Abstract
Background and Aims
Inflammatory bowel disease [IBD] is a chronic relapsing disorder of the gastrointestinal tract, which generally manifests as Crohn’s disease [CD] or ulcerative colitis [UC]. These subtypes are heterogeneous in terms of disease location and histological features, while sharing common clinical presentation, genetic associations and, thus, common immune regulatory pathways.
Methods
Using miRNA and mRNA coupled transcriptome profiling and systems biology approaches, we report a comprehensive analysis of blood transcriptomes from treatment-naïve [n = 110] and treatment-exposed [n = 177] IBD patients as well as symptomatic [n = 65] and healthy controls [n = 95].
Results
Broadly, the peripheral blood transcriptomes of CD and UC patients were similar. However, there was an extensive gene deregulation in the blood of IBD patients, while only a slight deregulation in symptomatic controls, when compared with healthy controls. The deregulated mRNAs and miRNAs are mainly involved in the innate immunity and are especially enriched in neutrophil activation-related pathways. Oxidative phosphorylation and neutrophil activation-related modules were found to be differentially co-expressed among treatment-naïve IBD as compared to healthy controls. In the deregulated neutrophil activation-related co-expression module, IL1B was identified as the central gene. Levels of co-expression among IL1B and chemosensing receptor [CXCR1/2 and FPR1/2] genes were reduced in the blood of IBD patients when compared with healthy controls.
Conclusions
Immune dysregulation seen in peripheral blood transcriptomes of treatment-naïve IBD patients is mainly driven by neutrophil activation.
Keywords: Inflammatory bowel disease, peripheral blood, gene expression
Graphical Abstract
1. Introduction
Beyond oxygenation and nutrition of tissues, circulating peripheral blood cells act as a surveillance system for damaged tissue, reflecting and providing information on pathological events occurring throughout the human body, especially those related to immune dysfunction.1 One such dysfunction is inflammatory bowel disease [IBD], an idiopathic disease probably caused by an inappropriate immune response against environmental factors, including luminal and microbial antigens, in genetically susceptible hosts.2,3 IBD encompasses two major subtypes, specifically Crohn’s disease [CD] and ulcerative colitis [UC], each showing heterogeneity in terms of disease location, histological features, as well as response to treatment,4 but both subtypes also show significant overlap in their clinical presentation and genetic predisposition.2,5,6 Various subtypes of blood circulating immune cells, including CD4+ and CD8+ T cells, CD14+ and CD16+ monocytes, as well as neutrophils, have been implicated in the pathogenesis of IBD and associated with clinical heterogeneity, including disease activity and treatment response.7–11 Associations between the observed heterogeneity of the IBD subtypes and variations in blood transcriptomes have also been described.12,13 Previous studies have specifically aimed to identify diagnostic biomarkers to discriminate between CD and UC using microRNA [miRNA] or messenger RNA [mRNA] expression data, but most of these studies used small clinical cohorts or only analysed a preselected group of candidate miRNAs or mRNAs. The results from these studies are rather inconsistent and may have been influenced by previous disease history, i.e. age of diagnosis, disease relapse frequency, comorbidities and especially systemic treatment. Imprecision in this information and unbalanced patient selection or missing cross-disease comparisons can easily lead to misinterpretation of newly identified biomarkers and predictive models. Also, the association of newly identified molecules to disease pathogenesis can be confounded, since blood-based biomarkers potentially reflect the secondary effects of the illness or treatment rather than pathophysiological factors.14
A comprehensive systemic network analysis of miRNA- and mRNA-coupled blood profiling in treatment-naïve IBD patients has not yet been performed. Here, we report an exploratory analysis of blood miRNA and mRNA transcriptomes from treatment-naïve and treatment-exposed IBD patients as well as control individuals using differential expression, gene set enrichment analysis and tensor decomposition of gene co-expression networks. By comparing transcriptomes among groups of interest, we describe differentially expressed transcripts and differentially co-expressed gene programmes as well as define biological pathways in which they are involved.
2. Materials and Methods
2.1. Patients and samples
Study participants were recruited in two cohorts, including 205 Swedish individuals and 242 German individuals. For all study participants, peripheral blood samples and clinical information were collected. The study was approved by the respective local ethics committees (PopGen 2.0 Network [P2N] and ethics committee of the Medical Faculty of the University Hospital Schleswig-Holstein, Kiel, Germany; Uppsala Regional Ethics Committee 2010/313). All participants provided written informed consent.
The German cohort comprised 65 healthy individuals and 177 patients diagnosed with IBD, including CD [n = 100] and ulcerative colitis UC [n = 77]. All German patients were systemically and/or topically treated with one or more of the following drugs: infliximab, adalimumab, methotrexate, azathioprine, mesalazine, sulfasalazine or corticosteroids [for details see Supplementary Table S1]. In contrast to the treatment-exposed German cohort, the Swedish cohort [Swedish Inception Cohort in IBD, SIC IBD] included 175 treatment-naïve patients, 17–78 years of age, referred to the gastroenterological unit at six Swedish hospitals, for suspected IBD. The presence of gastrointestinal symptoms, such as diarrhoea, abdominal pain and blood or mucus in stool for >2 weeks, indicative of IBD, was an inclusion criterion. The diagnosis of IBD was established according to internationally accepted criteria, following thorough clinical, microbiological, endoscopic, histological and radiological evaluation. The diagnoses comprise CD [n = 52] and UC [n = 58]. Patients with gastrointestinal symptoms with no endoscopic or histological signs of IBD-associated inflammation at inclusion, and no evidence of IBD during follow-up [for details see Supplementary Table S2] were considered as symptomatic controls [SC, n = 65]. In total, 30 healthy individuals were also included in the SIC IBD cohort.
Harvey–Bradshaw index [HBI]15 was used to classify disease activity in patients with CD and partial Mayo score16 in patients with UC. Activity groups [remission, mild, moderate and severe activity] were specified employing standard thresholds.16,17 In addition to medication and disease activity, patients were evaluated regarding common clinical parameters such as age, sex and smoking status. Furthermore, information on disease location, behaviour, extent [Montreal classification18] and serological markers (C-reactive protein [CRP], albumin) were collected. The summarized phenotypic and clinical information of the participants is provided in Table 1.
Table 1.
Clinical and phenotypic characteristics of the Swedish [treatment-naïve] and the German [treatment-exposed] study cohorts
| Swedish cohort | German cohort | ||||||
|---|---|---|---|---|---|---|---|
| Treatment- naïve CD, n = 52 | Treatment- naïve UC, n = 58 | Symptomatic controls, n = 65 | Healthy controls, n = 30 | Treatment-exposed CD, n = 100 | Treatment-exposed UC, n = 77 | Healthy controls, n = 65 | |
| Mean age, years [range] | 33.6 [17–76] | 35.5 [18–73] | 39.3 [18–78] | 47.4 [21–69] | 38.5 [15–61] | 39.4 [18–73] | 69.7 [56–82] |
| BMI [range] | 25 [16–92] | 27.1 [18.8–92] | |||||
| Sex female, n [%] | 26 [50] | 27 [46.6] | 39 [60] | 15 [50] | 61 [61] | 40 [51.9] | 25 [38.5] |
| Smoking, n [%] | |||||||
| Current | 9 [17.3] | 8 [13.8] | 9 [13.8] | 0 [0] | 21 [21] | 11 [14.3] | 33 [50.8] |
| Previous | 12 [23.1] | 16 [27.6] | 9 [13.8] | 0 [0] | 13 [13] | 5 [6.5] | 0 [0] |
| Never | 25 [48.1] | 31 [53.4] | 38 [58.5] | 0 [0] | 63 [63] | 56 [72.7] | 32 [49.2] |
| Unknown | 6 [11.5] | 3 [5.2] | 9 [13.8] | 30 [100] | 3 [3] | 5 [6.5] | 0 [0] |
| Location, n [%] | |||||||
| Ileal, L1 | 19 [38] | 12 [12] | |||||
| Colonic, L2 | 15 [30] | 44 [44] | |||||
| Ileocolonic, L3 | 16 [32] | 37 [37] | |||||
| Unknown | 7 [7] | ||||||
| Behaviour, n [%] | |||||||
| Inflammatory, B1 | 38 [76] | 52 [52] | |||||
| Stricturing, B2 | 6 [12] | 13 [13] | |||||
| Penetrating, B3 | 6 [12] | 30 [30] | |||||
| Unknown | 5 [5] | ||||||
| Perinanal disease | 5 [9.6] | 21 [21] | |||||
| Extent, n [%] | |||||||
| Proctitis, E1 | 19 [32.2] | 8 [10.4] | |||||
| Left-sided colitis, E2 | 16 [27.1] | 25 [32.5] | |||||
| Extensive colitis, E3 | 24 [40.7] | 33 [42.9] | |||||
| Unknown | 11 [14.3] | ||||||
| HBI, n [%] | |||||||
| Remission | 18 [36] | 23 [23] | |||||
| Mild | 7 [14] | 12 [12] | |||||
| Moderate | 11 [22] | 12 [12] | |||||
| Severe | 4 [8] | 2 [2] | |||||
| Unknown | 10 [20] | 51 [51] | |||||
| Partial Mayo Index, n [%] | |||||||
| Remission | 2 [3.4] | 15 [19.5] | |||||
| Mild | 17 [28.8] | 18 [23.4] | |||||
| Moderate | 28 [47.5] | 5 [6.5] | |||||
| Severe | 8 [13.6] | 7 [9.1] | |||||
| Unknown | 4 [6.8] | 32 [41.6] | |||||
| Mean albumin, g/L [range]a | 36.4 [24–45] | 39.2 [28–49] | 39.6 [30–48] | ||||
| Mean CRP, mg/L [range]b | 27.7 [0.89–242] | 7.6 [0.3–91] | 6.8 [0.3–87] | 8 [0–67.5] | 6.4 [0–70.2] | 3.6 [0–48.2] |
UC, ulcerative colitis; CD, Crohn’s diseases; BMI, body mass index; HBI, Harvey–Bradshaw index; CRP, C-reactive protein.
aInformation on albumin was missing in eight [4.6%] of Swedish cases [and symptomatic controls] and all German individuals.
bInformation on CRP was missing in eight [4.6%] of Swedish cases [and symptomatic controls] and 21 German individuals [8.7%].
2.2. RNA isolation
Peripheral blood samples were collected and stabilized using a PAXgene Blood miRNA System [Qiagen]. Total RNA was isolated using QIAcube automation with the PAXgene Blood RNA Kit [Qiagen] in accordance with the manufacturer’s instructions. Quality control and assessment of total RNA samples were performed using an Agilent 2200 TapeStation [Agilent Technologies].
2.3. Small RNA sequencing analysis for miRNA profiling
Small RNA libraries were prepared using a TruSeq Small RNA Sample Preparation Kit [Illumina] according to the manufacturer’s protocol with 1 μg of total RNA as an input per sample of Swedish [n = 205] and German [n = 242] cohorts. The generated small RNA libraries were quality-controlled using an Agilent 2200 TapeStation [Agilent Technologies] and sequenced using an Illumina HiSeq 2500 platform [1 × 50 bp SR, v3.0 or v4.0]. The obtained demultiplexed raw sequencing reads [.fastq] were processed using cutadapt v1.919 to remove adapters, low-quality bases [<Q20] and reads shorter than 18 nucleotides. Quality-controlled reads were mapped to miRNA reference sequences from miRBase release 2220 using mirAligner21 with default parameters. The R package isomiRs22 and its default parameters were used to generate the count matrix of miRNA reads per library. Samples with fewer than one million mapped reads, and those for which the number of detected miRNAs on a log2 scale fell below Q1−1.5 interquartile range [IQR], were removed from further analysis. Non-expressed miRNAs were excluded based on their average expression [>1 raw count] values in libraries per cohort. For each cohort, quality-controlled miRNA count data were then normalized using calcNormFactors followed by voomWithQualityWeights functions from the edgeR23 and the limma24 R packages, respectively. The generated quality-controlled counts and raw sequencing reads of Swedish and German cohorts have been deposited at NCBI Gene Expression Omnibus [GEO]25 under accession numbers GSE169569 and GSE169570, respectively.
2.4. BeadChip microarray analysis for transcriptomic profiling
The total RNA samples of the Swedish cohort [n = 205] were reverse transcribed and biotin-labelled using the TargetAmp-Nano Labeling Kit for Illumina Expression BeadChip [Epicentre] according to the manufacturer’s protocol. The labelled antisense RNA was hybridized to a Human HT-12 v4 BeadChip array [Illumina] following the standard hybridization protocol of the supplier. Array imaging was performed on an iScan system [Illumina] according to the manufacturer’s protocol. The obtained raw intensity information [.idat] and probe annotation [.bgx] files were used to generate a probe-intensity matrix of transcripts per sample using the read.idat function from the limma24 R package. Background correction followed by quantile normalization was performed using limma’s neqc function.26 The probes were annotated using the mappings from the illuminaHumanv4.db27 R package. Probes which were flagged as ‘bad’ or having ‘No match’ [without match to coding sequences or to any genomic region] were discarded. Non-expressed probes [in at least 10% of samples] were identified by limma’s detectionPValues function and removed from further analyses. Probes that were unannotated or mapped to ribosomal genes [those beginning with MRP, RPL and RPS] were also discarded from further analysis. Finally, the findLargest function from the R package genefilter28 was used to remove a set of quality-controlled probes that mapped to the same gene symbol, while retaining the probe with the highest median intensity value, resulting in 11 727 probes. The generated quality-controlled expression values and raw intensities have been deposited at NCBI GEO25 under accession number GSE169568.
2.5. Data analysis
The quality-controlled and normalized data of both miRNA and mRNA expression datasets were analysed using limma’s24 workflows for differential expression analysis including age and sex as covariates. Lists of significantly differentially expressed protein-coding genes or validated target genes [miRTarBase29] of significantly deregulated miRNAs were used for gene set enrichment analysis [GSEA] in Gene Ontology [GO] terms implemented in the clusterProfiler30 R package. Cell-type enrichment analysis was performed based on cell type-specific mRNA,31 and miRNA32 [Supplementary Table S3] signatures of main immune cell types were made using the single sample gene set enrichment analysis [ssGSEA] method.33 Co-expression networks [signed scale-free topology overlap matrices] of protein-coding genes were generated using the weighted gene correlation network analysis [WGCNA]34 workflow. Gene co-expression networks of each diagnosis [CD, UC, SC and HC] were assembled into third-order tensor and decomposed into ten latent components [R = 10]. Tensor decomposition was performed using the non-negative CP tensor decomposition by hierarchical alternating least squares [HALS] method35 implemented in the ncp_hals function from the TensorTools Python package.36 Knee point detection was applied to remove low scoring genes from each co-expression component [module] using the KneeLocator function implemented in the kneed package of Python.37 Co-expression modules were functionally annotated using GSEA. Experimentally validated protein-coding gene–gene interactions of the co-expression modules were retrieved from the STRING v11 database.38 Experimentally validated miRNA–target interactions [DIANA-TarBase39] were integrated into the networks using negative correlation. Correlations between clinical variables and a component’s eigengene40 were measured by Pearson’s correlation. Detailed descriptions of the data analyses performed are provided in the Supplementary Methods.
3. Results
3.1. Characterization of peripheral blood transcriptomes derived from IBD patients and controls
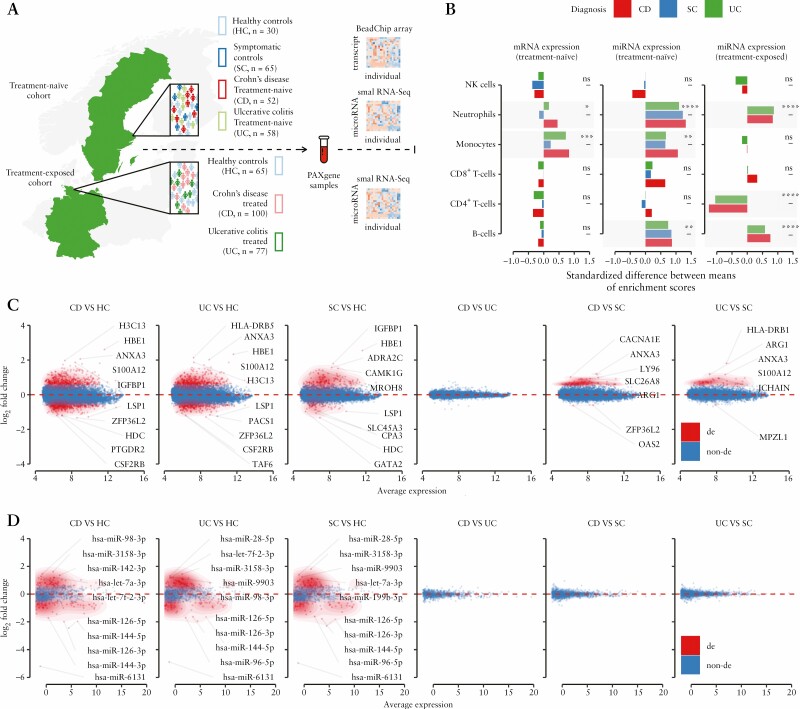
By including two independent cohorts of IBD patients and controls from Germany [treatment-exposed patients] and Sweden [treatment-naïve patients], we were able to investigate transcriptional profiles of IBD while considering the interference of medications via comparison of results from the two cohorts. For both cohorts, miRNA expression profiles were generated using small RNA-sequencing (RNA-seq), while mRNA expression profiles were generated using the BeadChip array only for the treatment-naïve cohort [Figure 1A].
Figure 1.
Characterization of peripheral blood miRNA/mRNA transcriptomes obtained from IBD treatment-naïve/-exposed patients and control individuals. [A] Schematic representation of the study design and included cohorts. The study consists of two independent cohorts – German and Swedish. The German cohort comprises treatment-exposed IBD patients [CD and UC subtypes] and healthy controls [HC], while the Swedish cohort comprises treatment-naïve patients [CD and UC], symptomatic [SC] and healthy controls [HC]. For both cohorts, miRNA expression profiles were generated using small RNA-seq, while the mRNA expression profiles were generated using the BeadChip array for the treatment-naïve cohort only. Significance levels: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant. [B] Immune cell type enrichment analysis based on cell-specific multi-marker gene expression. Peripheral blood transcriptomes show relative increase in B cells [on the miRNA level] and myeloid cells, including neutrophils and monocytes [on mRNA and miRNA levels] in the treatment-naïve IBD patients. While treatment-exposed IBD patients also show an increase in B cells and neutrophils, monocyte levels remain unchanged compared to healthy controls [HC]. The treatment-exposed IBD patients also show a relative decrease in CD4+ T cells, suggesting treatment effects on cellular blood composition. Coupled mRNA [C] and miRNA [D] differential gene expression analysis of treatment-naïve IBD patients [CD and UC] and control individuals [SC and HC]. While an extensive transcript deregulation [FC > 1.5 and pFDR < 0.05] was observed in the peripheral blood of inflammatory [CD and UC] and symptomatic traits [SC] when compared to healthy controls [HC], there were no significantly deregulated transcripts between the CD and UC subtypes of IBD. This observation was consistent on mRNA [treatment-naïve] and miRNA [treatment-naïve and treatment-exposed] expression levels [see Supplementary Figure 1B and Table S4 for results for the treatment-exposed cohort]. Top five up- and downregulated transcripts are annotated as gene symbols or miRNA names.
After data pre-processing and quality control, a total of 12 284 transcripts [including miRNAs] were found to be expressed in peripheral blood. For each generated dataset, normalized gene expression values were used to evaluate the global similarity structure of transcriptomes by utilizing multidimensional scaling analysis [MDS], which positions samples in two-dimensional space in relation to dissimilarity distances between them. In all three expression datasets, the results of MDS showed highly heterogeneous blood transcriptomes of the inflammatory and symptomatic traits [CD, UC and SC] when compared to healthy individuals [HC], who displayed lower within-group variability [Supplementary Figure S1A]. The analysis also revealed a high overlap [95% confidence ellipses for group centroids] among the subtypes of IBD [CD and UC] as well as symptomatic controls [SC], suggesting a considerable similarity of their blood transcriptomes. The overlap between CD and UC patients’ transcriptomes was consistent in both cohorts.
An elevated white blood cell count is common in IBD patients.41 To determine whether transcriptional changes reflect alterations of immune cell composition in peripheral blood, relative estimates of specific cell populations were evaluated using cell type enrichment analysis [see Methods]. The gene-based marker cell type estimates showed a consistent increase in relative abundance of neutrophils in the blood of IBD patients when compared to HC. This increase was independent of data type and treatment status [Figure 1B]. The relative increase of neutrophils was more pronounced in CD than in UC patients. Interestingly, when compared to HC, higher levels of monocytes were observed in the treatment-naïve IBD patients, but not in treatment-exposed patients. Based on miRNA data alone, the relative abundance of B cells was elevated in both cohorts.
3.2. Differential expression analysis reveals extensive gene deregulation between IBD patients and healthy controls, but not between the subtypes CD and UC
To further characterize the differences between IBD patients and healthy individuals, as well as the transcriptional differences between treatment-naïve and treatment-exposed IBD patients, differential gene expression analysis [DEA] was performed. To achieve this, the expression datasets (miRNA [i] and mRNA [ii] expression of treatment-naïve cohorts, and miRNA [iii] expression of treatment-exposed cohorts) were analysed separately and the results from the two cohorts were then compared.
The most profound deregulation of transcripts [FC > 1.5 and pFDR < 0.05] was observed in peripheral blood of inflammatory [and symptomatic] patients when compared to healthy controls. This effect was observed in both treatment-naïve [Figure 1C and D] and treatment-exposed cohorts [Supplementary Figure S1B] with a substantial overlap of differentially expressed transcripts among the pairwise comparisons. For example, in the treatment-naïve patients, transcripts encoded by HBE1, LSP1, PTGDR2, HIST2H3D and IGFBP1 were consistently among the most deregulated genes in the blood of IBD patients and symptomatic patients when compared to healthy controls [CD vs HC, UC vs HC and SC vs HC; Supplementary Table S4]. Additionally, hsa-mir-144, hsa-mir-618, hsa-mir-98 and hsa-mir-96 were among the most deregulated miRNAs in treatment-naïve, as well as in treatment-exposed, CD and UC patients compared with healthy controls. Overall, correlation analysis of fold change values showed significant concordance between miRNA differential expression in treatment-naïve and treatment-exposed IBD patients compared to healthy controls [r = 0.7 in CD vs HC; r = 0.6 in UC vs HC]. By contrast, several miRNAs [n = 12 in CD vs HC; n = 13 in UC vs HC] showed an opposite direction of deregulation in the treated IBD patients, suggesting their possible relation to the effects of medications [Supplementary Figure S1C].
Interestingly, none of the transcripts in the peripheral blood were found to be significantly differentially expressed between the subtypes of IBD [CD vs UC]. This observation was consistently observed on both the miRNA and mRNA levels and also independent of treatment status [Figure 1C and D and Supplementary Figure S1B], suggesting a similar pathological mechanism [at least secondary] between the subtypes. However, the comparisons of treatment-naïve IBD subtypes with symptomatic controls [SC] showed differential expression on the mRNA level but not on the miRNA level. Most of the differentially expressed genes [e.g. ANXA3, ARG1, S100A12, LY96, JCHAIN and SLC26A8] were overlapping in both of the comparisons [CD vs SC and UC vs SC], whereas some of the genes were found to be uniquely deregulated in one of the IBD subtypes. Genes such as HLA-DRB1 and CCL23 were found to be differentially expressed in UC alone, while genes including XIST and OAS2 were only deregulated in CD when compared to SC. These observations, however, should be interpreted with caution because the selected thresholds of DEA are always arbitrary [Figure 1C and D; Supplementary Table S4].
3.3. Differentially expressed mRNAs and miRNAs are mainly involved in neutrophil signalling pathways
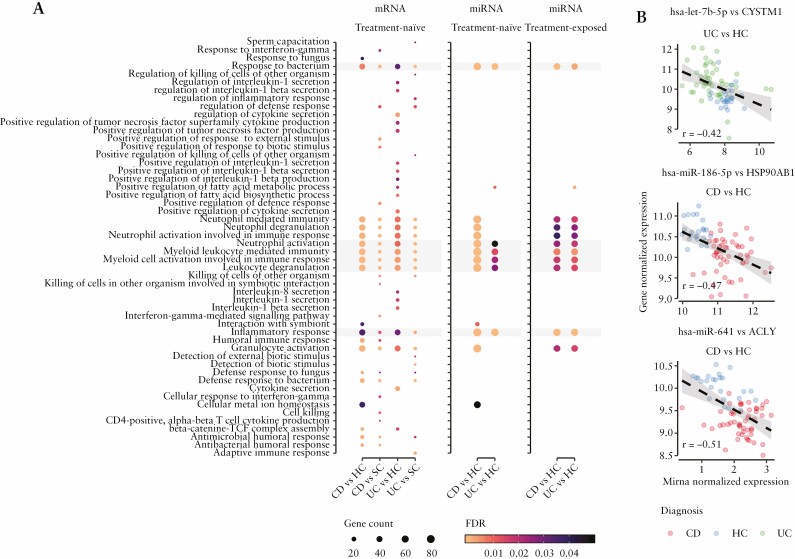
To place the differential expression results into a biological context, GSEA was performed for each pairwise comparison using lists of either significantly deregulated genes or validated target genes of significantly deregulated miRNAs [see Methods section].
Independent of data type and treatment status, the differentially expressed transcripts were recurrently enriched in pathways related to immune response and myeloid mediated immunity, especially neutrophil signalling [Figure 2A]. More precisely, GO terms including ‘neutrophil activation’, ‘neutrophil activation involved in immune response’, ‘neutrophil degranulation’ and ‘neutrophil mediated immunity’ were among the top ten most significant pathways in the comparisons between IBD subtypes and control groups of differentially expressed protein-coding genes [CD vs HC, UC vs HC, CD vs SC and UC vs SC; Supplementary Table S5]. To challenge whether these results are caused by the relative increase of the neutrophils in the blood of IBD patients [as shown in the cell type enrichment analysis], GSEA was performed separately on the lists of up- and downregulated genes. The results implied that although the majority of neutrophil activation-related genes were upregulated, a number of these genes [ACLY, HUWE1, PTPRC, CXCR1, CXCR2, etc.] were downregulated in comparisons between IBD and healthy controls [data not shown]. This supports the conjecture that the effect was not only driven by the quantitative increase in neutrophils, but to a certain extent rooted in a qualitative transcriptional change. Additionally, the reliability of miRNA–target enrichment analyses was assessed by Pearson’s correlation. The analysis was performed on normalized expression values of differentially expressed miRNAs and their known target genes that mapped to the neutrophil activation pathway [see Methods]. There were numerous [n = 912] unique negative correlations between miRNAs and their targets, reflecting the expected negative regulatory effect of miRNAs [Figure 2B]. Furthermore, the replicability of our results was evaluated by performing identical GSEA on the differentially expressed genes obtained from a study by Ostrowski et al.42 As in our data, neutrophil activation-related pathways were among the most significantly over-represented terms within differentially expressed genes comparing adult- as well as paediatric-onset IBD to their according controls [Supplementary Figure S2 and Table S6].
Figure 2.
Differentially expressed mRNAs as well as miRNAs are involved in inflammatory response and neutrophil activation signalling in the blood of IBD patients. [A] Gene set enrichment analysis [GSEA] of differentially expressed genes and validated target genes of significantly deregulated miRNAs. The figure displays the most significantly overrepresented biological pathways [GO biological process terms; y-axis] of differentially expressed protein-coding genes [mRNA] and significant terms that overlap with validated target genes of significantly deregulated miRNAs in each pairwise comparison [x-axis]. Dot size corresponds to the proportion of differentially expressed genes that overlap total genes of the particular pathway, while colour indicates statistical significance [FDR] of the pathway enrichment. Pathways highlighted in grey are overlapping in all pairwise comparisons of all three differential gene expression analyses (mRNA and miRNA results of treatment-naïve traits and miRNA results of treatment-exposed IBD patients compared to healthy controls [HC]). Complete results of GSEA are provided in Supplementary Table S5. [B] Example of negative correlations of miRNA and their target genes, which are involved in neutrophil activation pathways. The figure shows normalized miRNA expression on the x-axis, normalized expression values of its validated target gene on the y-axis and their regression line [see Methods]. Every data point corresponds to an individual whose diagnosis is indicated by colour. Overall, GSEA results of differentially expressed genes [and miRNAs] in blood of IBD patients show consistent overrepresentation of neutrophil activation pathways. These results are consistent independently of treatment status.
Collectively, the results show that differentially expressed genes, including miRNAs, in the blood of IBD patients are mainly enriched in pathways related to innate, rather than to adaptive, immunity.
3.4. Gene co-expression networks show disturbed connectivity in neutrophil activation pathways in patients with chronic inflammation
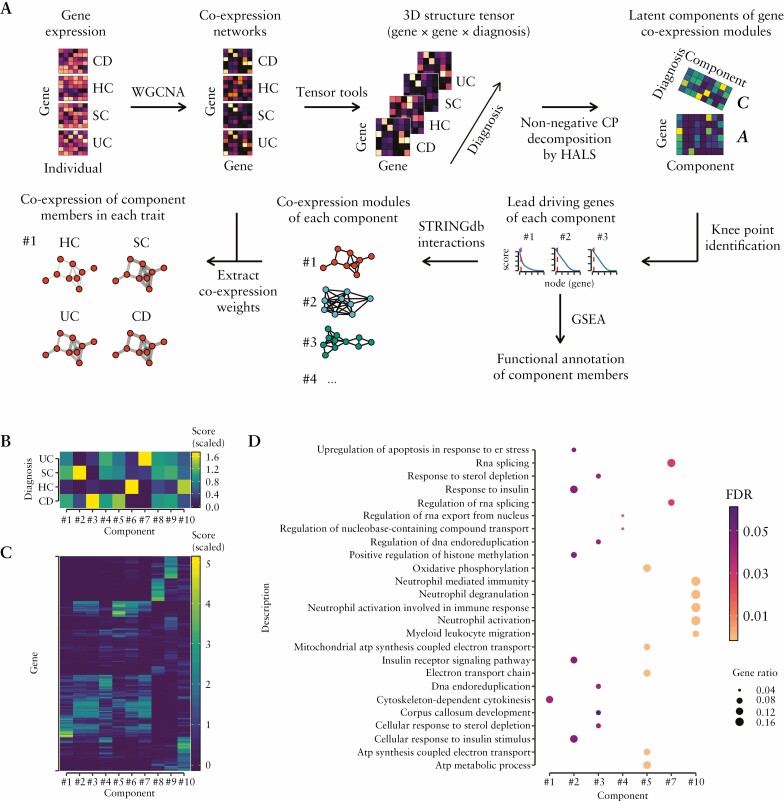
Together, the transcriptome analyses show higher heterogeneity, increased number of myeloid cells and deregulated innate immune-related pathways in the blood of individual IBD subtypes compared to healthy controls. To gain deeper insight into the complexity of blood transcriptomes and to extract differences in their modular structure [i.e. co-expressed gene programmes] among distinct diagnoses [CD, UC, SC and HC], a gene co-expression network analysis was conducted. First, co-expression networks for each diagnosis were constructed using weighted pairwise correlations of all included genes. Then, to capture differences of co-expressed gene programmes among traits, non-negative tensor decomposition and other downstream analyses were applied on the constructed co-expression networks [for detailed workflow see Figure 3A and Methods].
Figure 3.
Differences of gene co-expression patterns in blood among different diagnoses, including IBD. [A] To identify gene co-expression modules, activity of which is different across blood transcriptomes of treatment-naïve UC, CD, SC diagnoses and healthy controls [HC], the following strategy was used: [1] to identify co-expressed gene pairs, weighted gene correlation networks [using WGCNA] were generated for each trait [gene × gene]; [2] to determine gene co-expression modules, whose activity is different among diagnoses, diagnosis-wise co-expression networks were assembled into 3D tensor [gene × gene × diagnosis] and decomposed into latent matrices A and C, which represent the membership of each gene in each component [gene × component] as well as the membership of each diagnosis [CD, UC, SC and HC] in each component [diagnosis × component], which indicates the co-expression activity of a particular gene module in a given diagnosis; [3] to retain only the genes that are driving a particular co-expression component, knee point detection was used to remove low scoring genes; [4] to determine biological function, co-expression components were functionally characterized using gene set enrichment analysis [GSEA] and gene ontology terms; [5] to obtain biologically meaningful gene–gene interactions of each component’s network, the component-driving genes were mapped to the STRING database; [6] to visualize co-expression patterns of each component in different diagnoses, weighted correlation values were added to diagnosis-wise component networks. [B] The latent matrix C of the decomposed tensor of the co-expression networks. The score [indicated by colour intensity] shows activity of a particular co-expression component in a given diagnosis. [C] Similarly, this heatmap represents the latent matrix A, which contains the membership score [indicated by colour intensity] of each gene in each co-expression component. This score was used to identify the lead driving genes of each component. The lead driving genes mapped to the STRING database of each component are provided in Supplementary Figure S3. [D] Dotplot displaying statistically significant results of the component’s functional annotation using GSEA. Dot size corresponds to the proportion of a component’s [x-axis] genes that overlap the total genes of the particular GO term [y-axis], while colour indicates statistical significance [FDR] of the overrepresentation test.
The variability in gene co-expression among the diagnoses was captured in ten latent factors [components], which encode interactions between gene co-expression and diagnoses via membership scores of each diagnosis [Figure 3B] and each gene [Figure 3C] in a given component. The highest membership scores having genes [referred to as co-expression modules; Supplementary Figure S3] within each component were functionally annotated in silico using GSEA. Annotation of component’s co-expression modules showed that seven out of ten modules were significantly enriched [pFDR < 0.05] for at least one GO term [Figure 3D and Supplementary Table S7]. Some of the component’s co-expression modules were of particular interest, including neutrophil signalling- [component #10] and oxidative phosphorylation- [component #5] related modules [Figure 3D]. To get more detail on specific gene co-regulatory relationships, experimentally validated gene interactions of functionally annotated component module genes were retrieved [Supplementary Figure S4] and mapped to the previously generated co-expression networks [see Methods]. The co-expression level of the oxidative phosphorylation module network was highly pronounced in CD followed by UC, SC and lowest in HC [Supplementary Figure S5], showing its activation during gastrointestinal inflammation. The most interconnected genes, manifesting as central nodes of this co-expression network, were ATP5F1C, ATP5PO, COX7C and HINT1. This may indicate the respective importance of these nodes within this context. In contrast to the oxidative phosphorylation module, the co-expression level of the neutrophil signalling module was reduced in IBD. Co-expression among its member genes as well as their targeting miRNAs was lowest in the blood transcriptomes of CD followed by UC, and SC, when compared to HC [Figure 4A]. The most interconnected genes of this module included IL1B, CXCR1, CXCR2, FPR1 and FPR2. The two CXC chemokine receptors were downregulated, despite the fact that the genes IL1B and FPR1/2 were not significantly deregulated in inflammatory traits when compared to HC [Supplementary Table S4]. Negative correlation-based integration of miRNA and their known target mRNA expression revealed miR-10b-5p, miR-335-5p and let-7b-5p as being the most interconnected miRNAs in the module [Figure 4A], suggesting their possible regulatory function in neutrophil signalling.
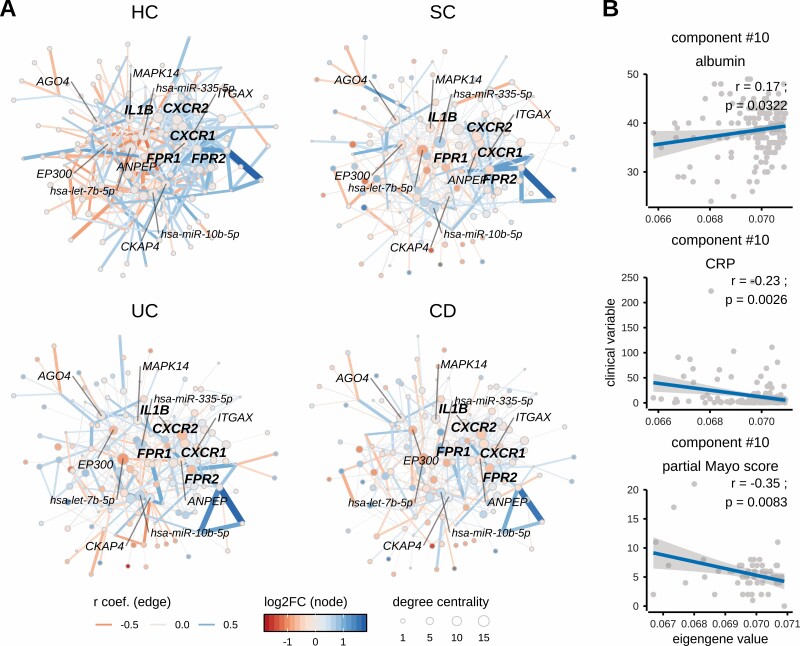
Figure 4.
Neutrophil activation-related co-expression module in different diagnoses and its correlation with clinical variables such as albumin and C reactive protein [CRP]. [A] Networks displaying co-expression module [component #10] activity among diagnoses [CD, UC, SC] and healthy controls [HC]. The neutrophil activation-related component module shows strong co-expression (note edge widths [thickness of line] between nodes) in healthy controls [HC], while co-expression of its member genes is reduced in inflammatory traits with the weakest co-expression in CD followed by UC and SC. The most central [hub] genes of this co-expression module are IL1B, CXCR1, CXCR2, FPR1 and FPR2 [highlighted in bold], whose differential expression during inflammation may disturb the co-expression of other member genes. Negative correlation-based integration of miRNA and their known target mRNA expression revealed miR-10b-5p, miR-335-5p and let-7b-5p as being the most interconnected miRNAs of co-expression module #10. The correlation coefficient [r] corresponds to co-expression activity [indicated by edge width], while the direction of correlation corresponds to edge colour. Nodes of the network represent genes [or miRNAs], the differential expression [log2FC] of which, compared to healthy controls [HC], is indicated by the colour gradient. The size of a node indicates its degree centrality, i.e. number of gene–gene interactions of a given gene [node]. The most central genes and miRNAs [hubs; having highest values of centrality degree] are annotated using gene or miRNA symbols. [B] Correlation of clinical variables [serum albumin concentration, serum and CRP concentration in CD and UC patients and partial Mayo score only in UC patients] and component [#10] eigengenes [summarized expression values, see Methods].
Further analysis of the clinical parameters showed that the eigengene [summarized expression value] of the neutrophil signalling module [component #10] was negatively correlated with CRP [r = −0.23; p = 0.003] and partial Mayo score [r = −0.35; p = 0.0083], but was positively correlated with albumin [r = 0.17; p = 0.032] levels of the treatment-naïve IBD patients [Figure 4B], but not with HBI score [data not shown]. Overall, this suggests a consequence of gastrointestinal inflammation rather than persistent and disease activity-independent deregulation of these signalling pathways.
4. Discussion
Using transcriptome profiling, we have generated the largest and most comprehensive investigation of combined mRNA and miRNA expression in peripheral blood of IBD to date. Our study comprised two independent IBD cohorts: a treatment-naïve cohort to study the combined miRNA and mRNA expression profiles without previous exposure to IBD medications, and a treatment-exposed cohort. The latter cohort was used to test if medications have an effect on miRNA expression profiles [Figure 1A]. As expected, we observed higher heterogeneity of blood transcriptomes in IBD patients and symptomatic controls [SC] than in healthy controls [HC] [Supplementary Figure S1A]. Using cell type enrichment analyses, we show that this observation was partially explained by a compositional shift towards innate immune cells [i.e. monocytes and neutrophils] in the peripheral blood of the treatment-naïve IBD patients [Figure 1B]. Based on miRNA cell markers, the blood of treatment-exposed IBD patients was also enriched in neutrophils, but not in monocytes. This may be explained by the effect of immunosuppressants, such as glucocorticoids, since their mechanism of action directly or indirectly affects the proliferation and migration of monocytes.7 Although we observed extensive gene deregulation between IBD and healthy controls, we did not identify any significantly differentially expressed transcripts [FC > 1.5 and pFDR < 0.05] between CD and UC [Figure 1Cand D; Supplementary Figure S1B]. Albeit in line with some previous studies, this observation also contradicts some of the earlier reported findings [including our own] on miRNA12,13,43,44 as well as mRNA levels.45,46 We believe that these inconsistencies may be due to small sample sizes in the previous studies, use of different profiling technologies or different measures of disease activity or treatment effects, since all previous studies have examined patients with ongoing or previous medical treatment. For example, Schaefer et al. suggested a panel of miRNAs to discriminate CD from UC.13 However, five of these six miRNAs [miR-21-3p, miR-31-5p, miR-101-3p, miR-375-3p and miR-146a-5p] are known to have anti-inflammatory properties and have been associated with immune response to infections and inflammation.47,48 This may indicate that the diagnostic capacity of this signature in the North American cohort was explained by differences in inflammatory activity, since the findings were not validated in an independent cohort. Intriguingly, we found the majority of these miRNAs, including miR-146a-5p, miR-21-3p, miR-31-5p and miR-375-3p, were differentially expressed in CD and/or UC compared with healthy controls, but not when comparing CD vs UC patients. Furthermore, miR-375-3p was upregulated in the blood of treatment-exposed patients [~30% of patients received corticosteroids], but was downregulated in the treatment-naïve IBD patients [Supplementary Figure 1C]. This observation is in accordance with the results by Lu et al., who showed reversibility of miR-375-3p downregulation by glucocorticoids in patients with eosinophilic oesophagitis.49 As observed in our data, the analogous effect for miR-210-3p clearly demonstrates the impact of treatment on miRNA expression levels in blood. Regarding protein-coding gene expression, Ostrowski et al. identified nine genes which showed moderate discriminative power [area under the curve = 0.81] for paediatric inactive IBD vs healthy controls.42 However, they were neither able to discriminate between active or inactive adult IBD patients from controls, nor to distinguish between active UC and active CD in paediatric or adult populations.42 We also observed dysregulation of seven out of these nine genes, including S100A12, ANXA3, CACNA1E, GALNT14, MMP9, OPLAH and ATP9A, comparing the IBD subtypes with healthy controls. Notably, ANXA3, CACNA1E and GALNT14 were also differentially expressed between IBD and SC. These genes are all highly expressed in neutrophils and have been previously reported to be differentially expressed in peripheral blood of patients with various inflammatory conditions, including chronic inflammatory diseases such as rheumatoid arthritis.50–53 Upregulation of these genes has also been associated with sepsis,54 and both ANXA3 and S100A12 have been identified as marker genes for bacterial infection50 in peripheral blood.
In general, the majority of differentially expressed coding genes between IBD and HC were enriched in pathways related to bacterial infection and innate immune response. This also includes neutrophil activation/degranulation signalling, which was deregulated based on miRNA differential expression [Figure 2A]. Investigating the miRNA–target gene relationships in the neutrophil activation pathway, we were able to capture known miRNA–target interactions in more detail [Figure 2B]. One such interaction was miR-641 and the ATP citrate lyase [ACLY] target gene. Expression of these transcripts was deregulated and negatively correlated in peripheral blood of IBD patients. Interestingly, Lauterbach et al. previously showed that ACLY is involved in TLR4 signalling and that its inhibition leads to decreased expression of the costimulatory molecule CD86 in the lipopolysaccharide-induced circulating neutrophils and inflammatory monocytes, indicating its importance in systemic immune response to bacterial infection.55 Our weighted gene correlation network analysis also revealed neutrophil activation-related module [component #10] as differentially co-regulated among traits. The co-expression level between gene members of this module was found to be reduced in the blood of IBD patients compared to healthy controls, showing loss of correlation among the module genes in IBD patients [Figure 3B–D]. The most interconnected [hub] genes of this module belong to the interleukin 1 beta [IL1B] signalling pathway, with IL1B itself being the most central gene in the co-expression module [Figure 4A]. The other hub genes of this co-expression module, including the CXC chemokine receptors CXCR1/2 and the formyl peptide receptors FPR1/2, had reduced co-expression as well as expression levels in IBD and symptomatic controls when compared to healthy individuals. These findings are in line with previous reports showing reduced CXCR1/2 expression via the IL1B/CXCL8/CXCR1/2 axis during neutrophil priming, activation and recruitment.56–58 Interestingly, not only can pro-inflammatory IL1B prime neutrophils,59 but primed neutrophils themselves can also activate expression of IL1B.60 Several genetic and functional studies have reported IL1B as being implicated in IBD pathogenesis.61,62 The chemokine receptors CXCR1/2 are known to be broadly co-expressed in immune cells such as neutrophils, T cells and mast cells. Inhibition of these receptors reduces neutrophil recruitment in vivo, suggesting a crucial role in mediating neutrophil migration to sites of acute inflammation.63 It has been previously shown that CXCR1/2 are consistently downregulated [as in our data] in response to bacterial infection in peripheral neutrophils, but are strongly upregulated in monocytes.64,65 This might be caused by either downregulation of CXCR1/2 genes in activated neutrophils, e.g. via CXCL8 and/or miRNAs, or depletion of high CXCR1/2-expressing peripheral neutrophils, due to migration towards the site of inflammation. For instance, miR-335-5p was upregulated in our data and has been previously shown to interact with CXCR1/2 mRNAs and might suppress their expression.66 On the other hand, the existence of heterogeneous neutrophil subtypes [including CXCR1/2low] was shown in peripheral blood of systemic lupus erythematosus patients,67 suggesting that a compositional shift of neutrophil populations in blood is also possible. Such a shift due to neutrophil migration is also supported by the observation of increased expression of CXCR1/2 and their ligand IL-8 in colonic tissues of newly diagnosed treatment-naïve IBD patients.68 This even may be caused by neutrophil priming [e.g. by chemoattractants, microbial products and inflammatory cytokines]59 once passing through inflammation site, which would stimulate the migration and infiltration of CXCR1/2high neutrophils and/or increased production of IL1B in CXCR1/2low neutrophils, which could further prime, or in the presence of other activating agents, activate other neutrophils in the circulation. Recently, Sudhakar et al. identified CXCR2 as a hub gene that was associated with monocyte gene expression modules, which were active across the CD phenotypes of disease behaviour or disease location. However, the fact that the authors did not include expression profiles of neutrophils69 limits the possibility to compare our findings with these previous data. Further genes we identified as hub genes of the neutrophil activation-related co-expression module, namely FPR1/2, are mainly expressed in neutrophils and monocytes and are well known as chemotactic receptors and pattern recognition receptors that interact with bacterial and mitochondria-derived formylated peptides.70 Several reports show that FPR1/2 are associated with IBD pathogenesis possibly via effects on neutrophil migration,71–73 perhaps in a similar fashion to the chemokine receptors. With respect to clinical parameters, our data revealed the summarized expression [eigengene] value of the neutrophil activation module was negatively correlated with CRP and positively correlated with albumin levels, suggesting that the genes of this co-expression module may contribute to the systemic burden of inflammation [Figure 4B]. Another interesting co-expression module [component #5], which we observed to be active in the blood of IBD patients, while inactive in healthy controls, was responsible for oxidative phosphorylation [Supplementary Figure S5]. Bao et al. reported that mitochondria regulate neutrophil activation by generating ATP for autocrine purinergic signalling.74 Purinergic signalling plays a key role in inflammatory processes and modulates immune responses against bacterial and eukaryotic parasites.75 Also, this pathway has been shown to be implicated in gastrointestinal inflammation as well as the pathogenesis of IBD.76
Together, the results of blood transcriptome analysis of treatment-naïve IBD patients presented here point to neutrophil-related innate immune activation, probably in response to bacterial antigens. However, it remains unclear whether these observations are disease-specific and explain the progression from subclinical disease to onset of symptoms, or if they only represent a secondary response to gastrointestinal inflammation. The comparisons with symptomatic controls, which include some patients diagnosed with bacterial infection [such as Clostridium difficile or Campylobacter], suggest that the latter scenario is more plausible, since expression on the miRNA level did not show any significantly deregulated miRNAs comparing IBD and SC. At this point, we also note that we did not apply depletion of highly abundant erythropoietic miRNAs77 in this study, and were therefore unable to capture the complete miRNA repertoire of peripheral blood. Thus, differential expression of some low-abundance miRNAs might be undetected. However, we have captured the signals coming from the most abundant cell types of blood, so the results are still representative. The ‘secondary response’ scenario is also supported by another study analysing blood transcriptomes from patients with primary biliary cholangitis [PBC], primary sclerosing cholangitis [PSC] and IBD.78 The results revealed commonly deregulated genes among the examined autoimmune inflammatory diseases when compared to healthy individuals. In support of this, Nikolaus et al. have shown that polymorphonuclear neutrophil granulocytes are primed to secrete enhanced amounts of pro-inflammatory cytokines [including IL1B] and that this observation is not specific for IBD but rather reflects intestinal inflammation in general.79 Therefore, technologies such as single-cell RNA-seq need to be used to analyse activated peripheral neutrophils coming from blood of various infectious and autoimmune diseases, including IBD, in order to fully answer this question.
To summarize, in this study we show a lack of broader differences in blood transcriptome profiles between the CD and UC subtypes of IBD. This observation was consistent on the mRNA, as well as miRNA, expression levels and also independent of treatment status. However, we cannot exclude that the application of technologies which achieve higher profiling resolution and depth might reveal subtle differences at the transcriptome level between CD and UC peripheral blood. In comparison with healthy controls, the differences in treatment-naïve IBD transcriptomes were highly pronounced and indicated neutrophil activation in peripheral blood. Gene co-expression network analysis showed that IL1B might be substantially involved in neutrophil activation during IBD, since it was identified as the central gene of the neutrophil-related co-expression module. Consistently, co-expression levels among IL1B and chemosensing receptor [CXCR1/2 and FPR1/2] genes were reduced in the blood of IBD patients when compared to healthy controls.
Supplementary Material
Contributor Information
Simonas Juzenas, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany; Institute of Biotechnology, Life Science Centre, Vilnius University, Vilnius, Lithuania.
Matthias Hübenthal, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany; Department of Dermatology, Quincke Research Center, University Hospital Schleswig-Holstein, Kiel, Germany.
Carl Mårten Lindqvist, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany; School of Medical Sciences, Faculty of Medicine and Health, Örebro University, Örebro, Sweden.
Robert Kruse, Department of Clinical Research Laboratory, Faculty of Medicine and Health, Örebro University, Örebro, Sweden; iRiSC – Inflammatory Response and Infection Susceptibility Centre, Faculty of Medicine and Health, Örebro University, Örebro, Sweden.
Tim Alexander Steiert, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany.
Frauke Degenhardt, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany.
Dominik Schulte, Division of Endocrinology, Diabetes and Clinical Nutrition, Department of Medicine I, University Hospital of Schleswig-Holstein , Kiel, Germany; Institute of Diabetes and Clinical Metabolic Research, Kiel University, Kiel, Germany.
Susanna Nikolaus, Department of Internal Medicine I, University Hospital Schleswig-Holstein, Kiel, Germany.
Sebastian Zeissig, Medical Department 1, University Hospital Dresden, Technische Universität Dresden (TU Dresden), Dresden, Germany; Center for Regenerative Therapies Dresden (CRTD), Technische Universität (TU) Dresden, Dresden, Germany.
Daniel Bergemalm, School of Medical Sciences, Faculty of Medicine and Health, Örebro University, Örebro, Sweden.
Sven Almer, Department of Medicine, Karolinska Institutet, Solna, and Division of Gastroenterology, Karolinska University Hospital, Stockholm, Sweden.
Henrik Hjortswang, Department of Gastroenterology and Hepatology, Linköping University, Linköping, and Department of Health, Medicine, and Caring Sciences, Linköping University, Linköping, Sweden.
Francesca Bresso, Department of Medicine, Karolinska Institutet, Solna, and Division of Gastroenterology, Karolinska University Hospital, Stockholm, Sweden.
Nina Strüning, Department of Internal Medicine I, University Hospital Schleswig-Holstein, Kiel, Germany.
Juozas Kupcinskas, Department of Gastroenterology and Institute for Digestive Research, Lithuanian University of Health Sciences, Kaunas, Lithuania.
Andreas Keller, Chair for Clinical Bioinformatics, Saarland University, Saarbrücken, Germany; Department of Neurology and Neurological Sciences, Stanford University School of Medicine, Stanford, CA, USA.
Wolfgang Lieb, Institute of Epidemiology, Christian-Albrechts-University of Kiel, Kiel, Germany.
Philip Rosenstiel, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany.
Stefan Schreiber, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany; Department of Internal Medicine I, University Hospital Schleswig-Holstein, Kiel, Germany.
Mauro D’Amato, Unit of Clinical Epidemiology, Department of Medicine Solna, Karolinska Institutet, Stockholm, Sweden; Gastrointestinal Genetics Lab, CIC bioGUNE – BRTA, Derio, Spain; Ikerbasque, Basque Foundation for Science, Bilbao, Spain.
Jonas Halfvarson, Department of Gastroenterology, Faculty of Medicine and Health, Örebro University, Örebro, Sweden.
Georg Hemmrich-Stanisak, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany.
Andre Franke, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany; University Hospital of Schleswig-Holstein (UKSH), Kiel Campus , Kiel, Germany.
Funding
This work was supported by the European Union’s Horizon 2020 SYSCID programme under the grant agreement No. 733100 and received infrastructure support from the German Research Foundation (DFG) Excellence Cluster ‘Precision Medicine in Chronic Inflammation’ [PMI, EXC 2167], the Swedish Foundation for Strategic Research [RB13-0160, BIO IBD] and the Swedish Research Council [grant number 2020-02021 to J.H.].
Conflicts of Interest
The authors declare no conflicts of interest.
Author Contributions
Study concept, design and joint supervision: A.F., G.H-S., J.H. Data analysis and interpretation: S.J., M.H., C.M.L., F.D. with contributions from R.K., A.K. Coordination of patient inclusion J.H., M.D’A., G.H-S., P.R., S.S. Sample and clinical data acquisition: D.S., S.N., S.Z., D.B., S.A., H.H., F.B., N.S., J.K., W.L. Drafting of the manuscript: S.J. with contributions from G.H-S., J.H., C.M.L., T.A.S. All authors revised and edited the manuscript for critical content and approved of the final version of the manuscript for publication.
Data Availability
The gene expression data are deposited at GEO under accession numbers GSE169568, GSE169569 and GSE169570. Scripts to reproduce the main downstream analyses of this study are deposited at https://github.com/ikmb/ibd-blood-reproducibility.
References
- 1. Martin P, Leibovich SJ. Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol 2005;15:599–607. [DOI] [PubMed] [Google Scholar]
- 2. de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol 2016;13:13–27. [DOI] [PubMed] [Google Scholar]
- 3. Franke A, Balschun T, Karlsen TH, et al. ; IBSEN study group. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet 2008;40:1319–23. [DOI] [PubMed] [Google Scholar]
- 4. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet 2017;389:1756–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doecke JD, Simms LA, Zhao ZZ, et al. Genetic susceptibility in IBD: overlap between ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis 2013;19:240–5. [DOI] [PubMed] [Google Scholar]
- 6. Ellinghaus D, Jostins L, Spain SL, et al. ; International IBD Genetics Consortium (IIBDGC); International Genetics of Ankylosing Spondylitis Consortium (IGAS); International PSC Study Group (IPSCSG); Genetic Analysis of Psoriasis Consortium (GAPC); Psoriasis Association Genetics Extension (PAGE). Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016;48:510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ai L, Ren Y, Zhu M, et al. Synbindin restrains proinflammatory macrophage activation against microbiota and mucosal inflammation during colitis. Gut 2021;70:2261–72. [DOI] [PubMed] [Google Scholar]
- 8. McCarthy DA, Rampton DS, Liu YC. Peripheral blood neutrophils in inflammatory bowel disease: morphological evidence of in vivo activation in active disease. Clin Exp Immunol 1991;86:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones GR, Bain CC, Fenton TM, et al. Dynamics of colon monocyte and macrophage activation during colitis. Front Immunol 2018;9:2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koch S, Kucharzik T, Heidemann J, Nusrat A, Luegering A. Investigating the role of proinflammatory CD16+ monocytes in the pathogenesis of inflammatory bowel disease. Clin Exp Immunol 2010;161:332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Funderburg NT, Stubblefield Park SR, Sung HC, et al. Circulating CD4+ and CD8+ T cells are activated in inflammatory bowel disease and are associated with plasma markers of inflammation. Immunology 2013;140:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu F, Guo NJ, Tian H, et al. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis 2011;17:241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schaefer JS, Attumi T, Opekun AR, et al. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol 2015;16:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haider BA, Baras AS, McCall MN, Hertel JA, Cornish TC, Halushka MK. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One 2014;9:e89565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harvey RF, Bradshaw JM. A simple index of Crohn’s-disease activity. Lancet 1980;1:514. [DOI] [PubMed] [Google Scholar]
- 16. Lewis JD, Chuai S, Nessel L, Lichtenstein GR, Aberra FN, Ellenberg JH. Use of the noninvasive components of the Mayo score to assess clinical response in ulcerative colitis. Inflamm Bowel Dis 2008;14:1660–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dhanda AD, Creed TJ, Greenwood R, Sands BE, Probert CS. Can endoscopy be avoided in the assessment of ulcerative colitis in clinical trials? Inflamm Bowel Dis 2012;18:2056–62. [DOI] [PubMed] [Google Scholar]
- 18. Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006;55:749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011;17:10–12. [Google Scholar]
- 20. Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019;47:D155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pantano L, Estivill X, Martí E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res 2010;38:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pantano L, Escaramis G. isomiRs: analyze isomiRs and miRNAs from small RNA-seq. R package version 1.16.2 ed, 2020. https://bioconductor.org/packages/isomiRs/. Accessed 20 January 2022.
- 23. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012;40:4288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi W, Oshlack A, Smyth GK. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res 2010;38:e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dunning M, Lynch A, Eldridge M. illuminaHumanv4.db: illumina HumanHT12v4 annotation data (chip illuminaHumanv4). R package version 1.26.0 ed, 2015. https://bioconductor.org/packages/illuminaHumanv4.db/. Accessed 20 January 2022.
- 28. Gentleman R, Carey V, Huber W, et al. genefilter: genefilter: methods for filtering genes from high-throughput experiments. R package version 1.70.0 ed , 2020. https://bioconductor.org/packages/genefilter/. Accessed 20 January 2022.
- 29. Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 2018;46:D296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Juzenas S, Venkatesh G, Hübenthal M, et al. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res 2017;45:9290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cichocki A, Zdunek R, Amari S. Hierarchical ALS algorithms for nonnegative matrix and 3D tensor factorization. In: Davies ME, James CJ, Abdallah SA, Plumbley MD, eds. Independent Component Analysis and Signal Separation. ICA 2007. Lecture Notes in Computer Science, Vol. 4666. Berlin: Springer; 2007. [Google Scholar]
- 36. Williams AH, Kim TH, Wang F, et al. Unsupervised discovery of demixed, low-dimensional neural dynamics across multiple timescales through tensor component analysis. Neuron 2018;98:1099–115.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Satopaa V, Albrecht J, Irwin D, et al. Finding a ‘Kneedle’ in a Haystack: detecting knee points in system behavior. In: 2011 31st International Conference on Distributed Computing Systems Workshops, 2011. https://ieeexplore.ieee.org/document/5961514. [Google Scholar]
- 38. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 2018;46:D239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci U S A 2000;97:10101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cappello M, Morreale GC. The role of laboratory tests in Crohn’s disease. Clin Med Insights Gastroenterol 2016;9:51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ostrowski J, Dabrowska M, Lazowska I, et al. Redefining the practical utility of blood transcriptome biomarkers in inflammatory bowel diseases. J Crohns Colitis 2019;13:626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mohammadi A, Kelly OB, Filice M, Kabakchiev B, Smith MI, Silverberg MS. Differential expression of microRNAs in peripheral blood mononuclear cells identifies autophagy and TGF-beta-related signatures aberrantly expressed in inflammatory bowel disease. J Crohns Colitis 2018;12:568–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hübenthal M, Hemmrich-Stanisak G, Degenhardt F, et al. Sparse modeling reveals miRNA signatures for diagnostics of inflammatory bowel disease. PLoS One 2015;10:e0140155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burakoff R, Pabby V, Onyewadume L, et al. Blood-based biomarkers used to predict disease activity in Crohn’s disease and ulcerative colitis. Inflamm Bowel Dis 2015;21:1132–40. [DOI] [PubMed] [Google Scholar]
- 46. Burakoff R, Chao S, Perencevich M, et al. Blood-based biomarkers can differentiate ulcerative colitis from Crohn’s disease and noninflammatory diarrhea. Inflamm Bowel Dis 2011;17:1719–25. [DOI] [PubMed] [Google Scholar]
- 47. Wu CP, Bi YJ, Liu DM, Wang LY. Hsa-miR-375 promotes the progression of inflammatory bowel disease by upregulating TLR4. Eur Rev Med Pharmacol Sci 2019;23:7543–9. [DOI] [PubMed] [Google Scholar]
- 48. Tahamtan A, Teymoori-Rad M, Nakstad B, Salimi V. Anti-inflammatory microRNAs and their potential for inflammatory diseases treatment. Front Immunol 2018;9:1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lu TX, Sherrill JD, Wen T, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol 2012;129:1064–75.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Song F, Qian Y, Peng X, et al. The frontline of immune response in peripheral blood. PLoS One 2017;12:e0182294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mesko B, Poliska S, Szegedi A, et al. Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets. BMC Med Genomics 2010;3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Juss JK, House D, Amour A, et al. Acute respiratory distress syndrome neutrophils have a distinct phenotype and are resistant to phosphoinositide 3-kinase inhibition. Am J Respir Crit Care Med 2016;194:961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tran TM, Guha R, Portugal S, et al. A molecular signature in blood reveals a role for p53 in regulating malaria-induced inflammation. Immunity 2019;51:750–65.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Toufiq M, Roelands J, Alfaki M, et al. Annexin A3 in sepsis: novel perspectives from an exploration of public transcriptome data. Immunology 2020;161:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lauterbach MA, Hanke JE, Serefidou M, et al. Toll-like receptor signaling rewires macrophage metabolism and promotes histone acetylation via ATP-citrate lyase. Immunity 2019;51:997–1011.e7. [DOI] [PubMed] [Google Scholar]
- 56. Khandaker MH, Xu L, Rahimpour R, et al. CXCR1 and CXCR2 are rapidly down-modulated by bacterial endotoxin through a unique agonist-independent, tyrosine kinase-dependent mechanism. J Immunol 1998;161:1930–8. [PubMed] [Google Scholar]
- 57. Hu N, Westra J, Rutgers A, et al. Decreased CXCR1 and CXCR2 expression on neutrophils in anti-neutrophil cytoplasmic autoantibody-associated vasculitides potentially increases neutrophil adhesion and impairs migration. Arthritis Res Ther 2011;13:R201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hauser CJ, Fekete Z, Goodman ER, Kleinstein E, Livingston DH, Deitch EA. CXCR2 stimulation primes CXCR1 [Ca2+]i responses to IL-8 in human neutrophils. Shock 1999;12:428–37. [DOI] [PubMed] [Google Scholar]
- 59. Summers C, Rankin SM, Condliffe AM, Singh N, Peters AM, Chilvers ER. Neutrophil kinetics in health and disease. Trends Immunol 2010;31:318–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yao Y, Matsushima H, Ohtola JA, Geng S, Lu R, Takashima A. Neutrophil priming occurs in a sequential manner and can be visualized in living animals by monitoring IL-1β promoter activation. J Immunol 2015;194:1211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maeda S, Hsu LC, Liu H, et al. Nod2 mutation in Crohn’s disease potentiates NF-kappaB activity and IL-1beta processing. Science 2005;307:734–8. [DOI] [PubMed] [Google Scholar]
- 62. Shouval DS, Biswas A, Kang YH, et al. Interleukin 1β mediates intestinal inflammation in mice and patients with interleukin 10 receptor deficiency. Gastroenterology 2016;151:1100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chapman RW, Phillips JE, Hipkin RW, Curran AK, Lundell D, Fine JS. CXCR2 antagonists for the treatment of pulmonary disease. Pharmacol Ther 2009;121:55–68. [DOI] [PubMed] [Google Scholar]
- 64. Gomes NE, Brunialti MK, Mendes ME, Freudenberg M, Galanos C, Salomão R. Lipopolysaccharide-induced expression of cell surface receptors and cell activation of neutrophils and monocytes in whole human blood. Braz J Med Biol Res 2010;43:853–8. [DOI] [PubMed] [Google Scholar]
- 65. Tikhonov I, Doroshenko T, Chaly Y, Smolnikova V, Pauza CD, Voitenok N. Down-regulation of CXCR1 and CXCR2 expression on human neutrophils upon activation of whole blood by S. aureus is mediated by TNF-alpha. Clin Exp Immunol 2001;125:414–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hamberg M, Backes C, Fehlmann T, et al. MiRTargetLink–miRNAs, genes and interaction networks. Int J Mol Sci 2016;17:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mistry P, Nakabo S, O’Neil L, et al. Transcriptomic, epigenetic, and functional analyses implicate neutrophil diversity in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A 2019;116:25222–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kvedaraite E, Lourda M, Ideström M, et al. Tissue-infiltrating neutrophils represent the main source of IL-23 in the colon of patients with IBD. Gut 2016;65:1632–41. [DOI] [PubMed] [Google Scholar]
- 69. Sudhakar P, Verstockt B, Cremer J, et al. Understanding the molecular drivers of disease heterogeneity in Crohn’s disease using multi-omic data integration and network analysis. Inflamm Bowel Dis 2021;27:870–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jeong YS, Bae YS. Formyl peptide receptors in the mucosal immune system. Exp Mol Med 2020;52:1694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen K, Liu M, Liu Y, et al. Formylpeptide receptor-2 contributes to colonic epithelial homeostasis, inflammation, and tumorigenesis. J Clin Invest 2013;123:1694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Leoni G, Gripentrog J, Lord C, et al. Human neutrophil formyl peptide receptor phosphorylation and the mucosal inflammatory response. J Leukoc Biol 2015;97:87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Anton PA, Targan SR, Shanahan F. Increased neutrophil receptors for and response to the proinflammatory bacterial peptide formyl-methionyl-leucyl-phenylalanine in Crohn’s disease. Gastroenterology 1989;97:20–8. [DOI] [PubMed] [Google Scholar]
- 74. Bao Y, Ledderose C, Seier T, et al. Mitochondria regulate neutrophil activation by generating ATP for autocrine purinergic signaling. J Biol Chem 2014;289:26794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Savio LEB, Coutinho-Silva R. Purinergic signaling in infection and autoimmune disease. Biomed J 2016;39:304–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Longhi MS, Moss A, Jiang ZG, Robson SC. Purinergic signaling during intestinal inflammation. J Mol Med 2017;95:915–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Juzenas S, Lindqvist CM, Ito G, et al. Depletion of erythropoietic miR-486-5p and miR-451a improves detectability of rare microRNAs in peripheral blood-derived small RNA sequencing libraries. NAR Genom Bioinform 2020;2:lqaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ostrowski J, Goryca K, Lazowska I, et al. ; Polish PBC study Group; Polish IBD study Group. Common functional alterations identified in blood transcriptome of autoimmune cholestatic liver and inflammatory bowel diseases. Sci Rep 2019;9:7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nikolaus S, Bauditz J, Gionchetti P, Witt C, Lochs H, Schreiber S. Increased secretion of pro-inflammatory cytokines by circulating polymorphonuclear neutrophils and regulation by interleukin 10 during intestinal inflammation. Gut 1998;42:470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression data are deposited at GEO under accession numbers GSE169568, GSE169569 and GSE169570. Scripts to reproduce the main downstream analyses of this study are deposited at https://github.com/ikmb/ibd-blood-reproducibility.