome sliding arises from the additional flanking DNA that is released, we measured remodeling on 601 + 20 hexasomes. The use of 601 + 20 hexasomes results in effectively a similar length of flanking DNA to the 601 + 40 nucleosomes (Table 2). We found that 601 + 20 hexasomes are also remodeled ~60-fold faster than 601 + 40 nucleosomes, ruling out any effects from increased flanking DNA length.
Furthermore, the 601 + 20 hexasome and the 601 + 40 hexasome are remodeled only 5-fold slower than a 601 + 80 hexasome (Figure 2F). These results indicate that, although hexasome remodeling by INO80 shows a flanking DNA length dependence, this dependence is less steep than that for nucleosomes. As a result, the timescales for sliding 601 + 20 and 601 + 40 hexasomes (t1/2 ~ 2 min) are now more compatible with timescales of transcription elongation within yeast (García-Martínez et al., 2004; Pelechano et al., 2010).
Given that hexasomes are better substrates, we next asked if this preference is also reflected in their ability to stimulate the ATPase activity of INO80. We found that 601 + 80 hexasomes stimulate the ATPase activity of INO80 6.3-fold more than 601 + 80 nucleosomes (Figure 2G; Table 1). In the course of these studies, we noticed another major difference in how nucleosomes and hexasomes stimulate INO80’s ATPase activity. The hexasome-stimulated ATPase activity shows a much bigger dependence on flanking DNA length than the nucleosome-stimulated ATPase activity. Previously, we showed that the ATPase activity of INO80 on nucleosomes is not strongly dependent on flanking DNA length (Zhou et al., 2018). Consistent with these prior studies, we find that a 601 + 80 nucleosome stimulates the ATPase activity of INO80 only ~1.5-fold more than a 601 + 40 nucleosome (Figure S1B; Table 1). Additionally, a 601 + 100 nucleosome shows comparable ATPase stimulation as a 601 + 80 nucleosome (Figure S1; Table 1). In contrast, a 601 + 80 hexasome stimulates the ATPase activity of INO80 7.6-fold more than a 601 + 20 hexasome (Figure 2H). Finally, a larger population of hexasomes are remodeled in comparison with the corresponding nucleosomes (Figure 2E), suggesting more productive DNA translocation with hexasomes compared with nucleosomes.
Table 1.
Rate constants of ATPase assays
| Construct | WT INO80 kobs (min−1) | SEM | INO80(∆arp5) kobs (min−1) | SEM |
|---|---|---|---|---|
| +40 WT N | 133.2 | 14.6 | 14.1 | 2.1 |
| +60 WT N | 274.2 | 25.8 | 26.9 | 13.8 |
| +80 WT N | 231.9 | 29.0 | 38.5 | 12.0 |
| +100 WT N | 402.8 | 53.7 | – | – |
| +40 APM N | 111.7 | 13.6 | 0.59 | 2.1 |
| +80 APM N | 163.8 | 7.7 | 24.4 | 9.5 |
| +20 WT H | 192.6 | 40.6 | – | – |
| +40 WT H | 404.9 | 89.1 | – | – |
| +80 WT H | 1463.0 | 79.4 | 45.9 | 3.2 |
| +80 APM H | 1504.0 | 96.8 | – | – |
In principle, the faster observed remodeling of hexasomes could also be explained if INO80 irreversibly slides the hexasome in one direction but slides the nucleosome in both directions, thereby seeming to be less effective at sliding the nucleosome (Zhou et al., 2018). Comparisons of rate constants from the gel-based sliding assay, bulk FRET, and prior single-molecule FRET indicate that we are mainly capturing unidirectional sliding toward the longer flanking DNA for nucleosomes (Figure S4). Importantly, the higher ATPase activation by hexasomes further indicates that reversibility is not an issue.
These results establish that a hexasome is the preferred substrate for INO80 in terms of remodeling rates, remodeling extent, and ATPase stimulation. These findings further raised the possibility that the slower remodeling of nucleosomes arises from a nucleosome-specific rate-limiting step. To uncover this step, we studied the functional significance of the interactions made by INO80 with the proximal H2A-H2B dimer that is missing in the hexasomes tested above.
INO80 largely uses only one acidic patch within the nucleosome
The acidic patch on the H2A-H2B dimer of the nucleosome has been shown to be crucial for the activities of many remodelers including INO80 (Eustermann et al., 2018; Gamarra et al., 2018). It has been shown through structural studies that within an INO80-nucleosome complex, the Arp5-Ies6 module binds the entry site (proximal) dimer, and the Ies2 subunit binds the exit site (distal) dimer (Ayala et al., 2018; Eustermann et al., 2018). Both binding interactions involve contacts with the acidic patch on the respective H2A-H2B dimer (Ayala et al., 2018; Eustermann et al., 2018) (Figures 3A and 3B). Importantly, the hexasome that we used above is missing the dimer that is normally contacted by the Arp5 module. We, therefore, sought to determine the role of the interactions made by INO80 with the acidic patch on the proximal dimer. Previous studies have shown that mutating the acidic patches on both H2A-H2B dimers causes a large decrease in nucleosome sliding by INO80 (Eustermann et al., 2018; Gamarra et al., 2018). To determine the functional role of interactions with the proximal dimer’s acidic patch, we generated asymmetric nucleosomes, which had a mutated acidic patch on either the proximal or the distal dimer (Figure 3C) (Levendosky and Bowman, 2019).
Figure 3. The acidic patch that binds to Ies2 is dispensable for INO80 sliding.
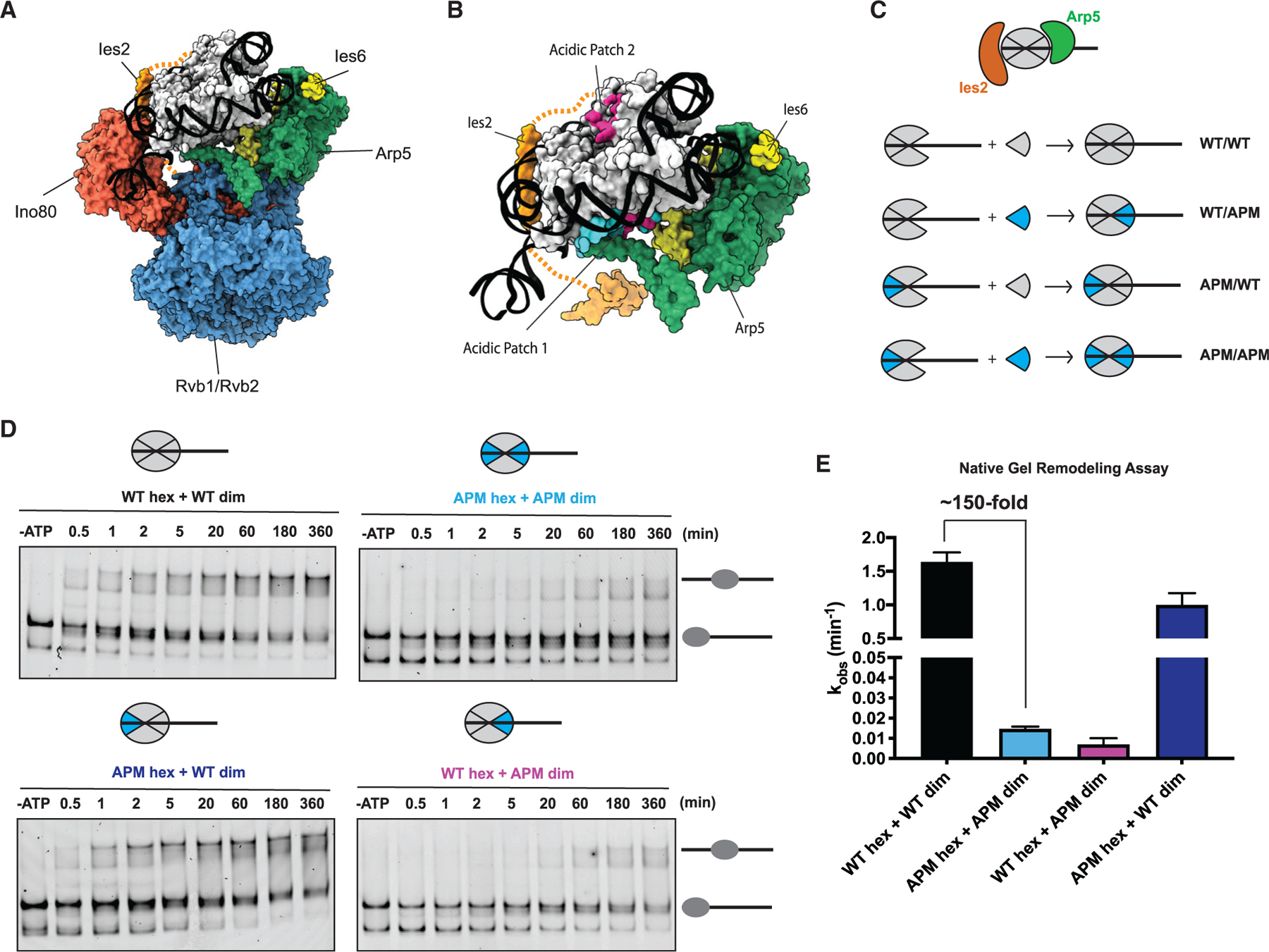
(A) Cryo-EM-based structure of the core INO80 components bound to the nucleosome (PDB: 6FML).
(B) Cryo-EM model from (A) showing only the Arp5 (green)-Ies6 (yellow) module and Ies2 (orange) bound to the nucleosome for clarity. Arp5-Ies6 and Ies2 bind to acidic patches (pink) on opposite sides of the nucleosome (PDB: 6FML).
(C) Schematic showing assembly of nucleosomes, with acidic patch mutations (APMs) depicted in blue. From (A), Arp5 module (green) binds dimer at the entry site, whereas Ies2 (orange) binds dimer at the exit site.
(D) Example native gels showing remodeling time courses for nucleosomes shown (C).
(E) Quantification of rate constants from multiple repeats of data, such as that shown in (D). Error bars represent SEM from n > 3.
Asymmetric nucleosomes are made by isolating hexasomes and adding in H2A-H2B dimers that are either WT or acidic patch mutant (APM) to reconstitute nucleosomes (Figure 3C). To confirm that this method of nucleosome assembly generates a nucleosome that can be remodeled, we mixed WT hexasomes with WT dimers and subjected the nucleosome to remodeling by INO80. Although this method of reconstitution led to more hexasomes in the starting substrate, the remodeling rate of the nucleosome substrate was comparable to canonically assembled nucleosomes (Figures S2A and S2C; Table 2). Furthermore, to ensure that excess dimer was not contributing to the effects seen in remodeling, we added comparable amounts of excess dimer to canonically assembled nucleosomes and did not see significant effects on remodeling kinetics (Figure S2B).
Unexpectedly, introducing a single APM dimer at the proximal location slowed nucleosome sliding by 200-fold, an effect that was comparable to the 150-fold defect of mutating both acidic patches (Figures 3D and 3E; Table 2). In contrast, introducing a single APM dimer at the distal location caused only a modest (<1.5-fold) remodeling defect (Figures 3D and 3E). Thus, despite recent EM structures showing contacts by INO80 with both acidic patches of the nucleosome, contacts with the acidic patch on the proximal dimer contribute significantly more to nucleosome remodeling. Furthermore, mutating the distal acidic patch in the context of hexasomes did not significantly change the remodeling rates (Figures S2E–S2G; Table 2). Together, these results indicate that the interactions made by the Arp5 module with the acidic patch of the proximal dimer play a major role in INO80 remodeling, whereas the interactions made between Ies2 and the acidic patch of the distal dimer do not make a large contribution to remodeling. We propose that, the Ies2-acidic patch contacts may be important for binding the nucleosome rather than for catalysis (Yao et al., 2015). Thus, the interaction between the H2A-H2B acidic patch and the Arp5 module may regulate a key step in nucleosome remodeling.
A differential role for the Arp5-Ies6 module in nucleosome versus hexasome remodeling
The results above suggested that the Arp5 module may play a bigger role in nucleosome sliding than in hexasome sliding because its contacts with the proximal acidic patch promote nucleosome remodeling. To test this possibility, we purified a mutant INO80 complex from yeast lacking Arp5, which we denote as INO80(Δarp5) (Figure S9). INO80(Δarp5) did not display large defects in the binding of hexasomes and nucleosomes (Figure S5). Previous studies have implied that deletion of the Arp5 module abrogates sliding by INO80 (Shen et al., 2003; Tosi et al., 2013; Yao et al., 2016). However, upon assaying for longer times, we found that saturating concentrations of INO80(Δarp5) display detectable nucleosome sliding activity (Figure 4A). Under excess and saturating enzyme conditions, we found that removing the Arp5 module still permits sliding on 601 + 80 nucleosomes, albeit 200-fold more slowly that WT INO80 (Figures 4A and 4B; Table 2). The products generated by INO80(Δarp5) align with the intermediate nucleosome positions generated by INO80(WT), indicating that these are INO80(Δarp5) sliding products and not supershifted bound bands (Figures 4A, 4C, and S7). In contrast, remodeling of a hexasome by INO80(Δarp5) is undetectable and is at least 800-fold slower than with that WT INO80 (Figures 4A and 4C). These results indicate that the Arp5 module plays a larger activating role in the context of a hexasome than a nucleosome.
Figure 4. The Arp5 module is a key regulatory component for remodeling.
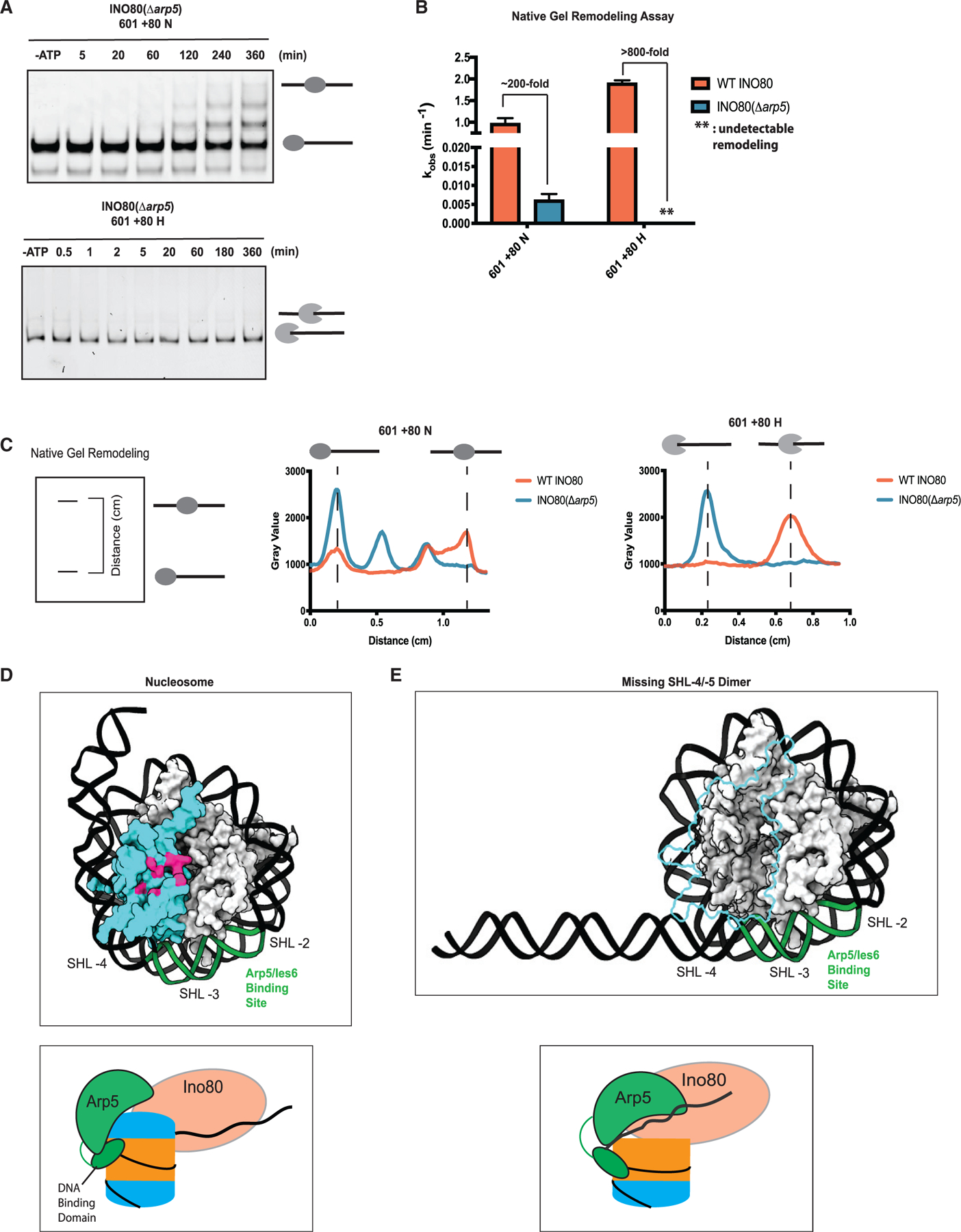
(A) Example time courses for remodeling of 601 + 80 N (top panel) and 601 + 80 H (bottom panel) by INO80(Δarp5) as assayed by native gel.
(B) Quantification of INO80(Δarp5) remodeling rate constants (blue) on 601 + 80 N and 601 + 80 H from repeats of data, such as in (A). For ease of comparison, the same WT INO80 data shown in Figure 2D are shown again here (coral).
(C) Left panel: schematic showing the distance parameter in the line-scan data depicted in the middle and right panels. Middle panel: line scan showing distribution of band intensity (gray value) on a native gel for the last time point for remodeling of 601 + 80 N by WT INO80 (coral, 360 min) and INO80(Δarp5) (blue, 360 min). Right panel: line scan showing the distribution of band intensity (gray value) on a native gel for the last time point for remodeling of 601 + 80 H by WT INO80 (coral, 360 min) and INO80(Δarp5) (blue, 360 min).
(D) Top panel: cryo-EM structure of the nucleosome (PDB: 6FML) showing the entry site dimer (cyan) and the Arp5 module’s DNA-binding site (green) at SHL-2/-3 (from PDB: 6FML). Lower panel: illustration of the Arp5 module binding the nucleosome (H2A-H2B dimers in cyan; H3-H4 tetramer in orange; DNA in black) at the designated binding site via a DNA-binding domain, based on PDB: 6FML.
(E) Top panel: model of the structure of a hexasome missing the entry site dimer. Model shows how the wrapping of the DNA may change upon proximal dimer loss to make Arp5 module’s binding site at SHL-2/-3 (in green) more accessible. Lower panel: illustrated model of the Arp5 module binding the hexasome in a different conformation due to the absence of the dimer. Error bars represent SEM from n > 3.
Superficially, this was a counterintuitive result as the hexasomes used here lack the dimer that the Arp5 module contacts in a nucleosome. However, in addition to contacting the acidic patch, the Arp5 module also contacts nucleosomal DNA at the internal location of SHL-2/-3 through the DNA-binding domain of the Arp5 subunit and through parts of the Ies6 subunit (Figure 4D) (Eustermann et al., 2018). A consequence of removing the proximal H2A-H2B dimer is that nucleosomal DNA at this location is released, increasing the length and changing the flexibility of the flanking DNA (Figure 4E). Importantly, such a change would make the internal DNA location of SHL-2/-3 more accessible to binding the Arp5 module. We, therefore, hypothesized that the hexasome is a better substrate in part because it allows INO80 to more productively engage the DNA at SHL-2/-3, resulting in unimpeded translocation of DNA. We further found that at saturating enzyme concentrations, the ATPase activity of INO80(Δarp5) on hexasomes is ~30-fold slower than that of INO80(WT) (Figure S1; Table 1). These results are consistent with the possibility that the Arp5 module contributes to the proper positioning of the Ino80 ATPase on hexasomal DNA, thereby explaining Arp5’s bigger activating role in hexasome remodeling.
Based on these results, we propose that the Arp5 module uses the acidic patch interactions to loosen the contacts between the H2A-H2B dimer and the H3-H4 tetramer at SHL-4/-5, making the substrate transiently resemble a hexasome. To further test this model, we next investigated which additional steps in nucleosome remodeling rely on the Arp5-acidic patch interaction.
Interactions between Arp5 and acidic patch prime the nucleosome for DNA translocation
Our previous work has suggested that the INO80-nucleosome complex forms an intermediate upon addition of ATP, prior to sliding (Zhou et al., 2018). The formation of this intermediate was uncovered by measuring the accessibility of nucleosomal DNA 18 bps into nucleosome from the exit DNA near the distal dimer, by incorporating a PstI restriction site at this location and measuring the rate of cutting by PstI (Figures 5A and S6) (Zhou et al., 2018). Using this restriction enzyme accessibility (REA) assay, we found that a PstI accessible INO80-nucleosome intermediate was formed in the presence of ATP that was independent of nucleosome sliding (Zhou et al., 2018). This experiment was conducted with 601 + 40 nucleosomes to assess the formation of the intermediate in the context of very slow or no sliding. To assess the role of the proximal dimer’s acidic patch, we used nucleosomes containing acidic patch mutations in both dimers because these nucleosomes show comparable rates of sliding as nucleosomes with only the proximal dimer mutated (Figures 3D and 3E). These double APM nucleosomes were 10.8-fold slower at generating the REA-accessible intermediate compared with a WT nucleosome (Figure 5B). These results suggest that the acidic patch is used in the formation of the intermediate. We next tested the effects of deleting the Arp5 module. With WT nucleosomes, INO80(Δarp5) was ~5-fold slower in generating the REA-accessible intermediate compared with INO80(WT) (Figure 5B). On APM nucleosomes, INO80(Δarp5) showed only a modest (~1.5-fold, within error) further decrease in the generation of the REA-accessible intermediate (Figure 5B). These results suggest that the direct interaction between the Arp5 module and the acidic patch is essential to form the intermediate.
Figure 5. The Arp5 module interacts with the acidic patch to regulate INO80 sliding.
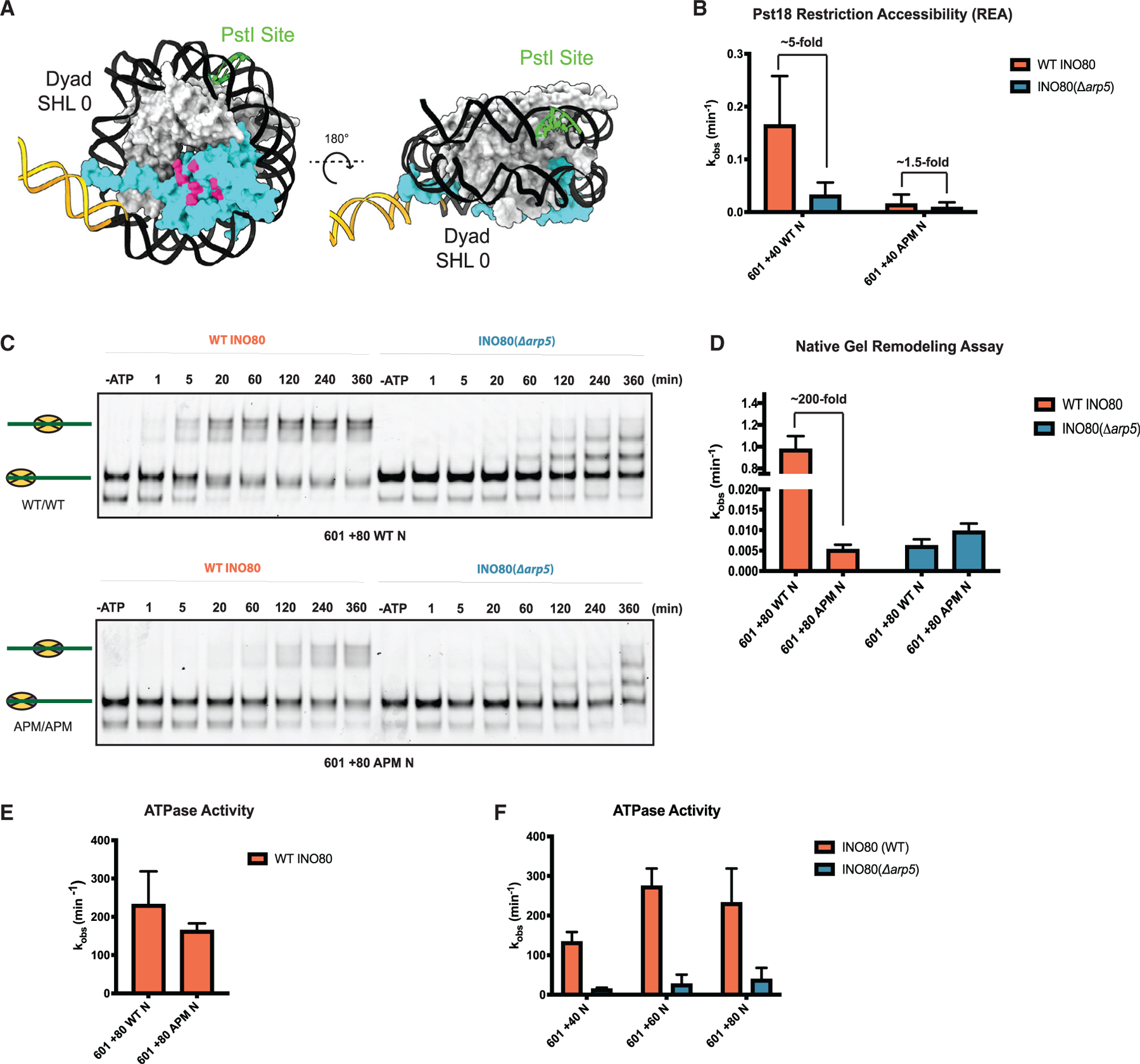
(A) Model of the nucleosome (PDB: 1KX5) from different angles with location of the engineered PstI restriction site in green. Proximal H2A-H2B dimer is in cyan.
(B) Rate constants of cutting by PstI for WT and double APM 601 + 40 N, with saturating ATP and saturating WT INO80 (coral) or INO80(Δarp5) (blue).
(C) Example native gels showing remodeling of WT (top panel) and double APM (bottom panel) 601 + 80 N with WT INO80 (coral) and INO80(Δarp5) (blue).
(D) Quantification of rate constants from multiple repeats of data, such as that shown in (C).
(E) Rate constants for INO80 ATPase activity with WT and double APM, 601 + 80 N. All ATPase assays were performed under the same conditions as native gel remodeling.
(F) Observed rate constants for ATPase activity of WT INO80 (coral) and INO80(Δarp5) (blue) on 601 + 40 N, 601 + 60 N, and 601 + 80 N. Error bars represent SEM from n > 3.
To test if the Arp5 module-acidic patch interactions are important for other remodeling steps, we used gel-based assays to measure sliding rates for APM nucleosomes with INO80(Δarp5). INO80(Δarp5) slid APM nucleosomes with similar rates to WT nucleosomes (Figures 5C and 5D), suggesting that the acidic patch does not contribute to INO80 remodeling in the absence of the Arp5 module. This result is consistent with the effects observed in the context of the REA assay (Figure 5B). Notably, mutating the acidic patch of 601 + 80 nucleosomes slows nucleosome sliding by 200-fold but does not affect WT INO80’s ability to center nucleosomes (Figures 5C and 5D). In comparison, as noted in Figures 4A and 4C, deleting the Arp5 module alone results in off-centered products (Figure 5C). This comparison suggests that in the context of a nucleosome, the Arp5 module has an additional role in regulating the extent of nucleosome sliding, which is independent of the acidic patch interaction.
The results above most simply suggest that the Arp5-acidic patch interaction promotes intermediate formation, which then allows nucleosome sliding to proceed efficiently. Given the large stimulatory effects of the Arp5-acidic patch interaction for the overall reaction, we tested if this interaction is coupled to ATP hydrolysis. Surprisingly, we found that mutating the acidic patch residues has small effects on ATPase activity (Figure 5E). In contrast, consistent with previous work, we find that deleting the Arp5 module decreases nucleosome-stimulated ATP hydrolysis by at least 10-fold (Shen et al., 2003; Tosi et al., 2013) (Figure 5F). Similar to the conclusions from Figures 4A, 4C, and 5B, these results suggest that the Arp5 module plays additional roles during nucleosome remodeling that are independent of the acidic patch interaction.
DISCUSSION
How INO80 achieves its many biological functions is poorly understood (Morrison and Shen, 2009). We have found here that INO80 preferentially slides hexasomes over nucleosomes. Our findings explain how subnucleosomal particles are repositioned in cells and point to a sophisticated remodeling mechanism that regulates the extent of INO80’s preference for hexasomes in a genomic context. Below, we discuss the mechanistic and biological implications of these findings.
Mechanistic explanation for how INO80 acts on hexasomes and nucleosomes
The faster remodeling of hexasomes raises the possibility that nucleosomes may be remodeled via a hexasome-like intermediate, which is mediated by the Arp5 module. This intermediate could involve transient loss of either the proximal or distal dimer. For the reasons discussed below, we suggest that transient loss of the proximal dimer through interaction with the Arp5 module is the more parsimonious model. However, we cannot rule out alternative models that involve transient loss of the distal dimer and different roles for the Arp5 module.
We propose that the proximal H2A-H2B dimer inhibits movement of DNA that is translocated from the entry site by the INO80 ATPase. In this model, the inhibition is relieved through transient dislodging of the H2A-H2B dimer by the Arp5 module through interactions with the acidic patch. Such dislodging may then allow unimpeded movement of the DNA translocated by the Ino80 motor (Figure 6A). This model is consistent with previous evidence for a nucleosomal intermediate that displays increased DNA accessibility and previous cross-linking studies, showing disruption of the H2A-H2B-DNA contacts by INO80 (Brahma et al., 2017; Zhou et al., 2018). Such a model is also consistent with prior single-molecule findings showing an ATP-dependent pause preceding a rapid DNA translocation step (Zhou et al., 2018). We suggest that the ATP-dependent pause represents the time taken to dislodge the proximal dimer.
Figure 6. Model of the INO80 remodeling mechanism.
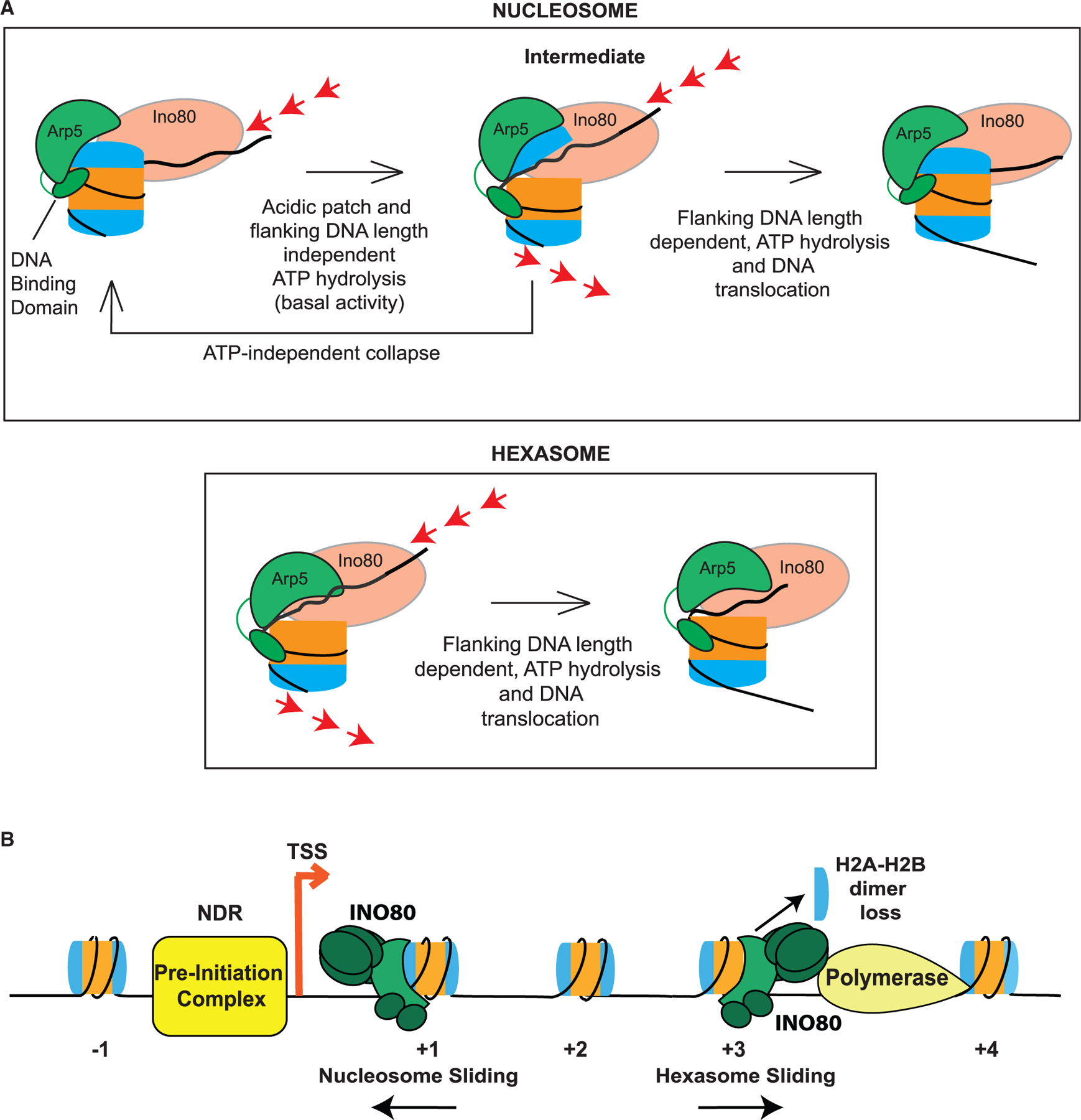
(A) Illustration of the model for INO80 action on a nucleosome and a hexasome. Nucleosome binding stimulates a basal level of ATP hydrolysis and the Ino80 motor pumps DNA into the nucleosome. This ATPase activity is independent of flanking DNA length and the acidic patch. The torsional strain caused by the pumping of the DNA is partially relieved through transient dislodging of the H2A-H2B dimer. Such dislodging is speculated to occur by the Arp5 module through contacts with the acidic patch and DNA at SHL-2/-3. This transition results in the formation of an intermediate, which can either collapse back in an ATP-independent manner, or transition forward in an ATP-dependent manner to translocate DNA across the nucleosome. Translocation is dictated by flanking DNA length and requires flanking DNA-length-dependent ATP hydrolysis. In comparison, for a hexasome, because the dimer is absent and does not inhibit INO80 remodeling, translocation occurs more readily.
(B) Schematic of INO80 participating in chromatin remodeling at sites of transcription. At the +1 location, INO80 helps position the nucleosome. During elongation as RNA Pol II actively removes the H2A-H2B dimer distal to the promoter, INO80 can act on these subnucleosomal particles to restore proper positioning and help prevent aberrant transcription initiation.
The absence of the proximal dimer will also increase the accessibility of DNA at SHL-2/-3, the region contacted by the DNA-binding domain of Arp5 and by Ies6 (Figure 4E). We, therefore, speculate that Arp5 is able to better engage the DNA at SHL-2/-3 in hexasomes, thereby enabling more efficient passage of the translocated DNA. In a nucleosome, such productive engagement of the DNA at SHL-2/-3 would occur after the proximal dimer is transiently dislodged, adding an unfavorable step to the overall reaction. Prior cryo-EM studies showed that the Arp5 module can adopt two different conformations on a nucleosome, one that appears inhibitory for DNA translocation and one that appears permissive for DNA translocation (Eustermann et al., 2018). Additionally, action of the Ino80 ATPase has been proposed to introduce torsional strain at the proximal H2A-H2B dimer in a nucleosome (Brahma et al., 2017). We, therefore, propose that in response to such strain, the Arp5 module switches between the inhibitory and permissive conformations, thereby transiently dislodging the proximal dimer to more effectively engage the DNA at SHL-2/-3.
To date, the H2A-H2B acidic patch has been shown to serve largely a binding purpose (McGinty and Tan, 2015), and, in the context of the human ISWI enzyme, SNF2h also has an allosteric activating role (Gamarra et al., 2018). Our studies suggest a third type of role, wherein the interactions made with the acidic patch transiently displace an H2A-H2B dimer to promote nucleosome remodeling.
It has been debated whether INO80 exchanges an H2AZ/H2B dimer with an H2A-H2B dimer (Brahma et al., 2017; Watanabe et al., 2013; Wang et al., 2016). Our results suggest that the exchange may be a side reaction. As INO80 transiently dislodges the dimer, it may fall off under certain reaction conditions and be replaced by free dimers in solution. If H2AZ destabilizes nucleosomes, this may preferentially dissociate H2AZ/H2B. Consistent with this possibility, the fraction of exchanged H2A-H2B dimers is less than slid nucleosomes (Papamichos-Chronakis et al., 2011).
Different flanking DNA length dependencies for hexasome versus nucleosome remodeling
Previous work has shown that the Arp8 module binds flanking DNA (Brahma et al., 2018). Using our model, we propose that productive engagement of the DNA at SHL-2/-3 by the Arp5 module is necessary to allow appropriate engagement of flanking DNA by the Arp8 module. Such engagement of flanking DNA by the Arp8 module could then be coupled to activation of the Ino80 ATPase in a manner that depends on the length of flanking DNA bound by Arp8. This proposal is consistent with prior work showing that deletion of the Arp8 module changes the nucleosome contacts made by the Arp5 module (Brahma et al., 2018). In a nucleosome, the DNA at SHL-2/-3 is partially occluded through interactions with the H2A-H2B dimer potentially inhibiting the productive engagement of the DNA by Arp5’s DNA-binding domain. In turn, such inhibition would reduce positive cooperation between the Arp5 and Arp8 module, resulting in a basal-nucleosome-stimulated ATPase rate that is not strongly dependent on the flanking DNA length. With a hexasome, given the greater accessibility of DNA at SHL-2/-3, the Arp5 and Arp8 modules would more effectively cooperate, resulting in higher ATPase activity that is also more strongly dependent on flanking DNA length.
Interestingly, unlike ATPase activity, remodeling of nucleosomes by INO80 is strongly dependent on flanking DNA length. Within the framework described here, the flanking DNA length dependence of nucleosome sliding may arise from two steps: (1) the DNA translocation steps carried out by the Ino80 subunit in the intermediate state and (2) ATP-independent collapse of the intermediate to the starting conformation (Figure 6A). The DNA translocation steps would get faster with longer flanking DNA reflecting productive engagement by Arp8 and Arp5. In contrast, the collapse step would get faster with shorter flanking DNA, reflecting the increased instability of the intermediate due to weaker cooperation between Arp5 and Arp8. We note that the flanking DNA length dependence of the collapse step proposed here draws from a model we had proposed previously (Zhou et al., 2018). In a hexasome, the flanking DNA length dependence would then primarily arise from the DNA translocation step, as the starting state for a hexasome resembles the intermediate for a nucleosome.
Roles for the Arp5 module in nucleosome and hexasome sliding
Our results uncovered two unexpected effects of deleting the Arp5 module. First, deleting the Arp5 module causes greater than an ~800-fold defect in hexasome sliding compared with an ~200-fold defect in nucleosome sliding, indicating that this module makes a larger energetic contribution to hexasome sliding. We propose that in a hexasome, the interaction made by the Arp5 module with the DNA at SHL-2/-3 is substantially stronger than the corresponding interaction made in the nucleosomal intermediate due to the complete absence of the proximal dimer. Second, with 601 + 80 nucleosomes, where sliding was detectable upon deleting the Arp5 module, the nucleosome was moved substantially less far compared with the ~40-bp movement observed with WT INO80. We propose that without the Arp5 module, the intermediate would have a reduced life-time, resulting in the movement of less DNA.
INO80 regulates positions of nucleosomes and subnucleosome particles at genes
Most previous work has focused on INO80’s role in establishing the +1 nucleosome at TSSs (Klein-Brill et al., 2019; Krietenstein et al., 2016; Yao et al., 2016; Yen et al., 2013). However, prior work has also implicated INO80 in regulating elongation by RNA polymerase II as well as nucleosome spacing in gene bodies independent of RNA Pol II (Klopf et al., 2009; Lafon et al., 2015; Poli et al., 2016; Singh et al., 2021). Our new findings that INO80 regulates the locations of both hexasomes and nucleosomes within the gene body provide some mechanistic basis for these latter set of studies.
Our findings also synergize with emerging evidence indicating an elevated prevalence of subnucleosomal particles at highly transcribed genes (Ramachandran et al., 2017; Rhee et al., 2014). Many of these subnucleosomal particles have DNA footprints consistent with hexasomes. In these studies, it has been suggested that hexasomes may arise during transcription as RNA polymerase navigates through a nucleosome. Indeed, careful biochemical studies have shown that RNA polymerases can dislodge an H2A-H2B dimer during transcription through a nucleosome (Kireeva et al., 2002). In this context, our findings provide one mechanistic explanation for how hexasome positions are regulated in vivo.
INO80’s role in regulating the position of the +1 nucleosome, where the average length of the NDR is ~140 bps (Mavrich et al., 2008), is compatible with INO80’s in vitro activity of rapidly (order of minutes) sliding nucleosomes with flanking DNA greater than ~60 bps. However, within gene bodies in yeast, the average linker DNA length is ~18 bps (Mavrich et al., 2008), and INO80 slides nucleosomes very poorly in vitro (order of hours) when the flanking DNA is less than 40 bp. Interestingly, we find that INO80 slides hexasomes with 20 bp of flanking DNA on the order of minutes. We, therefore, propose that INO80’s effect on nucleosome positions within gene bodies is through its mobilization of hexasomes, some of which are then converted to nucleosomes with the aid of histone chaperones, such as Nap1 and FACT (Belotserkovskaya et al., 2003; Kuryan et al., 2012). Consistent with this possibility, genes showing the most changes in nucleosome positioning upon Ino80 deletion also show mispositioning of both nucleosomes and hexasomes.
Restoration of hexasome positions by INO80 could regulate cryptic transcription. Indeed, INO80 is a key repressor of antisense transcription of long non-coding RNAs (Alcid and Tsukiyama, 2014). Because inter-nucleosomal spacing is largely regular in yeast gene bodies, losing a dimer would promote directional sliding of hexasomes by INO80. Indeed our in vivo results suggest that INO80 moves subnucleosomal particles away from the promoter. However, we cannot exclude that misregulation of the +1 nucleosome impairs RNA Pol II elongation, also contributing to defects in hexasome positions. Interestingly, genes showing large changes in nucleosome and hexasome positions in Δino80 cells are enriched for metabolic functions (Figure 1H). Metabolism regulating genes can show rapid changes in expression (Slavov et al., 2014). Restoring nucleosomes and hexasome positions at such genes during transcription could be a critical function of INO80.
The S. cerevisiae Chd1 remodeler was also shown to remodel hexasomes (Levendosky et al., 2016). However, unlike with INO80 where the loss of an H2A-H2B dimer stimulates sliding, with Chd1, the loss of an H2A-H2B dimer inhibits sliding in one direction, resulting in unidirectional sliding (Levendosky et al., 2016). These results demonstrate that although other remodelers, such as Chd1, can recognize both hexasomes and nucleosomes, their specific mechanisms differ. In the future, determining how other remodelers, such as those from the ISWI and SWI/SNF families, act on subnucleosomal particles will provide more insights into how these particles are regulated in vivo.
Broader implications
The model proposed here provides a means to imagine how histone variants or PTMs could regulate INO80 activity. For example, histone variants or PTMs that destabilize the H2A-H2B dimer-H3-H4 interface could specifically promote remodeling of nucleosomes. Altering interactions with the H2A-H2B acidic patch could also regulate INO80 activity. Indeed, replacing H2A with H2AZ or mutating H2A to possess an acidic patch more similar to H2A.Z results in faster remodeling (Brahma et al., 2017; Eustermann et al., 2018). Additionally, the ability of INO80 to tune its preference for hexasomes versus nucleosomes based on linker DNA length provides INO80 the versatility to act in different genomic contexts that vary in nucleosome density. Such versality could explain INO80’s global role in DNA repair, where rapid movement of nucleosomes as well as subnucleosomal particles may be needed for the repair machinery to access the damaged DNA. Finally, our results showing that deletion of the Arp5-Ies6 module changes the outcome and speed of nucleosome remodeling raises the possibility that in cells, INO80 activity can be tuned to carry out defined tasks through loss or modification of specific modules. Indeed, prior work has suggested the presence of INO80 subcomplexes in yeast (Yao et al., 2016). INO80 has also been shown to be post-translationally modified by other enzymes (Morrison et al., 2007). Overall, the ability to precisely tune its activities through both its subunits and substrates could allow INO80 to adopt multiple roles and explain INO80’s involvement in diverse chromatin processes. Moving forward, it will be important to understand the different types of chromatin or ‘‘chromatin-like’’ structures INO80 encounters in cells and how INO80 uniquely acts at these sites to facilitate and regulate essential DNA-based transactions.
Limitations of the study
Although our work provides an initial model for how INO80 acts on hexasomes, additional biochemical and structural work is needed to understand exactly how the different subunits of INO80 act together to slide hexasomes. In particular, the proposed role of the Arp5 module is a speculation based on its engagement of the acidic patch of the proximal dimer and the large defects from mutating this acidic patch. This role needs to be more directly tested with further mutagenesis and cryo-EM structural analysis of reaction intermediates.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
All inquiries for further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Geeta J. Narlikar (narlikar@ucsf.edu).
Materials availability
All plasmids and cell-lines generated in this study are found in the key resources table including those uniquely made for this study. Additional information about the materials used can provided upon requests made to lead contact.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| ATP | GE Healthcare | 27-2056-01 |
| Pst1 | NEB | R0140 |
| γ-32P-ATP | Perkin Elmer | Blu002Z250uC |
| Adenosine 5′-diphosphate sodium salt | Sigma-Aldrich | A2754-1G |
| Micrococcal Nuclease | Worthington | LS004797 |
|
Critical commercial assays | ||
| Ovation® Ultralow V2 DNA-Seq Library Preparation Kit | Nugen | 0344NB-A01 |
|
Deposited data | ||
| Raw data from MNase Seq experiments | NCBI’s Gene Expression Omnibus | GSE168700 |
| Code to analyze MNase data | Ramani et al., 2019 | PMID30811994 |
|
Experimental models: Cell lines | ||
| S. cerevisiae: INO80-FLAG: s288c INO80-FLAG | Shen, 2004 | N/A |
| S cerevisiae: INO80-FLAG(ΔArp5): s288c INO80-FLAG ΔArp5::KanMX | This paper | N/A |
| S cerevisiae: INO80-FLAG(Δino80): s288c INO80-FLAG ΔINO80::KanMX | Yao, 2016 | N/A |
|
Oligonucleotides | ||
| 0/40 601 Pst1-18 (Pst1 cut site bolded): CTGGAGAATCCCGGTCTGCAGGCCG CTCAATTGGTCGTAGACAGCTCTAGC ACCGCTTAAACGCACGTACGCGCTGT CCCCCGCGTTTTAACCGCCAAGGGGA TTACTCCCTAGTCTCCAGGCACGTGTC AGATATATACATCCTGTGCATGTATTG AACAGCGACCTTGCCGGTGCCAGTCG GATA |
Zhou et al., 2018 | N/A |
| 0/100 601 DNA sequence:CTGGAGA ATCCCGGTGCCGAGGCCGCTCAAT TGGTCGTAGACAGCTCTAGCACCG CTTAAACGCACGTACGCGCTGTCC CCCGCGTTTTAACCGCCAAGGGGA TTACTCCCTAGTCTCCAGGCACGTG TCAGATATATACATCCTGTGCATGT ATTGAACAGCGACCTTGCCGGTGC CAGTCGGATAGTGTTCCGAGCTCC CACTCTAGAGGATCCCCGGGTACC GAGCTCGAATTCGCCCTATA |
Zhou et al., 2018 | N/A |
|
Recombinant DNA | ||
| 601 plasmid | Lowary and Widom, 1998 | N/A |
| Pet3a_H2A (Xenopus laevis) | Yang et al., 2006 | N/A |
| Pet3a_H2B (Xenopus laevis) | Yang et al., 2006 | N/A |
| Pet3a_H3 (Xenopus laevis) | Yang et al., 2006 | N/A |
| Pet3a_H4 (Xenopus laevis) | Yang et al., 2006 | N/A |
|
Software and algorithms | ||
| Graphpad Prism 8/9 | Graphing and modeling software | https://www.graphpad.com |
Data code and availability
The data reported in this paper is available on NCBI’s Gene Expression Omnibus database with the accession number GSE168700, as listed in the key resources table.
The code used in this study originated from Ramani et al. (PMID30811994).
Any additional information required to reanalyze the data in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains
All yeast strains are MATa and in the s288c background. The Δino80 strain was a gift from the Ashby lab (Yao et al., 2016). The Δarp5 strain was generated by PCR knock-in of the KanMX cassette at the Arp5 locus. The yeast strains used in this study are listed in the key resources table. For protein purification, all yeast strains were grown at 30 degrees. Yeast were first inoculated in YPD from frozen glycerol stock then transferred to 18–20L of YPD. Yeast was grown until saturation, 50 g/L of YPD was added and cells were harvested after 1–2 overnight incubations at 30 degrees.
METHOD DETAILS
MNase-seq
Yeast chromatin was digested with MNase as previously described (Rodriguez et al., 2014). A MNase titration was performed on the chromatin samples, and run on a 1% agarose gel (Figure S8). Similarly digested chromatin among the different strains were chosen, and one replicate was size selected for mononucleosomes via gel extraction, while the other replicate was not size selected. Libraries were prepped using the Ovation Ultralow System V2 for DNA-Seq Kit (Nugen). Libraries were sequenced on a NextSeq.
MNase-seq data analysis & acquisition
Read processing and alignment
MNase-seq libraries were sequenced with paired end 80 bp reads on a NextSeq 500 instrument. Raw reads were clipped using SeqPrep and aligned to SacCer3 using bwa mem with default parameters. Aligned BAM files were sorted, deduplicated, and indexed using samtools and the PySAM API. The scripts and pipelines for mapping have been previously described (Ramani et al., 2019).
Normalized enrichment heatmaps
The nucleosome and subnucleosomal particle dyads were derived from chemical cleavage mapping data. A custom Python script calling the PySAM API was used to create arrays mapping midpoints of all reads +/− 1000 bp from an established +1 dyad position in the yeast genome (Ramani et al., 2019). An annotated list of 4,116 yeast +1 dyads mapped by chemical cleavage was used (Brogaard et al., 2012). Fragment midpoint enrichment was calculated as a simple z-score for each +1 nucleosome.
Line plots
Normalized enrichment was averaged across all genes and plotted using ggplot2 in R.
Expression sorting
Normalized enrichment arrays were sorted based on gene TPM count determined from RNA-Seq reads in wild type yeast (Voichek et al., 2018).The arrays for the top 20% and bottom 20% of genes were averaged and visualized as described above.
Heatmaps
Normalized enrichment arrays were visualized as heatmaps using matplotlib. Maximum and minimum color intensity values were set to the 90th and 10th percentiles of data, respectively. Data above and below these values were also displayed.
Correlation heatmaps
Normalized mutant data of fragment size 130–300 bp (nucleosome sized fragments) were correlated with normalized wild type arrays of the same fragment size on a gene-by-gene basis using Spearman’s r. Arrays were sorted based on correlation and genes with r < 0.33 were visualized using the same parameters as other heatmaps. Arrays of fragment size 90–109 were also sorted based on the correlation of the nucleosome sized fragments and visualized using the same parameters.
iPAGE analysis
Correlations were calculated for each gene as described above. iPAGE (Goodarzi et al., 2009) was used to sort genes based on correlation (binned into 11 categories) and calculate significantly enriched GO groups using mutual information content.
Purification of INO80 complexes
FLAG-tagged Ino80 strains were grown at 30°C in YPD to saturation and harvested for purification. INO80 was purified using FLAG immunoprecipitation, as described previously (Zhou et al., 2018), with a minor modification. A secondary elution for 15 minutes at 4°C was performed to increase yield.
Assembly of nucleosomes, asymmetric nucleosomes, and hexasomes
Recombinant Xenopus laevis histones were purified from bacteria as previously described. DNA constructs were generated using a plasmid containing 601 DNA. Large scale PCR was performed and the resulting DNA was separated on an 8% polyacrylamide gel, cut out, crushed and soaked into 1X TE. The gel particles were filtered through a 0.45 μm filter, and the DNA was ethanol precipitated and resuspended in 1X TE. To label the DNA, primers with a Cy3 fluorophore modification were used (IDT). Nucleosomes and hexasomes were assembled using salt gradient dialysis. Nucleosomes were purified by ultracentrifugation with a 10–30% glycerol gradient. Hexasomes were purified using the Mini PrepCell (BioRad) with a 7% acrylamide gel as described previously (Levendosky and Bowman, 2019). Briefly, this method by Levendosky and Bowman takes advantage of the asymmetry of the 601 nucleosomes positioning sequence. The asymmetry results in weaker affinity of one side of the DNA sequence for H2A/H2B dimers compared to the other side. Placing the flanking DNA adjacent to the side of the 601 sequence that binds dimers more weakly then allows assembly of hexasomes missing the dimer that is proximal (i.e. adjacent) to the longer flanking DNA. The hexasomes are further purified over a prep-cell as described by Levendosky and Bowman, to separate out hexasomes from small amounts of nucleosomes. Purified hexasomes were mixed with 2.5–4X excess dimer to assemble the asymmetric nucleosomes.
Native gel remodeling assay
Remodeling reactions were done under single turnover, saturating INO80 and saturating ATP conditions. The reaction conditions contained 10 nM nucleosomes, 30 or 60 nM INO80, 40 mM Tris pH7.5, 50 mM KCl, 1.1mM MgCl2, 0.02% NP-40, 1 mM ATP●MgCl2, and 3% glycerol, and reactions were carried out at 30°C. INO80 was pre-bound to nucleosomes for 10 minutes at 30°C before starting the reaction with the addition of ATP●Mg. The no ATP control was taken at the last time point of the reaction. Time points taken from the reaction were quenched with 0.6 mg/ml non-601 plasmid DNA, 4nM ADP, and 18% glycerol. Samples were loaded on a 6% polyacrylamide gel and resolved for 4 hours at 125V. Gels were scanned on a Typhoon Imager (GE Life Sciences). The fraction unremodeled was quantified over time using ImageJ. Undetectable remodeling was defined as reactions that displayed no change in the fraction unremodeled compared to the no ATP control after 6 hours. A limit for remodeling was determined by fitting the data to a 6 hour end point where ~5% of substrate was remodeled. The data were fit to a single exponential decay as indicated by Equation 1, using GraphPad:
| (Equation 1) |
EMSA assay
The reactions contained 2, 5 or 75 nM 601+80 nucleosomes (or hexasomes), varying concentrations of INO80(WT) and INO80(ArpΔ) as noted on each figure, 40 mM Tris pH7.5, 50 mM KCl, 1.1mM MgCl2, 0.02% NP-40, and 3% glycerol. Binding reactions were carried out for 30 minutes at 30°C. The bound and unbound fractions were separated by electrophoresis on a 4% acrylamide, 0.5X TBE native gel.
ATPase assay
ATPase assays performed under conditions that closely mimicked remodeling assays. All ATPase assays were performed under multiple turnover conditions (ATP in excess of INO80). Experimentally we that determined 320 μM ATP●MgCl2 was saturating for both 60 nM WT INO80 and 90 nM INO80(Δarp5). Reactions were performed with 15 nM nucleosomes (or hexasomes), 60 nM WT INO80 (or 90 nM INO80(Δarp5)), 40 mM Tris pH 7.5, 50 mM KCl, 320 μM ATP●MgCl2, 0.5 mM MgCl2, and trace amounts of γ-32P-ATP, at 30°C. Each reaction was started with the addition of ATP●MgCl2 after a 10-minute preincubation at 30°C. The no ATP control was taken at the last time point of the reaction. Samples of the reactions were quenched at various timepoints with equal volumes of 50 mM Trips pH 7.5, 3% SDS, and 100 mM EDTA. Inorganic phosphate was separated from ATP●MgCl2 on PEI-cellulose TLC plates with 0.5 M LiCl, 1 M formic acid. These plates were exposed overnight on a phosphoscreen and scanned on a Typhoon Imager (GE Life Sciences). To determine the rate constant, we measured the fraction of inorganic phosphate using ImageJ, and fit the initial 10% hydrolyzed.
Restriction enzyme accessibility (REA) assay
REA assays were performed under single turnover conditions (INO80 in excess of nucleosome). Experimentally, we determined that 60 nM WT and 90 nM INO80(Δarp5) was saturating under the following reaction conditions: INO80, 15 nM nucleosome, 40 mM Tris pH 7.5, 60 mM KCl, 1 mM ATP●MgCl2, 5 mM MgCl2, 0.01% NP40, 0.5 mg/mL FLAG peptide, and 3 U/μL Pst1 at 30°C. Each reaction was started with the addition of ATP●MgCl2 after a 10-minute preincubation at 30°C. The no ATP control was taken at the last time point of the reaction. Samples of the reactions were quenched at various timepoints with equal volumes of 20 mM Tris pH 7.5, 2% SDS, and 70 mM EDTA, 20% glycerol, 0.2 mg/mL xylene cyanole, and 0.2 mg/mL bromophenol blue. All quenched timepoints were incubated with 4 mg/mL of Proteinase K for 20 minutes at 50°C to digest all proteins. The DNA (cut and uncut fragments) were resolved for each timepoint with a native PAGE (10% acrylamide, 1X TBE) and scanned on a Typhoon Imager (GE Life Sciences). To determine the rate constant, we measured the fraction of DNA cut using ImageJ and fit the data to a single exponential decay using Prism 7 (GraphPad) (Equation 1).
QUANTIFICATION AND STATISTICAL ANALYSIS
Error estimation for ensemble measurements
All ensemble measurements of rate constants are reported as the mean of three or more experimental replicates and standard error of the mean (SEM). These values are reported in the figure legends. Graphing and statistical analyses were done using GraphPad Prism 8 or 9.
Supplementary Material
Highlights.
Locations of subnucleosome-sized particles are regulated by INO80 in yeast cells
INO80 slides hexasomes up to 60-fold faster than nucleosomes in vitro
Sliding of hexasomes is less sensitive to flanking DNA length than that of nucleosomes
The Arp5-acidic patch interaction plays a large role in nucleosome remodeling
ACKNOWLEDGMENTS
We thank Julia Tretyakova for the histone purifications. We thank Coral Zhou, Hiten Madhani, John Gross, Serena Sanulli, and Emily Wong for helpful comments on the manuscript. We thank the Narlikar Lab for stimulating discussions during the development of this work. We thank the Morrison Lab for the Δino80 yeast strain used in this study. We thank the Bowman Lab for their guidance with the assembly of hexasomes and asymmetric nucleosomes. This research was funded by grants from the NIH (R35 GM127020) to G.J.N., by a Ruth L. Kirschstein National Research Service Award (5F31GM136187-02) to M.A.G., and by an American Cancer Society—Roaring Fork Valley Research Fund Postdoctoral Fellowship (PF-18-155-01-DMC) to L.J.H.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2022.04.026.
DECLARATION OF INTERESTS
Geeta Narlikar is on the Molecular Cell advisory board.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
REFERENCES
- Alcid EA, and Tsukiyama T (2014). ATP-dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev 28, 2348–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala R, Willhoft O, Aramayo RJ, Wilkinson M, McCormack EA, Ocloo L, Wigley DB, and Zhang X (2018). Structure and regulation of the human INO80-nucleosome complex. Nature 556, 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, and Reinberg D (2003). FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093. [DOI] [PubMed] [Google Scholar]
- Brahma S, Udugama MI, Kim J, Hada A, Bhardwaj SK, Hailu SG, Lee T-H, and Bartholomew B (2017). INO80 exchanges H2a.Z for H2A by translocating on DNA proximal to histone dimers. Nat. Commun 8, 15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahma S, Ngubo M, Paul S, Udugama M, and Bartholomew B (2018). The Arp8 and Arp4 module acts as a DNA sensor controlling INO80 chromatin remodeling. Nat. Commun 9, 3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogaard K, Xi L, Wang J-P, and Widom J (2012). A map of nucleosome positions in yeast at base-pair resolution. Nature 486, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao HT, Dul BE, Dann GP, Liszczak GP, and Muir TW (2020). A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing. Nat. Chem. Biol 16, 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, and Hopfner K-P (2018). Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 556, 386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarra N, Johnson SL, Trnka MJ, Burlingame AL, and Narlikar GJ (2018). The nucleosomal acidic patch relieves auto-inhibition by the ISWI remodeler SNF2h. eLife 7, e35322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martínez J, Aranda A, and Pérez-Ortín JE (2004). Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell 15, 303–313. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Elemento O, and Tavazoie S (2009). Revealing global regulatory perturbations across human cancers. Mol. Cell 36, 900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S (2016). Mechanisms of nucleosome dynamics in vivo. Cold Spring Harb. Perspect. Med 6, a026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, and Studitsky VM (2002). Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9, 541–552. [DOI] [PubMed] [Google Scholar]
- Klein-Brill A, Joseph-Strauss D, Appleboim A, and Friedman N (2019). Dynamics of chromatin and transcription during transient depletion of the RSC chromatin remodeling complex. Cell Rep 26, 279–292.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopf E, Paskova L, Solé C, Mas G, Petryshyn A, Posas F, Wintersberger U, Ammerer G, and Schüller C (2009). Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol 29, 4994–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, and Korber P (2016). Genomic nucleosome organization reconstituted with pure proteins. Cell 167, 709–721.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, and Studitsky VM (2009). Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol 16, 1272–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryan BG, Kim J, Tran NNH, Lombardo SR, Venkatesh S, Workman JL, and Carey M (2012). Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc. Natl. Acad. Sci. USA 109, 1931–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon A, Taranum S, Pietrocola F, Dingli F, Loew D, Brahma S, Bartholomew B, and Papamichos-Chronakis M (2015). INO80 chromatin remodeler facilitates release of RNA polymerase II from chromatin for ubiquitin-mediated proteasomal degradation. Mol. Cell 60, 784–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levendosky RF, and Bowman GD (2019). Asymmetry between the two acidic patches dictates the direction of nucleosome sliding by the ISWI chromatin remodeler. eLife 8, 856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levendosky RF, Sabantsev A, Deindl S, and Bowman GD (2016). The Chd1 chromatin remodeler shifts hexasomes unidirectionally. eLife 5, 3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, and Widom J (1998). New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol 276, 19–42. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, and Pugh BF (2008). A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18, 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, and Tan S (2015). Nucleosome structure and function. Chem. Rev 115, 2255–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, Kim JA, Person MD, Highland J, Xiao J, Wehr TS, Hensley S, Bao Y, Shen J, Collins SR, et al. (2007). Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 130, 499–511. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, and Shen X (2009). Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol 10, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Watanabe S, Rando OJ, and Peterson CL (2011). Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Chávez S, and Pérez-Ortín JE (2010). A complete set of nascent transcription rates for yeast genes. PLoS One 5, e15442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J, Gerhold C-B, Tosi A, Hustedt N, Seeber A, Sack R, Herzog F, Pasero P, Shimada K, Hopfner K-P, and Gasser SM (2016). Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev 30, 337–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Levendosky RF, Chakravarthy S, Patel A, Bowman GD, and Myong S (2017). The Chd1 chromatin remodeler shifts nucleosomal DNA bidirectionally as a monomer. Mol. Cell 68, 76–88.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, and Rando OJ (2010). Nucleosome positioning: how is it established, and why does it matter? Dev. Biol 339, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Ahmad K, and Henikoff S (2017). Transcription and remodeling produce asymmetrically unwrapped nucleosomal intermediates. Mol. Cell 68, 1038–1053.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V, Qiu R, and Shendure J (2019). High sensitivity profiling of chromatin structure by MNase-SSP. Cell Rep 26, 2465–2476.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Bataille AR, Zhang L, and Pugh BF (2014). Subnucleosomal structures and nucleosome asymmetry across a genome. Cell 159, 1377–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, McKnight JN, and Tsukiyama T (2014). Genome-wide analysis of nucleosome positions, occupancy, and accessibility in yeast: nucleosome mapping, high-resolution histone ChIP, and NCAM. Curr. Protoc. Mol. Biol 108, 21.28.1–21.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang J-PZ, and Widom J (2006). A genomic code for nucleosome positioning. Nature 442, 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X (2004). Preparation and analysis of the INO80 complex. Methods Enzymol 377, 401–412. [DOI] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, and Wu C (2000). A chromatin remodelling complex involved in transcription and DNA processing. Nature. 406, 541–544. [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, and Wu C (2003). Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12, 147–155. [DOI] [PubMed] [Google Scholar]
- Singh AK, Schauer T, Pfaller L, Straub T, and Mueller-Planitz F (2021). The biogenesis and function of nucleosome arrays. Nat. Commun 12, 7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Budnik BA, Schwab D, Airoldi EM, and van Oudenaarden A (2014). Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis. Cell Rep 7, 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, and Hopfner KP (2013). Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154, 1207–1219. [DOI] [PubMed] [Google Scholar]
- Valencia AM, Collings CK, Dao HT, St Pierre R, Cheng Y-C, Huang J, Sun Z-Y, Seo H-S, Mashtalir N, Comstock DE, et al. (2019). Recurrent SMARCB1 mutations reveal a nucleosome acidic patch interaction site that potentiates mSWI/SNF complex chromatin remodeling. Cell 179, 1342–1356.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voichek Y, Mittelman K, Gordon Y, Bar-Ziv R, Lifshitz Smit D, Shenhav R, and Barkai N (2018). Epigenetic control of expression homeostasis during replication is stabilized by the replication checkpoint. Mol. Cell 70, 1121–1133.e9. [DOI] [PubMed] [Google Scholar]
- Wang F, Ranjan A, Wei D, and Wu C (2016). Comment on “A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme”. Science 353, 358. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Radman-Livaja M, Rando OJ, and Peterson CL (2013). A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science 340, 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JG, Madrid TS, Sevastopoulos E, and Narlikar GJ (2006). The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat. Struct. Mol. Biol 13, 1078–1083. [DOI] [PubMed] [Google Scholar]
- Yao W, Beckwith SL, Zheng T, Young T, Dinh VT, Ranjan A, and Morrison AJ (2015). Assembly of the Arp5 (actin-related protein) subunit involved in distinct INO80 chromatin remodeling activities. J. Biol. Chem 290, 25700–25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, King DA, Beckwith SL, Gowans GJ, Yen K, Zhou C, and Morrison AJ (2016). The INO80 complex requires the Arp5-Ies6 subcomplex for chromatin remodeling and metabolic regulation. Mol. Cell. Biol 36, 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, and Pugh BF (2013). SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154, 1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CY, Johnson SL, Gamarra NI, and Narlikar GJ (2016). Mechanisms of ATP-dependent chromatin remodeling motors. Annu. Rev. Biophys 45, 153–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CY, Johnson SL, Lee LJ, Longhurst AD, Beckwith SL, Johnson MJ, Morrison AJ, and Narlikar GJ (2018). The yeast INO80 complex operates as a tunable DNA length-sensitive switch to regulate nucleosome sliding. Mol. Cell 69, 677–688.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


