Abstract
SARS-CoV-2 infection is initiated by binding of the receptor-binding domain (RBD) of its spike glycoprotein to the peptidase domain (PD) of angiotensin-converting enzyme 2 (ACE2) receptors in host cells. Recently detected Omicron variant of SARS-CoV-2 (B.1.1.529) is heavily mutated on RBD. First the BA.1 and later the BA.2 variant became the most dominant strains of the Omicron variant. To investigate how the mutations of these strains affect RBD-PD interactions, we performed all-atom molecular dynamics simulations of the BA.1 and BA.2 RBD-PD in the presence of full-length glycans, explicit water, and ions. Simulations revealed that RBDs of BA.1 and BA.2 variants exhibit a more dispersed interaction network and make an increased number of salt bridges and hydrophobic interactions with PD compared to wild-type RBD. Although BA.1 and BA.2 differ in two residues at the RBD-ACE2 interface, no major difference in RBD-PD interactions and binding strengths were observed between these variants. Using the conformations sampled in each trajectory, the Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) method estimated ∼34% and ∼51% stronger binding free energies to PD for BA.1 and BA.2 RBD, respectively, than wild-type RBD, which may result in higher binding efficiency of the Omicron variant to infect host cells.
Keywords: ACE2 receptor, MMPBSA, Molecular dynamics simulations, Omicron variant, SARS-CoV-2, Spike glycoprotein
Graphical abstract
1. Introduction
The recent appearance and the rapid rate of infection of a heavily mutated B.1.1.529 variant of SARS-CoV-2, named Omicron, have raised concerns around the world, with many countries temporarily limiting their international travel. World Health Organization has designated the Omicron variant as a variant of concern (VOC) [1]. Currently, the Omicron variant has five major sub-lineages, namely BA.1, BA.2, BA.3, BA.4 and BA.5 [2]. BA.1 became the first dominant Omicron variant, while writing this manuscript BA.2 was the most observed SARS-CoV-2 variants, at its peak accounting for 83% of all new SARS-CoV-2 cases globally [3]. Currently, the BA.2 variant accounts for more than 20% of all new SARS-CoV-2 cases, while BA.5 accounts for more than 40% of all new cases [3]. The BA.1 variant comprises 30 mutations on the spike glycoprotein (S), while the BA.2 variant comprises 28. Remarkably, 15 and 16 of these mutations are located on the receptor-binding domain (RBD) of the BA.1 and BA.2 variants, respectively. Among these RBD mutations, 12 (G339D, S373P, S375F, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, and Y505H) are shared among the BA.1 and BA.2 variants (Fig. 1 ).
Fig. 1.
Location of RBD mutations for the Omicron variant. Mutations found on both BA.1 and BA.2 are highlighted with red beads, while the mutations specific to BA.1 and BA.2 variants are highlighted with blue and turquoise colored beads, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
RBD interacts with the peptidase domain (PD) of angiotensin-converting enzyme 2 (ACE2) receptors and plays a critical role in the host cell entry of the virus. RBD is a critical antibody and drug target, and all the available vaccines produce antibodies that neutralize the RBD-PD interaction. Mutations on both BA.1 RBD (RBDBA.1) and BA.2 RBD (RBDBA.2) are surface-exposed and being targeted by various antibodies (Fig. S1) and nanobodies. In addition, for BA.1, 11 of these 15 mutations are located on the ACE2 binding interface, while for BA.2 nine of these are located on the ACE2 binding interface (Fig. 1). For both BA.1 and BA.2 four hydrophilic residues mutated to positively charged residues (N440K, T478K, Q493R, and Q498R), one negatively charged residue mutated to hydrophobic residue (E484A), one positively charged residue mutated to hydrophilic residue (K417N), and three hydrophilic residues are mutated to again hydrophilic residues (S477N, N501Y, and Y505H) at RBD's PD binding interface. In addition, to these mutations, two neutral residues mutated to hydrophilic residues (G446S and G496S) in BA.1. Thus, both RBDBA.1's and RBDBA.2's PD binding interfaces are more positively charged than RBDWT. Furthermore, the PD binding interface of RBDBA.1 comprises more hydrophilic residues than RBDBA.2. Our previous all-atom Molecular Dynamics (MD) simulations [4] showed that 5 of these mutated residues form pairwise interactions between wild-type (WT) S and ACE2 (salt bridges between K417-D30 and E484-K31, and hydrogen bonding between Q493-E35, Q498-Q42, Q498-K353, and Y505-E37). It is still unclear how BA.2 Omicron mutations affect the binding strength of RBD to ACE2 and the ability of existing SARS-CoV-2 antibodies to neutralize RBD-ACE2 interaction. Furthermore, the difference in binding characteristics and strength of Omicron BA.1 and BA.2 variants remains to be explored.
In order to explore the effect of various Omicron variant mutations on RBD-ACE2 interactions, we performed an extensive set of MD simulations of the RBD-PD complex for the Omicron variants BA.1 and BA.2. Our simulations totaling 3 μs in length revealed that both RBDBA.1 and RBDBA.2 exhibit a more dispersed interaction network on the RBD-ACE2 interaction surface compared to WT RBD (RBDWT). Furthermore, an increased number of salt bridges and hydrophobic interactions of RBDBA.1 and RBDBA.2 with PD were observed. Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) method estimated ∼34% and ∼51% stronger binding free energy for RBDBA.1 and RBDBA.2, respectively, compared to RBDWT.
2. Methods
2.1. MD simulations system preparation
Most computational studies in the literature [[4], [5], [6], [7], [8], [9], [10]], including ours, have focused on the RBD-PD systems instead of the full-length S-ACE2 system due to the relevance of RBD-PD systems in providing insight to the S-ACE2 interactions and the computational difficulty of simulating the full-length S-ACE2 complex in the presence of explicit solvent and membrane, which goes up to 2 million atoms. Furthermore, a wide range of experimental studies [7,[11], [12], [13], [14], [15], [16]] have also focused on the RBD-PD systems. In our current study, systems were prepared in VMD [17] as we previously performed RBDWT-PD, RBDAlpha-PD, RBDBeta-PD, and RBDDelta-PD MD simulations [[4], [5], [6]]. The RBDBA.1-PD structure [11] did not exist when the preprint of this study was published. Thus, the structure of SARS-CoV-2 S protein RBD bound with ACE2 (PDB ID: 6M0J [12]) was used as a starting structure for MD simulations of the RBDBA.1-PD complex. Omicron BA.1 variant RBD structure was modeled by introducing the 15 mutations located at the RBD of the SARS-CoV-2 Omicron variant using the Mutator plugin of VMD [17] onto the WT RBD structure. Chloride ion, zinc ion, and water molecules in the structures were kept. Since full-length glycans are not visible in the crystal structure, we used glycan models [18]. RBDBA.1-PD was solvated into a water box with 25 Å cushion in each direction using TIP3P model water molecules. Ions were added to neutralize the system and set the NaCl concentration to 150 mM. The RBDBA.1-PD (PDB ID: 7T9L [11]) structure was published recently. Using this structure an additional solvated RBDBA.1-PD system was constructed.
For the RBDBA.2-PD simulations, solvated RBDBA.2-PD systems were modeled by introducing L371F, T376A, D405N, and R408S mutations and reversing G446S and G496S Mutations in the RBDBA.1-PD structure, and subsequent solvating it and adding ions to system. S proteins are coated by the glycans that shield its surface to thwart the host immune response [18]. Full-length glycans are not visible in the RBD-PD [11,12] structures that were used in our WT, BA.1 and BA.2 simulations. Thus, we used the glycan models [18,19] that were built based on the glycomics data [20,21] to obtain RBD and PD structures with complete glycosylation profiles. Glycan models were superimposed onto the partially visible glycans in the RBD-PD structures and partial glycan structures were replaced with full-length glycan models.
2.2. MD simulations
Two sets of conventional MD simulations were performed for the RBDWT-PD complex (MD 1–2), while four sets of simulations were performed for each of the RBDBA.1-PD complex (MD 3–6) and RBDBA.2-PD complex (MD7-10). Three of the RBDBA.1-PD simulations were initiated from the RBDWT-PD based RBDBA.1-PD model (MD3-5) and one was initiated from the RBDBA.1-PD structure (MD6). All MD simulations were performed in the presence of explicit water molecules, ions, and also full-length glycans (MD7-10). Prior to production simulations, each system was minimized for 10,000 steps and then equilibrated for 2 ns by keeping the protein fixed. Subsequently, system was minimized for an additional 10,000 steps without fixing the protein, which is followed by 4 ns of equilibration with harmonic constraints applied on Cα atoms. All constraints were removed from the system and an additional 4 ns of MD simulations were performed; finalizing the minimization and equilibration steps prior to production runs. Production runs for each set of MD simulations were of 300 ns length. Thus, a total of 3 μs of production simulations were performed.
MD simulations were performed in NAMD-2.14, [22] for MMPBSA calculations and system minimizations and equilibrations, and NAMD3 [22] for all production simulations under N, P, T conditions. CHARMM36 [23] force field and a time step of 2 fs were used in the simulations. Pressure was kept at 1 atm using the Langevin Nosé-Hoover method with an oscillation period of 100 fs and a damping time scale of 50 fs. Temperature was maintained at 310 K using Langevin dynamics with a damping coefficient of 1 ps−1. Periodic boundary conditions were applied in simulations and the Particle-mesh Ewald method was used for long-range electrostatic interactions. 12 Å cutoff distance was used for van der Waals interactions.
2.3. Criteria for interaction analysis
To determine salt bridge formation in MD simulations, a cutoff distance of 6 Å between the basic nitrogen and acidic oxygen was used [24], while for hydrophobic interactions, a cutoff distance of 8 Å between the side chain carbon atoms was used [[25], [26], [27]]. A cutoff distance of 3.5 Å between hydrogen bond donor and acceptor, and a 30° angle between the hydrogen atom, the donor heavy atom and the acceptor heavy atom was used to determine hydrogen bond formation [28]. Those interaction pairs that satisfied the hydrogen bonding distance criterion but did not satisfy the angle criterion, were classified as electrostatic interactions. As was performed in our previous studies [[4], [5], [6]], observation frequencies of interactions sampled from MD simulations were classified as high and moderate for interactions that occur in 49% and above and between 15 and 48% of the total trajectory, respectively. Pairwise interactions with observation frequencies below 15% were excluded from further analysis.
2.4. Binding free energy predictions via MMPBSA method
For each set of simulations, 3,000 snapshots separated by 0.1 ns were selected from the simulations. The binding free energies were predicted for the RBD-PD complexes using the MMPBSA method [29,30], which was conducted via VMD [17] plugin CaFE [31]. Entropy change during binding was neglected in calculations, consistent with previous MMPBSA calculations for RBD-PD interactions [32,33]. Default parameters were used in CaFE [31] calculations.
3. Results
3.1. RBDBA.1-PD interactions
We performed all-atom MD simulations of the RBDBA.1-PD in the presence of explicit water and ions, and full-length glycans on both S RBD and ACE2 PD [18,19] (∼200k atoms in total). Four sets of MD simulations each of 300 ns in length were performed using the parameters of our previous RBD-PD simulations for the WT [4], Alpha, and Beta variants [5]. These four sets of simulations were combined into a single 1.2 μs long trajectory to investigate the RBDBA.1-PD interactions. Simulations revealed a more extensive interaction network for RBDBA.1-PD with PD compared to RBDWT. We detected five salt bridges between RBDBA.1 and PD; one of them (K440-E329) medium and four (R403-E37, R493-E35, R493-D38, and R498-D38) with high frequency (Fig. 2 ). In comparison only 2 high frequency salt bridges existed between RBDWT and PD [4] (Fig. 2 and Table S1) and both of those disappeared in the BA.1 variant (Fig. 2 and Table S2). The RBDBA.1 forms all of the 10 high frequency hydrophobic interactions that were observed for RBDWT-PD and an additional high frequency hydrophobic interaction between Y501–Y41. Compared to eight hydrogen bonds between RBDWT and PD (three high and five medium frequency), six hydrogen bonds were observed between RBDBA.1 and PD (three high and three medium frequency). Only two of these interactions were also observed for the WT, while other four are newly formed (Fig. 2). Collectively, the total number of salt bridges, hydrophobic interactions, and hydrogen bonds at the S-ACE2 interface changed by 150%, 10%, and −25%, respectively.
Fig. 2.
(A) Interactions between RBDWT, RBDBA.1 and RBD BA.2 of the SARS-CoV-2 S protein and the PD of human ACE2. Representative snapshots of the all-atom MD simulations highlight salt bridges, hydrophobic interactions, and hydrogen bonding between RBDWT-PD, RBDBA.1-PD and RBDBA.2-PD. The interaction surface is divided into three distinct regions (CR1-3) [4,34] (B) Normalized distributions of the distances between the amino-acid pairs that form salt bridges (orange), hydrophobic interactions (red), and hydrogen bonds (purple) between RBDWT, RBDBA.1 and RBD BA.2 and PD. Newly formed interactions due to mutations are marked with an asterisk. Solid lines represent the minimal threshold distance between these residues to form each class of pairwise interactions. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Our simulations also revealed a change in the spatial distribution of RBD-PD interactions along the interaction surface due to the mutations in the BA.1 variant, which are mostly consistent with recently reported RBDBA.1-ACE2 structures [11,[14], [15], [16]]. Between RBDWT and PD, salt bridges are concentrated at the interface of contact region 1 (CR1) and CR2, while hydrogen bonding and hydrophobic interactions are concentrated in CR3 and CR1, respectively (Fig. 2A) [4]. In comparison, RBDBA.1 exhibits a more dispersed interaction network along the RBD-PD interaction surface (Fig. 2). RBDBA.1 mutations result in two additional interactions (hydrogen bonds) in CR1, four additional interactions (three salt bridges and one hydrogen bond) in CR2, and five additional interactions (two salt bridges, two hydrogen bonds, and one hydrophobic interaction) in CR3. Furthermore, RBDBA.1 mutations result in the loss of one interaction (one salt bridge) in CR1, three interactions (one salt bridge and two hydrogen bonds) in CR2, and five interactions (five hydrogen bonds) in CR3. This may result in an altered binding mechanism and negatively impact the current inhibition mechanism by neutralizing antibodies and nanobodies.
3.2. RBDBA.2-PD interactions
For RBDBA.2-PD, four sets of all-atom MD simulations, each of 300 ns in length, were performed in the presence of explicit water and ions, and full-length glycans. These simulations were combined into a single MD trajectory of 1.2 μs length to investigate RBDBA.2-PD Interactions. As was the case for RBDBA.1, RBDBA.2 showed a more extensive interaction network with PD compared to RBDWT. Between RBDBA.2 and PD a total of five salt-bridges (two high and three medium frequency), 11 hydrophobic interactions (all high frequency), and six hydrogen bonds (five high and one medium frequency) (Fig. 2 and Table S2) were observed. This corresponds to a 150%, 10% and −25% change in salt-bridges, hydrophobic interactions, and hydrogen bonds, respectively, compared to those observed for RBDWT-PD. Similar to BA.1, RBDBA.2 exhibits a dispersed interaction network for each type of interaction type along the RBD-PD interaction surface (Fig. 2).
The difference in the RBD-PD interface for BA.1 and BA.2 variants are the two residues located at residue positions 446 and 496, which are S446 and S496 for BA.1 and G446 and G496 for BA.2. Yet, the interacting RBD-PD residue pairs for BA.1 and BA.2 were identical. Comparing the frequencies of RBDBA.2-PD interactions with RBDBA.1-PD shows that the total number of high frequency hydrogen bonds increased by three, while high frequency salt bridges decreased by two for BA.2 with respect to BA.1. Conclusively, the binding interactions network for RBDBA.1–PD and RBDBA.2–PD share similar features, differing only in the observation frequencies in six out of 22 interactions.
3.3. Effect of Omicron BA.1 and BA.2 mutations on RBD, PD, and their interface fluctuations
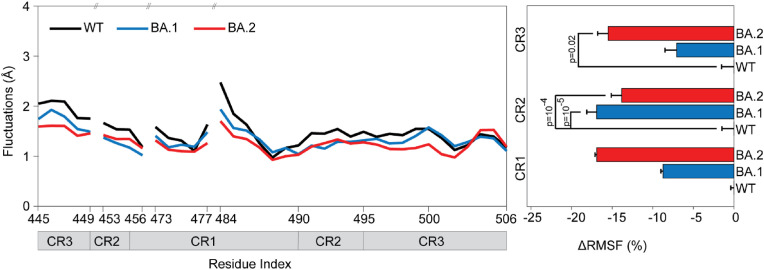
To investigate the effect of Omicron mutations on the RBD binding dynamics, we quantified the Root Mean Square Fluctuations (RMSF) of the Cα atoms of the RBD residues located on the PD binding surface for the RBD-PD complexes [4]. All sets of MD simulations performed for WT, BA.1, and BA.2 were combined into single trajectories of 600 ns, 1.2 μs, and 1.2 μs lengths, respectively. The rigid body motions were eliminated for each trajectory by aligning the RBD interacting surface of PD for each conformer with their starting crystal structures. Both BA.1 and BA.2 mutants of RBD had lower residue fluctuations at the interface suggesting that tighter and more rigid binding compared to WT (Fig. 3 ). At CR1 and CR3, BA.2 mutations caused a significantly larger decrease in fluctuations compared to BA.1, while in CR2 the opposite was observed (Fig. 3).
Fig. 3.
(Left) Effect of Omicron mutations on RBD and PD fluctuations. (Right) RMSF of RBD residues located on the PD binding surface of WT, BA.1 and BA.2 variants. P values were calculated using a two-tailed Student's t-test. P values larger than 0.05 are not shown. Error bars represent standard deviation (s.d.).
We aligned each trajectory with the RBDWT –PD crystal structure by using all of the RBD Cα atoms and also by using only the RBD beta sheet and helix Cα atoms. The RMSD of complete structure of RBDBA.1 and RBDBA.2 to RBDWT resulted in trajectory average values of 1.25 Å and 1.30 Å, respectively, while the RMSD of the beta sheet and helical regions of the RBDBA.1 and RBDBA.2 to RBDWT resulted in trajectory average values of 0.82 Å and 0.79 Å (Fig. S2). MD simulations generally deviate to some degree from their starting structures, especially if thermodynamic conditions of the structure and MD simulations differ [35]. Thus, we also compared the average RBDBA.1 and RBDBA.2 Cα atom coordinates obtained from MD simulation trajectories (Fig. S3) with those of RBDWT MD simulations. The average RBDBA.1 and RBDBA.2 conformations differed from the average RBDWT conformation by only 0.60 Å and 0.68 Å, respectively, while the RMSD between average RBDBA.1 and RBDBA.2 average conformations was only 0.51 Å. Thus, BA.1 and BA.2 mutations did not affect the RBD structure significantly, showing practically identical structures with WT.
3.4. Binding free energies for RBDBA.1-PD and RBDBA.2-PD
Binding free energies between RBD and PD were calculated from two sets of RBDWT-PD, four sets of RBDBA.1-PD, and four sets of RBDBA.2-PD simulations via the MMPBSA method [29,30] using the VMD [17] plugin CaFE [31] (Table S3). MMPBSA calculations estimated 34% stronger binding free energy (−40 ± 9.7 kcal/mol, mean ± s.d., N = 4 sets) between RBDBA.1 and PD, compared to the binding free energy between RBDWT and PD (−29.9 ± 7.3 kcal/mol, N = 2 sets). For RBDBA.2, MMPBSA calculations estimated 51% stronger binding free energy (−45.3 ± 9.1 kcal/mol, N = 4 sets) to PD than the binding free energy of RBDWT to PD (Fig. 4 ). Considering that BA.1 and BA.2 induced small changes in the total percentage in the number of hydrophobic interactions and hydrogen bonds, while their effect on the total number of salt bridges was considerably large, we conclude that the increase in the number of salt bridges in the S-ACE2 interface resulted in this higher binding strength of RBDBA.1 and RBDBA.2 to PD, which may result in a higher efficiency of the SARS-CoV-2 virus to infect host cells.
Fig. 4.
Binding free energies of RBDs to PD. (A) Distribution of the binding free energies of RBDWT, RBDBA.1, and RBDBA.2. (B) Mean binding free energy values of the RBDWT, RBDBA.1, and RBDBA.2 to PD. Error bars represent s.d. P values were calculated using a two-tailed Student's t-test.
4. Conclusion
An extensive set of MD simulations totaling 3 μs in length were performed to investigate the effect of the two most common Omicron variants BA.1 and BA.2 in RBD-PD interactions. The preprint of this study was the first in the literature to show via all-atom MD simulations the effect of the Omicron BA.1 mutations on RBD-PD interactions and binding strength. Our findings have been supported with recent computational [8,9] and experimental studies [7,11,13,14]. There is currently no consensus regarding the exact binding energy of the BA.1 Omicron variant S protein to ACE2. Binding free energies ranging from −107.04 to −635.32 kcal/mol were reported by post-processing all-atom MD trajectories [[8], [9], [10]]. Furthermore, KD values ranging 0.3–38.9 nM were experimentally reported for the binding of RBDBA.1 to PD [7,11,13,14]. While these approaches report different KD values, consistent with our findings, they all estimate a higher binding strengths for RBDBA.1 compared to the RBDWT. Affinity constant Kaff was measured experimentally as 6.01 × 10−7 and 0.37 × 10−7 for WT and BA.1, respectively, to PD [10]. However, these Kaff measurements were not statistically significantly different [10]. Yet, effect of BA.2 mutations on ACE2 binding have not yet been reported in the literature. The binding free energies between RBD and PD that we estimated via MMPBSA method exhibits a 34% and 51% increase in binding strength for RBDBA.1 and RBDBA.2 compared to RBDWT. The analysis of the pairwise interactions between RBD and PD provided a detailed insight into this increased binding strength of the Omicron variants. The most striking change induced by BA.1 and BA.2 mutations was the net change of three additional salt bridges. Both RBDBA.1 and RBDBA.2 mutations were shown to decrease the fluctuation of RBD residues at the ACE2 binding interface. Collectively our result highlight that both Omicron variants result in a more extensive interaction network, and a stronger and tighter binding. Our MD simulations also revealed differences in observation frequencies, which result in a stronger and tighter binding for BA.2 compared to BA.1.
RBDBA.1 and RBDBA.2 mutations may also affect the binding affinity and neutralizing capability of SARS-CoV-2 S antibodies and nanobodies. We investigated the binding surface of 160 antibody structures that were resolved in complex with RBD in the PDB, and determined if these surfaces overlap with the BA.1 and BA.2 mutations (Fig. S1). Mutations introduced by both BA.1 and BA.2 variants overlap with binding interface for all of these 160 antibodies. In the literature a categorization protocol for neutralizing nanobodies was introduced, and 24 of the 160 neutralizing antibodies investigated in our study were categorized into four different classes according to their binding regions and mechanisms (Fig. 5 ) [36]. Based on these categorization, mutations shared by both BA.1 and BA.2 appear to be more concentrated on class 2 antibodies than the other classes. For example, K417N, N501Y, and Y505H mutations are expected to eliminate salt bridges K417-E99 and K417-E96 and hydrogen bonds K417–Y52, N501-G29, Y505-E99 between RBD and the class 1 antibody C105 (Fig. 5). Similarly, these mutations are expected to disrupt RBD-antibody interactions involving residues 493 and 484 for the remaining class 1 antibodies C102, B38, CB6, and REGN10933. E484A, Q493R, Q498R, N501Y, and Y505H BA.1 and BA.2 mutations are expected to disrupt class 2 interactions, and G339D, S373P, N440K, Q498R N501Y, and Y505H mutations are expected to disrupt class 3 interactions, while S373P, S375F, N501Y, and Y505H mutations are expected to disrupt class 4 interactions. For example, E484A mutation disrupts interaction of E484-R112 and E484-Y34 (class 2). N440K and Q498R mutations disrupt interactions of N440–Y42, N440–Y102, N400-D103, and Q498-Y59 (class 3). S375F mutation disrupts interactions of class 4 antibody S2A4 S375-D95 and S375–S96 (Fig. 5). IC50 values were evaluated for a subset of 15 antibodies of the 160 antibodies investigated in our study. These 15 antibodies were also categorized into four classes and all of them showed higher IC50 values compared to WT when either BA.1 or BA.2 are introduced. Furthermore, binding free energies of launched monoclonal antibodies Etesevimab (class 2) and Bamlanivimab (class 1) to RBDBA.1 were computationally predicted to be weaker than to RBDWT [10]. Location of BA.1 and BA.2 mutations are also expected to affect nanobodies, which are single domain antibodies [37]. For example, we expect E484A mutation to eliminate E484-R52 salt bridge and E484-S57 hydrogen bonds in H11–H4 and H11-D4 nanobodies, and E484-N56, and E484-Y335 hydrogen bonds in Ty1 nanobody [5,38,39]. Additionally, Q493R mutation would eliminate the hydrogen bonds Q493-Y104 and Q493-S104 in H11–H4, and H11-D4, respectively.
Fig. 5.
Class 1–4 Antibody binding poses on RBD. (Upper Left) Binding poses of antibodies C105, P2B–2F6, REGN10987, and S2A4 categorized as class 1–4 antibodies, respectively, are shown [36]. Locations of RBD mutations of the Omicron variant are shown in red. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Funding
This work is supported by COVID-19 HPC Consortium (Grant numbers: TG-BIO200053 and TG-BIO210181).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmgm.2022.108286.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.WHO Classification of omicron (B.1.1.529): SARS-CoV-2 variant of concern. 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- 2.Viana R., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022 doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadfield J., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taka E., et al. Critical interactions between the SARS-CoV-2 spike glycoprotein and the human ACE2 receptor. J. Phys. Chem. B. 2021;125:5537–5548. doi: 10.1021/acs.jpcb.1c02048. [DOI] [PubMed] [Google Scholar]
- 5.Golcuk M., Hacisuleyman A., Erman B., Yildiz A., Gur M. Binding mechanism of neutralizing nanobodies targeting SARS-CoV-2 spike glycoprotein. J. Chem. Inf. Model. 2021;61:5152–5160. doi: 10.1021/acs.jcim.1c00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golcuk M., et al. SARS-Cov-2 delta variant decreases nanobody binding and ACE2 blocking effectivity. J. Chem. Inf. Model. 2022;62(10):2490–2498. doi: 10.1021/acs.jcim.1c01523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y., et al. Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA. 2 variant compared to BA. 1 and their possible mouse origins. Cell Res. 2022:1–12. doi: 10.1038/s41422-022-00672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupala C.S., Ye Y., Chen H., Su X.-D., Liu H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun. 2022;590:34–41. doi: 10.1016/j.bbrc.2021.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rath S.L., Padhi A.K., Mandal N. Scanning the RBD-ACE2 molecular interactions in Omicron variant. Biochem. Biophys. Res. Commun. 2022;592:18–23. doi: 10.1016/j.bbrc.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu L., et al. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Targeted Ther. 2022;7:1–3. doi: 10.1038/s41392-021-00863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mannar D., et al. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022;375:760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 13.Cameroni E., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin W., et al. Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science. 2022;375:1048–1053. doi: 10.1126/science.abn8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCallum, M. et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science 0, eabn8652, doi:doi:10.1126/science.abn8652. [DOI] [PMC free article] [PubMed]
- 16.Han P., et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell. 2022;185(4):630–640. doi: 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 18.Casalino L., et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barros E.P., et al. The flexibility of ACE2 in the context of SARS-CoV-2 infection. Biophys. J. 2021;120:1072–1084. doi: 10.1016/j.bpj.2020.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shajahan A., et al. Comprehensive characterization of N-and O-glycosylation of SARS-CoV-2 human receptor angiotensin converting enzyme 2. Glycobiology. 2021;31:410–424. doi: 10.1093/glycob/cwaa101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips J.C., et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020;153 doi: 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Best R.B., et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theor. Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckstein O., Denning E.J., Perilla J.R., Woolf T.B. Zipping and unzipping of adenylate kinase: atomistic insights into the ensemble of open↔ closed transitions. J. Mol. Biol. 2009;394:160–176. doi: 10.1016/j.jmb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stock P., Utzig T., Valtiner M. Direct and quantitative AFM measurements of the concentration and temperature dependence of the hydrophobic force law at nanoscopic contacts. J. Colloid Interface Sci. 2015;446:244–251. doi: 10.1016/j.jcis.2015.01.032. [DOI] [PubMed] [Google Scholar]
- 26.Manavalan P., Ponnuswamy P. A study of the preferred environment of amino acid residues in globular proteins. Arch. Biochem. Biophys. 1977;184:476–487. doi: 10.1016/0003-9861(77)90457-x. [DOI] [PubMed] [Google Scholar]
- 27.Stavrakoudis A., Tsoulos I.G., Shenkarev Z.O., Ovchinnikova T.V. Molecular dynamics simulation of antimicrobial peptide arenicin-2: b-hairpin stabilization by noncovalent interactions. Biopolymers. 2009;92:143–155. doi: 10.1002/bip.21149. [DOI] [PubMed] [Google Scholar]
- 28.Durrant J.D., McCammon J.A. HBonanza: a computer algorithm for molecular-dynamics-trajectory hydrogen-bond analysis. J. Mol. Graph. Model. 2011;31:5–9. doi: 10.1016/j.jmgm.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kollman P.A., et al. Calculating structures and free energies of complex molecules: combining molecular Mechanics and continuum models. Accounts Chem. Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan J., Cheatham T.E., Cieplak P., Kollman P.A., Case D.A. Continuum solvent studies of the stability of DNA, RNA, and Phosphoramidate−DNA helices. J. Am. Chem. Soc. 1998;120:9401–9409. doi: 10.1021/ja981844+. [DOI] [Google Scholar]
- 31.Liu H., Hou T.CaFE. A tool for binding affinity prediction using end-point free energy methods. Bioinformatics. 2016;32:2216–2218. doi: 10.1093/bioinformatics/btw215. [DOI] [PubMed] [Google Scholar]
- 32.Ghorbani M., Brooks B.R., Klauda J.B. Critical sequence hotspots for binding of novel coronavirus to angiotensin converter enzyme as evaluated by molecular simulations. J. Phys. Chem. B. 2020;124:10034–10047. doi: 10.1021/acs.jpcb.0c05994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aggarwal A., et al. Mechanistic insights into the effects of key mutations on SARS-CoV-2 RBD–ACE2 binding. Phys. Chem. Chem. Phys. 2021;23:26451–26458. doi: 10.1039/D1CP04005G. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Liu M., Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc. Natl. Acad. Sci. USA. 2020;117:13967–13974. doi: 10.1073/pnas.2008209117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pullara F., Wenzhi M., Gür M. Why protein conformers in molecular dynamics simulations differ from their crystal structures: a thermodynamic insight. Turk. J. Chem. 2019;43:394–403. [Google Scholar]
- 36.Barnes C.O., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bannas P., Hambach J., Koch-Nolte F. Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics. Front. Immunol. 2017;8:1603. doi: 10.3389/fimmu.2017.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huo J., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 39.Hanke L., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.