Abstract
Dual leucine zipper kinase (DLK) plays a pivotal role in the development, degeneration, and regeneration of neurons. DLK can regulate gene expression post-transcriptionally, but the underlying mechanism remains poorly understood. The Drosophila DLK, Wallenda (Wnd), regulates the expression of Down syndrome cell adhesion molecule (Dscam) to control presynaptic arbor growth. This regulation is mediated by the 3′ untranslated region (3′UTR) of Dscam mRNA, which suggests that RNA binding proteins (RBPs) mediate DLK function. We performed a genome-wide cell-based RNAi screen of RBPs and identified the cytoplasmic poly(A)-binding protein, pAbp, as an RBP that mediates Wnd-induced increase in Dscam expression. Genetic analysis shows that Wnd requires pAbp for promoting presynaptic arbor growth and for enhancing Dscam expression. Our analysis revealed that Dscam mRNAs harbor short poly(A) tails. We identified a region in Dscam 3′UTR that specifically interacts with pAbp. Removing this region significantly reduced Wnd-induced increase in Dscam expression. These suggest that a noncanonical interaction of PABP with the 3′UTR of target transcripts is essential for DLK functions.
SIGNIFICANCE STATEMENT The kinase DLK plays key roles in a multitude of neuronal responses, including axon development, neurodegeneration, and nerve injury. Previous studies show that DLK acts via mRNAs to regulate protein synthesis, but how DLK does so is poorly understood. This study demonstrates that DLK regulates the synthesis of Dscam through the poly(A)-binding protein PABP-C. Whereas PABP-C is known as a general translational activator, our study shows that DLK-mediated Dscam expression involves a noncanonical interaction between PABP-C and the Dscam mRNA, which leads to a selective regulation of Dscam translation by PABP-C. Thus, our study provides novel insights into the mechanisms that underlie the function of DLK and regulation of gene expression of PABP-C.
Keywords: axon growth, DLK, Dscam, PAPB, post-transcriptional regulation, RNAi screen
Introduction
The evolutionarily conserved dual leucine zipper kinase (DLK) is a neuronal protein kinase that plays central roles in axon development, synapse maintenance, axon regeneration and degeneration, and neuronal cell death (Nakata et al., 2005; Collins et al., 2006; Xiong et al., 2010; Ghosh et al., 2011; Shin et al., 2012; Klinedinst et al., 2013; Blondeau et al., 2016; Larhammar et al., 2017; Le Pichon et al., 2017). DLK is an upstream mitogen-activated protein kinase (MAP kinase), MAP3K, that activates MAP kinases, including p38 and JNK (Nakata et al., 2005; Collins et al., 2006). The expression changes in genes downstream of these MAP kinases are believed to be central to DLK functions (Collins et al., 2006; Wlaschin et al., 2018; Shin et al., 2019). Interestingly, DLK functions seem to rely on post-transcriptional regulations in different settings. In Caenorhabditis elegans, DLK promotes axonal regeneration through stabilizing CEBP mRNA in injured axons (Yan et al., 2009). DLK-JNK signal activates an integrated stress response kinase, PERK in mouse, which results in translational activation of ATF4 for DLK-mediated neuronal cell death (Larhammar et al., 2017). In Drosophila, the DLK ortholog Wallenda (Wnd) promotes presynaptic arbor growth by upregulating Down syndrome cell adhesion molecule (Dscam) protein levels (Kim et al., 2013).
Dscam plays critical roles in neuronal development, including the self-avoidance of dendrites in Drosophila and mammalian retinal neurons (Fuerst et al., 2008, 2009; Schmucker and Chen, 2009; Zipursky and Sanes, 2010; Schramm et al., 2012) and axon guidance and growth (Kim et al., 2013; Alavi et al., 2016; Bruce et al., 2017; Santos et al., 2018). Dscam has also been implicated in various brain disorders (Amano et al., 2008; Shen et al., 2011; O'Roak et al., 2014). Thus, the regulation of Dscam expression is important for understanding neuronal development and brain disorders. Our previous study discovered that Wnd does not change the abundance of Dscam mRNA but requires the 3′ untranslated region (3′UTR) of Dscam for this regulation (Kim et al., 2013). This suggests that Wnd enhances the mRNA translation of Dscam. As a protein kinase, DLK likely requires an RNA-binding protein (RBP) to control protein translation through 3′UTR. However, the identity of the RBP is unknown.
Through an unbiased RNAi screen, we have identified pAbp, Drosophila ortholog of the cytoplasmic Poly(A)-binding protein (PABP-C) as an RBP that is required for Wnd-induced upregulation of Dscam expression as well as for Wnd-induced axon arborization. Although PABP-C is considered as a general activator of protein translation through poly(A) tail interaction (Munroe and Jacobson, 1990); it also performs gene-specific post-transcriptional regulations (Wu and Bag, 1998; Lyabin et al., 2011; Vazquez-Pianzola et al., 2011; Eliseeva et al., 2012; Iwakawa et al., 2012; Smith et al., 2017). We identified an adenine nucleotide rich (A-rich) region in the 3′UTR of Dscam that mediates pAbp interaction. Deletion of this region significantly reduced Wnd-induced Dscam expression both in vitro and in vivo. We further show that Dscam mRNA has significantly short poly(A) tails. Our study demonstrates that DLK function relies on the noncanonical mRNA interaction of PABP-C. It identifies not only a novel mechanism that mediates DLK signaling but also one that underlies the post-transcriptional regulation of Dscam expression.
Materials and Methods
Drosophila melanogaster strains
Drosophila strains were kept under standard condition at 25°C in a humidified chamber. The following strains were used in this study: w1118 (stock #3605, Bloomington Drosophila Stock Center), ppk-GAL4 (Grueber et al., 2007), UAS-Dscam[TM2]::GFP-Dscam-3′UTR (ectodomain 3.36.25) and UAS-Dscam[TM2]::GFP (ectodomain 3.36.25; Kim et al., 2013), hiwΔN (Wu et al., 2005), pAbpEP310 (stock #17261, Bloomington Drosophila Stock Center) and pAbpK10109 (stock #10970, Bloomington Drosophila Stock Center; Sigrist et al., 2000), UAS-FRT-rCD2-STOP-FRT-mCD8::GFP (Yang et al., 2014), and UAS-pAbp (stock #914, Bloomington Drosophila Stock Center).
FRTG13, UAS-mCD8::GFP, pAbpK10109 was generated through recombination. The primers used for genotyping pAbpK10109 were pAbpK10109-top (GAGGGCGCAACGCACGACAA) and pAbpK10109-bottom (ATATAATCATGTGTGTGTGTGTACACACTGGC).
Axon arborization analysis
The mosaic analysis with a repressible cell marker (MARCM) technique for presynaptic arbor analysis (Kim et al., 2013) and the quantitation of axon arborization by counting the axonal connecting fibers (Sterne et al., 2015) were done as previously described.
DNA constructs and Drosophila transgenic flies and Schneider 2 cell transfection
The enhanced green fluorescent protein (EGFP) reporters for RNAi screen were reported previously (Kim et al., 2013). To generate Wnd-HSL (histone-stem-loop), the Wnd-Wnd-3′UTR sequence without polyadenylation signal (PAS) was recovered from UAS-Wnd (Collins et al., 2006) via PCR and inserted proximal to HSL sequence into pAC5.1-Rn-Luc-HSL (Weidmann et al., 2014) using the NdeI and StuI restriction sites. Wnd-Wnd-3′UTR-HSL sequence was then transferred into pUAST-HA using EagI and XhoI sites, followed by transferring HA-Wnd-Wnd-3′UTR-HSL into the pAttB vector (Bischof et al., 2007) using NotI and XbaI sites. UAS-Dscam-5′UTR-Dscam-Dendra2-SV40-3′UTR was generated in pUASTattB (Bischof et al., 2007). UAS-Dscam-5′UTR-Dscam-Dendra2-Dscam-3′UTR and UAS-Dscam-5′UTR-Dscam-Dendra2-Dscam-3′UTRΔpAbp were generated in pAttB (Bischof et al., 2007). The 5′UTR and 3′'UTR of Dscam were obtained from w1118 genomic DNA by PCR. The Dscam coding sequence was from UAS-Dscam[TM2]::GFP (ectodomain 3.36.25). Dscam-3′UTRΔpAbp was generated by a site-directed mutagenesis. The resulting PCR products were ligated together using an In-Fusion Cloning Kit (Clontech) following instructions of the manufacturer. UAS-DSCAM-SV40-3′UTR with the A-rich region from Dscam-3′UTR was generated in pUASTattB (Bischof et al., 2007). The 123-bp A-rich sequence was recovered from UAS-Dscam-UTR-Dscam-Dendra2-Dscam-3′UTR via PCR and inserted at 148 bp upstream of the polyadenylation signal in SV40-3′UTR using an In-Fusion Cloning Kit (Clontech) following directions of the manufacturer. Transgenic flies were generated by PhiC31-mediated germline transformation (Bischof et al., 2007).
Schneider 2 cell culture, transfection, and Western blot analysis
Schneider 2 (S2) cells were maintained in Drosophila Schneider's Medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Sigma-Aldrich) at 27°C in a humidified incubator. Cells were transfected with indicated DNA constructs along with tubulin-Gal4 (Lee and Luo, 2001) using Lipofectamine 2000 (Life Technologies) following instructions from the manufacturer. For Western blot analysis, S2 cells were washed two times with PBS. The whole lysates were resolved on 8% SDS-PAGE gels and subjected to Western blot analysis as previously described (Kim et al., 2013). Primary antibodies used were rabbit polyclonal anti-Wnd (Collins et al., 2006), rabbit polyclonal anti-pAbp (Antic et al., 2015), rabbit polyclonal anti-Dendra2 (Antibodies-online), mouse monoclonal anti-tubulin (DM1A, Sigma-Aldrich), mouse monoclonal anti-puromycin (12D10, Sigma-Aldrich), Rat Anti-Elav (catalog #7E8A10, Developmental Studies Hybridoma Bank).
The validation of Rabbit polyclonal anti-pAbp antibody was done using S2 cells that were treated with pAbp dsRNA (7.5 µg) in a 24 well plate for 4 d. Cells were harvested. Total cell lysates were resolved in SDS-PABP before Western analysis (Fig. 6D).
Figure 6.
pAbp physically interacts with an adenine-rich region in Dscam 3′UTR. A, Dscam-3′UTR contains a region that is enriched in pAbp-binding sites. A bioinformatic analysis revealed a region in Dscam-3′UTR (shaded in gray) that contains five potential pAbp binding sites (underlined). The region between the two gray arrows in Dscam-3′UTR was selected for the pull-down experiment. B, C, pAbp interacts with Dscam-3′UTR. An RNA sequence in Dscam-3′UTR (Dscam) and SV40-3′UTR (SV40) was synthesized and biotinylated in vitro. Dscam-ΔpAbp refers to a deletion mutant of Dscam-3′UTR that lacks the potential pAbp binding sites (gray highlighted region in A). Streptavidin beads were coupled with indicated RNA and incubated with S2 cell lysates. Bound pAbp proteins were visualized by Western blot analysis (B). A representative image from five independent experiments is shown. Blots were quantified, normalized by bead-bound RNA (C). Dscam-3′UTR-bound pAbp was expressed either by SV40-3′UTR or by Dscam-ΔpAbp as a control. Left, SV40- and Dscam-bound pAbp were normalized by Dscam-ΔpAbp -bound pAbp. Right, Dscam- and Dscam-ΔpAbp-bound pAbp were normalized by SV40-bound pAbp. The data were expressed as mean ± SEM (SV40 vs Dscam: t = 9.849, df = 7.342, p < 0.0001, 2-tailed Welch's t test; Dscam vs Dscam-ΔpAbp: t = 9.923, df = 6.643, p < 0.0001, 2-tailed Welch's t test); sample n = 5. D, Validation of anti-pAbp antibody. Drosophila S2 cells were treated with the dsRNA targeting either GFP or pAbp for 3 d. Total lysates were subjected to Western blot analysis using anti-pAbp antibody. A representative image from five replicate experiments is shown. Actin blot was used as a loading control. The data were expressed as mean ± SD (t = 11.45, df = 4.737, p < 0.0001, 2-tailed Welch's t test); sample n = 5 ***p < 0.001 and ****p < 0.0001.
RNAi screening
The RBP-RNAi library (Mohr et al., 2015) was obtained from the Drosophila RNAi Screening Center (DRSC). The screening was done in a duplicate, nonadherent 384-well plate format. S2 cells were plated directly in the wells that contain individual dsRNAs (0.25 µg) and cultured for 24 h before transfecting with UAS-Wnd along with EGFP-Dscam-3′UTR reporter. Plates were inverted and kept at 27°C in a humidified incubator to prevent excessive culture media evaporation. After 48 h post-transfection, the average intensity of green fluorescence from individual cells was measured using a high-throughput flow cytometry, the HyperCyt System (Intellicyt). A lacZ dsRNA was used as a negative control. The lacZ dsRNAs were placed in seven wells that were scattered through each 384-well plate. The average GFP intensity values from the seven lacZ dsRNA-treated wells were used to generate an average GFP intensity (AVE-GFPlacZ) and an SD (SDlacZ). The initial hits were selected if GFPRBP < AVE-GFPlacZ – 1.5 × SDlacZ, where GFPRBP refers to the average GFP intensity from the cells that were treated with individual RBP dsRNA. The hits were finalized only when the duplicate wells for a given RBP dsRNA were both selected by the criteria.
For the secondary screening, the dsRNAs from the initial hits were synthesized from the DRSC DNA templates using TranscriptionAid T7 High Yield Transcription Kit (Thermo Fisher Scientific) and purified using Quick-RNA Miniprep Kit (Zymo Research). S2 cells were bath incubated with individual dsRNAs for 24 h before transfecting with UAS-Wnd along with EGFP-SV40-3′UTR or EGFP-Dscam-3′UTR. The GFP intensity was measured 48 h after transfection.
RNP pulldown
The biotinylated, single-stranded Dscam-3′UTR, SV40-3′UTR, and Dscam-3′UTRΔpAbp RNA were generated using TranscriptionAid T7 High Yield Transcription Kit (Thermo Fisher Scientific) with Biotin-16-UTP (Sigma-Aldrich). The T7 sequence was used for in vitro transcription reaction. RNAs were purified using Quick-RNA Miniprep Kit (Zymo Research). All RNAs contain S1 aptamer sequence (Srisawat and Engelke, 2001) at their 5′-end for better Streptavidin bead coupling. RNAs were incubated with Streptavidin agarose (Thermo Fisher Scientific) in binding buffer (50 mm HEPES, pH7.4, 300 mm NaCl, 0.5% NP-40, 2 mm MgCl2, 1 mm DTT) in the presence of RNaseOUT (Invitrogen) for 1 h at 4°C. Unbound RNA was removed by washing the beads three times with binding buffer. An aliquot of RNA-bound Streptavidin agarose was transferred to a new tube that contained 100 ng firefly luciferase RNA. The agarose bead-bound RNA was extracted using Trizol (Thermo Fisher Scientific), resolved, and quantified on 1.5% formaldehyde-denaturing agarose gel using firefly luciferase RNA as an extraction and a loading control.
S2 cells were washed two times with PBS before being lysed for 30 min in ice-cold binding buffer containing EDTA-free protease inhibitor cocktail (Research Products International). The crude lysates were centrifuged at 1000 × g, 4°C, for 5 min. The resulting postnuclear supernatant was supplemented with SUPERase-In (Thermo Fisher Scientific) and further centrifuged at 22,000 × g, 4°C, for 20 min. The resulting lysates were precleared with Streptavidin agarose for 1 h at 4°C, incubated with the RNA-coupled Streptavidin agarose for 2 h at 4°C. The RNP complexes were washed five times with binding buffer before resolved on 8% SDS-PAGE gel and subsequent Western blot analysis.
Immunostaining
Drosophila third instar larvae were prepared as previously described (Kim et al., 2013). Primary antibodies used were chicken polyclonal anti-GFP (Aves Labs), rabbit polyclonal anti-Dendra2 (Antibodies-online), and rabbit polyclonal anti-RFP (Rockland Immunochemicals). The secondary antibodies used were Cy2- or Cy5-conjugated goat anti-chicken and Cy2- or Cy5-conjugated goat anti-rabbit (Jackson ImmunoResearch). The imaging was done with a Leica SP5 confocal system or a custom-built spinning disk confocal microscope equipped with a 63× oil-immersion objective with a 0.3 µm step size. The resulting 3D images were projected into 2D images using a maximum projection method in ImageJ software. A region of interest was drawn in the cell body of C4da neurons, and the mean fluorescence intensity was measured using ImageJ software to quantify UAS-Dscam transgene expression levels. Because the activity of GAL4 varies among individual neurons, the mean fluorescence intensity of UAS-mCD8::GFP or UAS-mCD8::mRFP transgene expression was measured from the same region of interest and used as a normalization control.
Puromycylation assay
The batches of 20 brains were harvested from third instar larvae in ice-cold PBS. PBS was removed after centrifugation at 4°C, and 1 ml HL3 (hemolymph-like) solution was added containing the following (in mm): 5 HEPES, pH 7.2, 70 NaCl, 5 KCl, 0.5 CaCl2, 20 MgCl2, 5 trehalose, and 115 sucrose containing 10 µg/ml puromycin (Thermo Fisher Scientific). Larval samples were then incubated in a nutator for 15 min at room temperature (RT). Following incubation, HL3 solution was removed after a brief centrifugation at RT. Larval brains were washed three times with ice-cold PBS before resolved on 10% SDS-PAGE gel and subsequent Western blot analysis.
Poly(A) tail measurement
The G/I (guanosine/inosine) tailing takes advantage of a yeast poly(A) polymerase that incorporates additional nucleotides at the very 3′-end of RNAs, and thus preserves the 3′-end of RNA. After the addition of repeating G and I nucleotides, cDNA is prepared using a universal reverse primer. PCR is performed with the PCR primers that anneal to the 3′UTR from the genes of interest to detect the gene-specific as well as the gene-specific poly(A) products. The PCR products were analyzed by Agilent 2100 Bioanalyzer high-resolution capillary gel electrophoresis. Total RNA was extracted using Quick-RNA Microprep Kit (Zymo Research). The quality and quantity of isolated RNA was monitored by denaturing 1.5% agarose gel electrophoresis and spectrophotometry. Guanosine and inosine residues were added in the 3′-end of the poly(A)-containing RNAs followed by cDNA synthesis using the newly added G/I tails as priming sequence using the Poly(A) Tail-Length Assay Kit (Thermo Fisher Scientific) following instructions of the manufacturer. A gene-specific forward/reverse primer set right upstream of the polyadenylation site was used to generate a PCR product. A primer set with a gene-specific forward and the universal reverse primer was used to generate a product that includes the poly(A) tails of the gene of interest. The poly(A) tail lengths of the gene of interest are the sizes of poly(A) PCR products minus the calculated length of the gene-specific forward primer to the putative polyadenylation site. The sizes of the PCR products were analyzed by Agilent 2100 Bioanalyzer high resolution capillary gel electrophoresis with Agilent 2100 software.
The primers used are the following: Dscam-forward (CGCAGCCACAACAATTGAATG), Dscam-reverse (AAAATAAAATCAAAATCATATATTTAGCAACTTATGAAC), GAPDH2-forward (CACTTCAGAAACGGCCTGAAAATGGC), GAPDH2-reverse (AAATATTTAAATGCTTATGAGTCGGCATTTTTAAAACTAC), and the universal-reverse (GGTAATACGACTCACTATAGCGAGACCCCCCCCCCTT).
Experimental design and statistical analysis
All statistical analysis was performed as two-tailed using GraphPad Prism software version 7.04. The Mann–Whitney test was used for presynaptic arbor analysis (see Fig. 2) and immunostaining results (see Figs. 3, 9). An unpaired Student's t-test was used for S2 cell RNAi (Fig. 1), poly(A) length measurement (see Fig. 5), and Western blot analysis (see Figs. 4, 6, 7, 8). For multigroup comparisons, one-way ANOVA was used followed by post hoc Tukey's multiple comparisons test. A p value smaller than 0.05 was considered statistically significant. All p values are indicated as not significant; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Figure 2.
pAbp is required for Wnd-induced exuberant presynaptic arborization. A, The axon terminals from C4da neurons that correspond to abdominal segments 4 and 5 were visualized with mCD8::RFP using ppk-Gal4 (left). Hemizygous male hiw mutant (hiwΔN) and male wild-type (w1118; ppk-GAL4, UAS-mCD8::RFP/+) animals were used. The average number of neuropil-connecting fibers was counted from each animal (abdominal segments 4–6) and expressed as median ± 95% CI (right). One-way ANOVA (F(4,66) = 114.6, p < 0.0001) followed by post hoc Tukey's multiple comparison test. The p values from Tukey's test are indicated in the graph. Sample numbers were wt (w1118; ppk-GAL4, UAS-mCD8::RFP/+) (n = 10), ppk > PABP (w1118; ppk-GAL4, UAS-mCD8::RFP/UAS-PABP) (n = 9), pAbpK10109/EP310 (w1118; pAbpK10109/pAbpEP310; ppk-GAL4, UAS-mCD8::RFP/+) (n = 19), hiwΔN (hiwΔN;; ppk-GAL4, UAS-mCD8::RFP/+) (n = 17), hiwΔN, pAbpK10109/EP31 (hiwΔN; pAbpK10109/pAbpEP310; ppk-GAL4, UAS-mCD8::RFP/+) (n = 16). wt, Wild type. Scale bar, 10 µm. B, Schematic of Wnd-HSL. Wnd-HSL contains HSL instead of PAS in its 3′UTR, which makes Wnd protein expression independent of poly(A) tail. The MARCM analysis was performed to visualize the presynaptic arbors from individual C4da neurons. Transgenes were expressed using a C4da-specific ppk-GAL4. Presynaptic arbors were visualized with a membrane marker mCD8::GFP transgene. Overexpressing Wnd-HSL caused a dramatic increase in presynaptic arbor length in wt C4da neurons, but not in pAbpK10109 C4da neurons. The pAbp mutation alone did not significantly affect presynaptic arborization. Sample numbers were wt (hs-FLP, y, w1118/w1118; FRTG13, UAS-mCD8::GFP/FRTG13, GAL80; ppk-GAL4, UAS-mCD8::GFP/+) (n = 14), Wnd-HSL (hs-FLP, y, w1118/w1118; FRTG13, UAS-mCD8::GFP/FRTG13, GAL80; ppk-GAL4, UAS-mCD8::GFP/UAS-Wnd-HSL) (n = 12), pAbpK10109 (hs-FLP, y, w1118/w1118; FRTG13, UAS-mCD8::GFP, pAbpK10109/FRTG13, GAL80; ppk-GAL4, UAS-mCD8::GFP/+) (n = 12), Wnd-HSL in pAbpK10109 (hs-FLP, y, w1118/w1118; FRTG13, UAS-mCD8::GFP, pAbpK10109/FRTG13, GAL80; ppk-GAL4, UAS-mCD8::GFP/UAS-Wnd-HSL) (n = 5). The data were expressed as median ± 95% CI. One-way ANOVA (F(3,36) = 21.91, p < 0.0001) followed by post hoc Tukey's multiple comparison test. The p values from Tukey's test are indicated in the graph *p < 0.05, ****p < 0.0001, ns = not significant. Scale bar, 10 µm.
Figure 3.
pAbp is required for Wnd-induced Dscam expression. A, Wnd-HSL or a control transgene, CD2, was expressed in the larval C4da neurons along with Dscam::GFP-SV40-3′UTR or Dscam::GFP-Dscam-3′UTR in wild-type (wt) control (w1118) or in pAbp mutants (pAbpk10109/EP310). The cell bodies of C4da neurons are shown, Dscam::GFP (green) and mCD8::RFP (magenta). Scale bar, 10 µm. Expression of Dscam in Dscam::GFP-SV40-3′UTR not shown. B, The expression levels of Dscam::GFP were measured from the individual C4da cell bodies. An mCD8::RFP (RFP) transgene was used to normalize Dscam::GFP intensity. The data were expressed as median ± 95% CI. Note that the increase in Dscam::GFP expression from Dscam::GFP-Dscam-3′UTR is significantly higher in Wnd-HSL than in Wnd-HSL, pAbpK10109/EP310; one-way ANOVA (F(7,203) = 12.03, p < 0.0001) followed by post hoc Tukey's multiple comparison test. Sample numbers were Dscam::GFP-SV40-3′UTR and CD2 (w1118; GAL44-77/UAS-Dscam::GFP-SV40-3′UTR; UAS-FRT-rCD2-STOP-FRT-mCD8::GFP/+) (n = 24), Dscam::GFP-SV40-3′UTR and Wnd-HSL (w1118; GAL44-77/UAS-Dscam::GFP-SV40-3′UTR; UAS-Wnd-HSL/+) (n = 29), Dscam::GFP-SV40-3′UTR and CD2 in pAbpK10109/EP310 (w1118; GAL44-77, pAbpK10109/pAbpEP310, UAS-Dscam::GFP-SV40-3′UTR; UAS-FRT-rCD2-STOP-FRT-mCD8::GFP/+) (n = 29), Dscam::GFP-SV40-3′UTR and Wnd-HSL in pAbpK10109/EP310 (w1118; GAL44-77, pAbpK10109/pAbpEP310, UAS-Dscam::GFP-SV40-3′UTR; UAS-Wnd-HSL/+) (n = 32), Dscam::GFP-Dscam-3′UTR and CD2 (w1118; GAL44-77/UAS-Dscam::GFP-Dscam-3′UTR; UAS-FRT-rCD2-STOP-FRT-mCD8::GFP/+) (n = 30), Dscam::GFP-Dscam-3′UTR and Wnd-HSL (w1118; GAL44-77/UAS-Dscam::GFP-Dscam-3′UTR; UAS-Wnd-HSL/+) (n = 28), Dscam::GFP-Dscam-3′UTR and CD2 in pAbpK10109/EP310 (w1118; GAL44-77, pAbpK10109/pAbpEP310, UAS-Dscam::GFP-Dscam-3′UTR; UAS-FRT-rCD2-STOP-FRT-mCD8::GFP/+) (n = 33), Dscam::GFP-Dscam-3′UTR and Wnd-HSL in pAbpK10109/EP310 (w1118; GAL44-77, pAbpK10109/pAbpEP310, UAS-Dscam::GFP-Dscam-3′UTR; UAS-Wnd-HSL/+) (n = 30). The p values from Tukey's test are indicated in the graph **p < 0.01, ****p < 0.0001, ns = not significant.
Figure 9.
pAbp binding to Dscam-3′UTR is required for Wnd-induced Dscam translation in vivo. A, The Dscam::Dendra2 transgenes, Dscam-5′UTR-Dscam::Dendra2-SV40-3′UTR (Dscam-SV40), Dscam-5′UTR-Dscam::Dendra2-Dscam-3′UTR (Dscam-3′UTR), and Dscam-5′UTR-Dscam::Dendra2-Dscam-3′UTRΔpAbp (Dscam-3′UTRΔpAbp) were expressed in C4da neurons with ppk-GAL4 either in wild-type control (w1118, wt) or in hiw mutant (hiwΔN) animals. The expression levels of Dscam::Dendra2 were measured from the individual C4da cell bodies. Dscam::Dendra2 expression levels were visualized with anti-Dendra2 antibody (magenta). A membrane marker, mCD8::GFP, was used as a normalizing control (green). Hemizygous male hiw mutant (hiwΔN) and male wild-type (w1118) animals were used. Scale bar, 10 µm. B, Dscam::Dendra2 fluorescence intensity was normalized by mCD8::GFP fluorescence intensity and expressed as a fold change of a corresponding control (hiwΔN/wt) from Dscam-5′UTR-Dscam::Dendra2-SV40-3′UTR (Dscam-SV40), Dscam-5′UTR-Dscam::Dendra2-Dscam-3′UTR (Dscam-3′UTR), and Dscam-5′UTR-Dscam::Dendra2-Dscam-3′UTRΔpAbp (Dscam-3′UTRΔpAbp). The data were expressed as median ± 95% CI. One-way ANOVA (F(5,255) = 14.40, p < 0.0001) followed by post hoc Tukey's multiple comparison test. The p values from Tukey's test are indicated in the graph. Sample numbers were Dscam-SV40 in wt (w1118; UAS-Dscam-5′UTR-Dscam::Dendra2-SV40-3′UTR/+; ppk-GAL4, UAS-mCD8::GFP/+) (n = 60), Dscam-SV40 in hiwΔN (hiwΔN; UAS-Dscam-5′UTR-Dscam::Dendra2-SV40-3′UTR/+; ppk-GAL4, UAS-mCD8::GFP/+) (n = 58), Dscam-3′UTR in wt (w1118; UAS-Dscam-5′UTR-Dscam::Dendra2-Dscam-3′UTR/+; ppk-GAL4, UAS-mCD8::GFP/+) (n = 44), Dscam-3′UTR in hiwΔN (hiwΔN; UAS-Dscam-5′UTR-Dscam::Dendra2-Dscam-3′UTR/+; ppk-GAL4, UAS-mCD8::GFP/+) (n = 41), Dscam-3′UTRΔpAbp in wt (w1118; UAS-Dscam-5′UTR-Dscam::Dendra2-Dscam-3′UTRΔpAbp/+; ppk-GAL4, UAS-mCD8::GFP/+) (n = 35), and Dscam-3′UTRΔpAbp in hiwΔN (hiwΔN; UAS-Dscam-5′UTR-Dscam::Dendra2-Dscam-3′UTRΔpAbp/+; ppk-GAL4, UAS-mCD8::GFP/+) (n = 23) **p < 0.01, ****p < 0.0001, and ns = not significant.
Figure 1.

Genome-wide cell-based RNAi screen identifies pAbp as an RBP that mediates Wnd-induced increase in Dscam expression. A, Cell-based RNAi screen. S2 cells were treated with the dsRNA library against Drosophila RBP genes and transfected with Wnd and EGFP-Dscam-3′UTR reporter constructs, followed by high-throughput flow cytometry analysis. Bottom, The results of the secondary screen. dsRNA-treated cells were transfected with EGFP-Dscam-3′UTR reporter constructs along with an empty vector (back bar) or with Wnd construct (gray bar). The empty vector control was used to normalize values for each dsRNA (n = 4). LacZ dsRNA was used as a control. The data shown are an example from the screening. Mean ± SEM; *p < 0.05, **p < 0.01, two-tailed, unpaired t test. B, pAbp RNAi does not reduce Wnd expression in S2 cells. Cell lysates obtained from the secondary screen was tested by Western blot analysis. Note that Wnd expression was largely decreased in SF2 and Hel25E RNAi-treated cells. pAbp RNAi did not affect Wnd expression, although it did reduce GFP expression from EGFP-Dscam-3′UTR reporter. Tubulin blot was used as a loading control. C, pAbp RNAi blunts Wnd-induced increase in EGFP-Dscam-3′UTR reporter. S2 cells were treated with pAbp dsRNA or with LacZ dsRNA before transfecting with EGFP-Dscam-3′UTR construct along with an empty vector (Vec) or Wnd construct (n = 4). The average GFP intensity was measured using flow cytometry. The data were expressed as mean ± SEM. One-way ANOVA (F(3,12) = 11.25, p = 0.0008) followed by post hoc Tukey's multiple comparison test. The p values from Tukey's test are indicated in the graph *p < 0.05, **p < 0.01, ns = not significant.
Figure 5.
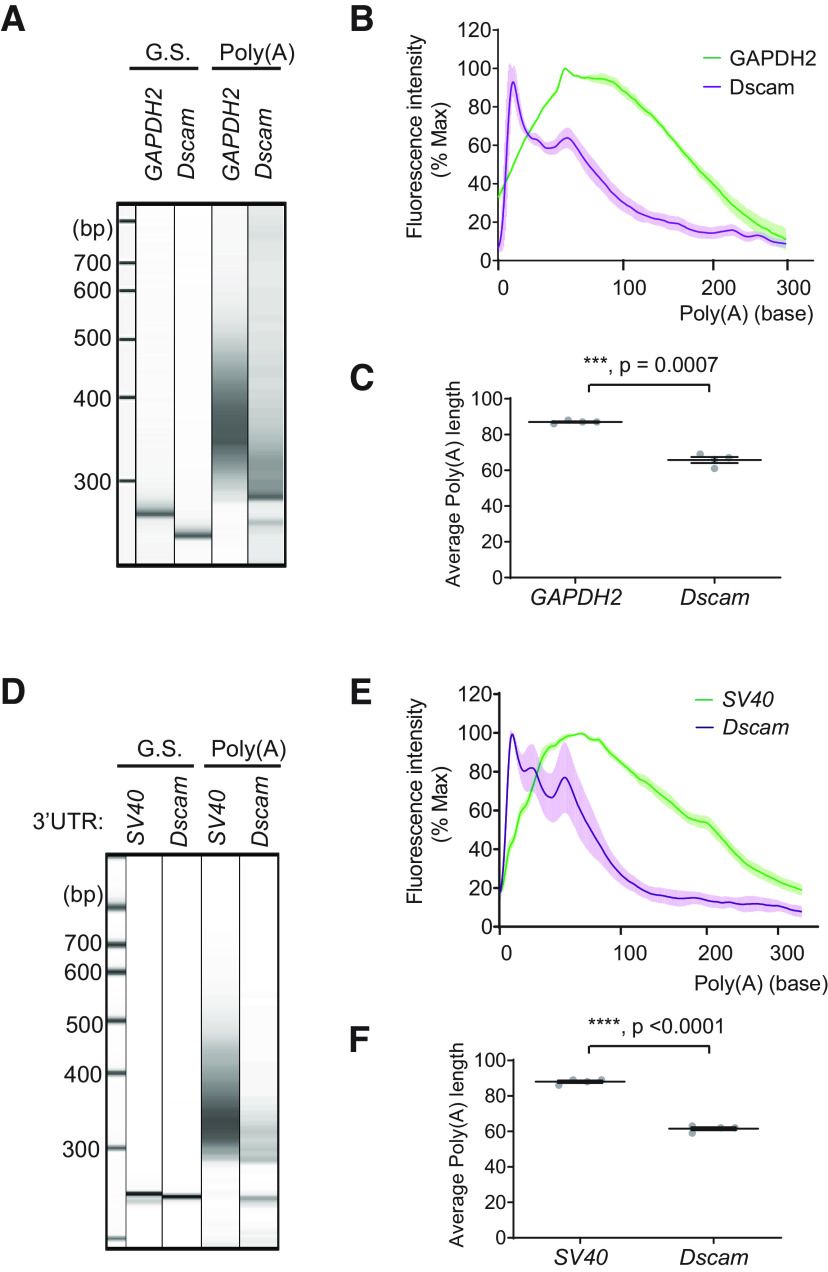
Dscam mRNA contains short poly(A) tails. A, Total RNA was extracted from the third instar larva brains from a wild-type control (w1118) and subjected to the poly(A) length analysis. Capillary gel electrophoresis images were shown for the gene-specific (G.S.) PCR product as a discrete band (the first and second lane) for GAPDH2 and Dscam. The PCR product using universal reverse primer exhibits a smear pattern (the third and fourth lane), which indicates the mRNAs with a range of poly(A) tail. B, The poly(A) length from GAPDH (green) and Dscam (magenta) was calculated from the capillary gel electrophoresis (see above, Materials and Methods), and the distribution was plotted from four technical replicates (mean ± SD). Note a difference in peak poly(A) length. C, Mean poly(A)-tail lengths were calculated and are displayed (mean ± SEM, t = 12.14, df = 3.344, p = 0.0007, 2-tailed Welch's t test); sample n = 4. D, Drosophila S2 cells were cotransfected with the Dscam constructs that contain Dscam-5′UTR, the coding region of Dscam, and the 3′UTR of either Dscam or SV40 and subjected to the poly(A) length analysis. Capillary gel electrophoresis images were shown for the gene-specific (G.S.) PCR product as a discrete band (the first and second lane) for Dscam-5′UTR-Dscam-SV40-3′UTR (SV40) and Dscam-5′UTR-Dscam-Dscam-3′UTR (Dscam). The PCR product using universal reverse primer exhibits a smear pattern (the third and fourth lane), which indicates the mRNAs with a range of poly(A) tail. E, The poly(A) length from Dscam-5′UTR-Dscam-SV40-3′UTR (SV40, green) and Dscam-5′UTR-Dscam-Dscam-3′UTR (Dscam, magenta) was calculated from the capillary gel electrophoresis, and the distribution was plotted from four technical replicates (mean ± SD). F, Mean poly(A)-tail lengths were calculated and displayed (mean ± SEM, t = 23.70, df = 5.769, p < 0.0001, 2-tailed Welch's t test); sample n = 4 ***p < 0.001 and ****p < 0.0001.
Figure 4.
Characterization of pAbpK10109/EP310. A, Total RNAs extracted from the wondering third instar larval brains from a wild-type (wt) control (w1118) and pAbpK10109/EP310 were subjected to RT-qPCR to measure pAbp mRNA abundance. The pAbp mRNA levels were normalized to GAPDH1 mRNA levels via the 2ΔΔCT method. The heterozygote combination of pAbpK10109 and pAbpEP310 alleles reduced by ∼65% pAbp mRNA abundance. A similar result was obtained using an independent qPCR primer pair. The data were expressed as mean ± SEM of five technical replicates (t = 13.47, df = 4.383, p < 0.0001, 2-tailed Welch's t test). B, Puromycylation to measure protein synthesis rate. The wondering third instar larval brains from a wt control (w1118) and pAbpK10109/EP310 were briefly incubated in puromycin and subjected to Western blot analysis with anti-puromycin antibody. Anti-Elav blot was used as a loading control. The incorporation of puromycin was displayed as mean ± SEM of five biological replicates (t = 3.167, df = 8, p = 0.022, 2-tailed Welch's t test) *p < 0.05, ****p < 0.0001.
Figure 7.
pAbp binding to Dscam-3′UTR is required for Wnd-induced Dscam translation in S2 cells. A, Schematics of Dscam-5′UTR-Dscam::Dendra2-SV40-3′UTR (5-Dscam-SV40), Dscam-5′UTR-Dscam::Dendra2-Dscam-3′UTR (5-Dscam-3UTR), and Dscam-5′UTR-Dscam::Dendra2- Dscam-3′UTRΔpAbp (5-Dscam-3UTRΔpAbp) are shown. All Dscam-3′UTR DNA constructs contains endogenous PAS from Dscam. B, S2 cells were transfected with indicated Dscam constructs along with either an empty vector (Vec) or Wnd-expressing construct. Total cell lysates were subjected to Western blot analysis using anti-Dendra2 antibody to detect Dscam::Dendra2. Tubulin blot was used as a loading control. A representative image from four replicate experiments is shown. C, Dendra2 blots were normalized with tubulin blot and expressed as a fold change of a corresponding control (Wnd/Vec) from Dscam-SV40, Dscam-3′UTR, and Dscam-3′UTRΔpAbp. The data were expressed as mean ± SEM; one-way ANOVA (F(5,18) = 9.899, p = 0.0001) followed by post hoc Tukey's multiple comparison test. The p values from Tukey's test are indicated in the graph; sample n = 4 *p < 0.05, ***p < 0.001, and ns = not significant.
Figure 8.
The A-rich region of Dscam is not sufficient for Wnd regulation. The 123-bp A-rich sequence from Dscam-3′UTR was inserted at the 148 bp upstream of the polyadenylation signal in Dscam-5′UTR-Dscam::Dendra2-SV40-3′UTR (Dscam-A-rich-SV40) (top) and expressed in Drosophila S2 cells in the presence of an empty vector (Vec) or Wnd expressing plasmid. Total lysates were subjected to Western blot with anti-Dendra2 to quantify Dscam::Dendra2 (Dscam). Tubulin blot was used as a loading control. The expression of Dscam-5′UTR-Dscam::Dendra2-SV40-3′UTR (Dscam-SV40) was shown as a comparison. The data were expressed as mean ± SEM. The two-tailed Welch's t test (t = 0.06,013, df = 7.289 for Dscam-SV40 and t = 0.6723, df = 5.937 for Dscam-A-rich-SV40); sample n = 5; *ns = not significant.
Results
Genome-wide cell-based RNAi screen identifies pAbp as an RBP that mediates Wnd-induced increase in Dscam expression
Our previous work has shown that Wnd post-transcriptionally regulates the expression of Dscam through the 3′UTR of Dscam without affecting the abundance of Dscam mRNA, which suggests the role of Wnd in Dscam translation (Kim et al., 2013). The study also suggested that the increased Dscam expression mediates the effect of Wnd on increased presynaptic axon arbors (Kim et al., 2013). However, how Wnd controls Dscam translation was unclear. Wnd may require an RBP for this regulation as Wnd, as a protein kinase, unlikely affects the 3′UTR of mRNAs directly. In search for such RBPs, an unbiased cell-based RNAi screen was performed in cultured Drosophila S2 cells. We took advantage of an EGFP reporter that recapitulated Wnd-mediated Dscam expression regulation in S2 cells (Kim et al., 2013). We used a dsRNA library for all annotated Drosophila RBPs (Mohr et al., 2015). In total, 820 individual dsRNAs against 427 Drosophila RBP genes were screened. S2 cells were plated in a microplate and bath incubated with individual dsRNAs to knock down RBP genes before transfecting an EGFP-Dscam 3′UTR construct either with a Wnd expression construct or an empty DNA construct. A dsRNA against lacZ was used as a negative control. The expression levels of EGFP were assessed 48 h post-transfection by a high-throughput flow cytometry analysis (Fig. 1A). The initial screen generated multiple hits in 18 RBP genes. We then performed a secondary screening using a control EGFP reporter that contains the 3′UTR from SV40 (Fig. 1A, bottom) and narrowed down to nine candidates. An RBP RNAi may decrease reporter expression by inhibiting plasmid gene expression or by reducing Wnd expression. To rule out these possibilities, we performed an additional test using Western blotting with an anti-Wnd antibody and obtained four RBPs as the final candidates, namely, pAbp, Int6, Rpb7, and CG14414. CG14414 has neither a known function nor is orthologous to any known mammalian genes. Three final hits have known roles in translation initiation. Int6 encodes the eukaryotic translation initiation factor 3 subunit E, eIF3e, which is involved in translation initiation (Akiyoshi et al., 2001). Rpb7 encodes a nonessential component of RNA-polymerase II that is involved in diverse functions including translation initiation (Harel-Sharvit et al., 2010). pAbp is a Drosophila ortholog of PABP-C. We selected pAbp for further study because of its well-characterized role in translation initiation (Munroe and Jacobson, 1990). Moreover, pAbp RNAi did not reduce Wnd expression (Fig. 1B) but significantly reduced Wnd-triggered increase in EGFP expression from the EGFP-Dscam-3′UTR reporter (Fig. 1C).
pAbp is required for Wnd-induced Dscam translation in vivo
Wnd increases the size of presynaptic axon arbors by enhancing Dscam expression in Drosophila larval class IV dendritic arborization (C4da) neurons (Kim et al., 2013). To evaluate the functional role of pAbp in this process, we first examined the axonal arborization of C4da neurons in pAbp overexpression (Fig. 2A). The C4da neuron axon arborization was measured by counting the number of fibers connecting the segmentally repeated neurophils. The method was successfully used before (Wang et al., 2013; Sterne et al., 2015). We did not observe any changes, which suggests that pAbp is expressed sufficiently under normal condition. We then tested the effect of highwire (hiw)-induced axon arborization by pAbp mutations. Hiw normally degrades Wnd expression to suppress axonal arborization (Collins et al., 2006). Thus, increased Wnd expression causes excessive axon arborization in an hiw loss-of-function allele (hiwΔN). Because strong loss-of-function alleles of pAbp caused larval lethality, we used a transheterozygous combination of two pAbp hypomorphic alleles—partial loss of function pAbpk10109 and pAbpEP310—to study the effects of loss of pAbp. The mutation of hiw caused a dramatic increase in axon arborization, which was mildly but significantly reduced by pAbp mutations (21% rescue; Fig. 2A).
PABP-C binds to the poly(A) tails of mRNAs and interacts with mRNA translation initiation complex (Wells et al., 1998). As virtually all cellular mRNAs contain a poly-(A) tail, PABP-C is considered as a general mRNA translation activator. A poly(A) tail addition is triggered by a cis-element at mRNA 3′UTR, also known as PAS. Histone mRNA is an exception because it does not contain a PAS. Instead, HSL sequence marks the transcriptional termination and recruits stem loop binding protein (SLBP). Through binding to HSL, SLBP plays a similar role as PABP-C to histone mRNA (Sànchez and Marzluff, 2002; Marzluff et al., 2008). To examine the role of pAbp in Wnd-mediated Dscam regulation in vivo, we generated a Wnd transgene to dissociate its expression from the pAbp regulation by replacing the PAS of Wnd mRNA to HSL sequences (Fig. 2B). In this transgene, Wnd-HSL, Wnd expression is through the HSL mechanism and is thus independent of the canonical pAbp function via poly(A) tail interaction. We performed the mosaic analysis with a repressible cell marker (MARCM; Lee and Luo, 2001) in wild-type and pAbp mutant larval C4da neurons that are expressing Wnd-HSL. MARCM allowed us to use a strong loss-of-function allele of pAbp, pAbpk10109, without causing larval lethality. As shown in Figure 2B, the expression of Wnd-HSL induced a dramatic increase in presynaptic arbor size, which was almost completely abolished by pAbp mutations. These results strongly suggest that pAbp mediates Wnd-induced axon terminal growth, which is through Dscam (Kim et al., 2013).
Having a tool for directly testing a role of pAbp in Wnd-induced Dscam regulation, we then asked whether Wnd controls Dscam expression through pAbp. The Wnd-HSL transgene was expressed by using the GAL4-UAS bipartite expression system in the C4da neurons with GAL44-77 in wild-type and in pAbp mutant (pAbpk10109/EP310) animals. As expected, Wnd-HSL increased expression of a Dscam transgene that contains Dscam-3′UTR but not the one with SV40-3′UTR (Fig. 3A,B). Importantly, this effect was significantly abolished by the loss of pAbp, which suggests that pAbp is required for Wnd-induced Dscam translation in vivo.
Dscam mRNA contains short poly(A) tails
How does PABP as a general translation activator exhibit selectivity for Dscam? And, what is the extent of this selectivity? To answer these questions, we first measured how much pAbp function is lost in the hypomorphic transheterozygote of the pAbpK10109 and pAbpEP310 alleles. We performed quantitative real-time PCR on the larval brains from wild type and pAbpK10109/EP310 and found a dramatic reduction (67%) in pAbp mRNA abundance (Fig. 4A). Although this clearly demonstrates a reduction in pAbp mRNA levels, pAbp proteins may exist in excess, and their functions be consequently unaffected by in pAbpK10109/EP310. Therefore, we decided to measure general protein synthesis, a proxy for pAbp function, using the puromycylation assay. Puromycylation takes advantage of puromycin, an amino-nucleoside antibiotic. Puromycin resembles a charged-tRNA and causes termination of mRNA translation by incorporating itself into a growing polypeptide chain from ribosomes (Pestka, 1971). Specific antibodies against puromycin allow the evaluation of the puromycin incorporation rate, which linearly correlates to the mRNA translation rate (Eggers et al., 1997). The larval brains from wild type and pAbpK10109/EP310 were isolated and briefly treated with puromycin at a low concentration. Western blot analysis was performed on the brain lysates using anti-puromycin antibody. The result showed a 50% reduction in the general protein synthesis rate in pAbpK10109/EP310 (Fig. 4B). Thus, pAbp proteins are not expressed in excess and can be a limiting factor for general translation initiation. This indicates that Dscam translation is more sensitive to a reduction in pAbp protein levels than other canonical targets.
This relative sensitivity suggests that Dscam mRNA may not provide a high-affinity binding site as other cellular mRNAs via poly(A) tail. Certain mRNAs undergo a process known as the cytoplasmic deadenylation, a form of post-transcriptional translational regulation (Richter, 1999). Interestingly, some isoforms of mouse Dscam mRNAs reportedly contain very short poly(A) tails of only 5–10 adenine residues (Alves-Sampaio et al., 2010). Efficient PABP-C recruitment to a poly(A) tail requires ∼27 adenine nucleotides (Deo et al., 1999). Thus, we decided to measure the lengths of the poly(A) tails of Dscam mRNA. To this end, we used a poly(A) tail analysis known as the G/I tailing (Kusov et al., 2001). To measure the poly(A) lengths of Dscam, total RNA was extracted from the larval brains from a wild-type control (w1118) and subjected to the poly(A) length analysis. GAPDH2 was used as a control. Polyadenylation is not a defined modification; therefore, the PCR product generates a smear pattern (Fig. 5A). The identity of the smear PCR band from GAPDH2 and Dscam were verified by sequencing.
The sizes of poly(A) PCR products and the gene-specific PCR product were used to calculate the poly(A) tail lengths of the gene of interest (Fig. 5B; see above, Materials and Methods). The result showed that Dscam has significantly shorter poly(A) tails than those from GAPDH2 (Fig. 5C). Unlike a bell-shape distribution of GAPDH2 poly(A) tails, Dscam poly(A) tails showed a sharp peak. The peak poly(A) tail length of Dscam was 5.75 ± 2.36 bases compared with 47.25 ± 1.26 bases of GAPDH2 (Fig. 5B). Wnd regulation on Dscam expression requires Dscam-3′UTR (Fig. 3). Therefore, we wondered whether the short poly(A) tails of Dscam mRNA is dependent on Dscam-3′UTR. We coexpressed the Dscam DNA constructs that contains either SV40-3′UTR or Dscam-3′UTR in Drosophila S2 cells. Total RNA was prepared from the S2 cell lysates and subjected to subsequent poly(A) length analysis. The result showed that the poly(A) tail distributions from Dscam-SV40-3′UTR were almost identical as those from GAPDH2, whereas those from Dscam-Dscam-3′UTR showed a similar pattern as those from endogenous Dscam (Fig. 5B,C,E,F). This clearly demonstrates that Dscam mRNAs contain short poly(A) tails, which is dependent on Dscam-3′UTR and further suggests a presence of pAbp binding site outside the poly(A) tail of Dscam mRNA.
pAbp Physically interacts with an adenine-rich region in the Dscam 3′UTR
Interestingly, PABP can interact with RNA sequences outside of poly(A) tail but rich in adenine nucleotides (Skabkina et al., 2003; Sladic et al., 2004; Vazquez-Pianzola et al., 2011; Kini et al., 2016; Smith et al., 2017). We reasoned that such binding likely takes place in the 3′UTR of Dscam because Wnd regulation of Dscam requires it. To test the possibility that the Dscam-3′UTR contains pAbp binding sites, bioinformatic analysis was performed using an RBP binding prediction tool (Paz et al., 2014). We selected the motifs that are shared by both Drosophila pAbp and human PABP-C1. The analysis revealed the five potential pAbp-binding sites in a 123-nucleotide region (Fig. 5A, shading) that is enriched with adenines (A; Fig. 6A, underline). To test whether this Dscam-3′UTR region can recruit pAbp, we performed an in vitro binding assay. Biotinylated RNA that contains the 123-nucleotide region was generated via in vitro transcription using the forward and reverse primers as indicated in Figure 6A (Dscam region). A 267-nucleotide region from SV40-3′UTR was generated as a control (SV40 region). We further generated an RNA that does not contain the 123-nucleotide sequences from Dscam-3′UTR (Dscam-ΔpAbp; Fig. 6A). The RNAs were incubated with S2 cell lysates. Bound pAbp was detected using Western blot analysis (Fig. 6B) following the quantitation of either using SV40 or Dscam-ΔpAbp as a control (Fig. 6C). In either quantitation, Dscam region showed an increased pAbp-binding (∼2.5-fold) than the controls. The specificity of the pAbp antibody was verified using the S2 cell lysates that were treated with pAbp dsRNA (Fig. 6D). These results demonstrate that pAbp physically interacts with Dscam-3′UTR, which is dependent on the 123-nucleotide A-rich region.
The interaction between pAbp and Dscam 3′UTR mediates Wnd-induced increase in dscam expression
Next, we determined whether this noncanonical interaction of pAbp with Dscam mRNA is required for Wnd-mediated Dscam upregulation. We generated Dscam-expressing DNA constructs that contain SV40-3′UTR, Dscam-3′UTR, or a Dscam-3′UTR without the A-rich region (Dscam-3′UTRΔpAbp; Fig. 7A). The transgenic Dscam proteins were tagged with the fluorescent protein Dendra2. We coexpressed a Wnd construct along with the Dscam constructs in S2 cells. Western blot analysis showed that although Wnd dramatically increased the expression of Dscam from Dscam::Dendra2-Dscam-3′UTR, it only mildly increased Dscam expression from Dscam::Dendra2-Dscam-3′UTRΔpAbp (Fig. 7B,C). There was about a one-fold difference in Wnd-induced Dscam increase between Dscam::Dendra2-Dscam-3′UTR and Dscam::Dendra2-Dscam-3′UTRΔpAbp (Fig. 7C). Next, we tested whether the A-rich region is sufficient to confer Wnd-mediated upregulation. We inserted the A-rich region from Dscam-3′UTR in SV40-3′UTR just before the PAS (Fig. 8, top). Dscam was expressed under the modified SV40-3′UTR (A-rich-SV40) in the presence of Wnd in S2 cells. We found no changes in Dscam expression (Fig. 8), which suggests the presence of an A-rich region per se is not sufficient for Wnd-mediated regulation.
To determine whether the interaction between pAbp and Dscam 3′UTR is essential for Dscam expression in vivo, we expressed the Dscam transgenes in Drosophila C4da neurons using a ppk-GAL4 driver in wild-type and highwire (hiw) mutants. We found that loss of hiw increased the expression of Dscam from Dscam::Dendra2-Dscam-3′UTR, but not that from Dscam::Dendra2-SV40-3′UTR (Fig. 9). Consistent with our in vitro result (Fig. 7B,C), loss of hiw caused a significantly smaller increase in Dscam expression from Dscam::Dendra2-Dscam-3′UTRΔpAbp than from Dscam::Dendra2-Dscam3'UTR (Fig. 9B). These results demonstrate that the interaction between pAbp and Dscam-3′UTR is essential for Wnd-induced enhancement of Dscam expression.
Discussion
DLK plays essential roles in multiple biological processes in the nervous system, some of which require post-transcriptional regulations of molecules in the DLK pathway. One of the molecules downstream of DLK is Dscam, whose altered expression is associated with multiple brain disorders. In this study, we identified pAbp, the Drosophila ortholog of PABP-C, as an RBP that mediates Wnd-induced post-transcriptional upregulation of Dscam. Our study revealed that the noncanonical recruitment of PABP-C to the 3′UTR of Dscam is essential in the process. Thus, our study uncovers a novel mechanism of DLK-mediated post-transcriptional gene regulation.
We showed that pAbp is required for Wnd-induced Dscam upregulation (Figs. 1, 3) as well as Wnd-induced increase in presynaptic arbor size (Fig. 2). How does pAbp, as a general mRNA translation activator, affect Dscam expression? It is known that simultaneous interaction between PABP-C and poly(A) tails and that between PABP-C and the mRNA translation initiation complex at the 5′-end of an mRNA underlies the role of PABP-C in the activation of mRNA translation (Munroe and Jacobson, 1990; Wells et al., 1998). However, recent studies have shown that PABP-C also can exert mRNA-specific post-transcriptional regulation through interacting the 5′ or 3′UTR of target mRNAs (Wu and Bag, 1998; Lyabin et al., 2011; Eliseeva et al., 2012; Iwakawa et al., 2012; Casper et al., 2013; Smith et al., 2017). These interactions lead to either a suppression or an activation of target translation, depending on target mRNA. In a conventional model, PABP-C bridges the 3′- and 5′-end of an mRNA for generating a closed-loop structure for efficient mRNA translation (Munroe and Jacobson, 1990; Wells et al., 1998). Interestingly, the recruitment of PABP-C to the 3′UTR of target mRNAs leads to translational activation (Lyabin et al., 2011; Eliseeva et al., 2012; Iwakawa et al., 2012; Smith et al., 2017). Consistently, our result strongly suggests that Dscam-3′UTR contains an A-rich region, which can recruit pAbp (Fig. 6). These suggest that the noncanonical PABP-C association near the end of 3′-UTR may involve a similar mechanism as the closed-loop model for translational activation.
Our result showed that pAbp knock-down selectively suppressed Wnd-induced upregulation of Dscam-3′UTR reporter but not that of SV40-3′UTR reporter or Wnd expression (Fig. 1). PABP controls general protein synthesis. How does pAbp RNAi selectively affect Dscam translation? PABP has a strong binding affinity toward the poly(A) tail and shows significantly lower affinity toward nonpoly(A) RNA sequences (Sladic et al., 2004). Our result indicates that Drosophila Dscam mRNAs have short poly(A) tails (Fig. 5), which is consistent with a previous report in mouse Dscam (Alves-Sampaio et al., 2010). Given that efficient PABP-C recruitment requires ∼27 consecutive adenine nucleotides (Deo et al., 1999), the absence of strong PABC-C binding through a poly(A) tail likely renders Dscam mRNA more dependent on the A-rich region in its 3′UTR. In this scenario, the noncanonical PABP binding requires a higher PABP concentration than that of canonical poly(A) tail interaction, which would make noncanonical interaction more sensitive to a reduction in cytoplasmic PABP protein levels. PABP is known to regulate its own protein homeostasis through binding its own 5′UTR and translational suppression (Wu and Bag, 1998). This autoregulation might not be as strong as it is believed as our analysis of transheterozygous pAbp mutants showed ∼50% reduction in general mRNA translation (Fig. 4). Nevertheless, our result showed that pAbp mutations caused more notable suppression of Dscam expression compared with that of a mCD8::RFP, a normalizing experimental control that contains SV40-3′UTR (Fig. 3). To further rule out a possibility of general mRNA translation suppression by pAbp mutations, we used Wnd-HSL in our study (Figs. 2B, 3). The translation of histone mRNAs employs a unique mechanism because these are the only metazoan mRNAs that do not possess poly(A) tails. The HSL at the 3′-end of histone mRNA recruits SLBP instead of PABP, which promotes the translation initiation of histone mRNA (Ling et al., 2002; Sànchez and Marzluff, 2002). Together, we envision that pAbp exerts a relative selectivity toward Dscam translation via a differential affinity toward canonical and noncanonical RNA interactions.
Our study shows that Wnd requires pAbp for enhancing Dscam expression (Figs. 1, 3) and for increasing presynaptic arbor growth (Fig. 2). This suggests that Wnd/DLK regulates PABP activity. Wnd is an upstream kinase in the MAP kinase pathway. It is possible that Wnd or downstream MAP kinases phosphorylate PABP and modify PABP affinity toward the noncanonical A-rich sequence in Dscam-3′UTR. For example, PABP is phosphorylated by CRK1, which promotes association with the poly(A) sequence, self-interaction, and interaction with eIF4E in Trypanosoma brucei (An et al., 2018). However, although human PABP-C is highly modified post-translationally via methylation and acetylation (Brook et al., 2012), PABP-C phosphorylation has not been detected in most animals. Then, how might Wnd modify PABP activity? Interestingly, A-rich sequences are not rare in the UTR sequences. In fact, the SV40-3′UTR segment that was used in our study (Fig. 6) also contains four putative noncanonical PABP binding sites, although it did not show a specific pAbp binding (Fig. 6) or being regulated by Wnd expression or a loss of hiw (Figs. 3, 7, 9). This raises an interesting possibility that these A-rich sequences may require additional factors for recruiting PABP-C. Consistent with this idea, inserting the A-rich region from Dscam-3′UTR in SV40-3′UTR did not affect reporter expression in the presence of Wnd (Fig. 8). The DLK/Wnd pathway may phosphorylate an unknown cofactor, which increases PABP-C recruitment to the A-rich region in the target 3′UTR. RNAi is known to have false negatives (Booker et al., 2011). Our RNAi screen may have simply missed the cofactor. It will be important to identify this cofactor to gain a complete understanding of DLK signaling in future studies.
Although our analyses showed that pAbp is critically required for Wnd-mediated presynaptic arborization (Fig. 2B) and Dscam expression (Fig. 3), the hypomorphic mutant pAbpK10109/EP310 showed only a mild effect in presynaptic arborization by hiw mutations (Fig. 2A). Complete loss of pAbp triggers cell lethality because pAbp is required for general protein synthesis. We estimate that ∼50% of pAbp function is lost in pAbpK10109/EP310 (Fig. 4), which may explain a partial rescue in hiw-induced axon arborization. Wnd-enhanced Dscam expression was reduced by ∼61% in pAbpK10109/EP310 mutants (Fig. 3B). Conversely, this may suggest that Hiw uses additional, Wnd-independent pathway for axonal arborization in C4da neurons. Consistently, ∼30% of hiw-enhanced Dscam expression remained in Dscam::Dendra2-Dscam-3′UTRΔpAbp (Fig. 9B). Although statistically insignificant, deleting the A-rich region from Dscam-3′UTR did not completely abolish Wnd-enhanced Dscam expression both in S2 cells and C4da neurons (Figs. 7, 9). Thus, this may suggest an existence of an independent RBP pathway downstream of Wnd in Dscam expression regulation.
Many functions of DLK are conserved from worms to mice, some of which rely on post-transcriptional gene regulation. Here, we show that the Drosophila ortholog of PABP-C is involved in presynaptic arbor growth through mediating post-transcriptional regulation of Dscam. The DLK homolog in C. elegans promotes mRNA stability of CEBP in response to axonal injury (Yan et al., 2009). A noncanonical PABP-C interaction is known for mRNA stability and localization of osk mRNA in Drosophila (Vazquez-Pianzola et al., 2011). Thus, it is conceivable that PABP-C may mediate multiple DLK functions. Interestingly, recent studies suggest a novel role of DLK and PABP-C in neuropathic pain (Barragán-lglesias et al., 2018; Wlaschin et al., 2018; Hu et al., 2019; Ma et al., 2021). It is possible that DLK and PABP-C target the same set of mRNAs in neuropathic pain. What factors or features would make these mRNAs under DLK-PABP control? Our results showed that Dscam-3′UTR has an A-rich region for direct pAbp recruitment (Fig. 6). In addition, Dscam mRNA has short poly(A) tails, which are dependent on Dscam-3′UTR (Fig. 5). The length of poly(A) tails is under the control of multiple pathways (Richter, 1999; Meijer et al., 2007; Nicholson and Pasquinelli, 2019). These may be the common features of DLK-PABP-targeted mRNAs. DLK is expressed in nonneuronal cell types and may function in these cell types (Jin and Zheng, 2019). Determining the roles of PABP-C and common mRNA targets downstream of various DLK functions will be an important research direction for future studies.
Footnotes
This study was supported by National Institute of Neurological Disorders and Stroke Grant R01NS116463 and National Institute of General Medical Sciences Grant P20GM103440 (Nevada IDeA Network of Biomedical Research Excellence) to J.H.K., National Institute of Mental Health Grant R01MH112669 and National Institute of Neurological Disorders and Stroke Grant R01NS104299 to B.Y., and a grant from the Protein Folding Disease Initiative of the University of Michigan to B.Y. We thank Drs. Catherine Colins, Aaron Goldstrohm, and Silke Dorner for sharing antibodies and DNA constructs; Drs. John Kim and Mallory Freeberg for the initial bioinformatic identification of RNA binding proteins (RBP) in Drosophila genome; Dr. Stephanie Mohr and the Drosophila RNAi Screening Center for constructing the RBP dsRNA library; the Center for Chemical Genomics at the University of Michigan–Ann Arbor for technical and financial support for the RNAi screen; and the Cellular and Molecular Imaging Core facility at the University of Nevada, Reno, which was supported by National Institutes of Health Grant P20GM103650 and used for research reported in this study.
Authors declare no competing financial interests.
References
- Akiyoshi Y, Clayton J, Phan L, Yamamoto M, Hinnebusch AG, Watanabe Y, Asano K (2001) Fission yeast homolog of murine Int-6 protein, encoded by mouse mammary tumor virus integration site, is associated with the conserved core subunits of eukaryotic translation initiation factor 3. J Biol Chem 276:10056–10062. 10.1074/jbc.M010188200 [DOI] [PubMed] [Google Scholar]
- Alavi M, Song M, King GLA, Gillis T, Propst R, Lamanuzzi M, Bousum A, Miller A, Allen R, Kidd T (2016) Dscam1 forms a complex with Robo1 and the N-terminal fragment of slit to promote the growth of longitudinal axons. PLoS Biol 14:e1002560. 10.1371/journal.pbio.1002560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Sampaio A, Troca-Marin JA, Montesinos ML (2010) NMDA-mediated regulation of DSCAM dendritic local translation is lost in a mouse model of Down's syndrome. J Neurosci 30:13537–13548. 10.1523/JNEUROSCI.3457-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano K, Yamada K, Iwayama Y, Detera-Wadleigh SD, Hattori E, Toyota T, Tokunaga K, Yoshikawa T, Yamakawa K (2008) Association study between the Down syndrome cell adhesion molecule (DSCAM) gene and bipolar disorder. Psychiatr Genet 18:1–10. 10.1097/YPG.0b013e3281ac238e [DOI] [PubMed] [Google Scholar]
- An T, Liu Y, Gourguechon S, Wang CC, Li Z (2018) CDK phosphorylation of translation initiation factors couples protein translation with cell-cycle transition. Cell Rep 25:3204–3214. 10.1016/j.celrep.2018.11.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antic S, Wolfinger MT, Skucha A, Hosiner S, Dorner S (2015) General and microRNA-mediated mRNA degradation occurs on ribosome complexes in Drosophila cells. Mol Cell Biol 35:2309–2320. 10.1128/MCB.01346-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragán-Iglesias P, Lou TF, Bhat VD, Megat S, Burton MD, Price TJ, Campbell ZT (2018) Inhibition of Poly(A)-binding protein with a synthetic RNA mimic reduces pain sensitization in mice. Nat Commun 9:10. 10.1038/s41467-017-02449-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A 104:3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondeau A, Lucier J-F, Matteau D, Dumont L, Rodrigue S, Jacques P-É, Blouin R (2016) Dual leucine zipper kinase regulates expression of axon guidance genes in mouse neuronal cells. Neural Dev 11:13. 10.1186/s13064-016-0068-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker M, Samsonova AA, Kwon Y, Flockhart I, Mohr SE, Perrimon N (2011) False negative rates in Drosophila cell-based RNAi screens: a case study. BMC Genomics 12:50. 10.1186/1471-2164-12-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook M, McCracken L, Reddington JP, Lu ZL, Morrice NA, Gray NK (2012) The multifunctional poly(A)-binding protein (PABP) 1 is subject to extensive dynamic post-translational modification, which molecular modelling suggests plays an important role in co-ordinating its activities. Biochem J 441:803–812. 10.1042/BJ20111474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce FM, Brown S, Smith JN, Fuerst PG, Erskine L (2017) DSCAM promotes axon fasciculation and growth in the developing optic pathway. Proc Natl Acad Sci U S A 114:1702–1707. 10.1073/pnas.1618606114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper I, Nowag S, Koch K, Hubrich T, Bollmann F, Henke J, Schmitz K, Kleinert H, Pautz A (2013) Post-transcriptional regulation of the human inducible nitric oxide synthase (iNOS) expression by the cytosolic poly(A)-binding protein (PABP). Nitric Oxide 33:6–17. 10.1016/j.niox.2013.05.002 [DOI] [PubMed] [Google Scholar]
- Collins CA, Wairkar YP, Johnson SL, DiAntonio A (2006) Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron 51:57–69. 10.1016/j.neuron.2006.05.026 [DOI] [PubMed] [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK (1999) Recognition of polyadenylate RNA by the poly (A)-binding protein. Cell 98:835–845. 10.1016/S0092-8674(00)81517-2 [DOI] [PubMed] [Google Scholar]
- Eggers DK, Welch WJ, Hansen WJ (1997) Complexes between nascent polypeptides and their molecular chaperones in the cytosol of mammalian cells. Mol Biol Cell 8:1559–1573. 10.1091/mbc.8.8.1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliseeva IA, Ovchinnikov LP, Lyabin DN (2012) Specific PABP effect on translation of YB-1 mRNA is neutralized by polyadenylation through a “mini-loop” at 3′ UTR. RNA Biol 9:1473–1487. 10.4161/rna.22711 [DOI] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW (2008) Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature 451:470–474. 10.1038/nature06514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW (2009) DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron 64:484–497. 10.1016/j.neuron.2009.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AS, Wang B, Pozniak CD, Chen M, Watts RJ, Lewcock JW (2011) DLK induces developmental neuronal degeneration via selective regulation of proapoptotic JNK activity. J Cell Biol 194:751–764. 10.1083/jcb.201103153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Yang C-H, Younger S, Borden K, Jan LY, Jan Y-N (2007) Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development 134:55–64. 10.1242/dev.02666 [DOI] [PubMed] [Google Scholar]
- Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M (2010) RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143:552–563. 10.1016/j.cell.2010.10.033 [DOI] [PubMed] [Google Scholar]
- Hu Z, Deng N, Liu K, Zeng W (2019) DLK mediates the neuronal intrinsic immune response and regulates glial reaction and neuropathic pain. Exp Neurol 322:1–13. [DOI] [PubMed] [Google Scholar]
- Iwakawa H.-o, Tajima Y, Taniguchi T, Kaido M, Mise K, Tomari Y, Taniguchi H, Okuno T (2012) Poly(A)-binding protein facilitates translation of an uncapped/nonpolyadenylated viral RNA by binding to the 3′ untranslated region. J Virol 86:7836–7849. 10.1128/JVI.00538-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Zheng B (2019) Multitasking: dual leucine zipper-bearing kinases in neuronal development and stress management. Annu Rev Cell Dev Biol 35:501–521. 10.1146/annurev-cellbio-100617-062644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Wang X, Coolon R, Ye B (2013) Dscam expression levels determine presynaptic arbor sizes in drosophila sensory neurons. Neuron 78:827–838. 10.1016/j.neuron.2013.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini HK, Silverman IM, Ji X, Gregory BD, Liebhaber SA (2016) Cytoplasmic poly(A) binding protein-1 binds to genomically encoded sequences within mammalian mRNAs. RNA 22:61–74. 10.1261/rna.053447.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinedinst S, Wang X, Xiong X, Haenfler JM, Collins CA (2013) Independent pathways downstream of the Wnd/DLK MAPKKK regulate synaptic structure, axonal transport, and injury signaling. J Neurosci 33:12764–12778. 10.1523/JNEUROSCI.5160-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusov YY, Shatirishvili G, Dzagurov G, Gauss-Müller V (2001) A new G-tailing method for the determination of the poly(A) tail length applied to hepatitis A virus RNA. Nucleic Acids Res 29: e57–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larhammar M, Huntwork-Rodriguez S, Jiang Z, Solanoy H, Ghosh AS, Wang B, Kaminker JS, Huang K, Eastham-Anderson J, Siu M, Modrusan Z, Farley MM, Tessier-Lavigne M, Lewcock JW, Watkins TA (2017) Dual leucine zipper kinase-dependent PERK activation contributes to neuronal degeneration following insult. Elife 6:e20725. 10.7554/eLife.20725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pichon CE, et al. (2017) Loss of dual leucine zipper kinase signaling is protective in animal models of neurodegenerative disease. Sci Transl Med 9:eaag0394. 10.1126/scitranslmed.aag0394 [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L (2001) Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24:251–254. 10.1016/S0166-2236(00)01791-4 [DOI] [PubMed] [Google Scholar]
- Ling J, Morley SJ, Pain VM, Marzluff WF, Gallie DR (2002) The histone 3′-terminal stem-loop-binding protein enhances translation through a functional and physical interaction with eukaryotic initiation factor 4G (eIF4G) and eIF3. Mol Cell Biol 22:7853–7867. 10.1128/MCB.22.22.7853-7867.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyabin DN, Eliseeva IA, Skabkina OV, Ovchinnikov LP (2011) Interplay between Y-box-binding protein 1 (YB-1) and poly(A) binding protein (PABP) in specific regulation of YB-1 mRNA translation. RNA Biol 8:883–892. 10.4161/rna.8.5.16022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Goodwani S, Acton PJ, Buggia-Prevot V, Kesler SR, Jamal I, Mahant ID, Liu Z, Mseeh F, Roth BL, Chakraborty C, Peng B, Wu Q, Jiang Y, Le K, Soth MJ, Jones P, Kavelaars A, Ray WJ, Heijnen CJ (2021) Inhibition of dual leucine zipper kinase prevents chemotherapy-induced peripheral neuropathy and cognitive impairments. Pain 162:2599–2612. 10.1097/j.pain.0000000000002256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ (2008) Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9:843–854. 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, de Moor CH (2007) A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res 35:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr SE, Hu Y, Rudd K, Buckner M, Gilly Q, Foster B, Sierzputowska K, Comjean A, Ye B, Perrimon N (2015) Reagent and data resources for investigation of RNA binding protein functions in Drosophila melanogaster cultured cells. G3 (Bethesda) 5:1919–1924. 10.1534/g3.115.019364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe D, Jacobson A (1990) mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol 10:3441–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Abrams B, Grill B, Goncharov A, Huang X, Chisholm AD, Jin Y (2005) Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell 120:407–420. 10.1016/j.cell.2004.12.017 [DOI] [PubMed] [Google Scholar]
- Nicholson AL, Pasquinelli AE (2019) Tales of detailed poly(A) tails. Trends Cell Biol 29:191–200. 10.1016/j.tcb.2018.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Stessman HA, Boyle EA, Witherspoon KT, Martin B, Lee C, Vives L, Baker C, Hiatt JB, Nickerson DA, Bernier R, Shendure J, Eichler EE (2014) Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun 5:5595. 10.1038/ncomms6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I, Kosti I, Ares M, Cline M, Mandel-Gutfreund Y (2014) RBPmap: a Web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res 42:W361–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S (1971) Inhibitors of ribosome functions. Annu Rev Microbiol 25:487–562. 10.1146/annurev.mi.25.100171.002415 [DOI] [PubMed] [Google Scholar]
- Richter JD (1999) Cytoplasmic polyadenylation in development and beyond. Microbiol Mol Biol Rev 63:446–456. 10.1128/MMBR.63.2.446-456.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sànchez R, Marzluff WF (2002) The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol Cell Biol 22:7093–7104. 10.1128/MCB.22.20.7093-7104.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RA, Fuertes AJC, Short G, Donohue KC, Shao H, Quintanilla J, Malakzadeh P, Cohen-Cory S (2018) DSCAM differentially modulates pre- and postsynaptic structural and functional central connectivity during visual system wiring. Neural Dev 13:22. 10.1186/s13064-018-0118-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Chen B (2009) Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev 23:147–156. 10.1101/gad.1752909 [DOI] [PubMed] [Google Scholar]
- Schramm RD, Li S, Harris BS, Rounds RP, Burgess RW, Ytreberg FM, Fuerst PG (2012) A novel mouse Dscam mutation inhibits localization and shedding of DSCAM. PLoS One 7:e52652. 10.1371/journal.pone.0052652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Xiao Z, Pan Y, Fang M, Li C, Chen D, Wang L, Xi Z, Xiao F, Wang X (2011) Altered expression of Dscam in temporal lobe tissue from human and experimental animals. Synapse 65:975–982. 10.1002/syn.20924 [DOI] [PubMed] [Google Scholar]
- Shin JE, Cho Y, Beirowski B, Milbrandt J, Cavalli V, DiAntonio A (2012) Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron 74:1015–1022. 10.1016/j.neuron.2012.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Ha H, Kim YK, Cho Y, DiAntonio A (2019) DLK regulates a distinctive transcriptional regeneration program after peripheral nerve injury. Neurobiol Dis 127:178–192. 10.1016/j.nbd.2019.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Thiel PR, Reiff DF, Lachance PED, Lasko P, Schuster CM (2000) Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature 405:1062–1065. 10.1038/35016598 [DOI] [PubMed] [Google Scholar]
- Skabkina OV, Skabkin MA, Popova NV, Lyabin DN, Penalva LO, Ovchinnikov LP (2003) Poly(A)-binding protein positively affects YB-1 mRNA translation through specific interaction with YB-1 mRNA. J Biol Chem 278:18191–18198. 10.1074/jbc.M209073200 [DOI] [PubMed] [Google Scholar]
- Sladic RT, Lagnado CA, Bagley CJ, Goodall GJ (2004) Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur J Biochem 271:450–457. 10.1046/j.1432-1033.2003.03945.x [DOI] [PubMed] [Google Scholar]
- Smith RWP, Anderson RC, Larralde O, Smith JWS, Gorgoni B, Richardson WA, Malik P, Graham SV, Gray NK (2017) Viral and cellular mRNA-specific activators harness PABP and eIF4G to promote translation initiation downstream of cap binding. Proc Natl Acad Sci U S A 114:6310–6315. 10.1073/pnas.1610417114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C, Engelke DR (2001) Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA 7:632–641. 10.1017/s135583820100245x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne GR, Kim JH, Ye B (2015) Dysregulated Dscam levels act through Abelson tyrosine kinase to enlarge presynaptic arbors. Elife 4:e05196. 10.7554/eLife.05196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Pianzola P, Urlaub H, Suter B (2011) Pabp binds to the osk 3′UTR and specifically contributes to osk mRNA stability and oocyte accumulation. Dev Biol 357:404–418. 10.1016/j.ydbio.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Wang X, Kim JH, Bazzi M, Robinson S, Collins CA, Ye B (2013) Bimodal control of dendritic and axonal growth by the dual leucine zipper kinase pathway. PloS Biol 11:e1001572. 10.1371/journal.pbio.1001572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann CA, Raynard NA, Blewett NH, Van Etten J, Goldstrohm AC (2014) The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation. Rna 20:1298–1319. 10.1261/rna.046029.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2:135–140. [DOI] [PubMed] [Google Scholar]
- Wlaschin JJ, Gluski JM, Nguyen E, Silberberg H, Thompson JH, Chesler AT, Le Pichon CE (2018) Dual leucine zipper kinase is required for mechanical allodynia and microgliosis after nerve injury. Elife 7:e33910. 10.7554/eLife.33910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bag J (1998) Negative control of the poly(A)-binding protein mRNA translation is mediated by the adenine-rich region of its 5′-untranslated region. J Biol Chem 273:34535–34542. 10.1074/jbc.273.51.34535 [DOI] [PubMed] [Google Scholar]
- Wu C, Wairkar YP, Collins CA, DiAntonio A (2005) Highwire function at the Drosophila neuromuscular junction: spatial, structural, and temporal requirements. J Neurosci 25:9557–9566. 10.1523/JNEUROSCI.2532-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang X, Ewanek R, Bhat P, DiAntonio A, Collins CA (2010) Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J Cell Biol 191:211–223. 10.1083/jcb.201006039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wu Z, Chisholm AD, Jin Y (2009) The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell 138:1005–1018. 10.1016/j.cell.2009.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li R, Kaneko T, Takle K, Morikawa RK, Essex L, Wang X, Zhou J, Emoto K, Xiang Y, Ye B (2014) Trim9 regulates activity-dependent fine-scale topography in Drosophila. Curr Biol 24:1024–1030. 10.1016/j.cub.2014.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky SL, Sanes JR (2010) Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell 143:343–353. 10.1016/j.cell.2010.10.009 [DOI] [PubMed] [Google Scholar]