Abstract
Objective
Gut dysbiosis contributes to multiple autoimmune diseases, including ankylosing spondylitis, which is commonly treated with tumor necrosis factor (TNF)-α inhibitors (TNFis). Because host TNF-α levels are considered to interact with gut microbiota, we aimed to systematically investigate the microbiota profile of ankylosing spondylitis patients with anti-TNF-α-based treatment and identify potential key bacteria.
Methods
Fecal samples were collected from 11 healthy controls and 24 ankylosing spondylitis patients before/after anti-TNF-α treatment, the microbiota profiles of which were evaluated by 16S ribosomal DNA amplicon sequencing and subsequent bioinformatic analysis.
Results
Significantly different microbial compositions were observed in samples from ankylosing spondylitis patients compared with healthy controls, characterized by a lower abundance of short-chain fatty acid (SCFA)-producing bacteria. All patients exhibited a positive response after anti-TNF-α treatment, accompanied by a trend of restoration in the microbiota compositions and functional profile of ankylosing spondylitis patients to healthy controls. In particular, the abundance of SCFA-producing bacteria (e.g. Megamonsa and Lachnoclostridium) was not only significantly lower in ankylosing spondylitis patients than in healthy controls and restored after anti-TNF-α treatment but also negatively correlated with disease severity (e.g. cor = -0.52, P = 8 × 10−5 for Megamonsa). In contrast, Bacilli and Haemophilus may contribute to ankylosing spondylitis onset and severity.
Conclusions
Microbiota dysbiosis in ankylosing spondylitis patients can be restored after anti-TNF-α treatment, possibly by impacting SCFA-producing bacteria.
Keywords: 16S sequencing, ankylosing spondylitis, gut microbiota, short-chain fatty acid, TNF-α inhibitor
Introduction
Ankylosing spondylitis, characterized as a chronic rheumatic disease with unclear etiology, is a prototype of spondylarthritis that can affect the spine and sacroiliac joints and is commonly accompanied by extra-articular manifestations (e.g. dermatitis, uveitis and colitis) [1]. As a consequence, ankylosing spondylitis has a serious negative impact on patients in terms of physical function, quality of life and work ability, and imposes a severe burden on society and families due to the high probability of disability [2]. Epidemiologically, ankylosing spondylitis predominantly affects young adults, with an overall incidence ranging from 0.2 to 0.54% in China [3,4]. Although a variety of factors may contribute to ankylosing spondylitis risk, including inherited predispositions (e.g. haplotype of HLA-B27) [5,6], the pathogenesis of autoimmunity in ankylosing spondylitis has not been fully elucidated.
Clinically, more than 50% of ankylosing spondylitis patients experience subclinical gut inflammation, 10% of whom may further develop inflammatory bowel disease (IBD) [7–9]. Accumulating evidence suggests that similar genetic risk factors and etiopathogenesis are shared by ankylosing spondylitis and IBD patients [10–12]. On the other hand, complicated interactions between gut microbiota and the function of host immune systems have been well revealed [13], including a demonstration in patients with IBD [14,15]. Not surprisingly, gut dysbiosis was also recently observed in patients with ankylosing spondylitis, illustrating significant differences in the composition and functional spectrum of gut microbiota in ankylosing spondylitis patients compared with healthy individuals [7,16,17]. Consistently, germ-free conditions can reduce the risk of ankylosing spondylitis development in animal models [18], further supporting the causal status of disturbed microbiota for ankylosing spondylitis. Mechanistically, the gut may be the first site of antigen exposure, which then activates the pathogenic mechanisms within the joint [19]. Rheumatologists speculated that gut bacterial antigens may invade sacroiliac and spine joints through lymphatic vessels due to the imbalance of gut microbiota and accompanying damage to the intestinal mucosa, thereby triggering immune responses and inflammation in these locations [20]. In addition, previous studies have pointed out that the genetic background of the host can affect gut microbes [21]. As the most common genetic susceptibility factor of ankylosing spondylitis, the pathogenicity of HLA-B27 is likely to induce ankylosing spondylitis by influencing the gut microbiome [22–24]. Based on this evidence, the gut microbiota is considered to play a critical role in ankylosing spondylitis development.
Recommendations for the use of tumor necrosis factor (TNF)-α inhibitors (TNFis) in patients with ankylosing spondylitis were proposed by several international guidelines [25,26]. TNFis have been proven to be effective in reducing disease activity, improving physical function and slowing radiographic progression; therefore, TNFis have revolutionized the treatment of ankylosing spondylitis patients who are inadequate or resistant to nonsteroidal anti-inflammatory drugs and conventional disease-modifying anti-rheumatic drugs (DMARDs) [27–30]. Mechanistically, TNF-α is a cytokine mainly produced by macrophages, as well as other immune cells, including CD4+ lymphocytes, NK cells, neutrophils, mast cells and eosinophils, thus triggering and aggravating inflammation [31]. Investigation of human sacroiliac joint specimens provided the earliest evidence, observing abundant TNF-α in patients with early sacroiliac arthritis and suggesting a possible association of spondylarthritis with TNF-α [32]. Clinical studies on TNFi further supported the role of TNF-α in the pathogenesis of ankylosing spondylitis [33,34]. Intriguingly, in addition to the impact of inherited predisposition on the anti-TNF-α treatment response [35], the correlation of TNF-α with gut microbiota has also been revealed. For instance, some patients with ulcerative colitis were not sensitive to TNFi and exhibited significantly different baseline gut microbiota compared with patients who were sensitive to TNFi [36], and the TNF-α level of the host can also influence the gut microbiota composition [37]. All these studies indicated the possible interaction between TNF-α and gut microbiota.
Because a number of studies have indicated the potentially important role of microbiota in drug or treatment response, we aimed to investigate the profile of microbiota and the involvement of specific gut microbes in the treatment of ankylosing spondylitis with TNFi in this study.
Materials and methods
Study subjects and sample collection
Anti-TNF-α treatment-naive patients with recent-onset ankylosing spondylitis and healthy controls were enrolled. Ankylosing spondylitis patients were treated with TNFi through injection, and individuals who used antibiotics in the last 3 months were excluded. Finally, 24 ankylosing spondylitis patients and 11 healthy controls were included in this study. The activity of ankylosing spondylitis was measured based on the Bath ankylosing spondylitis disease activity index (BASDAI). A reduction in the BASDAI score by at least 2 points or 50% from baseline was considered an indicator of clinical remission. Fecal samples were collected once from healthy controls and twice from ankylosing spondylitis patients (i.e. 1–3 days before anti-TNF-α treatment and ~1 month after treatment, except for the time point at which the patient experienced clinical remission for five patients). Fresh fecal samples were collected and stored in tightly closed tubes and immediately preserved at −80 °C, within 1 h from defecation to storage. Clinical information was obtained from the electronic system of West China Hospital as described previously [38–40]. This study was approved by the Ethics Committee of West China Hospital, Sichuan University [2020 (1151)].
Sequencing and bioinformatic analysis
DNA was extracted from frozen fecal samples (200 mg each) using the QIAamp Fast DNA Stool Mini Kit (Qiagen, #51604). The DNA quality was measured with a NanoDrop and agarose gel electrophoresis. The gut microbiota in all samples was determined via 16S rDNA sequencing at Novogene Bioinformatics Technology Institute (Sichuan, China) with the Illumina HiSeq X10 platform (paired-end 150 bp) and analyzed with the optimized pipeline based on which we have described previously [41,42]. Briefly, the universal forward primer (5′-GTGCCAGCMGCCGCGGTAA-3′) and the reverse primer (5′- GGACTACHVGGGTWTCTAAT-3′) were used to amplify the V3–V4 hypervariable regions. The sequences were processed with a USEARCH (http://www.drive5.com/usearch/) (VSEARCH) pipeline, clustered into operational taxonomic units (OTUs) based on a 97% similarity threshold, and then normalized according to the fewest number of OTUs in all samples for analysis of alpha diversity. The Ribosomal Database Project Classifier (https://www.drive5.com/sintax/) was used for taxonomic assignment of all OTUs, and the relative abundance was calculated using the annotation results in R software. Principal coordinates analysis was conducted on Bray–Curtis matrices using QIIME1 software (v.1.9.1). Linear discriminant analysis (LDA) effect size analysis (LefSe) was employed to identify features with significantly different abundances as unique bacteria between the indicated groups based on annotation results assigned by the Silva reference database (http://www.arb-silva.de/). A log LDA score of >3 was the threshold for discriminating between the indicated groups. The sequences were assigned to the Greengenes reference database for PICRUSt functional prediction.
Statistical analysis
R (version 4.0.3) software was used for statistical analyses. Statistical significance was calculated using the Wilcoxon rank-sum test or paired T-test for pairwise comparisons (e.g. α diversity, LefSe analysis, PICRUSt functional prediction). β diversity analysis was performed on Bray Curtis distances and statistical significance was calculated using the Adonis test. Differential OTUs in all groups were identified using the Kruskal–Wallis test. Spearman’s rank correlation test was used to assess correlations between the abundance of unique bacteria and the disease activity index. The results were considered to be statistically significant at P < 0.05.
Results
Overall intestinal microbial profile of ankylosing spondylitis patients and healthy controls
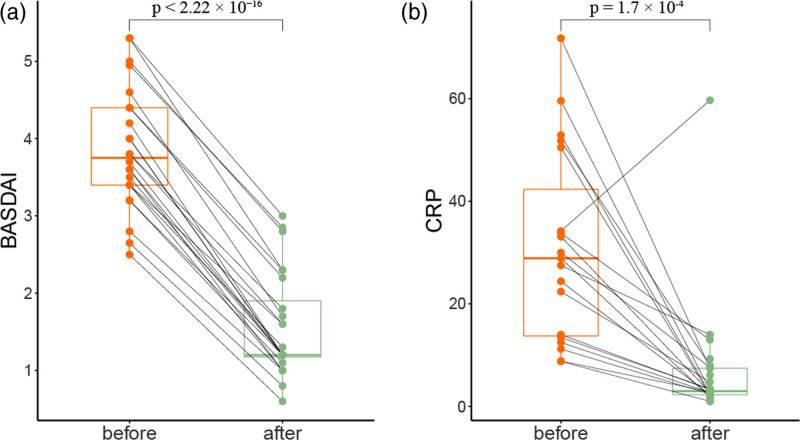
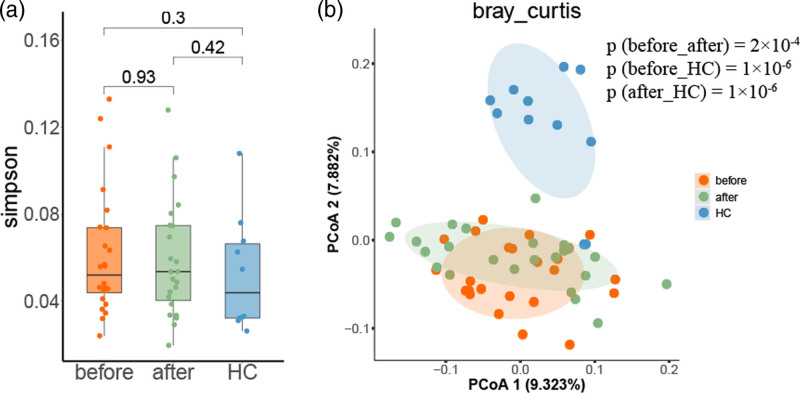
We collected fecal samples from 11 age/sex-matched healthy controls and 24 ankylosing spondylitis patients (Table 1 and Supplementary Table 1, Supplemental digital content 1, http://links.lww.com/FPC/B422). All ankylosing spondylitis patients had a significantly positive response to anti-TNF-α treatment in terms of decreased disease activity score BASDAI and C-reactive protein (CRP) level (Fig. 1 and Table 1), indicating satisfactory therapeutic effect of TNFi on ankylosing spondylitis. Subsequently, 16S ribosomal DNA amplicon sequencing was conducted to profile the gut microbiota with adequate sequencing depth (Supplementary Figure 1, Supplemental digital content 2, http://links.lww.com/FPC/B423). A total of 4 384 401 unique sequences of all samples were obtained and clustered into 1246 OTUs based on 97% similarity. Although the species richness of microbial communities exhibited no significant difference among groups in terms of α-diversity (Fig. 2a), the compositions of the gut microbiome of three sample groups were clustered separately in terms of β-diversity (Fig. 2b), and anti-TNF-α treatment shifted the microbial compositions of ankylosing spondylitis patients toward those of healthy controls.
Table 1.
Clinical characteristics of ankylosing spondylitis patients and healthy controls
| Feature | Ankylosing spondylitis patients | Control | P value |
|---|---|---|---|
| (n = 24) | (n = 11) | ||
| Mean ± SD | |||
| Age (years) at recruitment | 32.3 ± 10.5 | 35.1 ± 11.1 | 0.48 |
| Collection interval (days) | 41.0 ± 17.8 | NA | NA |
| BASDAI level | |||
| Pretreatment | 3.86 ± 0.78 | NA | <0.0001 |
| Post-treatment | 1.56 ± 0.68 | ||
| CRP level (mg/L) | |||
| Pretreatment | 31.0 ± 18.6 | NA | 0.0002 |
| Post-treatment | 6.8 ± 11.8 | ||
| No. (%) of patients | |||
| Gender | |||
| Female | 2 (8.3) | 2 (18.2) | 0.57 |
| Male | 22 (91.7) | 9 (81.8) | |
| Treatment strategy | |||
| Home | 12 (50) | NA | |
| Clinic | 12 (50) | NA | |
| Drug | |||
| Antibody | 7 (29.2) | NA | |
| Recombinant fusion protein | 17 (70.8) | NA | |
P value was estimated by comparing AS patients with controls or pretreatment with posttreatment.
BASDAI, Bath AS disease activity index; CRP, C-reactive protein; NA, not applicable.
Fig. 1.
Impact of anti-TNF-α treatment on the disease activity of ankylosing spondylitis. (a) BASDAI and (b) CRP scores were significantly downregulated after anti-TNF-α treatment. A paired t-test was performed to estimate the statistical significance. BASDAI, Bath AS disease activity index; CRP, C-reactive protein; TNF, tumor necrosis factor.
Fig. 2.
Microbiota diversity of ankylosing spondylitis patients and healthy controls. (a) α-diversity of microbiota was illustrated by the Simpson index; (b) β-diversity of microbiota calculated by Bray-Curtis was illustrated in three groups. HC, healthy control; before and after, before and after anti-TNF-α treatment. TNF, tumor necrosis factor.
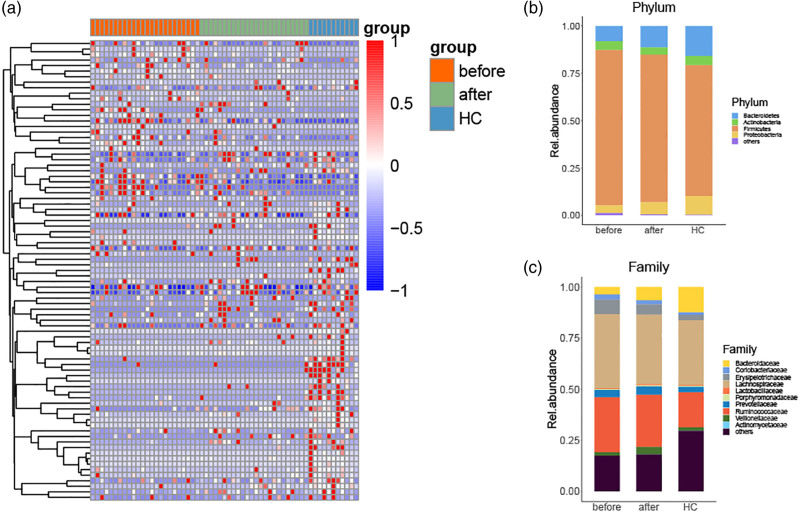
Furthermore, we estimated the significantly altered components of the gut microbiota among the three groups, identifying a total of 330 differently distributed OTUs. Considering the possible involvement of microbiota in ankylosing spondylitis development and treatment response, we focused on the 83 OTUs that exhibited gradually changed compositions in the order of pretreatment, post-treatment of ankylosing spondylitis patients and healthy controls (Fig. 3a). After annotation with the reference sequence of bacteria, we observed that anti-TNF-α treatment restored the relative abundance of the microbiota profile, including increased Bacteroidetes and Proteobacteria at the phylum level and decreased Erysipelotrichaceae, Lachnospiraceae and Ruminococcaceae at the family level (Fig. 3b,c).
Fig. 3.
Different components and taxonomic analysis of fecal microbiota in ankylosing spondylitis patients. (a) Heatmap of significant differential OTUs among the three groups. (b and c) Overall composition of bacterial microbiota at the phylum and family levels. OTU, operational taxonomic unit.
Specific microbiota components and pathways involved in ankylosing spondylitis onset and treatment outcomes
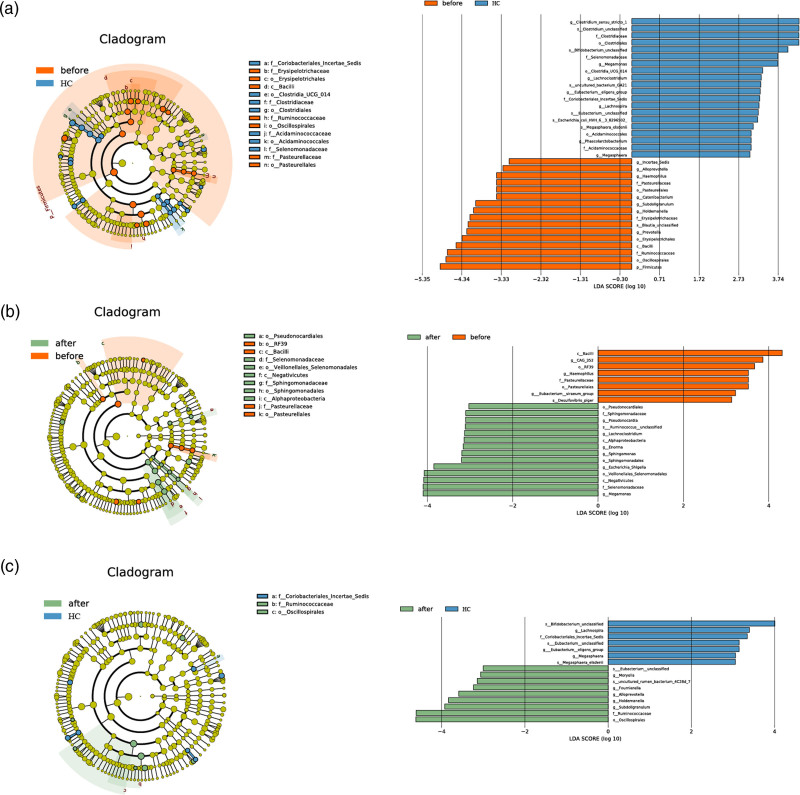
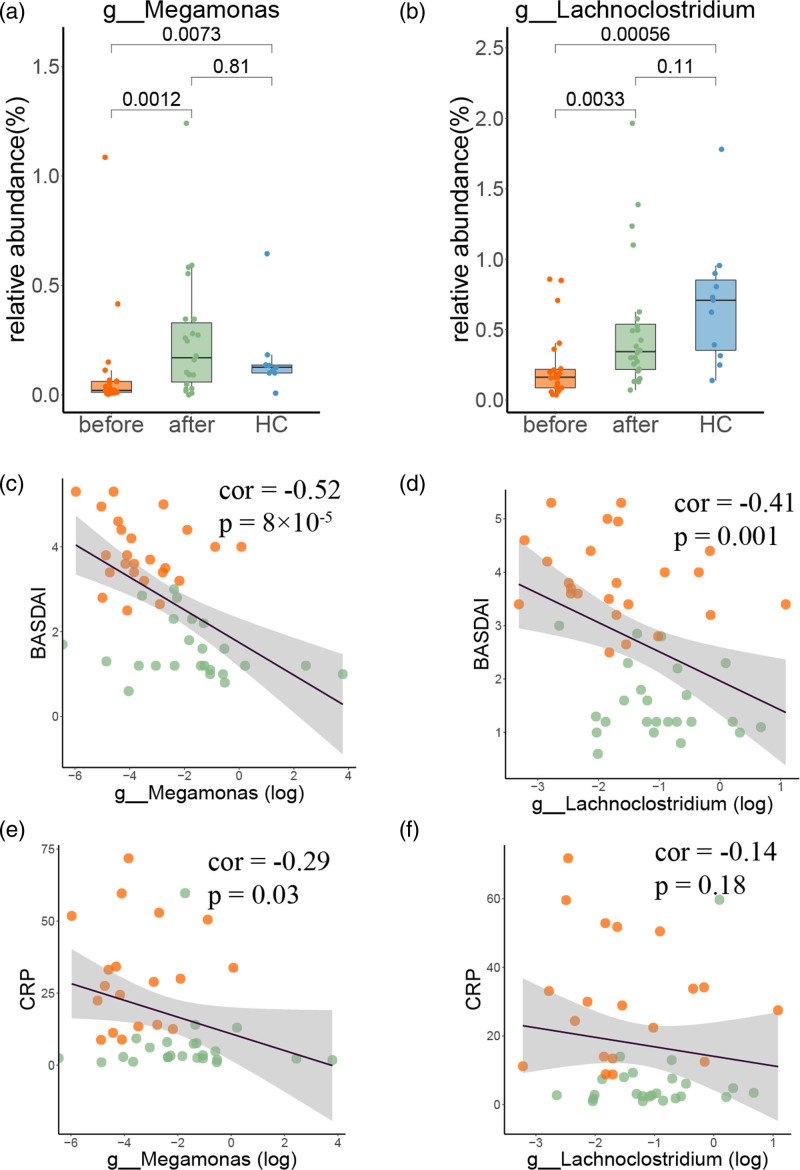
With LefSe discriminant analysis, we first compared the microbiota compositions of pretreated ankylosing spondylitis patients with those of healthy controls (before vs. healthy control), and subsequently, the differences in pre- and post-treatment groups were identified (before vs. after). Several unique bacteria (log LDA >3) in each group were identified. For instance, four species were filtered out by taking the intersection of two results of LefSe analysis: Bacilli phylum and Haemophilus genus were enriched in the pretreatment samples, and Megamonas and Lachnoclostridium genus were enriched in the post-treatment and healthy controls (Fig. 4). Intriguingly, the healthy controls were enriched in short-chain fatty acid (SCFA)-producing bacteria, such as Bifidobacterium, Megamonas, Lachnoclostridium, Lachnospira and Megasphaera (Fig. 4a and Supplementary Table 2, Supplemental digital content 3, http://links.lww.com/FPC/B424), and some of these bacteria (e.g. Megamonas and Lachnoclostridium) were significantly restored in post-treatment samples (Fig. 4b and Supplementary Table 3, Supplemental digital content 4, http://links.lww.com/FPC/B425), suggesting the possible involvement of SCFAs in the pathogenesis of ankylosing spondylitis as well as treatment outcomes. Not surprisingly, the SCFA-producing bacteria (i.e. Megamonas and Lachnoclostridium) no longer exhibited a significant difference between the post-treatment group and the healthy group (Fig. 4c and Supplementary Table 3, Supplemental digital content 4, http://links.lww.com/FPC/B425). These results illustrated that the microbiota dysbiosis of ankylosing spondylitis patients was restored to normal conditions after anti-TNF-α treatment, particularly SCFA-producing bacteria.
Fig. 4.
Microbiota differences among three groups. LefSe analysis illustrated the top differed microbiota components between pretreatment group (before) vs. healthy control (a), pretreatment group vs. posttreatment group (after) (b), and post-treatment group (after) vs. healthy control (c). log LDA >3 was defined as the cutoff for the identification of unique bacteria.
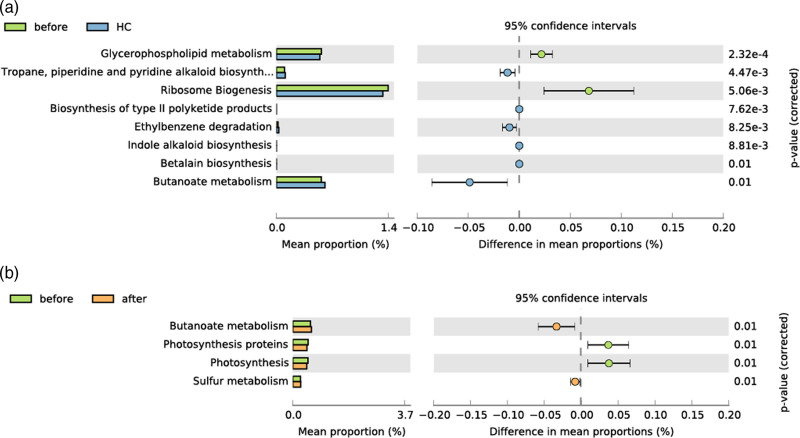
Next, we performed kyoto encyclopedia of genes and genomes (KEGG) pathway prediction with PICRUSt, revealing that a total of eight differential pathways were significantly altered in the pretreatment group vs. healthy controls, compared with four in the pretreatment group vs. the post-treatment group and 0 in the post-treatment group vs. healthy controls (Fig. 5), indicating that the functional profile of ankylosing spondylitis patients was restored to normal levels. In particular, the butyrate metabolism pathway overlapped between the two PICRUSt analysis results (before vs. after and before vs. healthy control) (Fig. 5), which is worthy of attention due to the strong effect of butyrate on immunomodulation and inflammation.
Fig. 5.
Functional prediction of altered pathways among the three groups. Pairwise PICRUSt analysis illustrated the top affected pathways between the (a) pretreatment group (before) vs. healthy control, (b) and pretreatment group vs. post-treatment group (after). The results were considered to be statistically significant with P < 0.01.
Correlation of microbiota components with disease activity
The correlation between the abundance of the unique microbiota components and disease activity was evaluated. In particular, the SCFA-producing bacteria described above (i.e. Megamonas and Lachnoclostridium genera), which were restored in post-treatment group (Fig. 6a,b), were significantly negatively correlated with the BASDAI score (Fig. 6c,d). The abundance of Megamonas may have a stronger effect than that of Lachnoclostridium, consistent with their association with the CRP score (Fig. 6e,f). Moreover, several microbiota components were enriched in ankylosing spondylitis patients and restored to healthy controls after anti-TNF-α treatment, including Bacilli and Haemophilus (Supplementary Figure 2A and B, Supplemental digital content 2, http://links.lww.com/FPC/B423). Intriguingly, Haemophilus was positively associated with disease activity (Supplementary Figure 2D and F, Supplemental digital content 2, http://links.lww.com/FPC/B423), suggesting that other gut microbiota-related factors may also be involved in ankylosing spondylitis onset and anti-TNF-α treatment outcomes.
Fig. 6.
Relative abundance of unique bacteria and their correlation with disease activity. (a and b) Relative abundances of Megamonsa and Lachnoclostridium in the three groups. Spearman’s rank correlation between the relative abundance of each bacterium with BASDAI (c and d) and CRP score (e and f). BASDAI, Bath ankylosing spondylitis disease activity index; CRP, C-reactive protein.
Discussion
Gut inflammation and damaged integrity of the intestinal mucosa are commonly observed in patients with ankylosing spondylitis. Proinflammatory factors in the gut, such as IL-17, IL-23 or pathogenic bacteria, can invade the blood circulation through the inflamed intestinal mucosa and stimulate the host immune system. Subsequently, activated immune cells can produce several inflammatory factors (e.g. TNF and IL-17), which accumulate in the joint cavity and trigger inflammation [5]. The imbalance of gut microbiota has been well reported in patients with ankylosing spondylitis [7,16,17,43], and there was an obvious comorbidity between ankylosing spondylitis and gut inflammation. TNF-α inhibitors, second-line clinical agents for ankylosing spondylitis, have been strongly effective in patients who were resistant or intolerant to standard treatment [25]. Although the TNF-α level of the host had a conversely regulatory effect on the gut microbiota [37], it is not clear whether TNFi could regulate the gut microbiota of ankylosing spondylitis patients or further exert a therapeutic effect by restoring the gut microbiota. Therefore, we aimed to link the relationship between the effect of TNFi on ankylosing spondylitis and the gut microbiota in this study. To the best of our knowledge, no other study has performed a longitudinal investigation on the correlation of anti-TNF-α treatment with the microbiota profile, particularly in ankylosing spondylitis patients.
After profiling the microbiota of healthy controls and patients before/after anti-TNF-α treatment, we noticed that the compositions of gut microbiota in ankylosing spondylitis patients were significantly different from those in healthy controls, which was consistent with previous reports [7,17]. The composition of some specific bacteria altered in ankylosing spondylitis patients can be restored to healthy controls after anti-TNF-α treatment, particularly SCFA-producing bacteria. For instance, Megamonas, a propionate-producing bacteria, was obviously upregulated after anti-TNF-α treatment and negatively correlated with the disease activity score BASDAI. Intriguingly, a significantly decreased abundance of Megamonas was also found in an animal model of IBD [44], which exhibited obvious clinical comorbidity with ankylosing spondylitis [7–9]. In addition, Megamonas belongs to the Negativicutes class, the abundance of which increased to normal levels after anti-TNF-α treatment (Supplementary Figure 3, Supplemental digital content 2, http://links.lww.com/FPC/B423) and was consistently downregulated in ankylosing spondylitis patients according to a previous report [16,45]. Taken together, Megamonas may be one of the key bacterial genera involved in the development of ankylosing spondylitis and the therapeutic efficacy of TNFi.
In particular, we noticed that SCFA-producing bacteria were enriched in healthy controls and ankylosing spondylitis patients after treatment. As one of the major bacterial metabolites, SCFAs play an indispensable role in host and gut immune homeostasis. For instance, SCFAs can induce the differentiation of regulatory T (Treg) cells [46] and increase the secretion of IL-10 family factors and other anti-inflammatory cytokines from several types of T cells, including Tregs and CD4+ Th cells [46,47]. Therefore, SCFAs may have a positive therapeutic effect on ankylosing spondylitis, which is an autoimmune inflammatory disease. SCFAs can attenuate HLA-B27-related inflammation [23], which is a risk factor for ankylosing spondylitis susceptibility [5,6], further suggesting the possible interaction between germline variants and microbiota on ankylosing spondylitis onset. In particular, propionate produced by Megamonas is an important member of the SCFA family and can reduce TNF-α levels in a dose-dependent manner [48]. A study has shown that propionate and butyrate can lower the abundance of osteoclasts in the joints of mice and alleviate arthritis symptoms [49], suggesting that Megamonas may play an essential role in ankylosing spondylitis through its metabolite propionate. In the present study, TNFi increased the abundance of SCFA-producing bacteria (e.g. Megamonas and Lachnoclostridium), and the abundance of these bacteria was negatively correlated with disease severity, which supported the possible impact of microbiota on ankylosing spondylitis through their metabolites (e.g. SCFAs). Speculatively, TNFi may upregulate SCFA levels by increasing the abundance of SCFA-producing bacteria, and SCFAs thus enter the blood circulation and induce immune cells to secrete more anti-inflammatory cytokines and finally alleviate the severity of ankylosing spondylitis.
Several limitations of our study should be clarified. First, only relatively small samples were included in our study due to the difficulty of obtaining longitudinal fecal samples from the same patients at specific time points, thus requiring validation in independent cohorts. Second, the fecal samples were all used to perform 16S sequencing, and SCFA levels could not be accurately evaluated with the leftover due to the rapid degradation of SCFAs. Therefore, more evidence is required to establish the crucial role of SCFAs in ankylosing spondylitis and determine whether TNFis can exhibit a therapeutic role in ankylosing spondylitis by regulating SCFA-producing bacteria.
Collectively, we observed dysfunction of the microbiota in ankylosing spondylitis patients, characterized by a reduced abundance of SCFA-producing bacteria. Meanwhile, TNFi can restore the microbiota dysbiosis of ankylosing spondylitis patients to healthy controls. The abundance of SCFA-producing bacteria, such as Megamonsa and Lachnoclostridium, was significantly increased and negatively correlated with disease severity. We have also proposed that TNFi may upregulate SCFA levels by increasing the abundance of SCFA-related gut bacteria, which in turn modulate host immunity and alleviate ankylosing spondylitis severity. Exploring the potential mechanistic role of gut microbiota in ankylosing spondylitis and corresponding anti-TNF-α treatment is largely required in the future.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81973408), National key research development program of China (No. 2021YFA1301203), Sichuan Science & Technology Program (No. 2020YJ0094, 2020YJ0105, 2021YJ0475) and 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (No. ZYJC21024, ZYYC20003, ZYJC18004, ZYJC18035, ZYYC20007).
H.X. designed the study; Q.H.D., X.Y.X., Y.S. and H.X. analyzed and interpreted the data; C.J.H., Y.P.H., Y.D.C., Y.W., Y.H.C. collected the clinical materials and information; W.Z., G.Y. and Q.B.X. provided technical and material support; H.X., G.Y., and Q.B.X. surprised this study. All authors contributed to the writing of the manuscript and final approval.
Most data supporting the findings of this study are available within the article and Supplementary Table, Supplement digital content 1, http://links.lww.com/FPC/B422. The raw sequencing data from this study have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, under accession numbers CRA005495 (BioProject: PRJCA007396) that can be accessed at https://bigd.big.ac.cn/gsa/browse/CRA005495.
Supplementary Material
Footnotes
Mr. Qinghong Dai, Dr. Xuyang Xia and Dr. Chenjia He contributed equally to the writing of this article.
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.pharmacogeneticsandgenomics.com.
Reference
- 1.Sieper J, Poddubnyy D. Axial spondyloarthritis. Lancet 2017; 390:73–84. [DOI] [PubMed] [Google Scholar]
- 2.Rashid T, Ebringer A. Ankylosing spondylitis is linked to Klebsiella–the evidence. Clin Rheumatol 2007; 26:858–864. [DOI] [PubMed] [Google Scholar]
- 3.Ng SC, Liao Z, Yu DT, Chan ES, Zhao L, Gu J. Epidemiology of spondyloarthritis in the People’s Republic of China: review of the literature and commentary. Semin Arthritis Rheum 2007; 37:39–47. [DOI] [PubMed] [Google Scholar]
- 4.Zeng QY, Chen R, Darmawan J, Xiao ZY, Chen SB, Wigley R, et al. Rheumatic diseases in China. Arthritis Res Ther 2008; 10:R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown MA, Kenna T, Wordsworth BP. Genetics of ankylosing spondylitis–insights into pathogenesis. Nat Rev Rheumatol 2016; 12:81–91. [DOI] [PubMed] [Google Scholar]
- 6.Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, Leo P, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet 2013; 45:730–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costello ME, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B, et al. Brief report: intestinal dysbiosis in ankylosing spondylitis. Arthritis Rheumatol 2015; 67:686–691. [DOI] [PubMed] [Google Scholar]
- 8.Ciccia F, Rizzo A, Triolo G. Subclinical gut inflammation in ankylosing spondylitis. Curr Opin Rheumatol 2016; 28:89–96. [DOI] [PubMed] [Google Scholar]
- 9.Van Praet L, Van den Bosch FE, Jacques P, Carron P, Jans L, Colman R, et al. Microscopic gut inflammation in axial spondyloarthritis: a multiparametric predictive model. Ann Rheum Dis 2013; 72:414–417. [DOI] [PubMed] [Google Scholar]
- 10.Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B, et al. ; International IBD Genetics Consortium (IIBDGC); International Genetics of Ankylosing Spondylitis Consortium (IGAS); International PSC Study Group (IPSCSG); Genetic Analysis of Psoriasis Consortium (GAPC); Psoriasis Association Genetics Extension (PAGE). Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016; 48:510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. ; International IBD Genetics Consortium (IIBDGC). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012; 491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 2013; 14:661–673. [DOI] [PubMed] [Google Scholar]
- 13.Pronovost GN, Hsiao EY. Perinatal Interactions between the Microbiome, Immunity, and Neurodevelopment. Immunity 2019; 50:18–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen B, Sun L, Zhang X. Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases. J Autoimmun 2017; 83:31–42. [DOI] [PubMed] [Google Scholar]
- 15.Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 2017; 152:327–339.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Dai B, Tang Y, Lei L, Li N, Liu C, et al. Altered bacterial-fungal interkingdom networks in the guts of ankylosing spondylitis patients. mSystems 2019; 4:e00176–e00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen C, Zheng Z, Shao T, Liu L, Xie Z, Le Chatelier E, et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol 2017; 18:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernández-Sueiro JL, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med 1994; 180:2359–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranganathan V, Gracey E, Brown MA, Inman RD, Haroon N. Pathogenesis of ankylosing spondylitis - recent advances and future directions. Nat Rev Rheumatol 2017; 13:359–367. [DOI] [PubMed] [Google Scholar]
- 20.Berthelot JM, Claudepierre P. Trafficking of antigens from gut to sacroiliac joints and spine in reactive arthritis and spondyloarthropathies: mainly through lymphatics? Joint Bone Spine 2016; 83:485–490. [DOI] [PubMed] [Google Scholar]
- 21.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 2011; 9:279–290. [DOI] [PubMed] [Google Scholar]
- 22.Rosenbaum JT, Davey MP. Time for a gut check: evidence for the hypothesis that HLA-B27 predisposes to ankylosing spondylitis by altering the microbiome. Arthritis Rheum 2011; 63:3195–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asquith M, Davin S, Stauffer P, Michell C, Janowitz C, Lin P, et al. Intestinal metabolites are profoundly altered in the context of HLA-B27 expression and functionally modulate disease in a rat model of spondyloarthritis. Arthritis Rheumatol 2017; 69:1984–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill T, Asquith M, Brooks SR, Rosenbaum JT, Colbert RA. Effects of HLA-B27 on gut microbiota in experimental spondyloarthritis implicate an ecological model of dysbiosis. Arthritis Rheumatol 2018; 70:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Heijde D, Ramiro S, Landewé R, Baraliakos X, Van den Bosch F, Sepriano A, et al. 2016 update of the ASAS-EULAR management recommendations for axial spondyloarthritis. Ann Rheum Dis 2017; 76:978–991. [DOI] [PubMed] [Google Scholar]
- 26.van der Heijde D, Sieper J, Maksymowych WP, Dougados M, Burgos-Vargas R, Landewé R, et al. ; Assessment of SpondyloArthritis international Society. 2010 Update of the international ASAS recommendations for the use of anti-TNF agents in patients with axial spondyloarthritis. Ann Rheum Dis 2011; 70:905–908. [DOI] [PubMed] [Google Scholar]
- 27.Baraliakos X, Listing J, Fritz C, Haibel H, Alten R, Burmester GR, et al. Persistent clinical efficacy and safety of infliximab in ankylosing spondylitis after 8 years–early clinical response predicts long-term outcome. Rheumatology (Oxford) 2011; 50:1690–1699. [DOI] [PubMed] [Google Scholar]
- 28.Sedger LM, McDermott MF. TNF and TNF-receptors: from mediators of cell death and inflammation to therapeutic giants - past, present and future. Cytokine Growth Factor Rev 2014; 25:453–472. [DOI] [PubMed] [Google Scholar]
- 29.Sfikakis PP. The first decade of biologic TNF antagonists in clinical practice: lessons learned, unresolved issues and future directions. Curr Dir Autoimmun 2010; 11:180–210. [DOI] [PubMed] [Google Scholar]
- 30.Ward MM, Deodhar A, Akl EA, Lui A, Ermann J, Gensler LS, et al. American College of Rheumatology/Spondylitis Association of America/Spondyloarthritis Research and Treatment Network 2015 Recommendations for the Treatment of Ankylosing Spondylitis and Nonradiographic Axial Spondyloarthritis. Arthritis Rheumatol 2016; 68:282–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 2001; 104:487–501. [DOI] [PubMed] [Google Scholar]
- 32.Braun J, Bollow M, Neure L, Seipelt E, Seyrekbasan F, Herbst H, et al. Use of immunohistologic and in situ hybridization techniques in the examination of sacroiliac joint biopsy specimens from patients with ankylosing spondylitis. Arthritis Rheum 1995; 38:499–505. [DOI] [PubMed] [Google Scholar]
- 33.Braun J, Brandt J, Listing J, Zink A, Alten R, Golder W, et al. Treatment of active ankylosing spondylitis with infliximab: a randomised controlled multicentre trial. Lancet 2002; 359:1187–1193. [DOI] [PubMed] [Google Scholar]
- 34.Redlich K, Görtz B, Hayer S, Zwerina J, Kollias G, Steiner G, et al. Overexpression of tumor necrosis factor causes bilateral sacroiliitis. Arthritis Rheum 2004; 50:1001–1005. [DOI] [PubMed] [Google Scholar]
- 35.De T, Zhang H, Alarcon C, Lec B, Avitia J, Smithberger E, et al. Genetic association of primary nonresponse to anti-TNFα therapy in patients with inflammatory bowel disease. Pharmacogenet Genomics 2022; 32:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magnusson MK, Strid H, Sapnara M, Lasson A, Bajor A, Ung KA, Öhman L. Anti-TNF therapy response in patients with ulcerative colitis is associated with colonic antimicrobial peptide expression and microbiota composition. J Crohns Colitis 2016; 10:943–952. [DOI] [PubMed] [Google Scholar]
- 37.Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe 2017; 21:455–466.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Ren Z, Su Z, Liu Y, Yang T, Cao M, et al. Novel recurrent altered genes in chinese patients with anaplastic thyroid cancer. J Clin Endocrinol Metab 2021; 106:988–998. [DOI] [PubMed] [Google Scholar]
- 39.Chen HN, Shu Y, Liao F, Liao X, Zhang H, Qin Y, et al. Genomic evolution and diverse models of systemic metastases in colorectal cancer. Gut 2022; 71:322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo H, Xia X, Kim GD, Liu Y, Xue Z, Zhang L, et al. Characterizing dedifferentiation of thyroid cancer by integrated analysis. Sci Adv 2021; 7:eabf3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou B, Xia X, Wang P, Chen S, Yu C, Huang R, et al. Induction and amelioration of methotrexate-induced gastrointestinal toxicity are related to immune response and gut microbiota. EBioMedicine 2018; 33:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu C, Zhou B, Xia X, Chen S, Deng Y, Wang Y, et al. Prevotella copri is associated with carboplatin-induced gut toxicity. Cell Death Dis 2019; 10:714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breban M, Tap J, Leboime A, Said-Nahal R, Langella P, Chiocchia G, et al. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis 2017; 76:1614–1622. [DOI] [PubMed] [Google Scholar]
- 44.Maldonado-Contreras A, Ferrer L, Cawley C, Crain S, Bhattarai S, Toscano J, et al. Dysbiosis in a canine model of human fistulizing Crohn’s disease. Gut Microbes 2020; 12:1785246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. New insights from uncultivated genomes of the global human gut microbiome. Nature 2019; 568:505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016; 165:1332–1345. [DOI] [PubMed] [Google Scholar]
- 47.Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun 2020; 11:4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vinolo MA, Rodrigues HG, Hatanaka E, Sato FT, Sampaio SC, Curi R. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J Nutr Biochem 2011; 22:849–855. [DOI] [PubMed] [Google Scholar]
- 49.Lucas S, Omata Y, Hofmann J, Böttcher M, Iljazovic A, Sarter K, et al. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat Commun 2018; 9:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.