Abstract
The interactions between trypsin and gallic acid (GA) were investigated by means of fluorescence spectroscopy, UV-vis absorption spectroscopy, resonance light scattering (RLS) spectroscopy, synchronous fluorescence spectroscopy, and enzymatic inhibition assay. It was found that GA can cause the fluorescence quenching of trypsin during the process of formation of GA-trypsin complex, resulting in inhibition of trypsin activity (IC50 = 3.9 × 10−6 mol/L). The fluorescence spectroscopic data showed that the quenching efficiency can reach about 80%. The binding constants were 1.9371 × 104 L/mol, 1.8192 × 104 L/mol, and 1.7465 × 104 L/mol at three temperatures, respectively. The thermodynamic parameters revealed that hydrogen bonds, van der Waals, hydrophobic, and electrostatic interactions were involved in the binding process of GA to trypsin. Molecular modeling studies illustrated a specific display of binding information and explained most of the experiment phenomena. The microenvironments of tryptophan and tyrosine residue in trypsin were changed by the GA. Results indicated that GA was a strong quencher and inhibitor of trypsin.
Keywords: gallic acid, interaction, spectroscopy, trypsin
1. Introduction
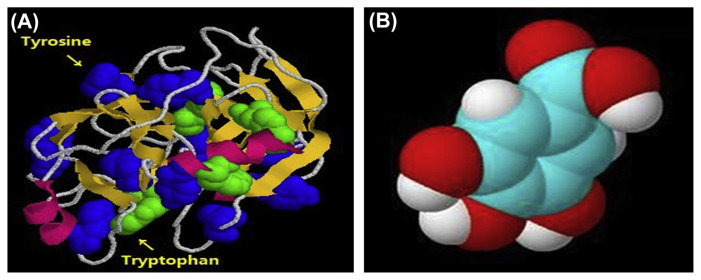
In recent years, there has been much attention to the interaction of small molecules with proteinases [1–3], which is related to understanding the medicine, chemistry, food science, toxicology, and biology [4]. Trypsin (EC 3.4.21.4) is an important protease in the human digestive system and plays an important role in many physiological processes [5]. Therefore, studies on the interactions of trypsin with small molecules have great significance in understanding the inhibition mechanisms of the small molecules [6]. The structure of trypsin is shown in Fig. 1A. Fruits, health drinks, and functional beverages are the main sources of the small molecules entering the human body, which influence the trypsin activity in vivo [7,8]. Several reports about interactions between trypsin and several proteinase inhibitors have been published, such as cinnamic acid [9], methotrexate [10], caffeine, and theophylline [11]. However, there was no report on the interactions between polyphenol and trypsin.
Fig. 1.
The molecule structure of trypsin (A) and gallic acid (GA) (B).
Gallic acid (3,4,5-trihydroxybenzoic acid, GA) is one of the natural occurring polyphenolic compounds in food, biological, pharmaceutical, chemical, and other fields [12]. The structure of the GA is shown in Fig. 1B. Reports confirmed the restoration of lysosomal damage, myocardial damage, and lipid peroxidation induced by isoproterenol during GA treatment [13]. GA shows cytotoxicity against cancer cells without damaging normal cells [14]. GA can be used to treat diabetes and albuminuria, and also exhibits a variety of pharmacologic properties such as antioxidant, antitumor, antiinflammatory, antibacterial, and antiviral activities [15–22]. However, there has not been enough attention paid to the interaction of GA with trypsin by spectroscopy.
The use of fluorescence quenching is simple and sensitive. Therefore, it is an important method to study the interaction between small molecules and proteinases [23–25]. Fluorescence spectroscopy can not only explain the result of the experiment, but also obtain dynamic information and special structure [26,27]. In this paper, the effects of GA on the structure and activity of trypsin and the interaction mechanism between GA and trypsin were studied by UV-vis absorption spectroscopy, fluorescence spectroscopy, synchronous fluorescence spectroscopy and resonance light scattering (RLS). The data provided information on the influence of the binding constant, binding distance, binding force, and thermodynamic aspects in the binding process.
2. Materials and methods
2.1. Apparatus
All the fluorescence spectra were recorded on an LS-50 fluorescence spectrophotometer (Perkinelmer Corp., London, UK) equipped with 3 cm quartz cell. UV-vis absorption spectra were measured at room temperature with an Ultrospec 2100 pro spectrophotometer (General Electric Company, London, UK) equipped with 3 cm quartz cell. The widths of the excitation and the emission slits of trypsin were 5.0 nm and 5.0 nm, respectively.
2.2. Reagents
The trypsin (Sigma Aldrich, New York, USA) was obtained from the USA and the GA (Guizhou Dida Biological Technolog Co., Ltd, Guiyang, China) was purchased from Guizhou Dida Biological Technology Co., Ltd. The trypsin solution (4 × 10−5 mol/L) was prepared in pH 7.40 phosphate buffer. GA solution was prepared in deionized water. Other reagents were analytical grade and deionized water was used throughout. All solution was stored at 4°C in a refrigerator.
2.3. Fluorescence measurements
The trypsin (2 mL, 4 × 10−5 mol/L) solution and 2 mL different concentration of GA were added into a 10 mL volumetric flask, and phosphate buffer was added to dilute the mixture to the scale mark. The fluorescence intensities of all solutions were measured (excitation at 280 nm and emission wavelengths of 300–500 nm) with a, LS-50 fluorescence spectrophotometer at three temperatures (25°C, 31°C, 37°C). Synchronous fluorescence spectra were recorded with λex = 280 nm. The D-value (Δλ) was 15 nm and 60 nm, respectively.
2.4. UV-vis absorbance measurement
The UV-vis absorbance spectra of GA and trypsin were recorded at 298 K. Trypsin of fixed concentration in the absence and presence of GA was added to the test tube.
2.5. Study of trypsin activity
The catalysis activity of trypsin was determined by the modified Lowry assay. Specific methods of operation were 0.5% casein, GA of different concentrations, and 4 × 10−5 mol/L trypsin solution preheated for 5 minutes. GA (2 mL) and casein (2 mL) solutions were added to 2 mL trypsin solution. The reaction kept for 10 minutes at 40°C (accurate to seconds). An amount of 4 ml 0.4 mol/L Trichlooacetic acid was added to the mixture to end the reaction. They were then kept for 20 minutes at 40°C. The precipitate was removed by filtration. Amounts of 1 mL supernatant, 5 mL 0.4 mol/L Na2CO3, and 1 mL Folin-phenol reagent were mixed and colored for 20 minutes at 40°C. The absorbance of the mixture was measured at a wavelength of 680 nm. No GA-mixed trypsin was taken as the blank group. Trichloroacetic acid solution was added to the trypsin solution to make the trypsin inactive. Then, the casein solution was added to the mixture as the control group. Each experiment group had a control group. The inhibition rate was calculated as follows:
| (1) |
where A and a were the absorbance of the blank group and the experimental group, respectively.
2.6. Molecular modeling study
The 3D structure of trypsin (PDB ID: 4I8G) was downloaded from Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). AutoDock Vina 1.1.2 was used for all docking calculations. The AutoDockTools 1.5.6 package (http://mgltools.scripps.edu) was employed to generate the docking input files. The search grid of trypsin was identified as center_x: −1.793, center_y: −14.925, and center_z: −16.941 with dimensions size_x: 16, size_y: 16, and size_z: 16. The value of exhaustiveness was set to 20. For Vina docking, the default parameters were used if it was not mentioned. The best-scoring pose as judged by the Vina docking score was chosen and visually analyzed using PyMOL (http://www.pymol.org/).
3. Results and discussion
3.1. UV-vis absorption spectra
UV-vis absorption is a simple and effective method to research the change of structure and the formation of a complex [28]. The change of microenvironment around the chromophore in trypsin can result in the change of absorption spectra about protein. So, the UV-vis absorption spectra of protein can be used for studying the changes of protein structure in solution. The absorption spectral changes of trypsin in the presence and absence of drugs are shown in Fig. 2. There was an absorption peak at 290 nm, which was related to the aromatic amino acid absorption. With the addition of GA, the peaks of trypsin increased and a red shift was observed. The results indicated the formation of a new complex between GA and trypsin. Fig. 2 shows different spectra between trypsin, GA, and GA-trypsin. If no interaction happened between trypsin and GA, the spectra of trypsin and (gallic acid + trypsin)-gallic acid should be the same. The results illustrated that there was a change in the conformation of trypsin and the microenvironment around the tryptophan residues when GA was mixed. GA influenced the functional groups and exposed the chromophore in the trypsin molecules to the surfaces of molecules.
Fig. 2.
The UV absorption spectra of gallic acid (GA) bound to trypsin at pH 7.4. C(GA) = 1 × 10−5 mol/L, C(trypsin) = 1.5 × 10−5 mol/L.
3.2. Fluorescence quenching of trypsin
The method of fluorescence can reveal some information about binding between small molecules and proteins [29]. Due to the fluorescent properties of tryptophan and tyrosine in trypsin, tryptophan, and tyrosine can be regarded as probes to detect the conformation change of trypsin. Tryptophan and tyrosine residues are excited when the excitation wavelength is at about 280 nm. The fluorescence intensity and position are related to the microenvironment of these residues [30].
Fluorescence quenching is a phenomenon of interaction between fluorescent substances and other molecules [31]. The mechanisms of fluorescence quenching include dynamic quenching and static quenching [32].
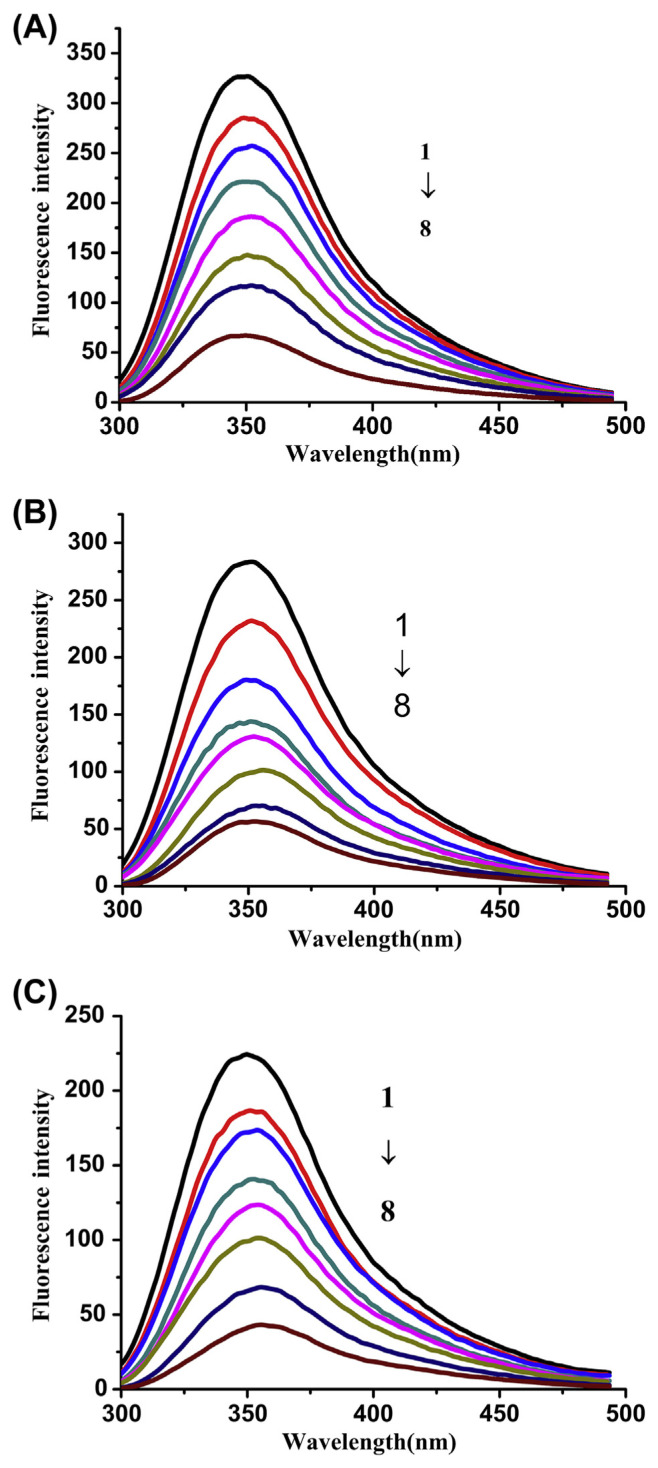
The influence of GA on fluorescence intensity of trypsin was investigated. The GA is nonfluorescent. The trypsin fluorescence peak was at about 350 nm. An obvious decrease of trypsin fluorescence intensity was observed when GA was added into the solution as shown in Fig. 3A, B, and C. The quenching efficiencies were 80.75%, 80.04%, and 79.48%, respectively. The data of fluorescence were analyzed by the Stern-Volmer equation:
Fig. 3.
Effect of gallic acid (GA) on fluorescence spectra of trypsin (pH =7.4, λex = 280 nm) at different temperatures (A: 25 °C, B: 31 °C, and C: 37 °C), with the trypsin concentration of 4 × 10−5 mol/L, and GA concentration of 0 mol/L, 1.0 × 10−5 mol/L, 2.0 × 10−5 mol/L, 3.0 × 10−5 mol/L, 4.0 × 10−5 mol/L, 5.0 × 10−5 mol/L, 6.0 × 10−5 mol/L, and 8.0 × 10−5 mol/L, respectively (from spectra 1 to spectra 8).
| (2) |
where F0 and F are the trypsin fluorescence intensities before and after GA is added into solution, respectively, Kq is the quenching rate constant of trypsin [33], τ0 is the fluorescent life of trypsin in the presence of quencher and taken as 10−8 [34], Ksv is the Stem-Volmer quenching constant, and [Q] is the concentration of GA. Kq and Ksv can be determined by mapping for F0/F and concentration of GA.
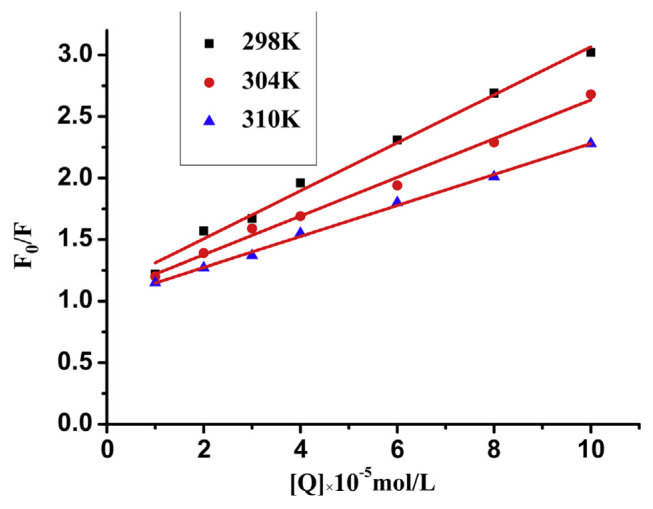
Fig. 4 shows the Stern-Volmer plots of the fluorescence quenching of trypsin by GA. Good linearity appeared in the curves. Linear Stern-Volmer plots may reveal the occurrence of a single type of quenching [35]. For dynamic quenching, the maximum scatter collision quenching rate constant of various quenchers is 2.0 × 1010 L/mol/s [36]. In this paper, Ksv and Kq values at different temperatures are shown in Table 1. In this table, Ksv was negatively correlated with temperature, which revealed that the quenching mechanism between trypsin and GA was due to the formation of a GA-trypsin complex. In addition, all the values of Kq are greater than the maximum collisional quenching constant of various quenchers for biological macromolecules (2.0 × 1010 L/mol/s). Therefore, these results indicated that the quenching mechanism of trypsin with GA was initiated by the formation of a GA-trypsin complex.
Fig. 4.
The Stern-Volmer plots for the fluorescence quenching of trypsin by gallic acid (GA). The concentration of trypsin is 4 × 10−5 mol/L, pH 7.4, λex = 280 nm.
Table 1.
Stern-Volmer quenching constants for the gallic acid (GA)-trypsin system at different temperatures.
| T (°C) | Equation | R2 | Ksv (105 L/mol) | Kq (1013 L/mol/s) | |
|---|---|---|---|---|---|
| Trypsin + GA | 25 °C | F0/F = 1.0165 + 0.2008 × 105 [Q] | 0.9906 | 0.2008 | 0.2008 |
| 31 °C | F0/F = 1.0413 + 0.1603 × 105 [Q] | 0.9927 | 0.1603 | 0.1603 | |
| 37 °C | F0/F = 1.0743 + 0.1269 × 105 [Q] | 0.9976 | 0.1269 | 0.1269 |
3.3. Binding constants
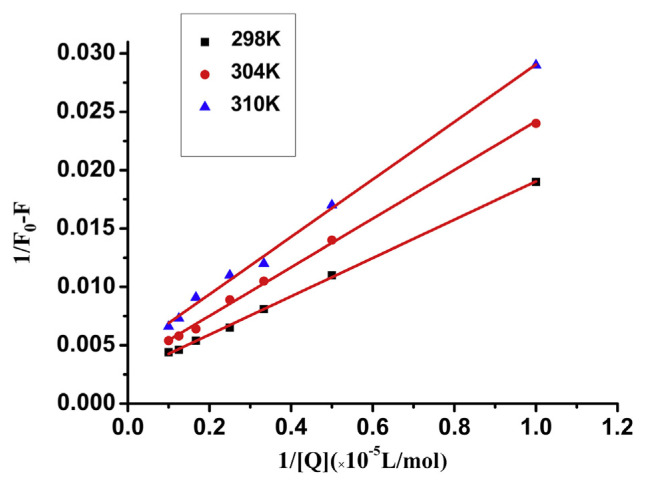
For the static quenching interaction, the binding constant (KA) between trypsin and GA can be obtained from the Lineweaver-Burk equation [37]:
| (3) |
where KA is the binding constant, which can be calculated from the slope and intercept of the curve from 1/F0-F vs. 1/[Q], as shown in Fig. 5. The KA values at different temperatures are shown in Table 2. The binding constant decreased with the increasing temperature, which indicated the formation of a low stability complex.
Fig. 5.
The Lineweaver-Burk curves for quenching of trypsin by gallic acid (GA) at different temperature.
Table 2.
The binding constants between gallic acid (GA) and trypsin by Lineweaver-Burk equation.
| T (°C) | Lineweaver-Burk equation | KA (104 L/mol) | R2 | |
|---|---|---|---|---|
| GA + trypsin | 25 °C | Y = 0.0030 + 0.0158 × 10−5 [Q]−1 | 1.9371 | 0.9969 |
| 31 °C | Y = 0.0035 + 0.0202 × 10−5 [Q]−1 | 1.8192 | 0.9949 | |
| 37 °C | Y = 0.0045 + 0.0245 × 10−5 [Q]−1 | 1.7465 | 0.9970 |
3.4. Thermodynamic parameters and binding forces
The interaction forces between biomolecules and small molecules include hydrogen bonds, electrostatic interactions, van der Waals interactions, hydrophobic forces, etc. [38]. Thermodynamic parameters are major evidence for certificating the binding force. If the effect of temperature is small in the reaction, the enthalpy change (ΔH) can be regarded as a constant. The values of enthalpy change (ΔH), entropy change (ΔS) and the free-energy change (ΔG) can be calculated using the Van’t Hoff and thermodynamic equations [39]:
| (4) |
| (5) |
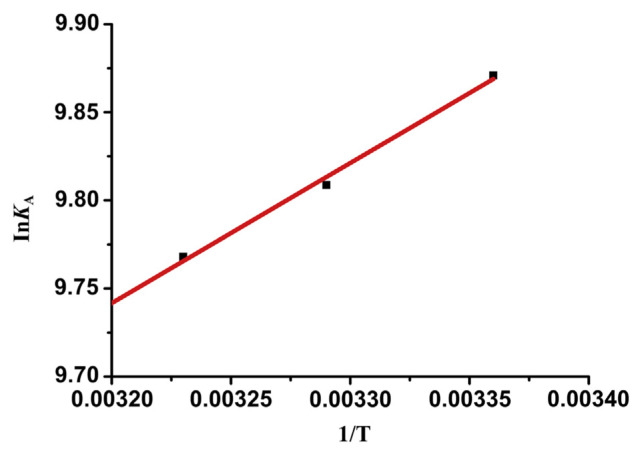
KA is the Lineweaver-Burk binding constant at different temperatures. R is the gas constant (R = 8.314 J/mol/K). ΔH and ΔS can be calculated by the linear relationship between InKA and 1/T based on Equation (4). The free energy change (ΔG) can be obtained from Equation (5). The Van’t Hoff curve is shown in Fig. 6. The fitting equation is InKA = 7.1977 + 795/T (R2 = 0.999). The values of ΔH and ΔS could be calculated from the intercept and slope of the curve, respectively. The values of ΔH, ΔS, and ΔG for GA binding to trypsin are presented in Table 3.
Fig. 6.
Van’t Hoff plot for gallic acid (GA) bound to trypsin.
Table 3.
Thermodynamic parameters of trypsingallic acid (GA) interaction at pH 7.4.
| T (°C) | ΔG (kJ/mol) | ΔH (kJ/mol) | ΔS (J/mol/K) | |
|---|---|---|---|---|
| Trypsin + GA | 25 °C | −24.28 | −6.61 | 59.84 |
| 31 °C | −4.80 | |||
| 37 °C | −25.16 |
There are several noncovalent interaction forces between small molecules and protein macromolecules according to Ross and Subramanian [40], such as hydrogen bonds, van der Waals forces, hydrophobic, and electrostatic interactions [41]. ΔG < 0 means that the binding of GA and trypsin is a process of spontaneous exothermic. ΔH < 0 and ΔS > 0 show that hydrophobic interactions are involved in the binding process. Electrostatic interactions are characterized by ΔH ≈ 0 and ΔS > 0. In addition, ΔH < 0 could also indicate van der Waals interactions. As shown in Table 3, it could be inferred that hydrophobic and electrostatic interactions were drawn into the binding process. In most cases, the reaction of small molecules and the proteins is the synergetic effect of a variety of forces. Three hydroxyl groups in GA molecules also indicate that hydrogen bonds may participate in the reaction. Therefore, it could be concluded that hydrogen bonds, van der Waals, hydrophobic, and electrostatic interactions were involved in the binding process of GA and trypsin.
3.5. Energy transfer between trypsin and GA
Fluorescence resonance energy transfer is an important technology, which has wide applications for interaction analysis between biological macromolecules and small molecules, cellular physiology, and immunity analysis [42]. According to Förster’s theory [43], the nonradiative energy transfer may occur if the compound met the following conditions: (1) the donor emits fluorescence; (2) there is enough overlap between the fluorescence emission spectrum of the donor and absorption spectrum of the receptor; and (3) the maximum distance between the donor and the receptor is <7 nm. The binding distance between the emission fluorescence groups can be determined between protein molecules and small molecules by the Förster theory.
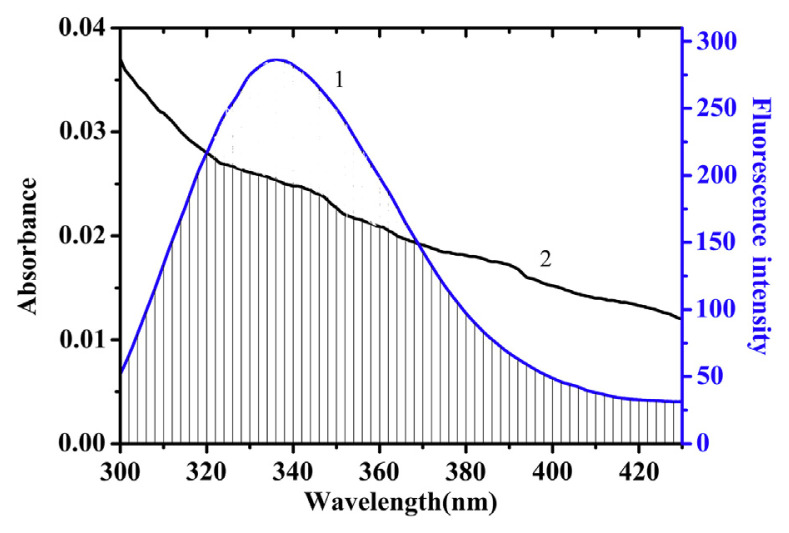
The overlap between UV-vis absorption spectrum of GA and the fluorescence emission spectrum of trypsin is shown in Fig. 7. The efficiency of energy transfer E can be described by the Förster nonradiative energy transfer theory in the following equation:
Fig. 7.
Overlap of fluorescence emission of trypsin with absorption spectra of gallic acid (GA). C(trypsin) = C(GA) = 1 × 10−5 mol/L. 1: The fluorescence emission spectrum of Trypsin. 2. The UV-vis absorption spectrum of GA.
| (6) |
where R0 is the Förster critical distance when the transfer efficiency is 50%, and r is the distance between the donor and the receptor. F and F0 are the fluorescence intensities of trypsin in the presence and absence of GA, respectively:
| (7) |
In Equation (7)K2 is the spatial orientation factor of the dipole related to the random distribution of the donor and the receptor. N is the refractive index of the medium. Φ is the fluorescence quantum yield of the donor and J is the overlap integral between the fluorescence emission spectra of the donor and the absorption spectra of the receptor, which can be calculated by the equation:
| (8) |
where F(λ) is the donor fluorescence intensity at the wavelength of λ, and ɛ is the molar absorption coefficient of the receptor at the wavelength of λ.
In the above equations, K2 = 2/3, N = 1.336, and Φ = 0.118. According to Equations (6–8), J, R0, E and r were calculated and are shown in Table 4. The binding distance was <7 nm and 0.5 R0 < r < 2.0 R0. The results showed that the nonradiative energy transfer occurred between GA and trypsin.
Table 4.
The distance parameters of gallic acid (GA)-trypsin interaction at pH 7.4.
| J (cm3·L·mol−1) | E | R0 (nm) | r (nm) | |
|---|---|---|---|---|
| GA-trypsin | 2.72 × 10−14 | 0.4169 | 2.31 | 2.82 |
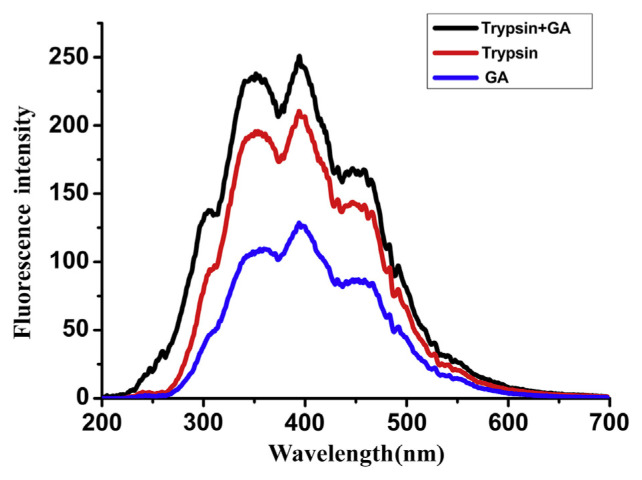
3.6. Characteristics of the RLS spectra
RLS is a new analysis to determine the scattering signal by the fluorescence spectrophotometer. The RLS analysis is simple, rapid, and highly sensitive. Therefore, it becomes more popular in life sciences, environmental science, nanomaterial analysis, and testing [44,45]. The RLS spectra of trypsin, GA, and GA-trypsin complex synchronously scanned from 200 nm to 600 nm with Δλ = 0 nm are recorded. The results are shown in Fig. 8. An obviously increased RLS was found when a little GA was added to trypsin solution. It was guessed from the results that trypsin might interact with GA in solution, forming a new GA-trypsin complex that could be expected to be an aggregate [46]. The size of GA-trypsin particles may be larger than those of trypsin, and the increased light scattering signal appeared under the given conditions.
Fig. 8.
The resonance light scattering (RLS) spectra of gallic acid (GA)-trypsin, trypsin, and GA. C(trypsin) = 1.5 × 10−5 mol/L, C (GA) = 1.0 × 10−5 mol/L.
3.7. Conformation investigation
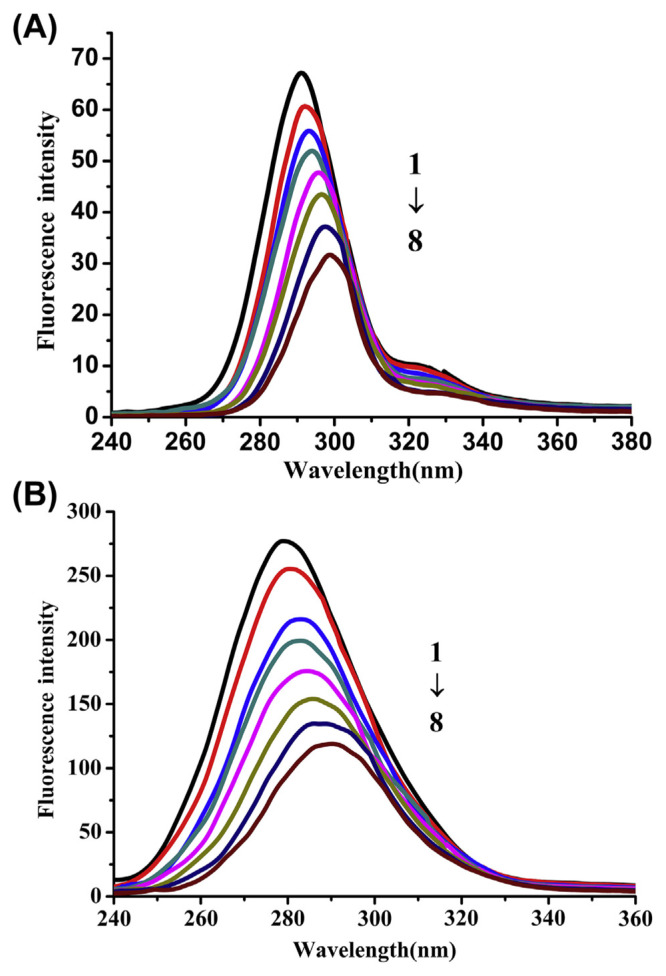
Synchronous fluorescence spectroscopy is a common method to provide information about the conformational changes near fluorophore functional groups [47]. Synchronous fluorescence spectroscopy is a useful method and tool to investigate the environment of amino acid residues and the influence of drug on the protein conformation. When the Δλ values between excitation wavelength and emission wavelength are set at 15 nm or 60 nm, the characteristic information of tryptophan and tyrosine residues are given by the synchronous fluorescence.
The influence of GA on trypsin synchronous fluorescence spectra is shown in Fig. 9. The synchronous fluorescence spectra gave the characteristic information of tyrosine (Fig. 9A) and tryptophan (Fig. 9B), respectively. The red shift indicated that the tryptophan and tyrosine residues in the nonpolar environment moved to a more polar environment. Intensities decreased of tyrosine and tryptophan residues and the red shift of peaks were observed when GA was gradually added into the trypsin solution. It can be concluded that GA may bind to trypsin. Tryptophan and tyrosine residues are sensitive to the microenvironment and show the change of trypsin tertiary structure by the synchronous spectra shift.
Fig. 9.
Influence of gallic acid (GA) on synchronous fluorescence spectra of trypsin, (A) Δλ = 15 nm, (B) Δλ = 60 nm. Both at pH = 7.4, T = 25 °C, with trypsin concentration of 4 × 10−5 mol/L, and GA concentration of 0 × 10−5 mol/L, 1.0 × 10−5 mol/L, 2.0 × 10−5 mol/L, 3.0 × 10−5 mol/L, 4.0 × 10−5 mol/L, 5.0 × 10−5 mol/L, 6.0 × 10−5 mol/L, and 8.0 × 10−5 mol/L, respectively (from spectra 1 to spectra 8).
3.8. Effect on trypsin activity
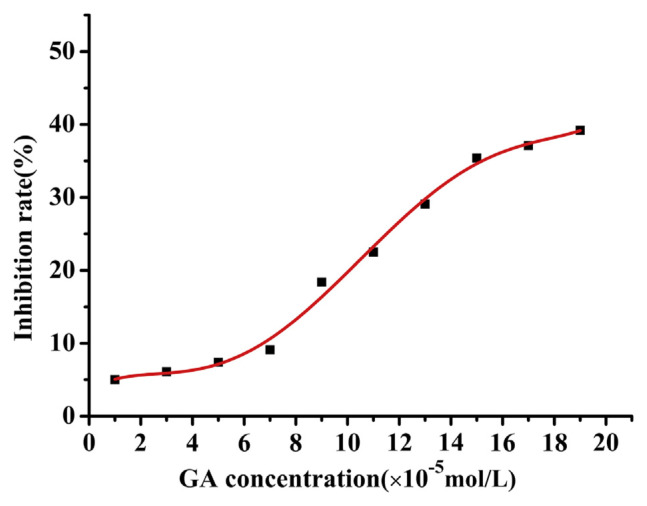
In order to investigate the influence on the enzymatic activity of trypsin, the inhibition effects of GA on the activity of trypsin are shown in Fig. 10. With casein as a substrate, the absorbance of trypsin in the presence of GA was determined. IC50 can be calculated from the fitting equation:
Fig. 10.
The inhibition rate of gallic acid (GA) to trypsin at different concentrations (pH = 7.4, T = 313 K), C(trypsin) = 4 × 10−5 mol/L.
| (9) |
where x is the concentration of GA, and Y is the inhibition rate. IC50 was 3.9 × 10−6 mol/L obtained from Equation (9). The results indicated that the inhibition rate of GA to trypsin was increased by the addition of GA, which was in accordance with previous reports.
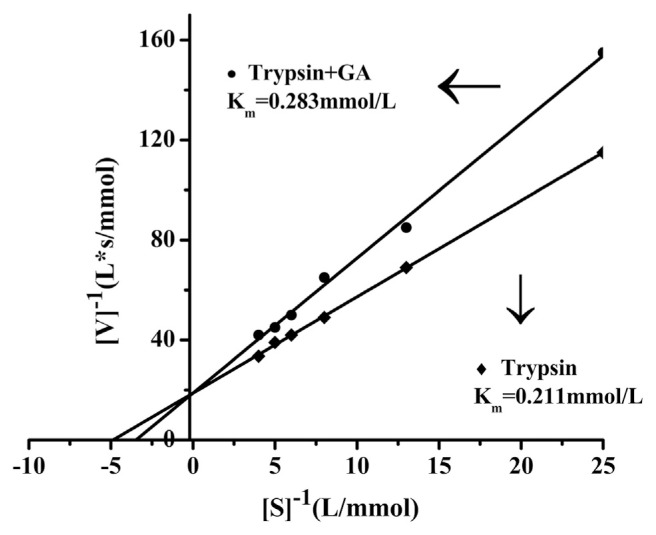
In order to confirm the inhibition mode, the Lineweaver-Burk plots of trypsin in the absence and presence of GA were drawn (Fig. 11). When GA was added, Km was 0.283 mmol/L, while without GA, Km was 0.211 mmol/L. From the above enzymatic activity experiment data, the results indicated that GA could interact with trypsin by the formation of a trypsin-GA complex, resulting in inhibition of trypsin activity.
Fig. 11.
The Lineweaver-Burk plots of trypsin in the absence and presence of gallic acid (GA). pH = 7.4, T = 313 K), C (trypsin) = 4 × 10−5 mol/L.
3.9. Computational modeling of the trypsin-GA
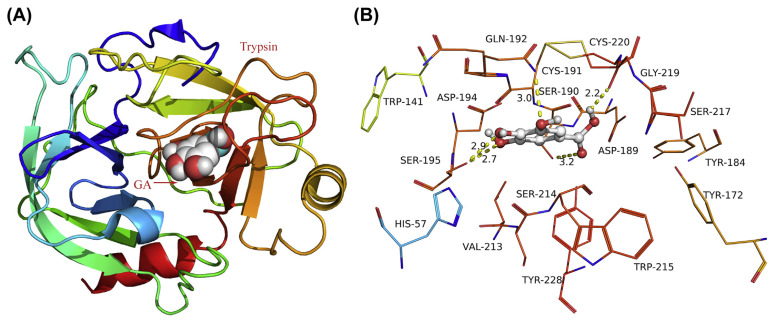
The molecular modeling method was originally performed in drug design and widely used to provide visible binding information between small molecules and proteins in recent years [48]. As shown in Fig. 12, GA fitted into the binding pocket of trypsin and produced a high density of van der Waals contacts with the residues. Importantly, one of the hydroxyl groups forms a hydrogen bond with residue Gln192, and the other two hydroxyl groups form hydrogen bonds with residue Ser195. Additionally, two hydrogen bonds were observed between the carboxy group and side chain of residues Ser190 and Gly219. From Fig. 12B, we could see that there were several tryptophan and tyrosine residues around GA and their microenvironment may be changed because of the addition of exogenous substance. All in all, the above molecular simulation gives us a rational explanation for the excellent activity of GA against trypsin, which provided valuable information for further study of trypsin inhibitors.
Fig. 12.
(A) The full view of gallic acid (GA) in trypsin. (B) Detailed illustration of the surroundings nearby GA in trypsin.
4. Conclusion
This paper shows spectroscopic studies on the interactions between trypsin and GA. The fluorescence spectroscopy results revealed that GA could bind to trypsin by a static quenching to form a new compound. Hydrogen bonds, van der Waals, hydrophobic, and electrostatic interactions were involved in the formation process of the GA-trypsin complex. The structural change of trypsin was shown by using UV-vis absorption spectra, synchronous fluorescence spectra, and RLS spectroscopy, when GA was added to the trypsin solution. The tryptophan and tyrosine residue microenvironments were changed by GA. The results indicated that GA was a strong quencher and inhibitor. The molecular docking results agreed well with the experiment data and offered more specific evidence for explaining the interaction mechanism. Finally, these results provided a new method to probe the inhibition effect of digestive enzymes induced by other small molecules and further advanced the interaction mechanism between the small molecules and proteinases.
Acknowledgments
We gratefully acknowledge financial support from the Fund for Science and Technology Project of Yunnan Province (2009EB081, 2011AB006) and Supporting Plan Issue of the Ministry of Science and Technology (2011BAD46B00).
Funding Statement
We gratefully acknowledge financial support from the Fund for Science and Technology Project of Yunnan Province (2009EB081, 2011AB006) and Supporting Plan Issue of the Ministry of Science and Technology (2011BAD46B00).
Footnotes
Conflicts of interest
All authors declare no conflicts of interest.
REFERENCES
- 1. Rufino FPS, Pedroso V, Araujo JN, França AF, Rabêlo LM, Migliolo L, Kiyota S, Santos EA, Franco OL, Oliveira AS. Inhibitory effects of a Kunitz-type inhibitor from Pithecellobium dumosum (Benth) seeds against insect-pests’ digestive proteinases. Plant Physiol Biochem. 2013;63:70–6. doi: 10.1016/j.plaphy.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 2. Rawdkuen S, Benjakul S, Visessanguan W, Lanier TC. Cysteine proteinase inhibitor from chicken plasma: fractionation, characterization and autolysis inhibition of fish myofibrillar proteins. Food Chem. 2007;101:1647–57. [Google Scholar]
- 3. Maltas E. Binding interactions of niclosamide with serum proteins. J Food Drug Anal. 2014;22:549–55. doi: 10.1016/j.jfda.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goncalves R, Mateus N, Pianet I, Laguerre M, de Freitaset V. Mechanisms of tannin-induced trypsin inhibition: a molecular approach. Langmuir. 2011;27:13122–9. doi: 10.1021/la202280c. [DOI] [PubMed] [Google Scholar]
- 5. Chen JM, Montier T, Férec C. Molecular pathology and evolutionary and physiological implications of pancreatitis-associated cationic trypsinogen mutations. Hum Genet. 2001;109:245–52. doi: 10.1007/s004390100580. [DOI] [PubMed] [Google Scholar]
- 6. Silva FP, Jr, De-Simone SG. S1 subsite in snake venom thrombin-like enzymes: can S1 subsite lipophilicity be used to sort binding affinities of trypsin-like enzymes to small-molecule inhibitors. Bioorg Med Chem. 2004;12:2571–87. doi: 10.1016/j.bmc.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 7. Zuo Y, Chen H, Deng Y. Simultaneous determination of catechins, caffeine and gallic acids in green, Oolong, black and puerh teas using HPLC with a photodiode array detector. Talanta. 2002;57:307–16. doi: 10.1016/s0039-9140(02)00030-9. [DOI] [PubMed] [Google Scholar]
- 8. Mämmelä P, Savolainen H, Lindroos L, Kangas J, Vartianen T. Analysis of oak tannins by liquid chromatography-electrospray ionisation mass spectrometry. J Chromatogr A. 2000;891:75–83. doi: 10.1016/s0021-9673(00)00624-5. [DOI] [PubMed] [Google Scholar]
- 9. Zhang H, Zhou Q, Cao J, Wang Y. Mechanism of cinnamic acid-induced trypsin inhibition: a multi-technique approach. Spectrochim Acta Part A. 2013;116:251–7. doi: 10.1016/j.saa.2013.07.035. [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, Zhang H, Cao J, Zhou Q. J Mol Struct. 2013:1051–78. [Google Scholar]
- 11. Wang R, Kang X, Wang R, Wang R, Dou H, Wu J, Song C, Chang J. Comparative study of the binding of trypsin to caffeine and theophylline by spectrofluorimetry. J Luminescence. 2013;138:258–66. [Google Scholar]
- 12. Mansouri MT, Naghizadeh B, Ghorbanzadeh B, Farbood Y, Sarkaki A, Bavarsad K. Gallic acid prevents memory deficits and oxidative stress induced by intracerebroventricular injection of streptozotocin in rats. Pharmacol Biochem Behav. 2013;111:90–6. doi: 10.1016/j.pbb.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 13. Do QD, Angkawijaya AE, Tran-Nguyen PL, Huynh LH, Soetaredjo FE, Ismadji S, Ju YH. Effect of extraction solvent on total phenol content, total flavonoids content, and antioxidant activity of Limnophila aromatica. J Food Drug Anal. 2013;22:296–302. doi: 10.1016/j.jfda.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Priscilla DH, Prince P. Cardioprotective effect of gallic acid on cardiac troponin-T, cardiac marker enzymes, lipid peroxidation products and antioxidants in experimentally induced myocardial infarction in Wistar rats. Chem Biol Interact. 2009;179:118–24. doi: 10.1016/j.cbi.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 15. de Mejía EG, Chandra S, Ramírez-Mares MV, Wang W. Catalytic inhibition of human DNA topoisomerase by phenolic compounds in Ardisia compressa extracts and their effect on human colon cancer cells. Food Chem Toxicol. 2006;44:1191–203. doi: 10.1016/j.fct.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 16. Luo JH, Li BL, Li NB, Luo HQ. Sensitive detection of gallic acid based on polyethyleneimine-functionalized graphene modified glassy carbon electrode. Sens Actuators B. 2013;186:84–9. [Google Scholar]
- 17. Wang X, Wang J, Yang N. Flow injection chemiluminescent detection of gallic acid in olive fruits. Food Chem. 2007;105:340–5. [Google Scholar]
- 18. Chuysinuan P, Chimnoi N, Techasakul S, Supaphol P. Gallic acid-loaded electrospun poly (L-lactic acid) fiber mats and their release characteristic. Macromol Chem Phys. 2009;210:814–22. [Google Scholar]
- 19. Neo YP, Ray S, Jin J, Gizdavic-Nikolaidis M, Nieuwoudt MK, Liu D, Quek SY. Encapsulation of food grade antioxidant in natural biopolymer by electrospinning technique: a physicochemical study based on zein–gallic acid system. Food Chem. 2013;136:1013–21. doi: 10.1016/j.foodchem.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 20. Lu Z, Nie G, Belton PS, Tang H, Zhao B. Structure-activity relationship analysis of antioxidant ability and neuroprotective effect of gallic acid derivatives. Neurochem Int. 2006;48:263–74. doi: 10.1016/j.neuint.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 21. Locatelli C, Filippin-Monteiro FB, Creczynski-Pasa TB. Alkyl esters of gallic acid as anticancer agents: A review. Eur J Med Chem. 2013;60:233–9. doi: 10.1016/j.ejmech.2012.10.056. [DOI] [PubMed] [Google Scholar]
- 22. Punithavathi VR, Prince PSM, Kumar R, Selvakumari J. Antihyperglycaemic, antilipid peroxidative and antioxidant effects of gallic acid on streptozotocin induced diabetic Wistar rats. Eur J Pharmacol. 2011;650:465–71. doi: 10.1016/j.ejphar.2010.08.059. [DOI] [PubMed] [Google Scholar]
- 23. Fang X, Sen A, Vicens M, Tan W. Synthetic DNA aptamers to detect protein molecular variants in a high-throughput fluorescence quenching assay. ChemBioChem. 2003;4:829–34. doi: 10.1002/cbic.200300615. [DOI] [PubMed] [Google Scholar]
- 24. Liu B, Tian H. A ratiometric fluorescent chemosensor for fluoride ions based on a proton transfer signaling mechanism. J Mater Chem. 2005;15:2681–6. [Google Scholar]
- 25. Han B, Yuan J, Wang E. Sensitive and selective sensor for biothiols in the cell based on the recovered fluorescence of the CdTe quantum dots− Hg (II) system. Anal Chem. 2009;81:5569–73. doi: 10.1021/ac900769h. [DOI] [PubMed] [Google Scholar]
- 26. Ohtaki H, Radnai T. Structure and dynamics of hydrated ions. Chem Rev. 1993;93:1157–204. [Google Scholar]
- 27. Molina-García L, Santos JLM, Ruiz-Medina A, Llorent-Martinez EJ. Determination of ketoprofen based on its quenching effect in the fluorescence of quantum dots. J Food Drug Anal. 2013;21:426–31. [Google Scholar]
- 28. Bi S, Song D, Tian Y, Zhou X, Liu Z, Zhang H. Molecular spectroscopic study on the interaction of tetracyclines with serum albumins. Spectrochim Acta Part A. 2005;61:629–36. doi: 10.1016/j.saa.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 29. Ni Y, Su S, Kokot S. Spectrometric studies on the interaction of fluoroquinolones and bovine serum albumin. Spectrochimica Acta Part A. 2010;75:547–52. doi: 10.1016/j.saa.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 30. Zhang HM, Wang YQ, Zhou QH. Investigation of the interactions of quercetin and morin with trypsin. Luminescence. 2009;24:355–62. doi: 10.1002/bio.1121. [DOI] [PubMed] [Google Scholar]
- 31. Szmacinski H, Ray K, Lakowicz JR. Metal-enhanced fluorescence of tryptophan residues in proteins: application toward label-free bioassays. Anal Biochem. 2009;385:358–64. doi: 10.1016/j.ab.2008.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fraiji LK, Hayes DM, Werner T. Static and dynamic fluorescence quenching experiments for the physical chemistry laboratory. J Chem Educa. 1992;69:424. [Google Scholar]
- 33. Gerbanowski A, Malabat C, Rabiller C, Guéguen J. Grafting of aliphatic and aromatic probes on rapeseed 2S and 12S proteins: influence on their structural and physicochemical properties. J Agric Food Chem. 1999;47:5218–26. doi: 10.1021/jf990226p. [DOI] [PubMed] [Google Scholar]
- 34. Hegde AH, Sandhya B, Seetharamappa J. Evaluation of binding and thermodynamic characteristics of interactions between a citrus flavonoid hesperitin with protein and effects of metal ions on binding. Mol Biol Rep. 2011;38:4921–9. doi: 10.1007/s11033-010-0634-9. [DOI] [PubMed] [Google Scholar]
- 35. Li Y, He WY, Liu H, Yao X, Hu Z. Daidzein interaction with human serum albumin studied using optical spectroscopy and molecular modeling methods. J Mol Struct. 2007;831:144–50. [Google Scholar]
- 36. Naveenraj S, Raj MR, Anandan S. Binding interaction between serum albumins and perylene-3, 4, 9, 10-tetracarboxylate–A spectroscopic investigation. Dyes Pigm. 2012;94:330–7. [Google Scholar]
- 37. Cui F, Zhang Q, Yan Y, Yao X, Qu G, Lu Y. Study of characterization and application on the binding between 5-iodouridine with HSA by spectroscopic and modeling. Carbohydr Polym. 2008;73:464–72. [Google Scholar]
- 38. Liang J, Cheng Y, Han H. Study on the interaction between bovine serum albumin and CdTe quantum dots with spectroscopic techniques. J Mol Struct. 2008;892:116–20. [Google Scholar]
- 39. Zhao H, Ge M, Zhang Z, Wang W, Wu G. Spectroscopic studies on the interaction between riboflavin and albumins. Spectrochim Acta Part A. 2006;65:811–7. doi: 10.1016/j.saa.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 40. Ross PD, Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochem. 1981;20:3096–102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 41. Reidenberg MM, Affrime M. Influence of disease on binding of drugs to plasma proteins. Ann N Y Acad Sci. 1973;226:115–26. doi: 10.1111/j.1749-6632.1973.tb20474.x. [DOI] [PubMed] [Google Scholar]
- 42. Haldar B, Mallick A, Chattopadhyay N. Photophysics of pyrene-end-capped poly (ethyleneoxide) in aqueous micellar environments. J Mol Liq. 2004;115:113–20. [Google Scholar]
- 43. Förster T. Zwischenmolekulare energiewanderung und fluoreszenz. Annalen der physik. 1948;437:55–75. [in German] [Google Scholar]
- 44. Zhu J. Determination of aniline and related mono-aromatic amines in indoor air in selected Canadian residences by a modified thermal desorption GC/MS method. Environ Int. 2004;30:135–43. doi: 10.1016/S0160-4120(03)00168-5. [DOI] [PubMed] [Google Scholar]
- 45. Collings PJ, Gibbs EJ, Starr TE, Vafek O, Yee C, Pomerance LA, Pasternack RF. Resonance light scattering and its application in determining the size, shape, and aggregation number for supramolecular assemblies of chromophores. J Phys Chem B. 1999;103:8474–81. [Google Scholar]
- 46. Xiao J, Shi J, Cao H, Wu S, Ren F, Xu M. Analysis of binding interaction between puerarin and bovine serum albumin by multi-spectroscopic method. J Pharm Biomed Anal. 2007;45:609–15. doi: 10.1016/j.jpba.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 47. Hu YJ, Liu Y, Pi ZB, Qu SS. Interaction of cromolyn sodium with human serum albumin: a fluorescence quenching study. Bioorg Med Chem. 2005;13:6609–14. doi: 10.1016/j.bmc.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 48. Lien GS, Chen CS, Chen WY, Huang SH, Cheng KT, Lin CM, Shao SH. Photoproducts of indomethacin exhibit decreased hydroxyl radical scavenging and xanthine oxidase inhibition activities. J Food Drug Anal. 2013;21:332–6. [Google Scholar]