Abstract
Most isolates of Salmonella enterica serovar Typhimurium contain a 90-kb virulence plasmid. This plasmid is reported to be mobilizable but nonconjugative. However, we have determined that the virulence plasmid of strains LT2, 14028, and SR-11 is indeed self-transmissible. The plasmid of strain SL1344 is not. Optimal conjugation frequency requires filter matings on M9 minimal glucose plates with a recipient strain lacking the virulence plasmid. These conditions result in a frequency of 2.9 × 10−4 transconjugants/donor. Matings on Luria-Bertani plates, liquid matings, or matings with a recipient strain carrying the virulence plasmid reduce the efficiency by up to 400-fold. Homologs of the F plasmid conjugation genes are physically located on the virulence plasmid and are required for the conjugative phenotype.
Salmonella enterica serovar Typhimurium (commonly referred to as Salmonella typhimurium) has been studied extensively, both as a model of fundamental genetic mechanisms and as a model of host-pathogen interactions. This bacterium is a broad-host-range pathogen that causes gastroenteritis in humans and cattle and typhoid-like disease in mice. Most S. typhimurium isolates (88%) carry a plasmid of 60 MDa (roughly 90 kb) (15). This plasmid has been given a variety of names, including pSLT, MP10, pRQ28, pSTV, the cryptic plasmid, and the virulence plasmid (7, 15, 25, 28, 30, 34). We will refer to this plasmid simply as the virulence plasmid of a particular host strain.
The most apparent consequence of virulence plasmid carriage is to enhance the growth rate of the bacterium during the systemic phase of disease (13). This phenotype is conferred by an 8-kb region of the plasmid that encodes the spv (Salmonella plasmid virulence) genes (11, 12). The product of the spv regulatory locus, SpvR, in combination with the alternative sigma factor RpoS, is required for the expression of the four structural genes spvABCD (35 and references therein). The molecular functions of SpvA, SpvB, SpvC, and SpvD have not been determined, although it appears that SpvD is secreted (8). Other loci on the plasmid include the pef (plasmid-encoded fimbriae) region, which has been implicated in bacterial adhesion to intestinal epithelial cells, and the srg region which includes rck (resistance to complement killing), a homolog of dsbA (disulfide bond isomerase), and a homolog of the AraC family of transcriptional regulators (1, 3, 10, 14). Another interesting gene on the virulence plasmid is tlpA, which encodes an apparent thermometer in that it regulates its own transcription according to temperature (17). The downstream targets of this regulator have not yet been identified.
Isolation of MudJ mutations on the virulence plasmid.
We recently performed a genetic screen to identify genes regulated by SirA, a transcriptional regulator of genes within SPI1 (Salmonella pathogenicity island 1) (2, 18). This screen resulted in the isolation of 74 transcriptional fusions (MudJ transposon insertions) that respond to sirA expressed from a multicopy plasmid. However, only 10 of the 74 fusions were found to respond to sirA in its natural chromosomal context. These 10 fusions were defined as class 1 and are definitively regulated by sirA. The class 1 fusions are located within SPI1, SPI4, and SPI5 (2). The remaining 64 fusions are probably not regulated directly by SirA but are expressed as an indirect effect of sirA overexpression. These 64 fusions were defined as class 2. By cloning and sequencing the genomic DNA flanking the class 2 insertions we discovered that many of the fusions are located on the virulence plasmid (Table 1). The known plasmid genes disrupted include pefC, tlpA, and spv.
TABLE 1.
Strains used in this study and their abilities to act as virulence plasmid conjugative donors
| Strain | Descriptiona | Construction, source, or referenceb | Donor abilityc |
|---|---|---|---|
| W3110 | E. coli K-12 | CGSCd | NA |
| BA769 | W3110 Nalr | Selection for spontaneous nalidixic acid resistance | NA |
| LT2 (ATCC23565) | S. typhimurium | ATCCd | NA |
| 14028 (ATCC14028) | S. typhimurium | ATCCd | NA |
| SR-11 | S. typhimurium | Susanne Lindgren | NA |
| SL1344 | S. typhimurium | 16, Cathy Lee | NA |
| IR715 | 14028 Nalr | 31 | NA |
| 14028 −pSLT | 14028, cured of the virulence plasmid | 3 | NA |
| BA770 | 14028 −pSLT Nalr | Selection for spontaneous nalidixic acid resistance | NA |
| BA901a,b,c | LT2 spv1541::MudJ | LT2 × P22/BA1541 | Yes |
| BA902a,b,c | SR-11 spv1541::MudJ | SR-11 × P22/BA1541 | Yes |
| BA903a,b,c | SL1344 spv1541::MudJ | SL1344 × P22/BA1541 | No |
| BA1504 | 14028 traC1504::MudJ (antisense) | 2, this study | No |
| BA1505 | 14028 traG1505::MudJ (antisense) | 2, this study | No |
| BA1523 | 14028 orfG1523::MudJ (antisense) | 2, this study | Noe |
| BA1527 | 14028 traW1527::MudJ (sense) | 2, this study | No |
| BA1532 | 14028 trbC1532::MudJ (antisense) | 2, this study | Noe |
| BA1533 | 14028 pefC1533::MudJ (sense) | 2, this study | Yes |
| BA1534 | 14028 traP1534::MudJ (sense) | 2, this study | No |
| BA1535 | 14028 traC1535::MudJ (antisense) | 2, this study | No |
| BA1539 | 14028 tra1539::MudJ (MudJ is between traR and traC; antisense) | 2, this study | No |
| BA1541 | 14028 spv1541::MudJ (sense) | 2, this study | Yes |
| BA1542 | 14028 orfG1542::MudJ (antisense) | 2, this study | Noe |
| BA1544 | 14028 traD1544::MudJ (antisense) | 2, this study | No |
| BA1545 | 14028 traD1545::MudJ (antisense) | 2, this study | No |
| BA1546 | 14028 traG1546::MudJ (antisense) | 2, this study | No |
| BA1550 | 14028 hilA1550::MudJ (sense) | 2 | NA |
| BA1569 | 14028 tlpA1569::MudJ (antisense) | 2, this study | Yes |
| BA1571 | 14028 trbI1571::MudJ (antisense) | 2, this study | No |
| BA1572 | 14028 traG1572::MudJ (antisense) | 2, this study | No |
| BA1576 | 14028 traW1576::MudJ (antisense) | 2, this study | No |
| BA1741 | 14028 spv1541::MudJ sirA::cam | 2 | Yesf |
The DNA surrounding the MudJ insertions was amplified by inverse PCR, cloned, and sequenced as described previously (1). Three MudJ insertions were within previously sequenced genes. The pefC1533 insertion is before nucleotide 4740 of GenBank accession no. L08613. The spv1541 insertion is upstream of spvA, before nucleotide 2440 of GenBank accession no. X56727. The tlpA1569 insertion is before nucleotide 216 of GenBank accession no. M88208. When the MudJ insertion is within a gene that has not been sequenced, the name of the closest homolog is given. The orientations of the promoterless lacZY genes of the MudJ element are listed with respect to the orientation of the gene disrupted (sense or antisense).
Transductions were performed with phage P22HTint, followed by streaking to isolation in the presence of 10 mM EGTA and confirming smooth lipopolysaccharide and lack of pseudolysogeny by cross-streaking transductants against P22vir on Evans blue-uranine plates (21). As a side note, we observed that P22 can be used to introduce genes into strain SR-11, but it cannot form plaques on SR-11. Therefore, it always appears rough in the Evans blue-uranine test, presumably because it is lysogenized with a phage conferring P22 immunity. We have found that phage KB1int-1 (22) works well for transducing markers out of SR-11.
Ability to act as a donor of the virulence plasmid in filter matings on M9 glucose plates is indicated by Yes, and failure to act as a donor is indicated by No. NA, not applicable.
CGSC, E. coli Genetic Stock Center; ATCC, American Type Culture Collection.
Donor ability was dramatically reduced but not completely eliminated by mutations in orfG or trbC.
Donor ability was slightly reduced (two- to threefold) by a sirA mutation.
The virulence plasmid contains genes homologous to the conjugation genes of F and R-100 plasmids.
We were able to sequence the genomic DNA surrounding 53 of the 64 MudJ mutations described above. Database searches indicated that 15 of the 53 mutations are within homologs of the F and R-100 plasmid conjugation genes (Table 1). Consistent with this, Boyd and Hartl reported that 50% of Salmonella isolates contain an F plasmid (5). More recently these authors determined that there is a correlation in Salmonella isolates between the presence of several virulence plasmid loci and the F plasmid locus, traD (6). Based on this correlation, they hypothesized that the F plasmid and the virulence plasmid might be one and the same.
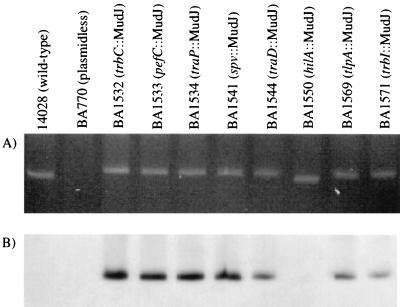
The F plasmid homologs could be located on the virulence plasmid, the chromosome, or on a second plasmid in the cell. To determine their physical location, pulsed-field gel electrophoresis was used to separate the virulence plasmid from the rest of the Salmonella genome, and Southern blotting was used to identify the MudJ insertions (20). Strains carrying MudJ mutations within known virulence plasmid loci were used as positive controls (spv, tlpA, and pefC). Negative controls were a plasmidless strain (BA770), a wild-type strain that does not contain a MudJ mutation (14028), and a strain that contains a MudJ mutation in the chromosome (hilA::MudJ). Ethidium bromide staining of the pulsed-field gel showed that the virulence plasmid was present in all strains except the plasmidless strain, BA770 (Fig. 1A). The plasmids of the wild type and the hilA mutant are slightly smaller than the others, consistent with the lack of a MudJ insertion. The identity of this plasmid was confirmed by using Southern hybridization with the kanamycin resistance gene of MudJ (Fig. 1B). The control strains that contain MudJ insertions in known virulence plasmid loci yield positive hybridization signals (pefC::MudJ, spv::MudJ, and tlpA::MudJ). The negative control strains do not (wild type, plasmidless, and hilA::MudJ). The virulence plasmid of strains containing MudJ insertions within F plasmid homologs (trbC::MudJ, traP::MudJ, traD::MudJ, and trbI::MudJ) also yields positive hybridization signals. This conclusively demonstrates that the F plasmid homologs are physically located on the virulence plasmid. It also suggests that the F plasmid detected by Boyd and Hartl in 50% of Salmonella isolates is actually the virulence plasmid of these strains (5).
FIG. 1.
Homologs of the F plasmid conjugation genes are physically located on the 90-kb virulence plasmid of S. typhimurium. DNA was prepared according to the methods described in reference 36. Pulsed-field gel electrophoresis utilized a 1% 0.5× Tris-borate-EDTA agarose gel with separation by a Bio-Rad CHEF DRII at 200 V. The separation was run for 5 h with a switching time that ramped from 6 to 150 s, 5 h at 6 to 12 s, and 10 h at 6 to 36 s. (A) The gel was stained with ethidium bromide. (B) Detection of the kanamycin resistance gene of MudJ by Southern hybridization after transfer to a positively charged nylon membrane obtained from Boehringer Mannheim. A random prime DNA labeling kit (Boehringer Mannheim) and [32P]dCTP were used to label the 1.2-kb SmaI fragment from pUC4K-KIXX (Pharmacia), which contains the kanamycin resistance gene.
The conjugation genes of the virulence plasmid are functional.
Previous reports have stated that the virulence plasmid of S. typhimurium is mobilizable but not self-transmissible (19, 26–29). However, these reports did not focus on the transmissibility of the virulence plasmid. This fact, coupled with the large number of conjugation genes located on the virulence plasmid, suggested that the virulence plasmid might be self-transmissible under certain conditions. To test this hypothesis, we assayed the ability of several strains to act as virulence plasmid donors in conjugation experiments (Table 1). In each case, the donor bacteria contained a MudJ insertion on the virulence plasmid. This served two purposes. First, recipients that had acquired the virulence plasmid could be selected by using the kanamycin resistance of the MudJ insertion. Second, using donors that had MudJ mutations in various genes of the virulence plasmid allowed us to determine which plasmid genes are required for conjugation. The recipient strain was BA770, which is resistant to nalidixic acid (allowing counterselection) and has been cured of the virulence plasmid (3). All donors containing a MudJ insertion in a conjugation gene homolog (traC, traD, traG, traW, traP, orfG, trbC, or trbI) failed to conjugate, either due to the direct disruption of the gene itself or polarity on downstream genes. Donors containing a MudJ insertion elsewhere on the virulence plasmid (spv, pefC, and tlpA) were conjugative according to a qualitative assay (all yielded conjugation frequencies of approximately 10−4 transconjugants/donor; Table 1).
A quantitative analysis of conjugation frequency under optimal conditions yielded a frequency of 2.9 × 10−4 transconjugants/donor (optimal conditions utilized filter matings on M9 minimal glucose plates with a recipient strain lacking the virulence plasmid; Table 2). This conjugation frequency is much lower than that of the Escherichia coli F plasmid but is reasonable considering that most natural self-transmissible plasmids yield frequencies of between 10−3 and 10−5 (23). The F plasmid of E. coli K-12 conjugates with an unusually high frequency of 0.2 to 1.0, because a repressor of the conjugation genes, finO, has been disrupted by an IS3 element (9). It is also possible that currently undiscovered conditions could result in higher virulence plasmid conjugation frequencies.
TABLE 2.
Conjugation frequencies of the S. typhimurium virulence plasmid under various conditions
| Donor | Recipientb | Condition | Conjugation frequency (SE)a |
|---|---|---|---|
| BA1541 | BA770 | LB plate | 4.2 × 10−6 (3.9 × 10−7) |
| BA1541 | BA770 | M9 plate | 2.9 × 10−4 (1.3 × 10−5) |
| BA1541 | BA770 | LB broth | 1.1 × 10−5 (2.5 × 10−6) |
| BA1541 | BA770 | M9 broth | 2.2 × 10−5 (1.6 × 10−6) |
| BA1541 | IR715 | M9 plate | 7.0 × 10−7 (6.8 × 10−8) |
| BA1741 | BA770 | LB plate | 1.4 × 10−6 (2.1 × 10−7) |
| BA1741 | BA770 | M9 plate | 1.7 × 10−4 (2.3 × 10−5) |
| BA1541 | None | LB plate | 9.0 × 10−8 |
| BA1541 | None | M9 plate | 1.1 × 10−7 |
Bacteria were grown as rolling overnight cultures at 37°C in LB medium. A total of 5 μl of donor strain and 50 μl of recipient strain were added to 5 ml of 10 mM MgSO4. This solution was filtered through a 0.45-μm-pore size filter, and the filter was placed on either an M9 minimal plate containing 0.2% glucose (recipe in reference 1) or an LB plate. After overnight incubation at 37°C, the filter was lifted from the plate, added to 5 ml of 10 mM MgSO4, and vortexted to remove the bacteria from the filter. Dilutions were plated on LB plates containing kanamycin (60 μg/ml) to select for donors or on LB plates containing kanamycin and nalidixic acid (50 μg/ml) to select for transconjugants. To perform liquid matings, bacteria were grown as rolling overnight cultures at 37°C in LB medium. A total of 5 μl of donor strain and 50 μl of recipient strain were added to either 1 ml of M9 minimal medium containing 0.2% glucose or 1 ml of LB medium in a 1.5-ml microcentrifuge tube. After a standing overnight incubation at 37°C, dilutions were plated on selective plates as described for filter matings. The conjugation frequency is reported as the number of transconjugants per donor (the number of Kanr Nalr colonies divided by the number of Kanr colonies). The conjugation frequency for each donor-recipient pair was determined by averaging the results of triplicate matings performed on three separate days (nine total matings). The error value is the standard error of the three daily means.
In cases where there is no recipient strain, the value represents the isolation frequency of spontaneously Nalr colonies. These experiments were performed exactly like the others, with triplicate matings on three separate days. However, most filters resulted in no spontaneously Nalr colonies, so an average was not calculated. The value given is the single highest frequency obtained from the nine individual filters.
Because the donor and recipient strains in these experiments were isogenic, it is possible that the transconjugants obtained were actually donors that had spontaneously acquired resistance to nalidixic acid. To eliminate this possibility, two types of control experiment were performed. The first control was to perform mock matings in the absence of a recipient strain. These experiments determined the frequency of spontaneous nalidixic acid resistance by the donors. These mock matings were performed exactly like actual matings, with three separate cultures on three separate days. Many of the mock matings resulted in no spontaneous nalidixic acid-resistant mutants, so an average frequency was not calculated (the detection limit for mutation frequency was 10−9). Instead, the highest frequency observed, during growth on either Luria-Bertani (LB) plates or M9 glucose, was reported (Table 2; 9.0 × 10−8 on LB and 1.1 × 10−7 on M9). This highest frequency of spontaneous nalidixic acid resistance is much lower than the observed conjugation frequencies (Table 2). Therefore, spontaneous drug resistance was not a factor in these experiments.
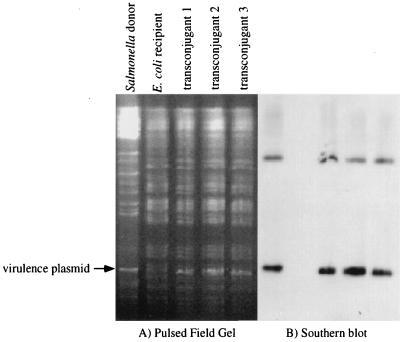
The second experiment was to physically observe the transferred virulence plasmid in the recipient strain. This requires the recipient to have a genome that is easily distinguished from that of the donor. Therefore, E. coli was chosen to be the recipient in matings with Salmonella. Filter matings were performed on M9 minimal glucose plates with the donor strain BA1541 (spv::MudJ) and recipient strain BA769 (E. coli W3110 Nalr). Transconjugants were obtained at a frequency of approximately 2 × 10−5 transconjugants/donor. Genomic DNA was isolated from the transconjugants, digested with XbaI, and examined by pulsed-field gel electrophoresis (Fig. 2A). Ethidium bromide staining of the pulsed-field gel shows that the restriction pattern of the transconjugant genomes matches that of the E. coli recipient, except for the acquisition of the virulence plasmid from the Salmonella donor. The identity of the virulence plasmid was confirmed by subsequent Southern hybridization with the kanamycin resistance gene of MudJ (Fig. 2B). Because the restriction patterns of the E. coli and Salmonella genomes are different, it is easily seen that the three transconjugants are E. coli bacteria that have obtained the virulence plasmid, rather than donors that have become spontaneously resistant to nalidixic acid. This is clear evidence that conjugation has occurred.
FIG. 2.
Visualization of virulence plasmid transfer from Salmonella to E. coli. (A) Pulsed-field gel stained with ethidium bromide. (B) Southern hybridization of the pulsed-field gel with a probe specific to the kanamycin resistance gene of MudJ. The virulence plasmid is present in the donor Salmonella strain, BA1541 (spv::MudJ), and the three transconjugants but is not present in the recipient E. coli strain, BA769. Pulsed-field gel electrophoresis and Southern blotting were performed as described in the legend to Fig. 1.
Factors affecting conjugation frequency.
Several factors were found to alter the conjugation frequency of the S. typhimurium virulence plasmid. First, the conjugation frequency is 70-fold lower when filter matings are performed on LB plates rather than on M9 minimal glucose plates (Table 2). LB plates contain a rich mixture of yeast components that can be used as biosynthetic precursors and as a carbon and energy source. M9 minimal glucose plates, on the other hand, contain only glucose as a carbon source and contain no biosynthetic precursors. This suggests that conjugation proficiency is regulated in response to environmental conditions. This could involve gene expression changes in the donor, the recipient, or both.
Second, liquid matings result in fewer transconjugants per donor than filter matings with M9 medium (Table 2). However, unlike the situation with filter matings, in which the use of M9 medium results in 70-fold higher efficiencies than the use of LB medium, liquid matings in either medium result in roughly the same frequency. This frequency is intermediate to those obtained with the filter matings on M9 glucose and LB plates (Table 2).
Third, the presence of a virulence plasmid in the recipient strain inhibits conjugation approximately 400-fold (Table 2; recipient strain IR715). This is probably due to a surface exclusion process mediated by the plasmid in the recipient strain. This phenomenon is common among plasmids utilizing an F plasmid-like conjugative apparatus (9). The traS and traT genes of the F plasmid are responsible for surface exclusion, and it is known that the S. typhimurium virulence plasmid encodes a homolog of traT (24, 32, 33).
The F plasmid homologs were originally isolated by screening for transcriptional fusions that respond to sirA carried on a multicopy plasmid (2). Because SirA regulates genes required for enteropathogenesis in the bovine model, we hypothesized that SirA may also activate conjugation functions in the intestine. This inherently makes sense, given the large numbers of potential mating partners present in the intestinal environment. Therefore, the effect of a sirA mutation on conjugation frequency was tested. We found a small, but statistically significant, decrease in conjugation efficiency for a sirA mutant donor on either LB or M9 minimal glucose plates (two- to threefold decrease; P = 0.003 on LB plates, P = 0.0096 on M9 glucose; Table 2). However, this is a very small effect, compared to the effects of varying the medium composition. Also, the SirA-regulated MudJ fusions in the virulence plasmid transfer region are found in both the sense and antisense orientations. Although the transfer region of the F plasmid is known to incorporate antisense transcripts (9), we conclude that the regulatory effect of sirA on these genes is probably indirect and not of significant consequence.
Strain background does not explain the previous literature.
Because the virulence plasmid has been reported to be nonconjugative, we wanted to determine why our results differed from those of previous studies (19, 26–29). One hypothesis was that there was a difference in strain background. We were utilizing a virulent isolate of S. typhimurium (14028), while historically Salmonella geneticists have used strain LT2. To test this hypothesis we compared the conjugation frequencies of virulence plasmids from several S. typhimurium strains (14028, LT2, SR-11, and SL1344). P22 phage transduction was used to move the spv::MudJ mutation from BA1541 (14028 spv::MudJ) into LT2, SL1344, and SR-11 (strain constructions and lineages are described in Table 1). The spv::MudJ mutation was chosen because it is located directly opposite the conjugation genes on the plasmid (4). Because the virulence plasmid is roughly 90 kb and P22 can package only 45 kb, this should prevent the cotransduction of the MudJ marker and the functional conjugation genes present on the plasmid of 14028. Three transductants from each strain construction were tested for conjugation proficiency. Surprisingly, all three spv::MudJ transductants of strains SR-11 and LT2 (BA901a,b,c and BA902a,b,c; Table 1) acted as virulence plasmid donors at a frequency similar to that of 14028, but all three isolates of strain SL1344 were nonconjugative (BA903a,b,c; Table 1). On a later occasion, all of these strains were constructed again, and the conjugation experiments were repeated. The result was the same: the virulence plasmids of strains LT2 and SR-11 are self-transmissible, but that of SL1344 is not.
Although it appears that strain background cannot explain the discrepancy between our results and those of previous studies, there are other potential explanations. One possibility is that previous investigators utilized conditions that are not optimal for virulence plasmid conjugation. Because conjugation was not the focus of these reports, all of the methods attempted were not listed. However, commonly utilized conditions for conjugation include liquid matings and filter matings on LB plates. Since we determined that conjugation frequency of the virulence plasmid is reduced by using either of these conditions, this is a likely explanation. It is also likely that the recipient strain in previous experiments contained a virulence plasmid. This greatly decreases the conjugation frequency. In addition, some investigators have tested the transmissibility of plasmids marked with Tn10 (19, 27). However, the location of Tn10 on the virulence plasmid was unknown. If the transposon disrupted the conjugation genes, this would also explain the lack of conjugation. In any case, there are several factors that could contribute to the previous lack of detected conjugation.
The demonstration that the virulence plasmid of S. typhimurium is self-transmissible provides an example of horizontal gene transfer. The fact that this genetic exchange involves virulence factors has implications for the evolution of enteric pathogens such as Salmonella. The horizontal transfer of genetic material by conjugation is likely to increase the evolutionary rate at which pathogens can test new virulence gene combinations.
Acknowledgments
This work was supported by grant AI22933 from the United States National Institutes of Health.
We thank Cindy Arvidson for a critical reading of the manuscript.
REFERENCES
- 1.Ahmer B M M, van Reeuwijk J, Timmers C D, Valentine P J, Heffron F. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol. 1998;180:1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmer B M M, van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 3.Bäumler A J, Tsolis R M, Bowe F A, Kusters J G, Hoffmann S, Heffron F. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect Immun. 1996;64:61–68. doi: 10.1128/iai.64.1.61-68.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäumler A J, Tsolis R M, Ficht T A, Adams L G. Evolution of host adaptation in Salmonella enterica. Infect Immun. 1998;66:4579–4587. doi: 10.1128/iai.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd E F, Hartl D L. Recent horizontal transmission of plasmids between natural populations of Escherichia coli and Salmonella enterica. J Bacteriol. 1997;179:1622–1627. doi: 10.1128/jb.179.5.1622-1627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd E F, Hartl D L. Salmonella virulence plasmid. Modular acquisition of the spv virulence region by an F-plasmid in Salmonella enterica subspecies I and insertion into the chromosome of subspecies II, IIIa, IV and VII isolates. Genetics. 1998;149:1183–1190. doi: 10.1093/genetics/149.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowman J E, Meynell G G. Pleiotropic effects of derepressed bacterial sex factors on colicinogeny and cell wall structure. Mol Gen Genet. 1970;109:57–68. doi: 10.1007/BF00334046. [DOI] [PubMed] [Google Scholar]
- 8.El-Gedaily A, Paesold G, Krause M. Expression profile and subcellular location of the plasmid-encoded virulence (Spv) proteins in wild-type Salmonella dublin. Infect Immun. 1997;65:3406–3411. doi: 10.1128/iai.65.8.3406-3411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firth N, Ippen-Ihler K, Skurray R A. Structure and function of the F factor and mechanism of conjugation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2377–2401. [Google Scholar]
- 10.Friedrich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 11.Guiney D G, Fang F C, Krause M, Libby S. Plasmid-mediated virulence genes in non-typhoid Salmonella serovars. FEMS Microbiol Lett. 1994;124:1–9. doi: 10.1111/j.1574-6968.1994.tb07253.x. [DOI] [PubMed] [Google Scholar]
- 12.Gulig P A, Danbara H, Guiney D G, Lax A J, Norel F, Rhen M. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol Microbiol. 1993;7:825–830. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 13.Gulig P A, Doyle T J. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect Immun. 1993;61:504–511. doi: 10.1128/iai.61.2.504-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heffernan E J, Reed S, Hackett J, Fierer J, Roudier C, Guiney D. Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella typhimurium virulence plasmid gene rck. J Clin Investig. 1992;90:953–964. doi: 10.1172/JCI115972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helmuth R, Stephan R, Bunge C, Hoog B, Steinbeck A, Bulling E. Epidemiology of virulence-associated plasmids and outer membrane protein patterns within seven common Salmonella serotypes. Infect Immun. 1985;48:175–182. doi: 10.1128/iai.48.1.175-182.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoiseth S K, Stocker B A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 17.Hurme R, Berndt K D, Normark S J, Rhen M. A proteinaceous gene regulatory thermometer in Salmonella. Cell. 1997;90:55–64. doi: 10.1016/s0092-8674(00)80313-x. [DOI] [PubMed] [Google Scholar]
- 18.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 19.Jones G W, Rabert D K, Svinarich D M, Whitfield H J. Association of adhesive, invasive, and virulent phenotypes of Salmonella typhimurium with autonomous 60-megadalton plasmids. Infect Immun. 1982;38:476–486. doi: 10.1128/iai.38.2.476-486.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S L, Sanderson K E. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J Bacteriol. 1992;174:1662–1672. doi: 10.1128/jb.174.5.1662-1672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 22.McIntire S A. Transduction with integration-defective mutants of Salmonella typhimurium bacteriophage KB1. J Bacteriol. 1974;117:907–908. doi: 10.1128/jb.117.2.907-908.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meynell E, Meynell G, Datta N. Phylogenetic relationships of drug-resistance factors and other transmissible bacterial plasmids. Bacteriol Rev. 1968;32:55–83. doi: 10.1128/br.32.1.55-83.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montenegro M A, Bitter-Suermann D, Timmis J K, Aguero M E, Cabello F C, Sanyal S C, Timmis K N. traT gene sequences, serum resistance and pathogenicity-related factors in clinical isolates of Escherichia coli and other Gram-negative bacteria. J Gen Microbiol. 1985;131:1511–1521. doi: 10.1099/00221287-131-6-1511. [DOI] [PubMed] [Google Scholar]
- 25.Ou J T. The 90 kilobase pair virulence plasmid of Salmonella serovar Typhimurium coexists in strains with a plasmid of the 23 incompatibility groups. Microb Pathog. 1993;15:237–242. doi: 10.1006/mpat.1993.1074. [DOI] [PubMed] [Google Scholar]
- 26.Ou J T, Baron L S, Dai X Y, Life C A. The virulence plasmids of Salmonella serovars typhimurium, choleraesuis, dublin, and enteritidis, and the cryptic plasmids of Salmonella serovars copenhagen and sendai belong to the same incompatibility group, but not those of Salmonella serovars durban, gallinarum, give, infantis, and pullorum. Microb Pathog. 1990;8:101–107. doi: 10.1016/0882-4010(90)90074-z. [DOI] [PubMed] [Google Scholar]
- 27.Pardon P, Popoff M Y, Coynault C, Marly J, Miras I. Virulence associated plasmids of Salmonella serotype Typhimurium in experimental murine infection. Ann Inst Pasteur Microbiol. 1986;137B:47–60. doi: 10.1016/s0769-2609(86)80093-x. [DOI] [PubMed] [Google Scholar]
- 28.Sanderson K E. F-mediated conjugation, F+ strains, and Hfr strains of Salmonella typhimurium and Salmonella abony. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2406–2412. [Google Scholar]
- 29.Sanderson K E, Kadam S K, MacLachlan P R. Derepression of F factor function in Salmonella typhimurium. Can J Microbiol. 1983;29:1205–1212. doi: 10.1139/m83-184. [DOI] [PubMed] [Google Scholar]
- 30.Smith H R, Humphreys G O, Grindley N D F, Grindley J N, Anderson E S. Molecular studies of an fi+ plasmid from strains of S. typhimurium. Mol Gen Genet. 1973;126:143–151. doi: 10.1007/BF00330989. [DOI] [PubMed] [Google Scholar]
- 31.Stojiljkovic I, Bäumler A J, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sukupolvi S, O’Connor C D. TraT lipoprotein, a plasmid-specified mediator of interactions between gram-negative bacteria and their environment. Microbiol Rev. 1990;54:331–341. doi: 10.1128/mr.54.4.331-341.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sukupolvi S, Vuorio R, Qi S Y, O’Connor D, Rhen M. Characterization of the traT gene and mutants that increase outer membrane permeability from the Salmonella typhimurium virulence plasmid. Mol Microbiol. 1990;4:47–57. doi: 10.1111/j.1365-2958.1990.tb02014.x. [DOI] [PubMed] [Google Scholar]
- 34.Tinge S A, Curtiss R., III Isolation of the replication and partitioning regions of the Salmonella typhimurium virulence plasmid and stabilization of heterologous replicons. J Bacteriol. 1990;172:5266–5277. doi: 10.1128/jb.172.9.5266-5277.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson J A, Doyle T J, Gulig P A. Exponential-phase expression of spvA of the Salmonella typhimurium virulence plasmid: induction in intracellular salts medium and intracellularly in mice and cultured mammalian cells. Microbiology. 1997;143:3827–3839. doi: 10.1099/00221287-143-12-3827. [DOI] [PubMed] [Google Scholar]
- 36.Wong K K, Cheng R, Saffer J D, Ralph D, Welsh J, McClelland M. DNA fingerprinting by arbitrarily primed PCR (RAPDs) In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 3.4.2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 1–14. [Google Scholar]