Abstract
The T4 head protein, gp2, promotes head-tail joining during phage morphogenesis and is also incorporated into the phage head. It protects the injected DNA from degradation by exonuclease V during the subsequent infection. In this study, we show that recombinant gp2, a very basic protein, rapidly kills the cells in which it is expressed. To further illustrate the protectiveness of gp2 for DNA termini, we compare the effect of gp2 expression on Red-mediated and Int-mediated recombination. Red-mediated recombination is nonspecific and requires the transient formation of double-stranded DNA termini. Int-mediated recombination, on the other hand, is site specific and does not require chromosomal termini. Red-mediated recombination is inhibited to a much greater extent than is Int-mediated recombination. We conclude from the results of these physiological and genetic experiments that T4 gp2 expression, like Mu Gam expression, kills bacteria by binding to double-stranded DNA termini, the most likely mode for its protection of entering phage DNA from exonuclease V.
The gp2 protein (10) of bacteriophage T4 gene2 (5) is associated with phage particles (25) and protects newly injected T4 DNA from degradation by exonuclease V (ExoV) (16, 17), an activity of the Escherichia coli holoenzyme composed of gpRecB, gpRecC, and gpRecD (12). Several aspects of this protection, as follows, have led to the proposal that T4 gp2 binds to and thereby mechanically shields the ends of injected T4 DNA. (i) T4 gp2− phage do not form plaques on Rec+ E. coli but do form plaques on ExoV− E. coli. The ExoV− mutation can suppress the T4 gene2 mutation whether the complete holoenzyme recombinase function is destroyed (recB or recC nulls [18]) or even if only the nuclease function is disabled (recD), while leaving other activities intact or enhanced (2, 13). (ii) Labeled DNA of T4 gp2 is degraded by ExoV upon injection (17). (iii) The inability of T4gp2 to grow on Rec+ E. coli can be suppressed by the simultaneous presence of certain alleles of gene23 (13). These gene23 alleles result in long phage heads filled with a long chromosome comprised of about three T4 genomes connected in tandem (4). When a T4 gene2 gene23 double-mutant phage injects its DNA, the same amount (but a lower fraction) of DNA is degraded as for a T4 gene2 mutant (13). Thus, the chromosome is left with more than one genome of T4 DNA by the time the anti-ExoV activity, which is encoded by the entering DNA and expressed early during infection (21), inhibits further degradation. (iv) T4 gp2 does not protect in trans. Simultaneous infection by gene 2 and gene2− bacteriophage results in the degradation of only the latter phage’s chromosome (16). The lack of protection in trans implies that RecBC activity is not inhibited by gp2 but that resistance to RecBC activity enters with the phage DNA. Protection of DNA ends, only in cis, contrasts with Gam protein protection of bacteriophage λ, which functions by inactivation of the ExoV enzyme (7). Although we prefer the mechanism whereby DNA is protected at its termini by bound gp2, we have not ruled out, for example, modification of DNA termini.
Various expression vectors for T4 gp2 with or without simultaneous expression of the contiguous T4 gene3 were constructed (10). The biochemical purification and analysis and in vitro assay of gp2 are reported elsewhere (26, 27). In this study we describe some in vivo observations on cells harboring these expression vectors (Table 1). The expression of gene2 and gene3 from vectors used in the studies described herein (10) was induced by a temperature shift to 42°C, which inactivates the bacteriophage lambda repressor CI857 controlling the pL promoter (19).
TABLE 1.
Plasmids, bacterial strains, and lambda phages
| Plasmid, bacterial strain, or lambda phage | Genotype or relevant property | Source or reference |
|---|---|---|
| Plasmids | ||
| pT713 | Expression vector | 19 |
| pKA23am | gene2+, gene3 amber | 10 |
| pKA2oc3 | gene2 ochre, gene3+ | 10 |
| pKA23 | gene2+, gene3+ | 10 |
| pGP1-2 | T7 RNApol, ts induction | 19 |
| Strains | ||
| K38 | Host for plasmids | 15 |
| 594 | Su− | 28 |
| 594d(recA-srlR)306::Tn10 | Su−recA | 24 |
| Lambda phages | ||
| Jam | J amber, Int+ | 24 |
| Rts | Rts, Int+ | 24 |
| Jam int4 | J amber, Int− | 24 |
| Rts int4 | Rts, Int− | 24 |
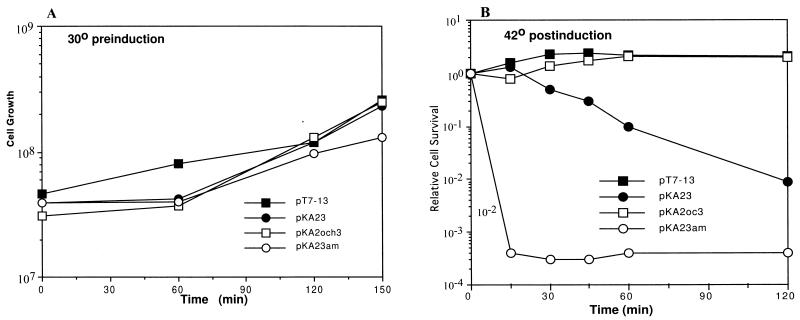
The expression of T4 gp2 was rapidly lethal to E. coli (Fig. 1). Fifteen minutes of expression, induced by incubation at 42°C, followed by plating under nonexpression conditions and colony counts, resulted in a loss of viability of greater than 1,000-fold. This lethality is even more rapid than that resulting from the endogenous activation of EcoRI nuclease in the absence of its cognate methylase (6). The lethal effect of T4 gp2 was not observed if the expression vector contained an ochre allele of gp2. The toxic effect therefore requires intact gp2 protein production. A similar strong effect on cell viability has been found for another putative end-binding protein, the Gam protein of phage Mu (1).
FIG. 1.
Kinetics of bacterial killing by gp2. E. coli K38/pGP1-2 cells containing the pT7-13 derivatives specified below were grown at 30°C (A) and switched after 2.5 h of cell growth to 42°C (B) to induce gp2 production. At the times indicated, the titers of samples of cells were determined on plates with ampicillin and kanamycin and grown overnight at 30°C. Ordinates represent viable cell titer (A) and viability relative to the preinduction cell titer at 150 min (B). Symbols: ■, pT7-13 containing no gene2 or gene3 insert; ●, pKA23 containing insert with intact gene2 and gene3; □, pKA2oc3 containing insert with ochre mutation in gene2 and intact gene3; ○, pKA23am containing insert with intact gene2 and an amber mutation in gene3.
The lethal effect of gp2 is alleviated by the simultaneous expression of gp3. This effect is not due to attenuation of the production of gp2. Biochemical assays showed that the quantity of gp2 synthesized in cells that make both gp2 and gp3 was the same as that in cells that make gp2 alone (data not shown). In addition, more recent work has shown that with the same promoter (27), gp2 is barely detectable at 15 min but comprises about 10 to 20% of total cell protein 2 h after induction. Therefore, the toxic effect of gp2 was not the result of a drain on the cells’ resources for protein synthesis but, more likely, the effect of the protein product on a vulnerable aspect of essential metabolism.
Our interpretation of the lethality of gp2 and its rescue by gp3 is that gp2 binds to a vulnerable and essential target in E. coli and that gp3 modulates this binding. G. R. Wang (25b) has shown that gp2 is a DNA binding protein and that gp3 can bind to gp2 under certain conditions. The work presented above as well as other evidence to be presented later in this article suggests that gp2 binds to double-stranded ends of DNA in vivo.
What in E. coli presents a double-stranded target for gp2 and why sequestering it should be lethal for the cell are not immediately obvious. It has been proposed (12) that a potentially vulnerable target in E. coli results from a prolapsing of the replication fork. According to this hypothesis, the leading and lagging strands of the replication fork occasionally pair and flare out, thereby presenting a double-stranded end. Other routes to double-chain breaks at the replication fork have been proposed to result from breakage at lesions (9). In these cases also, the double-stranded end is a necessary intermediate in repairing an otherwise lethal stall of the replication fork. We suppose that T4 gp2 can bind tightly to these double-chain ends and that trapping of this structure precludes repair of the fork. Alternatively, the protection of cell viability by gp3 may occur via modulation of inclusion body formation during overexpression rather than through a mechanism which directly reflects the interaction of gp2 and gp3 in the life cycle of T4.
The experiments shown in Fig. 1 involved enumerating colonies on plates with ampicillin (to select for the plasmid). gene2 may inhibit either the plasmid or the chromosomal replicon or both. We suppose that the reduction of cells is due to the stabilization of broken chromosomes rather than plasmids, since the chromosomal DNA provides a much greater target for breakage than the plasmids and killing would require only one stabilized break in the chromosome but as many stabilized breaks as there are plasmids. In fact, some evidence for each type was seen in the following experiments.
Plasmid-harboring strains were grown at different temperatures for 15 h, and then their titers were determined at 30°C. Table 2 shows that below 37°C, the temperature for T7 RNA polymerase induction, there is no toxicity (columns 1 and 2). Even at 37°C and higher, there is relatively little toxicity unless gp2 is expressed (compare lines 1 and 3 with lines 2 and 4). Table 2 also shows that gp3 alone has little effect on viability (line 3). Under conditions of continuous production, gp3 was able to protect only partially against the deleterious effects of gp2 (line 2). Table 3 shows that when there was no selection for plasmid maintenance (no drugs, columns 2 and 4), gp2 expression caused a 100-fold decrease in viability in the presence (line 2) or absence (line 4) of gp3 expression. However, when plasmid maintenance was selected for in the presence of gp2 expression, viability decreased a further 1,000-fold in the absence of gp3 expression (compare columns 1 and 2, row 4). Loss of viability at 42°C, when gp2 is expressed in the absence of antibiotic, implies toxicity to the chromosome. The further loss of viability in the presence of antibiotic (plasmid maintenance) implies additional toxicity to the plasmid replicon.
TABLE 2.
gene2 toxicity under conditions of continuous expressiona
| Plasmid | Titer (10−9) at indicated growth temperature
|
||||
|---|---|---|---|---|---|
| 30°C | 34°C | 37°C | 40°C | 42°C | |
| pT713 | 5.0 | 6 | 1 | 6 | 0.25 |
| pKA23 | 7.5 | 7.5 | 0.75 | 0.009 | 0.006 |
| pKA2oc3 | 8.0 | 7 | 1 | 0.4 | 0.8 |
| pKA23am | 7 | 7.5 | 0.08 | 0.004 | 0.00009 |
Small inocula of K38 were grown 15 h at various temperatures in Luria-Bertani medium with kanamycin and ampicillin, both at 40 μg/ml. The next day the inocula were diluted and plated on Luria-Bertani medium with antibiotics and incubated at 30°C. Titers were determined after another day of incubation.
TABLE 3.
T4 gp2 inhibits both plasmid and chromosomal repliconsa
| Plasmid | Titer at 42°C
|
Titer at 30°C
|
||
|---|---|---|---|---|
| Kan + Amp | No drugs | Kan + Amp | No drugs | |
| pT713 | 2.4 × 109 | 2.5 × 109 | 1.0 × 109 | 2.0 × 109 |
| pKA23 | <106 | 6.0 × 107 | 1.5 × 109 | 2.5 × 109 |
| pKA2oc3 | 2.5 × 109 | 3.3 × 109 | 1.8 × 109 | 3.5 × 109 |
| pKA23am | <104 | 9.0 × 107 | 1.0 × 109 | 2.1 × 109 |
One batch of K38 cells containing plasmids was grown to saturation in Luria-Bertani medium containing kanamycin and ampicillin. The cells were then diluted, and their titers were determined on Luria-Bertani medium with and without antibiotics at 30°C and 42°C.
The hypothesis that T4 gp2 acts in vivo by binding to double-stranded termini was supported further by a set of genetic crosses with bacteriophage lambda. Generalized recombination of wild-type bacteriophage lambda is mediated by the genes redα and redβ (14). Red-mediated recombination is stimulated by double-chain ends created by type II restriction enzymes (23, 25) and arguably, all normal Red-mediated recombination is dependent on double-chain ends of lambda that occur in the course of phage replication and packaging (22).
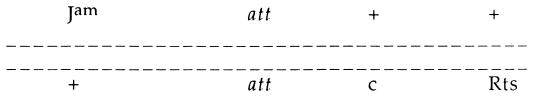
In contrast to Red, the site-specific system of Int-mediated recombination in lambda is not dependent upon either parent having chromosomal termini (8). The relative frequencies of Int- and Red-mediated recombination can be assayed by comparing the ratios of crossovers in each of two genetic intervals in crosses of the type shown in Fig. 2 (18), conducted with int+ and int phages, respectively. If am+ and ts+ progeny are selected from such a cross, Int-mediated recombination (which occurs only at att) can make only J+c+R+ (turbid) phage. Generalized (or Red-dependent) recombination from such a cross can make both J+c+R+ (turbid) and J+cR+ (clear) phage. If the removal of Int function has little effect on the c+/c ratio among J+ R+ recombinants, the generalized recombination activity is judged to be relatively high. However, if the Int+ cross gives a higher c+/c ratio among J+ R+ recombinants than in the Int− cross, generalized recombination activity is relatively low. Normalizing the ratio of c+/c− (obtained from normal crosses in which both Red- and Int-mediated recombination are active) to the same ratio obtained when only Red is active (i.e., in int crosses) allows the quantification of Int activity as follows (18): c+/c Int+ cross ÷ c+/c Int− cross.
FIG. 2.
Crosses conducted with int+ and int phages.
The results of these crosses (Table 4) show that the expression of T4 gp2 results in a diminished ratio of Red-mediated recombination (line 1, column 2) compared to Int-mediated recombination (line 1, column 1). Inhibition of Red recombination was dependent on the expression of the gene2 protein; no effect was seen from expression of the ochre fragment. Simultaneous expression of gene3 obviated the inhibitory effect of gene2 (row 3), supporting the idea that gene3 acts in vivo to modulate the activity of gp2 but does not affect synthesis of the protein.
TABLE 4.
T4 gp2 interferes with Red− recombination in E. coli recAa
| Bacterial strain/plasmid | +/c int+ cross | +/c int− cross | Int activity |
|---|---|---|---|
| 594recA/pKA23am | 2.92 | 1.19 | 2.45 |
| 594recA/pKA2oc3 | 1.63 | 1.47 | 1.11 |
| 594recA/pKA23 | 1.71 | 1.47 | 1.16 |
| 594/pKA23am | 1.42 | 1.42 | 1.00 |
| 594/pKA2oc3 | 1.45 | 1.32 | 1.01 |
| 594/pKA23 | 1.57 | 1.47 | 1.07 |
λ Crosses to assay the effect of T4 gp2, T4 gp3, and E. coli recA on Red recombination. Crosses were carried out exactly as described in previous work with Mu Gam protein (24). Cells harboring temperature-sensitive alleles of CI857, which controlled the expression of gene2 and gene3, were grown at 30°C to 2 × 108, at which time they were shifted to 41.5°C. After 15 min at 41.5°C, phage were added at a multiplicity of infection of 7 of each parent. Clear (c) and turbid (c+) recombinants in the yield were enumerated on E. coli 594 after incubation at 42°C.
The results with T4 gp2 on Red recombination are qualitatively similar but quantitatively smaller than the results of similar experiments conducted with another DNA binding protein purported to have a preference for double-chain termini, gpGam of bacteriophage Mu (24). Interestingly, the effect of gp2 on λ Red recombination was evident only when the host cells were recA. A similar effect of recA, suppressing the inhibitory effect of Mu Gam on Red-mediated recombination, was found (24a). The killing by T4 gp2 and Mu Gam appears entirely different from that mediated by bacteriophage Mu ligts and other kil functions (3). The latter has been shown only in the context of an abortive infection. The expression of gene2 was about 20% of total cellular protein (26, 27); therefore, it seems unlikely that the amount of protein was limiting for its in vivo activity. Thus, greater inhibition of Red by Mu Gam is more likely due to differences in their activities and to affinities for particular substrates.
There is a paradox or, at any rate, an incompleteness at this stage in the in vivo and in vitro characterizations of gp2. Biochemical experiments (25b) have shown T4 gp2 to be a DNA binding protein. However, no specific binding to DNA termini has been found in gel retardation assays (but see also reference 29). The in vivo evidence that gp2 binds to double-chain termini, which in molar terms represent a tiny fraction of the total intracellular DNA, is circumstantial but quite strong. Thus, there is reason to suspect the existence in vivo of an agent to guide or stabilize gp2 on double-chain termini. The neighboring gene3 is a candidate for an in vivo modulator of the binding of gp2. The creation, sequestration, and processing of DNA termini are among the most important and potentially dangerous aspects of genetic metabolism and appear to be under sophisticated controls in both procaryotes and eucaryotes (20, 25a).
Acknowledgments
K.A. and D.S.T. contributed equally to this work. The λ crosses were performed in the laboratory of Frank and Mary Stahl. We thank G. R. Wang for permission to cite his unpublished observations. D.T. thanks Fiona Doetsch and Joshua Lederberg for comments on the manuscript.
K.A. and E.B.G. were supported by NIH grant GM13511. D.T. acknowledges support from the Markey Trust.
REFERENCES
- 1.Akroyd J, Symonds N. Localization of the gam gene of bacteriophage mu and characterization of the gene product. Gene. 1986;49:273–282. doi: 10.1016/0378-1119(86)90288-x. [DOI] [PubMed] [Google Scholar]
- 2.Amundsen S K, Taylor A F, Chaudhury A M, Smith G R. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buu A, Ghelardini P, Monaci P, Paolozzi L. Analysis of the killing effect of Mu ligts mutants on host cells. FEMS Microbiol Lett. 1987;40:111–117. [Google Scholar]
- 4.Doermann A H, Eiserling F A, Boehner L. Genetic control of capsid length in bacteriophage T4. I. Isolation and preliminary description of four new mutants. J Virol. 1973;12:374–385. doi: 10.1128/jvi.12.2.374-385.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epstein R I, Bolle A, Steinberg C, Kellenberger E, Boy de la Tour E, Chevalley R, Edgar R S, Susman M, Denhardt C, Lielausis J. Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harbor Symp Quant Biol. 1964;28:375–392. [Google Scholar]
- 6.Heitman J, Zinder N D, Model P. Repair of the Escherichia coli chromosome after in vivo scission by the EcoRI endonuclease. Proc Natl Acad Sci USA. 1989;86:2281–2285. doi: 10.1073/pnas.86.7.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karu A E, Sakaki Y, Echols H, Linn S. The gamma protein specified by bacteriophage gamma. Structure and inhibitory activity for the recBC enzyme of Escherichia coli. J Biol Chem. 1975;250:7377–7387. [PubMed] [Google Scholar]
- 8.Kitts P, Richet E, Nash H A. Lambda integrative recombination: supercoiling, synapsis, and strand exchange. Cold Spring Harbor Symp Quant Biol. 1984;49:735–744. doi: 10.1101/sqb.1984.049.01.083. [DOI] [PubMed] [Google Scholar]
- 9.Kuzminov A. Instability of inhibited replication forks in E. coli. Bioessays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- 10.Lipinska B, Krishna Rao A S M, Bolten B M, Balakrishnan R, Goldberg E B. Cloning and identification of bacteriophage T4 gene 2 product gp2 and action of gp2 on infecting DNA in vivo. J Bacteriol. 1989;171:488–497. doi: 10.1128/jb.171.1.488-497.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louarn J-M, Louarn J, François V, Patte J. Analysis and possible role of hyperrecombination in the termination region of the Escherichia coli chromosome. J Bacteriol. 1991;173:5097–5104. doi: 10.1128/jb.173.16.5097-5104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers R S, Stahl F W. Chi and the RecBCD enzyme of Escherichia coli. Annu Rev Genet. 1994;28:49–70. doi: 10.1146/annurev.ge.28.120194.000405. [DOI] [PubMed] [Google Scholar]
- 13.Oliver D B, Goldberg E B. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J Mol Biol. 1977;116:877–881. doi: 10.1016/0022-2836(77)90276-5. [DOI] [PubMed] [Google Scholar]
- 14.Poteete A R, Fenton A C. Efficient double-strand break-stimulated recombination promoted by the general recombination systems of phages lambda and P22. Genetics. 1993;134:1013–1021. doi: 10.1093/genetics/134.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russel M, Model P. Replacement of the fip gene of Escherichia coli by an inactive gene cloned on a plasmid. J Bacteriol. 1984;159:1034–1039. doi: 10.1128/jb.159.3.1034-1039.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverstein J L, Goldberg E B. T4 DNA injection. I. Growth cycle of a gene 2 mutant. Virology. 1976;72:195–211. doi: 10.1016/0042-6822(76)90323-8. [DOI] [PubMed] [Google Scholar]
- 17.Silverstein J L, Goldberg E B. T4 DNA injection. II. Protection of entering DNA from host exonuclease V. Virology. 1976;72:212–223. doi: 10.1016/0042-6822(76)90324-x. [DOI] [PubMed] [Google Scholar]
- 18.Stahl F W, Stahl M M. Recombination pathway specificity of Chi. Genetics. 1977;86:715–725. doi: 10.1093/genetics/86.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taccioli G E, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A R, Alt R W, Jackson S P, Jeggo P A. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 21.Tanner D, Oishi M. The effect of bacteriophage T4 infection on an ATP-dependent deoxyribonuclease in Escherichia coli. Biochim Biophys Acta. 1971;228:767–769. doi: 10.1016/0005-2787(71)90747-7. [DOI] [PubMed] [Google Scholar]
- 22.Thaler D S, Stahl F W. DNA double-chain breaks in recombination of phage lambda and of yeast. Annu Rev Genet. 1988;22:169–197. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- 23.Thaler D S, Stahl M M, Stahl F W. Double-chain-cut sites are recombination hotspots in the Red pathway of phage lambda. J Mol Biol. 1987;195:75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- 24.Thaler D S, Stahl M M, Stahl F W. Evidence that the normal route of replication-allowed Red-mediated recombination involves double-chain ends. EMBO J. 1987;6:3171–3176. doi: 10.1002/j.1460-2075.1987.tb02628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Thaler, D. S., M. M. Stahl, and F. W. Stahl. Unpublished data.
- 25.Thaler D S, Stahl M M, Stahl F W. Tests of the double-strand-break repair model for Red-mediated recombination of phage λ and plasmid λ dv. Genetics. 1987;116:501–511. doi: 10.1093/genetics/116.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Van Steensel B, Smogorzewska A, De Lange T. TR F2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 25b.Wang, G. R. Unpublished results.
- 26.Wang, G.-R., and E. B. Goldberg. Unpublished data.
- 27.Wang, G.-R., A. Vianelli, and E. B. Goldberg. Unpublished data.
- 28.Weigle J. Assembly of phage lambda in vitro. Proc Natl Acad Sci USA. 1966;55:1462–1466. doi: 10.1073/pnas.55.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W-W, Yaneva M. On the mechanisms of Ku protein binding to DNA. Biochem Biophys Res Commun. 1992;186:574–579. doi: 10.1016/s0006-291x(05)80847-2. [DOI] [PubMed] [Google Scholar]