Abstract
Short-term rates of chromosome evolution were analyzed in experimental populations of Escherichia coli B that had been propagated for 2,000 generations under four thermal regimens. Chromosome alterations were monitored in 24 independent populations by pulsed-field gel electrophoresis of DNA treated with five rare-cutting restriction enzymes. A total of 11 changes, 8 affecting chromosome size and 3 altering restriction sites, were observed in these populations, with none occurring in strains cultured at 37°C. Considering the changes detected in these experimental populations, the rate of chromosome alteration of E. coli is estimated to be half of that observed in experimental populations of yeast.
Comparisons of the genetic maps of Escherichia coli K-12 and Salmonella enterica serovar Typhimurium LT2 have established that the chromosomes of these enteric species are evolutionarily well conserved (12). Yet, in natural populations of each of these species, chromosome size can differ by as much as 1 Mb–more than 20% of total chromosome length (5, 22).
How quickly do bacterial chromosomes evolve? Despite the observed variation in chromosome lengths, evidence from natural isolates of E. coli and S. enterica suggests that large-scale changes in the size and organization of bacterial genomes are relatively rare events on an evolutionary timescale. This contrasts with the high incidence of spontaneous duplications, deletions, and inversions (2, 3, 11, 20), with the ability of these species to acquire large regions by horizontal transfer (13), and with the finding that very closely related strains of S. enterica serovar Typhi display vast differences in chromosome size and gene arrangement (15, 22).
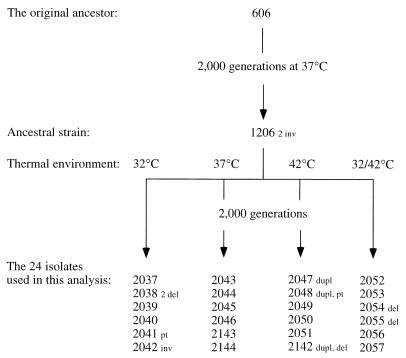
To investigate the short-term rates and patterns of chromosome evolution, we analyzed restriction fragment patterns in experimental populations of E. coli that were propagated for 2,000 generations. These strains represent 24 independent populations of E. coli B that were used in experiments by Bennett et al. (4) to study adaptation of E. coli to different thermal regimens and which were maintained at either 32, 37, or 42°C or alternating between 32 and 42°C (Fig. 1) (4).
FIG. 1.
Genealogical relationships and growth conditions of E. coli strains. Strains 2037 to 2042 were grown at 32°C; strains 2043 to 2046, 2143, and 2144 were grown at 37°C; and strains 2047 to 2051 and 2142 were grown at 42°C. Strains 2052 to 2057 were grown at temperatures that alternated daily between 32 and 42°C. Chromosomal changes occurring in these strains are denoted as follows: deletions, del; duplications, dupl; inversions, inv; point mutations, pt. Strains were grown in 10 ml of glucose limited minimal medium, and each day, 0.1 ml of culture was transferred to 9.9 ml of fresh medium (4). At 37°C, the population size of the ancestral strain in this medium is 4 × 108 cells in stationary phase (4). The common ancestor of all populations, strain 1206, was derived from a population, seeded with strain 606, that had been evolving for 2,000 generations at 37°C (14).
Restriction endonuclease digestion.
Agarose plugs containing intact genomic DNA were prepared as previously described (6). Five rare-cutting endonucleases were used to fractionate the genome: I-CeuI (NEB), which cuts in bacterial rRNA operons; BlnI (TaKaRa); NotI; SfiI, and XbaI (NEB). When digesting with I-CeuI, approximately 50 μl of each agarose plug (containing 10 ng of DNA/μl) was incubated overnight at 37°C with 0.3 U of I-CeuI in 50 μl of restriction enzyme buffer. For all other enzymes, 15 U of enzyme were added per 50-μl plug in 50 μl of the appropriate reaction buffer. Partial digests with I-CeuI were generated by overnight digestion with 0.05 U of enzyme per 50-μl plug in 50 μl of restriction enzyme buffer.
Pulsed-field gel electrophoresis.
Approximately 10 μl of an agarose plug (100 ng of DNA) was inserted into a 0.9% agarose gel and subjected to electrophoresis in 0.5× Tris-borate-EDTA buffer at 14°C in a CHEF-DR II pulsed-field gel box (Bio-Rad, Richmond, Calif.) for 24 h at 180 V with pulse times varying according to the intended range of resolution (6). To resolve I-CeuI restriction fragment A, which is typically 2,400 kb in these strains, samples were electrophoresed in a 0.7% agarose gel for 120 h at 60 V with pulse times ramped from 10 to 16 min over the course of the run. Gels were stained in 0.01% ethidium bromide and photographed under UV light. Lambda ladder, low-range pulsed-field gel marker (NEB), and chromosomes from Saccharomyces cerevisiae and Hanensula wingei (Bio-Rad) were used as molecular size standards.
Chromosome alterations in laboratory cultures.
A total of 11 changes were detected in the 24 strains, with none occurring in strains cultured at 37°C. Eight changes in six strains affected chromosome size, two strains acquired a restriction site, and one putative inversion was detected. The alterations in chromosome size usually involved regions of less than 50 kb and were as follows (Tables 1 and 2). (i) Isolates 2047 and 2048 each incurred a 25-kb duplication within I-CeuI fragment B, which spans 55′ to 73′ on the E. coli K-12 chromosome. In all likelihood, these duplications involve the same region because the restriction patterns produced by all five restriction enzymes are identical in the two strains. In digests of strain 2047, there is also a faint 700-kb band of equal size to the original I-CeuI fragment B, as well as the 725-kb fragment harboring the duplication; this is presumably due to instability of the duplication. (ii) Strain 2055 has a deletion of 20 to 30 kb in fragment I-CeuI fragment B, but at a different location from the duplications in strains 2047 and 2048. (iii) Strain 2038 has two deletions totaling 40 kb in I-CeuI fragment A, which spans 5′ to 55′ on the E. coli K-12 chromosome. (iv) Strain 2054 harbors a 20-kb deletion in I-CeuI fragment A. (v) Strain 2142 incurred two changes, a 30-kb deletion and a 50-kb duplication, in I-CeuI fragment A.
TABLE 1.
Sizes of I-CeuI restriction fragments and total chromosome size of isolates from experimental populations of E. coli
| Fragment | Size (kb) of I-CeuI restriction fragment in indicated E. coli strain from experimental populationa:
|
|||||
|---|---|---|---|---|---|---|
| 606 | 1206 | 2038 | 2047 | 2048 | 2055 | |
| I-CeuI A | 2,400 | 2,400 | 2,350 | 2,400 | 2,400 | 2,400 |
| I-CeuI B | 700 | 700 | 700 | 725 | 725 | 680 |
| I-CeuI C | 530 | 530 | 530 | 530 | 530 | 530 |
| I-CeuI D | 100 | 100 | 100 | 100 | 100 | 100 |
| I-CeuI E | 130 | 130 | 130 | 130 | 130 | 130 |
| I-CeuI F | 37 | 37 | 37 | 37 | 37 | 37 |
| I-CeuI G | 650 | 650 | 650 | 650 | 650 | 650 |
| Total size | 4,550 | 4,550 | 4,500 | 4,580 | 4,580 | 4,550 |
I-CeuI recognition sites occur in the seven rRNA operons. I-CeuI fragment A spans the region between 4.9′ and 58.5′ on the E. coli K-12 chromosome, B spans the region between 58.5′ to 73.7′, C spans 73.7′ to 84.9′, D spans 84.9′ to 86.9′, E spans 86.9′ to 89.7′, F spans 89.7′ to 90.6′, and G spans 90.6′ to 4.9′. Only strains exhibiting changes in fragment size relative to the ancestral strains 606 and 1206 are included in the table. Strain 2038 was grown at 32°C, strains 2047 and 2048 were grown at 42°C, and strain 2055 was grown at temperatures that alternated daily between 32 and 42°C.
TABLE 2.
Changes in sizes of restriction fragments and total chromosome size for experimental population using BlnI and NotI
| Restriction fragment | Size (kb) of restriction fragment in indicated E. coli strain from experimental populationa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1206 | 2038 | 2041 | 2042 | 2047 | 2048 | 2054 | 2055 | 2142 | |
| BlnI | 930 | 1,100 | 650 | 930 | 930 | 930 | 930 | 930 | 950 |
| 680 | 650 | 680 | 680 | 680 | 680 | 660 | 680 | 680 | |
| 350 | 350 | 350 | 350 | 380 | 380 | 350 | 350 | 350 | |
| 340 | 340 | 340 | 340 | 340 | 340 | 340 | 310 | 340 | |
| 330 | |||||||||
| 180 | 180 | 180 | 180 | 180 | 180 | 180 | 180 | ||
| NotI | 1,060 | 1,060 | 1,060 | 950 | 1,090 | 1,090 | 1,060 | 1,030 | 1,060 |
| 480 | 465 | 480 | 480 | 480 | 435 | 460 | 480 | 480 | |
| 410 | 410 | 410 | 410 | 410 | 410 | 410 | 410 | 460 | |
| 295 | 295 | 295 | 295 | 295 | 295 | 295 | 295 | 276 | |
| 250 | 250 | 250 | 362 | 250 | 250 | 250 | 250 | 250 | |
| 242 | 210 | 242 | 242 | 242 | 242 | 242 | 242 | 242 | |
| 40 | |||||||||
Only strains exhibiting changes relative to the ancestral strain 1206 are included in the table. Sizes of fragments that have changed are shown in boldface. Strains 2038, 2041, and 2042 were grown at 32°C; strains 2047, 2048, and 2142 were grown at 42°C; and strains 2054 and 2055 were grown at temperatures that alternated daily between 32 and 42°C.
In addition to changes in chromosome size, strain 2041 acquired a BlnI site, strain 2048 gained a NotI site, strain 2038 lost a NotI site, and strain 2142 lost an XbaI site (Table 2). New restriction sites were probably acquired through point mutations. However, losses of restriction sites may have resulted from deletions and were not counted as independent events. Strain 2042 incurred changes that are best explained by a single inversion: for three of the enzymes (NotI, SfiI, and XbaI) the changes involve a reduction and a commensurate increase in the sizes of two restriction fragments, and there were no changes in any I-CeuI and BlnI restriction fragments.
Comparisons of strain 1206, the common ancestor of the 24 independent populations (Fig. 1), with its parent strain (606) revealed changes in restriction patterns that are compatible with two inversion events. The I-CeuI digests are identical in strains 606 and 1206 (Table 1); however, digests with BlnI, NotI, SfiI, and XbaI each showed changes in the sizes of three fragments, i.e., the absence of three fragments in 1206 that are present in 606 along with the addition of three new fragments (Table 3). These changes could not have arisen from point mutations because it would require exactly the same number of gains and losses of sites for all four restriction enzymes.
TABLE 3.
Sizes of BlnI and NotI restriction fragments for strains 606 and 1206
| Size (kb) of restriction fragment in indicated E. coli straina:
| |||
|---|---|---|---|
|
BlnI
|
NotI
|
||
| 606 | 1206 | 606 | 1206 |
| 1,150 | 1,060 | 1,060 | |
| 930 | 480 | ||
| 630 | 410 | ||
| 660 | 660 | 410 | 410 |
| 550 | 380 | ||
| 545 | 380 | 380 | |
| 530 | 530 | 295 | |
| 445 | 265 | 265 | |
| 350 | 350 | 250 | 250 |
| 340 | 340 | 250 | 250 |
| 180 | 180 | 242 | 242 |
| 134 | 134 | 239 | |
| 105 | 105 | 230 | |
| 50 | 50 | 205 | 205 |
| 38 | 38 | 176 | 176 |
| 23 | 23 | 104 | 104 |
| 14 | 14 | 88 | 88 |
| 79 | 79 | ||
| 50 | 50 | ||
| 36 | 36 | ||
| 18 | 18 | ||
| 17 | 17 | ||
Sizes of fragments that have changed relative to these of the ancestor strain are shown in boldface. Total chromosome sizes (kilobases) of the restriction-digested strains were as follows: BlnI-digested 606, 4,569; BlnI-digested 1206, 4,529; NotI-digested 606, 4,635; NotI-digested 1206, 4,659.
Given the frequency of changes observed in these strains, and assuming that all changes resulted from independent events, the probability that the populations cultured at 37°C would have no changes is 2.5% [(13/24)6 = 0.025]. This suggests that strains propagated at temperatures other than 37°C either have higher mutation rates or incur changes that are adaptive under these nonstandard growth conditions. Although the fitness effects of the observed chromosomal rearrangements are not known, the parallel changes maintained in two independent lineages at 42°C, strains 2047 and 2048 harbor the same 25-kb duplication, might denote a beneficial mutation.
Based on the numbers of changes in restriction patterns observed for these five enzymes, these experimental populations have elevated mutation rates. E. coli typically incurs 5.4 × 10−10 point mutations per base pair per generation (9), and if all mutations are neutral, each of these experimental populations should have, on average, five point mutations after 2,000 generations. Omitting I-CeuI, which is targeted to rRNA operons, the four other rare-cutting endonucleases employed in this study survey a total of 588 nucleotides, yielding a probability of 0.13% for gaining or losing a restriction site due to a point mutation in 2,000 generations (2,000 × 2 × 588 × 5.4 × 10−10 = 1.27 × 10−3), assuming that the probability of gaining a restriction site is equal to that of losing a restriction site. This means that the probability of detecting a change in any of the 24 independent populations is only 3%. However, we observed at least two point mutational changes, implying that mutation rates in some of the strains may be elevated by nearly two orders of magnitude. Temperature is known to increase mutation rates and the transposition of some mobile elements (10, 18); however, mutator alleles could also be selected during periods of rapid adaptation (7, 8, 19, 21). These calculations are based on the assumption that the number of restriction sites in a genome is the result of an equilibrium between mutations leading to the gain or loss of a site. If the chances of gaining a restriction site were greater than that of losing one, the target size for creating sites would be greater, increasing the chances of observing a change in restriction fragments due to point mutations. This might be the case if there is selection against the presence of these rare restriction sites in the genome, as might be the case for XbaI.
Distribution of chromosome alterations.
All the deletions and duplications observed in these strains occurred in I-CeuI fragments A and B, which together span the region between 5′ and 73′ and encompass 68% of the chromosome. Since the probability that eight events would be limited to this portion of the chromosome by chance is 0.046, it appears that this region, which broadly spans the replication terminus, sustains most of the structural changes to the chromosome. This is consistent with the notion that bacterial chromosomes are less conserved, and contain fewer essential genes, closer to the replication terminus; and, in a comparison of low-resolution physical maps of E. coli K-12 derivatives, Perkins et al. (17) also noted that most of the differences between these laboratory strains occurred close to the terminus. The terminus region of the E. coli chromosome is also subject to higher rates of recombination than other parts of the chromosome (16), which should lead to clustering of chromosomal variation close to the terminus.
Unlike natural isolates, these experimental populations cannot augment their chromosomes by incorporating DNA from foreign sources and do not harbor plasmids or active phages that might facilitate DNA transfer. Instead, these strains can either lose DNA by deletions (which are irreversible) or gain DNA by duplications (which are generally unstable), so we might expect chromosome size in experimental populations to decrease with time. However, chromosome size in the strain of E. coli used to seed these experimental populations is only 4.6 Mb, close to a minimum size observed in E. coli, and it is likely that only small deletions can be tolerated.
Finally, it is tempting to compare the rates of chromosomal change in bacteria with those observed in eukaryotes. Adams et al. (1) detected a total of 17 large-scale rearrangements in 13 experimental populations of the fission yeast, Saccharomyces cerevisiae, after 1,000 generations, compared to the nine chromosomal rearrangements (i.e., duplications, deletions, and inversions) that we observed in 24 populations of E. coli after 2,000 generations. After correcting for genome size, the rate of chromosome alteration, measured as number of changes per megabase per generation, in experimental populations of yeast is about twofold higher than that observed in E. coli. Despite the high rate of chromosomal duplication and deletion measured in E. coli under laboratory conditions, most of the isolates analyzed here do not contain detectable chromosome rearrangements. If bacterial genomes are under selection to minimize replication time, it might be expected that the propagation of strains under stable and homogeneous laboratory conditions would favor a reduction in chromosome size. However, there is no evidence that these experimental populations are evolving smaller genomes to enhance their growth rates.
Acknowledgments
We thank Richard Lenski for providing the strains.
This work was supported by NIH grant GM56120 to H.O.
REFERENCES
- 1.Adams J, Puskas-Rozsa S, Simlar J, Wilke C M. Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Curr Genet. 1992;22:13–19. doi: 10.1007/BF00351736. [DOI] [PubMed] [Google Scholar]
- 2.Anderson P, Roth J. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc Natl Acad Sci USA. 1981;78:3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson R P, Roth J R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- 4.Bennett A F, Lenski R E, Mittler J E. Evolutionary adaptation to temperature. I. Fitness responses of Escherichia coli to changes in its thermal environment. Evolution. 1992;46:16–30. doi: 10.1111/j.1558-5646.1992.tb01981.x. [DOI] [PubMed] [Google Scholar]
- 5.Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:9–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 6.Bergthorsson U, Ochman H. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J Bacteriol. 1995;177:5784–5789. doi: 10.1128/jb.177.20.5784-5789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao L, Vargas C, Spear B B, Cox E C. Transposable elements as mutator genes in evolution. Nature. 1983;303:633–635. doi: 10.1038/303633a0. [DOI] [PubMed] [Google Scholar]
- 8.Cox E C, Gibson T C. Selection for high mutation rates in chemostats. Genetics. 1974;77:169–184. doi: 10.1093/genetics/77.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake J W. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haren L, Betermier M, Polard P, Chandler M. IS911-mediated intramolecular transposition is naturally temperature sensitive. Mol Microbiol. 1997;25:531–540. doi: 10.1046/j.1365-2958.1997.4951854.x. [DOI] [PubMed] [Google Scholar]
- 11.Heath J D, Weinstock G M. Tandem duplications of the lac region of the Escherichia coli chromosome. Biochimie. 1991;73:343–352. doi: 10.1016/0300-9084(91)90099-m. [DOI] [PubMed] [Google Scholar]
- 12.Krawiec S, Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990;54:502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence J G, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 14.Lenski R E, Rose M R, Simpson S C, Tadler S C. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- 15.Liu S L, Sanderson K E. Highly plastic chromosomal organization in Salmonella typhi. Proc Natl Acad Sci USA. 1996;93:10303–10308. doi: 10.1073/pnas.93.19.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louarn J, Cornet F, Francois V, Patte J, Louarn J-M. Hyperrecombination in the terminus region of the Escherichia coli chromosome: possible relation to nucleoid organization. J Bacteriol. 1994;176:7524–7531. doi: 10.1128/jb.176.24.7524-7531.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perkins J D, Heath J D, Sharma B R, Weinstock G M. XbaI and BlnI genomic cleavage maps of Escherichia coli K-12 strain MG1655 and comparative analysis of other strains. J Mol Biol. 1993;232:419–445. doi: 10.1006/jmbi.1993.1401. [DOI] [PubMed] [Google Scholar]
- 18.Savva D. Spontaneous mutation rates in continuous cultures: the effect of some environmental factors. Microbios. 1982;33:81–92. [PubMed] [Google Scholar]
- 19.Sniegowski P D, Gerrish P J, Lenski R E. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 20.Starlinger P. DNA rearrangements in prokaryotes. Annu Rev Genet. 1977;11:103–126. doi: 10.1146/annurev.ge.11.120177.000535. [DOI] [PubMed] [Google Scholar]
- 21.Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon P H, Godelle B. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 22.Thong K L, Puthucheary S D, Pang T. Genome size variation among recent human isolates of Salmonella typhi. Res Microbiol. 1997;148:229–235. doi: 10.1016/S0923-2508(97)85243-6. [DOI] [PubMed] [Google Scholar]