Abstract
The quantitation of the available antibody binding-site concentration of polyclonal antibodies in serum is critical in defining the efficacy of vaccines against substances of abuse. We have conceptualized an equilibrium dialysis (ED)-based approach coupled with fluorimetry (ED-fluorimetry) to measure the antibody binding-site concentration to the ligand in an aqueous environment. The measured binding-site concentrations in monoclonal antibody (mAb) and sera samples from TT-6-AmHap-immunized rats by ED-fluorimetry are in agreement with those determined by a more established equilibrium dialysis coupled with ultraperformance liquid chromatography tandem mass spectrometry (ED-UPLC-MS/MS). Importantly, we have shown that the measured antibody binding-site concentrations to the ligand by ED-fluorimetry were not influenced by the sample serum matrix; thus, this method is valid for determining the binding-site concentration of polyclonal antibodies in sera samples. Further, we have demonstrated that under appropriate analytical conditions, this method resolved the total binding-site concentrations on a nanomolar scale with good accuracy and repeatability within the microliter sample volumes. This simple, rapid, and sample preparation-free approach has the potential to reliably perform quantitative antibody binding-site screening in serum and other more complex biological fluids.
Introduction
Therapeutic vaccines that induce drug-sequestering antibodies have been recognized as potential treatment modalities for opioid use disorder (OUD).1−6 These antibodies negate the antinociceptive effects of the drugs by capturing and preventing them from permeating through the blood–brain barrier.7,8 In order for this vaccine to effectively elicit an immune response, a hapten that structurally resembles the target drug is conjugated to an immunogenic carrier protein to allow the presentation of the hapten to the immune cells.9,10 Hapten-based vaccines against opioids and other related drugs have been described in the literature.3,5,6,11−14
The efficacy of these vaccines relies not only on the binding affinity of the induced antibodies to the target drugs15−18 but also on the available antibody binding sites, otherwise known as the total antibody binding-site concentration.19 In most cases, the needed optimal concentration of induced antibodies in circulation is not defined and is expected to vary depending on the amount of drugs present in the patients.20 Thus, the monitoring of drug-sequestering antibody binding-site concentrations in serum and other biological fluids is not only a crucial requirement in defining vaccine efficacy but may also be beneficial in personalized treatment and precision medicine, as this can provide information on the appropriate boosting interval of the vaccine. Further, in the field of monoclonal antibody (mAb) immunotherapy, the quantitation of antibody concentration is also critical, particularly in establishing the pharmacokinetics of new immunotherapeutic drugs.21−23 While quantitative antibody analysis in mAb is relatively straightforward, the challenges and complications arise when conducted with polyclonal antibodies in complex matrices such as serum or cell lysates, which contain unidentified endogenous species. Thus, to address this limitation, it is imperative to develop simple and appropriate biophysical methods that can accurately and reliably quantify the available antibody binding sites in biological fluids. Traditionally, antibody binding-site concentrations were estimated from surface-based ligand-binding assays such as an enzyme-linked immunosorbent assay (ELISA)24−28 and surface plasmon resonance (SPR).24,29,30 These involve the surface immobilization of antigens that can potentially induce conformational changes, which have unpredictable influences on the results. This problem was minimized in solution-based ligand-binding assays such as radioimmunoassays (RIAs)31 and fluorescence-based approaches.32−34 However, most of these approaches pose significant disadvantages over the former solid-based ligand-binding assays due to the required sample preparation and purification steps or the need for radioactive labels. Over a couple of decades, there have been several reports on the utilization of liquid chromatography tandem mass spectrometry (LC-MS/MS) to directly quantify antibody binding-site concentrations in biological fluids.18,21,35
While LC-MS/MS-based methods can reliably quantify antibody binding sites, most of them suffer major experimental drawbacks such as the involvement of multiple sample preparation and purification steps, their dependence on the sensitivity of the developed method for quantification, and the integrity of the standards and/or the calibration curve. A previous report from our group described the use of ED-UPLC-MS/MS to indirectly determine the polyclonal antibody binding-site concentration in postimmune sera from their binding affinities (i.e., Kd values) to drugs such as 6-acetylmorphine (6-AM) and morphine.18 Although sample purification steps are not required in this approach, the determination of antibody binding-site concentration is a two-step process. The first step involves an equilibrium dialysis (ED) experiment that establishes the fraction of bound ligand in the absence of the competitor (b-value), followed by another set of an ED experiment that utilizes the optimized b-value from the first step to determine the Kd values. Hence, this approach involves long hours of dialysis and quantitation of drugs in the sample and buffer solutions by UPLC-MS/MS.
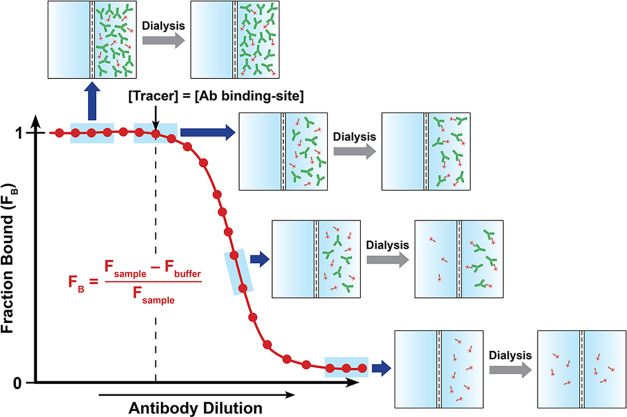
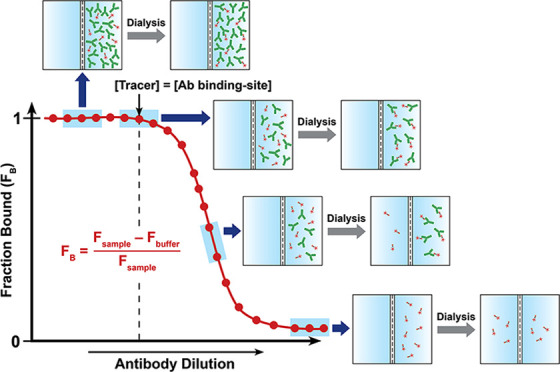
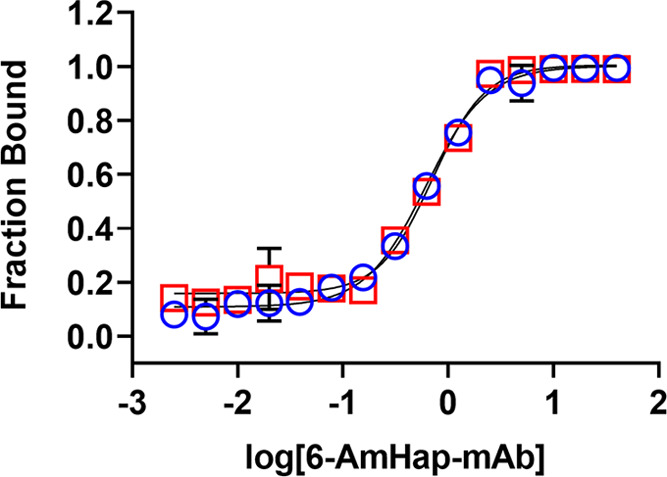
In the present work, we devised a simple ED-based approach coupled with nanodifferential scanning fluorimetry (ED-fluorimetry) to measure the antibody binding-site concentration to the ligand in sera from TT-6-AmHap-immunized rats14 using an easy-to-operate Monolith NT.115 system (NanoTemper). Unlike other analytical methods, this approach does not require sample preparation and immobilization; thus, measurements were done in the aqueous solution environment of the polyclonal antibodies. In this strategy, the antibody binding-site concentration was directly determined from the binding curve, thus requiring a shorter dialysis time (i.e., one-step dialysis). We have shown that the antibody binding-site concentration to the ligand can be fitted in the binding curve of fraction bound (FB) as a function of log[antibody] or serum dilution at the region where [tracer] = [antibody binding-site] (Figure 1). The proof-of-concept of this approach was first demonstrated in mAbs and further applied to sera samples. This method is promising in reliably measuring the antibody binding-site concentration on a nanomolar scale and has the potential to perform the quantitative antibody binding-site screening in biological fluids.
Figure 1.
Binding curve of the fraction bound ligand (FB) as a function of antibody dilutions/concentrations from ED experiments, highlighting the different regions of the curve. The antibody binding-site concentration was directly fitted at the equivalence point of the binding curve, where [tracer] = [antibody binding-site]. Fsample and Fbuffer are the fluorescence signals from the sample and buffer chambers, respectively.
Results and Discussion
Synthesis of 6-AmHap-Cy5 and 6-AmHap-acetamide Tracers
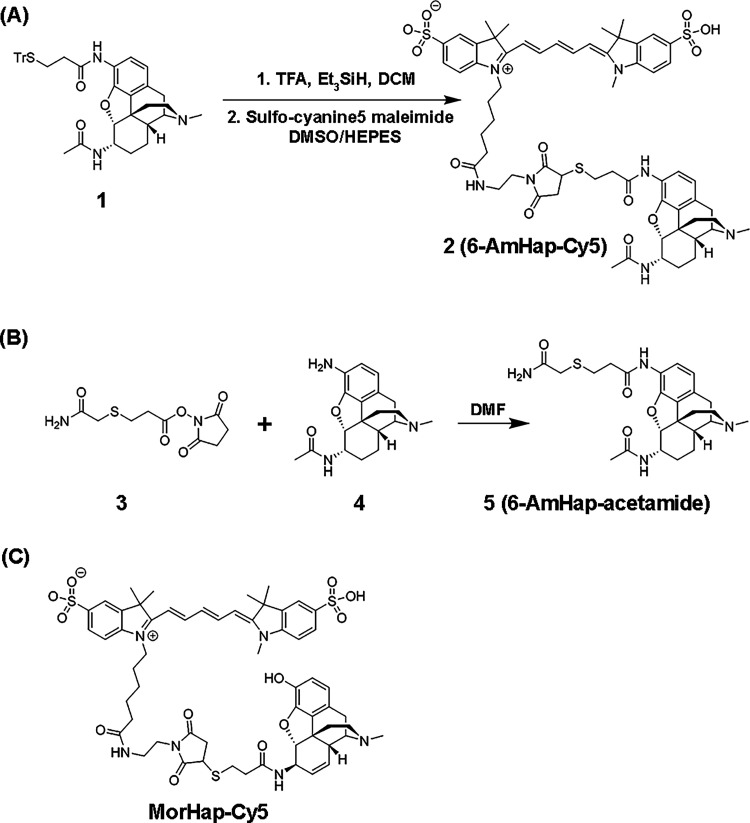
The hapten-fluorophore tracer, 6-AmHap-Cy5 (2; Scheme 1), was accessed using the previously prepared N-(7-acetamido-3-methyl-2,3,4,4a,5,6,7,7a-octahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinolin-9-yl)-3-mercaptopropanamide (1; Scheme 1a).14 The deprotection of the thiol group in 1 provided an activated sulfhydryl, which was subsequently coupled with the commercially available sulfo-cyanine5 in 30% DMSO in HEPES buffer to afford the desired product. The final purification by reversed-phase chromatography gave spectroscopically pure 6-AmHap-Cy5 in a 22% isolated yield and >98% purity based on HPLC. The formation of pure product 2 was confirmed by high-resolution mass spectrometry (HRMS-ESI) characterized by an m/z of 1180.4564 for [M + H]+ of C60H74N7O12S3 (calcd: 1180.4558).
Scheme 1. Preparation of (a) Sulfo-Cyanine5 Maleimide Conjugate of N-(7-Acetamido-3-methyl-2,3,4,4a,5,6,7,7a-octahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoliean-9-yl)-3-mercaptopropanamide (2), (b) N-(7-Acetamido-3-methyl-2,3,4,4a,5,6,7,7a-octahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinolin-9-yl)-3-((2-amino-2-oxoethyl)thio)propanamide (5), and (c) Chemical Structure of MorHap-Cy5.
The hapten-acetamide tracer, 6-AmHap-acetamide (5), was also prepared following the common coupling procedure. Briefly, 2,5-dioxopyrrolidin-1-yl 3-((2-amino-2-oxoethyl)thio)propanoate (3; Scheme 1b) was synthesized from the reaction of commercially available 3-[(carbamoylmethyl)sulfanyl]propanoic acid with N-hydroxysuccinimide (NHS) in DMF. The desired product 5 was accessed through the coupling of amine 4, an intermediate from our previously reported synthesis of DiAmHap,36 with the NHS ester 3. Column chromatography purification (2×) of the crude product generated spectroscopically pure product 5 in a 23% isolated yield. The identity and purity of 5 were established by 1H and 13C NMR spectroscopy (Figures S2 and S3), supported by HRMS-ESI and characterized by an m/z of 473.2229 for [M + H]+ of C60H74N7O12S3 (calcd: 473.2223).
Determination of Antibody Binding-Site Concentration: ED-UPLC-MS/MS vs ED-Fluorimetry
A simple strategy to measure the antibody binding-site concentration was proposed based on ED coupled with fluorimetry using an easy-to-operate Monolith NT.115 system (NanoTemper), which is equipped with an IR laser and a red fluorescence channel. In this approach, a red fluorescent dye, sulfo-cyanine5 maleimide, was conjugated to a sulfhydryl-containing ligand, 6-AmHap, to generate an appropriate fluorescent 6-AmHap-Cy5 tracer. This method is applicable to a wide range of fluorescent dyes; however, the readily available sulfo-cyanine5 was employed based on its ease of conjugation to the ligand and suitability of the analytical detection. The straightforward utilization of this NanoTemper technology was previously shown in the analysis of antibody binding affinities to drugs17 and in quantitative analysis of different biomolecular interactions37 by the microscale thermophoresis (MST) assay. This proposed method was compared with the previously reported ED-UPLC-MS/MS.18
The main concept in the present work involves fitting the antibody binding-site concentration to the ligand in the binding curve of FB as a function of log[antibody] or serum dilution from ED experiments. We proposed that the point where FB started to deviate from 1 estimates the antibody binding-site concentration to the ligand, i.e., [tracer] = [antibody binding-site] (the equivalence point). The [antibody binding-site] was extrapolated from the binding curve using two different methods (Figure S4): (i) best-fit by linear function at 0 < FB < 1 and (ii) second derivative plot of the 4-parameter logistic (4 PL) model of the binding curve. Both strategies of fitting the antibody binding-site concentrations from the binding curve provided comparable measured values. Extrapolation was done with caution due to each inherent limitation. In the case of the linear regression along the slope of the sigmoidal curve, we observed that reasonable values were obtained at R2 greater than 0.95. On the other hand, the second derivative method requires more data points (n > 5) at the top and bottom plateaus to have a defined minimum where the equivalence point is located. Unlike the previously reported ED-based approach coupled with UPLC-MS/MS,18 this method does not need the Kd values of antibodies to drugs to estimate the binding-site concentration; instead, it was directly fitted from the binding curve. The proof-of-concept of this approach was illustrated in ED experiments of 6-AmHap-mAb against 6-AmHap-acetamide, employing UPLC-MS/MS as a method of quantification.
Determination of Antibody Binding-Site Concentration to Hapten-Acetamide by ED-UPLC-MS/MS
A modified UPLC-MS/MS-based method was optimized to quantify the concentrations of 6-AmHap-acetamide tracer in the sample and buffer solutions from ED experiments. A traditional approach of an ED experiment that involves different mAb concentrations against a constant ligand concentration to establish a binding curve was employed.18 From the binding curve of FB as a function of log[6-AmHap-mAb] (Figure 2; red trace), the fitted antibody binding-site concentrations in mAb were 7.19 ± 0.02 nM (best-fit by linear function) and 7.91 ± 0.15 nM (second derivative method) (Table S1) with their corresponding errors of 44 and 58% (Figure 3A and Table S1) relative to the theoretical concentration (5 nM). To determine the effect of hapten-acetamide tracer concentration on the accuracy of measured antibody binding-site concentrations, we explored ED experiments against higher 6-AmHap-acetamide concentrations. In the case of the ED of 6-AmHap-mAb against 50 nM 6-AmHap-acetamide, measured binding-site concentrations were 53.46 ± 0.36 nM and 47.50 ± 0.33 nM from best-fit by linear function and second derivative method, respectively, relative to the theoretical concentration (50 nM).
Figure 2.
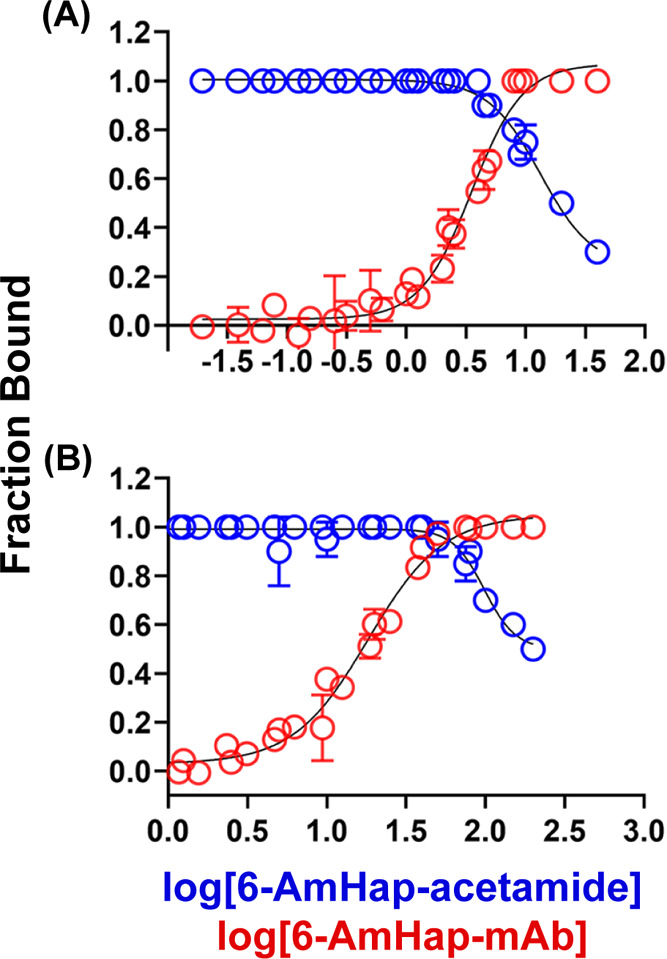
Binding curves of 6-AmHap-mAb against 6-AmHap-acetamide tracer by ED-UPLC-MS/MS. (a) Binding curves at low hapten-acetamide and mAb concentrations (5 nM). ED experiments of different mAb concentrations against 5 nM 6-AmHap-acetamide (red), and ED experiments of a series of 6-AmHap-acetamide concentrations against 5 nM mAb (blue). (b) Binding curves at high hapten-acetamide and mAb concentrations (50 nM). ED experiments of different mAb concentrations against 50 nM 6-AmHap-acetamide (red), and ED experiments of a series of 6-AmHap-acetamide concentrations against 50 nM mAb (blue).
Figure 3.
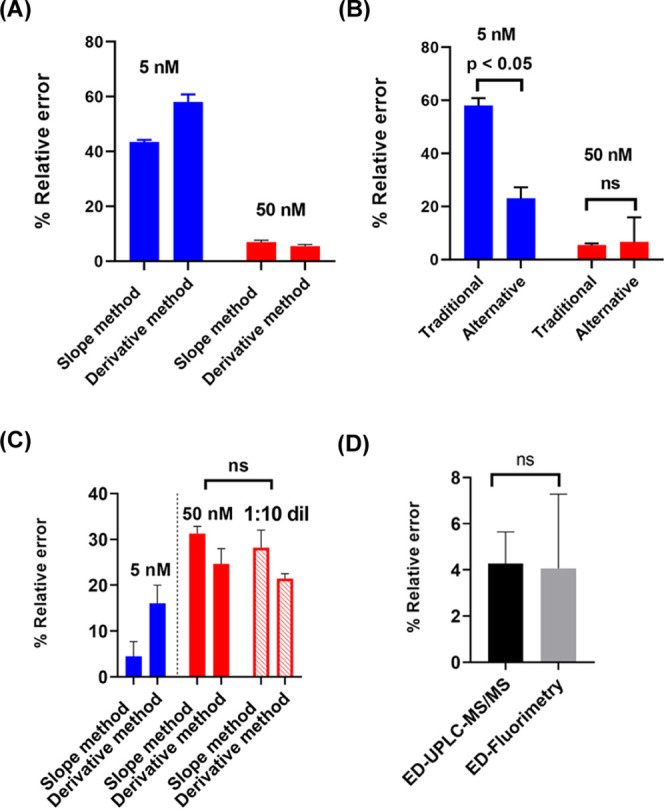
Statistical comparisons of % relative errors associated with determining the antibody binding-site concentration using ED-UPLC-MS/MS vs ED-fluorimetry. (A) Comparison of % relative errors in calculating the antibody binding-site concentration in 5 nM 6-AmHap-acetamide (blue, low hapten-acetamide tracer concentration) and 50 nM 6-AmHap-acetamide (red, high hapten-acetamide tracer concentration) using UPLC-MS/MS. (B) Comparison of % relative errors in the measured antibody binding-site concentrations using traditional vs alternative ED experiments at low and high hapten-acetamide tracers and mAb concentrations. (C) Comparison of % relative errors in 5 nM (blue, low hapten-fluorophore tracer concentration) vs 50 nM (red, high hapten-fluorophore tracer concentration) 6-AmHap-Cy5 by ED-fluorimetry (solid red: fluorescence data collected at low laser power; hollow red: fluorescence data collected at high laser power in a 1:10 dilution of ED samples/buffers). (D) Comparison of % relative errors associated with measuring the antibody binding-site concentration at 50 nM 6-AmHap-acetamide using UPLC-MS/MS (black) vs at 5 nM 6-AmHap-Cy5 using ED-fluorimetry (gray).
A large improvement in the measurement accuracy was attained at 50 nM 6-AmHap-acetamide with errors in the range of 5–7% (Table S1). Since the antibody binding-site concentrations were fitted in the binding curve and extrapolated at the point where the [6-AmHap-acetamide tracer] = [antibody binding-site], in this case the measurement reliability is dependent on the sensitivity of the method for quantifying 6-AmHap-acetamide. The developed UPLC-MS/MS-based method has a linear range of 1.25–160 nM with R2 > 0.99. It might be expected that with the ED at 5 nM 6-AmHap-acetamide, some of the buffer and sample solution points would have 6-AmHap-acetamide concentrations below the method detection limit, which impaired the behavior of the binding curve and thus influenced the measured antibody binding-site concentrations. The dependency on the developed quantification method for hapten-acetamide tracer shows that inflexibility is one of the drawbacks in utilizing the previously known ED-UPLC-MS/MS for the antibody binding-site quantitative analysis.
To further investigate the applicability of utilizing the binding curve from ED to directly measure the antibody binding-site concentrations in mAb and serum, we explored an alternative approach of an ED experiment. In this approach, ED was carried out at a constant mAb concentration against different ligand concentrations. This was demonstrated in the ED of 5 and 50 nM mAb against different 6-AmHap-acetamide tracer concentrations. In the case of ED at 5.0 nM 6-AmHap-mAb, antibody binding-site concentrations fitted to the binding curve (Figure 2A; blue traces) were 3.95 ± 0.07 nM (best-fit by linear function) and 3.85 ± 0.2 nM (second derivative method), with relative errors of 21 and 23%, respectively (Table S2). On the other hand, the ED experiment at 50 nM 6-AmHap-mAb resulted in the measured antibody binding-site concentrations of 53.46 ± 0.36 nM (best-fit by linear function) and 47.50 ± 0.33 nM (second derivative method) associated with their corresponding errors of 12 and 8% (Table S2). All measurements at different concentrations of mAb exhibit good repeatability with the % coefficient of variation (CV) in the range of 2–8%.
At high hapten-acetamide tracer and mAb concentrations (50 nM), there is no significant difference (p = not significant; paired T-test; Figure 3B) in the % relative errors associated with antibody binding-site concentrations measured by traditional (at constant hapten-acetamide concentration) vs alternative (at constant mAb concentration) ED approaches. However, it is apparent that at low hapten-acetamide and mAb concentrations (5 nM), there is a significant lowering (Figure 3B) of the % errors (Δ = 23 and 35%) observed in the alternative ED approach at 5 nM mAb, compared to that of traditional ED at 5 nM 6-AmHap-acetamide. Although the alternative ED experiment at a constant mAb concentration showed a better accuracy (lower % errors) in determining antibody binding-site concentrations compared to the traditional approach where constant [6-AmHap-acetamide] is dialyzed against different mAb concentrations, the alternative is less applicable in fluorimetry. ED experiments at constant mAb concentration against different hapten-fluorophore tracer concentrations will have a large concentration range of the fluorophore; this will pose problems during fluorescence measurements where high concentrations tend to cause detector saturation, while the lower end will be beyond the sensitivity of the detector. Thus, the traditional setting of the ED experiment, where a constant concentration of hapten-fluorophore is dialyzed against different mAb concentrations or serum dilution, was utilized to investigate the viability of the proposed ED-fluorimetry in determining the antibody binding-site concentration. Overall, the direct measurement of the antibody binding-site concentration from the binding curve has good accuracy and repeatability. This further demonstrated the reliability of directly extracting the concentration of the antibody binding site from the binding curve.
Determination of the Antibody Binding-Site Concentration to Hapten-Fluorophore by ED-Fluorimetry
ED-Fluorimetry of Monoclonal Antibodies: Accuracy, Precision, and Matrix Effect
The viability of using ED-fluorimetry in determining the antibody binding-site concentration was demonstrated using the in-house-generated 6-AmHap-mAb with hapten-fluorophore tracers, 6-AmHap-Cy5 and morHap-Cy5 (Scheme 1A,C). The binding affinities of 6-AmHap-mAb against 6-AmHap-Cy5 and morHap-Cy5 established by a conventional MST assay17 were found to be 0.65 ± 0.28 and 0.15 ± 0.03 pM, respectively. There is no significant difference (p = not significant; paired T-test) in the binding affinities of 6-AmHap-mAb to both hapten-fluorophore tracers. The observed cross-reactivities of 6-AmHap-mAb to both 6-AmHap-Cy5 and morHap-Cy5 are not unexpected as these hapten-fluorophore tracers share similar structural faces, which is consistent with the “facial recognition” hypothesis.38,39
In a similar manner as ED-UPLC-MS/MS, the antibody binding-site concentrations were fitted in the binding curve of FB vs log[mAb] or serum dilution from the ED experiment of mAb/serum against a known concentration of a hapten-fluorophore. The plot of FB as a function of log[mAb] from the ED of 6-AmHap-mAb against 5.0 nM 6-AmHap-Cy5 (Figure 4A) fitted the antibody binding-site concentrations in mAb to tracer at 4.8 ± 0.2 nM (best-fit by linear function) and 4.2 ± 0.2 nM (second derivative method) (Table S3). In comparison to the theoretical concentration (5 nM) of 6-AmHap-Cy5, these measured concentrations have % relative errors of 4 and 16%, respectively. Deterioration of the accuracy was observed at a higher concentration of 6-AmHap-fluorophore tracer. The measured antibody binding-site concentrations from the ED experiment of 6-AmHap-mAb against 50 nM 6-AmHap-Cy5 were 34.3 ± 0.8 and 37.7 ± 1.7 nM with errors of 25 and 34%, respectively, relative to the theoretical concentration (50 nM). All measurements at different tracer concentrations exhibit good repeatability with % CV in the range of 2–5% (Table S3). The observed increase in the measurement errors relative to the theoretical values at higher tracer concentrations can be ascribed, in part, to detector saturation due to higher concentrations of the fluorophore. In an attempt to circumvent this problem, we resorted to the dilution of samples prior to the measurement of fluorescence signals. However, at 50 nM 6-AmHap-Cy5, the dilution of each buffer and sample solutions by 1:10 prior to fluorescence measurement resulted only in a minor enhancement of the measurement accuracy (decrease in % error by 3%). In addition, it is apparent in Figure 4B that dilution has an effect on the behavior of the binding curve, particularly in the region where FB approaches zero.
Figure 4.
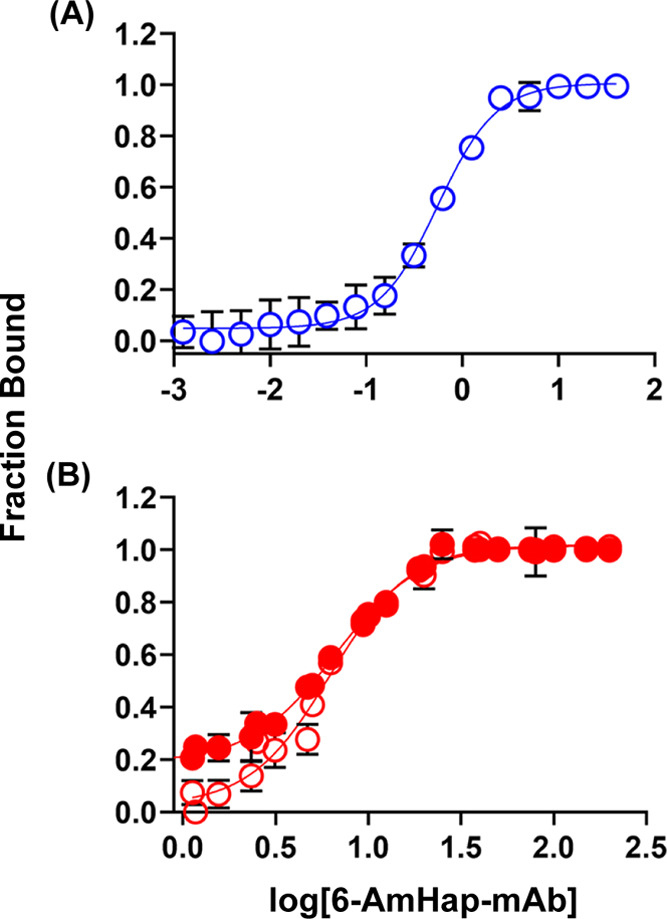
Binding curves of 6-AmHap-mAb against 6-AmHap-Cy5 by ED-fluorimetry. (A) Low hapten-fluorophore tracer concentration: ED of different mAb concentrations against 5 nM 6-AmHap-Cy5. (B) High hapten-fluorophore tracer concentration: ED of different mAb concentrations against 50 nM 6-AmHap-Cy5 (solid red: fluorescence measurement of ED sample/buffer solutions with 50 nM tracer at low laser power; hollow red: fluorescence measurements after a 1:10 dilution of 50 nM ED sample/buffer solutions at high laser power).
While % relative errors of the measured antibody binding-site concentrations by ED-UPLC-MS/MS decrease with an increasing concentration of 6-AmHap-acetamide (Figure 3A), the opposite was observed in ED-fluorimetry (Table S3 and Figure 3C). This is partly due to the different intrinsic limitations of each method of quantification. UPLC-MS/MS as an analytical tool for quantification is dependent on the overall sensitivity of the developed method, while fluorimetry has an inherent problem with detector saturation at a higher concentration of the fluorophore. Despite the differences and limitations of these two methods, they are strongly correlated based on the Pearson correlation analysis (r = 0.9654; p = 0.011) of the measured antibody binding-site concentrations in mAb (Figure 5A). The measurement errors (%) from the most accurate quantification condition in ED-UPLC-MS/MS (at 50 nM 6-AmHap-acetamide) are not significantly different (p = not significant; paired T-test) compared to that of ED-fluorimetry (at 5 nM 6-AmHap-Cy5) (Figure 3D).
Figure 5.
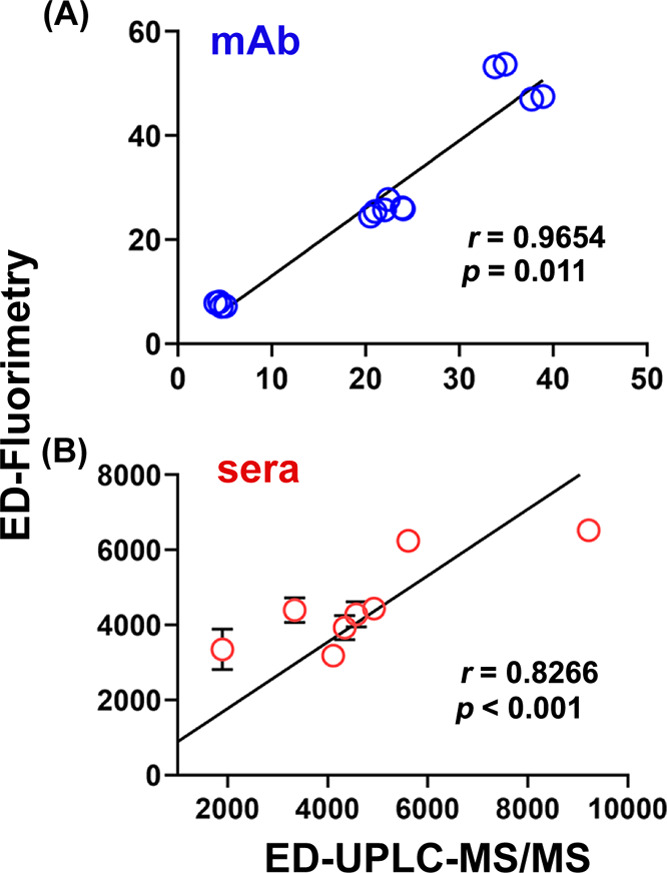
Correlation plots of antibody binding-site concentrations determined by ED-UPLC-MS/MS vs ED-fluorimetry, showing Pearson correlation coefficients (r) and p-values. (A) mAb samples with known concentrations and (B) sera samples.
Since the main goal of this method development was to measure the antibody binding-site concentrations in rat sera samples, we investigated the matrix effect in the presence of preimmune sera. As shown in Figure 6, the profile of the binding curve generated from the ED experiment of 6-AmHap-mAb with serum against 6-AmHap-Cy5 (red trace) is similar to that of the ED experiment in the absence of serum (blue trace). The fitted antibody binding-site concentration in mAb diluted with preimmune serum is 4.45 ± 0.85 nM with an error of 11% relative to the expected 5.0 nM. This measured binding-site concentration is not statistically different (p = not significant; paired T-test) from that determined from the ED experiment of mAb in the absence of serum (4.8 ± 0.2 nM). These observations confirmed that there was no appreciable matrix effect, and thus, ED-fluorimetry is a suitable method for determining the antibody binding-site concentration in rat sera samples.
Figure 6.
Matrix effect in the measured antibody binding-site concentration. Overlay of the binding curves from ED experiments; (blue) 6-AmHap-mAb against 6-AmHap-Cy5 and (red) 6-AmHap-mAb with preimmune serum (week 0) against 6-AmHap-Cy5.
ED-Fluorimetry of Sera Samples
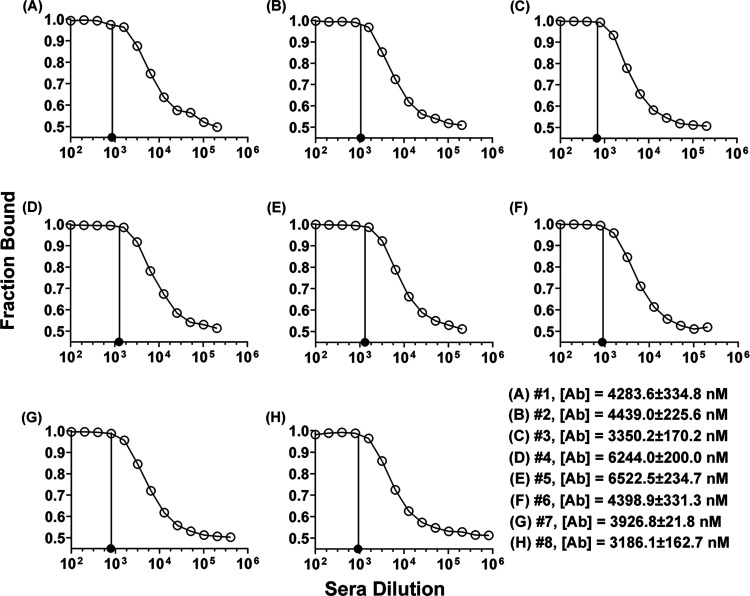
The use of ED-fluorimetry for measuring antibody binding-site concentrations was applied to sera samples from Sprague Dawley rats immunized with TT-6-AmHap.14 Since the accurate measurement of the antibody binding-site concentration in mAb was achieved at 5 nM hapten-fluorophore tracer, ED experiments in sera samples were performed with 5 nM 6-AmHap-Cy5. Under this experimental condition, the fitted antibody binding-site concentrations in sera samples by ED-fluorimetry range from 0.14 to 0.69 mg/mL (1899.6–9215.9 nM; Figure 7 and Table S5). These results were compared with the measured antibody binding-site concentrations in the range of 0.24–0.49 mg/mL (3186.1–6522.5 nM; Figure 7 and Table S5) by a more established ED-UPLC-MS/MS method based on the Kd values of antibodies against their native ligand 6-AM. The measured antibody binding-site concentrations in sera samples from different rats immunized with TT-6-AmHap were relatively sporadic with variances of 45 and 27% for ED-UPLC-MS/MS and ED-fluorimetry, respectively; the new technique was more precise than the ED-UPLC-MS/MS (Table S5). Further, we observed that under our analytical and experimental conditions, the measured antibody binding-site concentrations by both methods were not correlated with the corresponding measured binding affinities (Kd values in the nM range) of polyclonal antibodies in sera samples to 6-AmHap (Figure S5).
Figure 7.
Binding curves (A–H) from ED experiments of sera from TT-6-AmHap-immunized rats (I.D. #s 1–8) against 6-AmHap-Cy5 and their corresponding fitted antibody binding-site concentrations measured by ED-fluorimetry.
Consistent with the measured antibody binding-site concentrations in mAb at different tracer concentrations (vide supra; Figure 5A), the Pearson correlation analysis of measured binding-site concentrations in sera samples by ED-fluorimetry vs ED-UPLC-MS/MS revealed a correlation coefficient (r) of 0.8266 with p < 0.001 (vide supra; Figure 5B), suggesting a good statistical correlation between these two methods. Thus, ED-fluorimetry can be used as an alternative to ED-UPLC-MS/MS for measuring antibody binding-site concentrations in serum.
Summary
ED-fluorimetry is an improved method for measuring binding-site concentrations of polyclonal antibodies in serum. Unlike the current biophysical methods for determining the antibody binding-site concentration, ED-fluorimetry is simple and time-economical. It is immobilization-free, does not require radioactive labels, and does not involve tedious sample preparation and purification steps. In comparison with the previously reported ED-UPLC/MS/MS method,18 this approach does not require predetermined Kd values for estimating the binding-site concentration; thus, ED-fluorimetry involves a shorter dialysis and data collection time, and it is not dependent on the integrity of the standards and/or calibration curve.
Despite the ability of ED-fluorimetry to determine binding-site concentrations, this assay does have some limitations. The proposed approach based on the binding curve is not capable of establishing the specific antibody–ligand binding ratio. Further, the method requires the synthesis of fluorophores that can be attached to the haptens, which have tight binding affinities to the polyclonal antibodies.
Overall, ED-fluorimetry adapted in the experimental setting of the NanoTemper technology is a simple method that can be used to obtain the quantitative binding-site analysis of polyclonal antibodies in complex biological fluids. Our current efforts are aimed at improving and translating the method to enhance the throughput rate.
Materials and Methods
Commercially available reagents and solvents were used without further purification. Trifluoroacetic acid (TFA), iodoacetamide, triethylsilane (Et3SiH), dichloromethane (CH2Cl2), dimethylformamide (DMF), dimethylsulfoxide (DMSO), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and Tween 20 were purchased from Sigma-Aldrich (Saint Louis, MO). 3-[(Carbamoylmethyl)sulfanyl]propanoic acid was purchased from Enamine Ltd. (Monmouth Jct., NJ). The UPLC-MS/MS standard, 6-acetylmorphine-D3 (6-AM-D3), was purchased from Lipomed Inc. (Cambridge, MA), and sulfo-cyanine5 maleimide (≥95%; Cy5) was purchased from Lumiprobe Corporation (Hallandale Beach, FL). The newly synthesized compounds were characterized by 1H and 13C NMR spectroscopy (400 MHz Bruker spectrometer), HPLC, and high-resolution mass spectrometry (HRMS-ESI).
6-AmHap-mAb was produced from mice immunized with TT-6-AmHap.14 Spleens were removed and fused with P3X63/Ag8.653 cells. Monoclonal antibodies were produced using standard methods.40 The mAb were purified by protein G affinity chromatograph from hybridoma cell culture supernatants of cells grown in sera-free media. Sera samples used in this study were from Sprague Dawley rats immunized with 6-AmHap conjugated to tetanus toxoid with a polyethylene glycol linker and adjuvanted with army liposome formulation (ALF43) similar to those previously described.14 IgG concentrations of mAb were determined using nanodrop one (ThermoFisher) and bicinchoninic (BCA) assay. The molecular weight (MW) of mAb was established using the Axima MegaTOF (Shimadzu Scientific, MD).
All ED experiments were done in rapid equilibrium dialysis (RED) plates with 24 h incubation time, as described previously.18 Dulbecco’s phosphate buffer saline (DPBS, 10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, pH = 7.4) was purchased from Quality Biological Inc. (Gaithersburg, MD).
Quantification of 6-AmHap-acetamide tracer in the sample and buffer chambers of RED plates was done using a Thermo Scientific UPLC system coupled with a Q-Exactive quadrupole-orbitrap mass spectrometer. 6-AM-D3 from the equilibrium dialysis experiment of 6-AmHap-mAb against 6-AM/6-AM-D3 was quantified using the water’s LC-MS/MS instruments, as described.41 Optima LC/MS grade ammonium formate (NH4COOH), methanol (MeOH), and water (H2O) were purchased from Fisher Scientific (Suwanee, GA).
Fluorescence measurements of hapten-fluorophore tracers (e.g., 6-AmHap-Cy5, morHap-Cy5) were performed on standard treated glass capillary tubes using a Monolith NT.115 system (NanoTemper Technologies GmbH, Munich, Germany).
Hapten-Fluorophore Synthesis
Sulfo-Cyanine5 Maleimide Conjugate of N-(7-Acetamido-3-methyl-2,3,4,4a,5,6,7,7a-octahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinolin-9-yl)-3-mercaptopropanamide (2; 6-AmHap-Cy5)
To a solution of trityl-protected thiol 1 (21 mg, 32 μmol) and triethylsilane (7.7 μL, 48 μmol) in dichloromethane (640 μL) was added trifluoroacetic acid (24.5 μL, 320 μmol) at 0 °C under an atmosphere of argon. The reaction mixture was allowed to warm to room temperature and stirred continually for an additional 0.5 h. The solution was then concentrated under reduced pressure. The resulting residue was dissolved in a degassed 30% DMSO in 1 M HEPES buffer (pH 7.0–7.6; 500 μL) under an atmosphere of argon. This solution was protected from light by wrapping the reaction flask with aluminum foil. A solution of sulfo-cyanine5 maleimide (25 mg, 31 μmol) in a degassed 30% DMSO in 1 M HEPES buffer (500 μL) was then added to the above solution of the deprotected thiol 1. A 400 μL portion of 30% DMSO in 1 M HEPES buffer was used to wash the transfer vial containing Cy5 maleimide solution. The mixture was then stirred for 2.5 h at room temperature at which point no free thiol 1 was detectable using analytical HPLC, described below.
Analytical HPLC was carried out on an Agilent 1260 Infinity instrument equipped with a waters XBridge BEH C18 column (3.0 mm × 50 mm, 2.5 μm) and UV–vis detection at 220, 280, and 630 nm. The column flow rate and temperature were 0.8 mL/min and 30 °C, respectively. Mobile phase A: 0.1% TFA in H2O; mobile phase B: 0.1% TFA in acetonitrile; solvent gradient: isocratic 18% B in 9 min.
The hapten-fluorophore 2 was purified on an Agilent 1200 preparative HPLC equipped with an Agilent Prep-C18 column (21.2 mm × 100 mm, 5 mm) and UV–vis detection at 220 nm. The column flow rate was 20 mL/min. Mobile phase A was 0.1% TFA in H2O, and mobile phase B was 0.1% TFA in acetonitrile, with a gradient from 10 to 40% B in 14 min. The leading and trailing edges of the product peaks were discarded to collect a high purity material. Fractions containing product 2 were collected and subsequently lyophilized to yield a dark blue powder (8 mg, 22% isolated yield, >98% purity by HPLC). HRMS-ESI (m/z): [M + H]+ calcd for C60H74N7O12S3: 1180.4558; found: 1180.4564.
N-(7-Acetamido-3-methyl-2,3,4,4a,5,6,7,7a-octahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinolin-9-yl)-3-((2-amino-2-oxoethyl)thio)propanamide (5; 6-AmHap-acetamide)
To a solution of 3-(2-amino-2-oxoethyl)sulfanylpropanoic acid (3; 200.7 mg, 1.23 mmol) in DMF (3 mL) was added N-hydroxysuccinimide (226.5 mg, 1.97 mmol), followed by the addition of N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride (282.9 mg, 1.47 mmol) under an atmosphere of N2 and the mixture was stirred overnight. The reaction was diluted with H2O (5 mL) and extracted with EtOAc (3 mL × 5 mL). The organic layers were combined, washed with brine, and dried over anhydrous MgSO4. The solution was filtered, and the solvent was evaporated. The resulting residue was further dried under high vacuum overnight. To a solution of the above NHS ester 3 in DMF (2 mL) was added amine 4 (18 mg, 55 μmol) and the mixture was heated at 50 °C for 3 h under a N2 atmosphere. The reaction mixture was allowed to cool to room temperature. The mixture was then diluted with H2O (5 mL), and the organic product was extracted with CHCl3 (3 mL × 5 mL). The combined organic layers were washed with brine and dried over anhydrous MgSO4. The solution was then filtered and concentrated under reduced pressure. The residue was purified two times by column chromatography with 5% MeOH/28% NH4OH in CHCl3 yielding 6 mg of 5 (23% isolated yield).1H NMR (400 MHz, methanol-d4, δ = ppm) δ 7.35 (d, J = 8.1 Hz, 1H), 6.69 (d, J = 8.2 Hz, 1H), 4.67 (d, J = 4.6 Hz, 1H), 4.13 (dt, J = 12.1, 4.4 Hz, 1H), 3.25 (s, 2H), 3.18 (dd, J = 6.3, 2.9 Hz, 1H), 3.05 (d, J = 19.2 Hz, 1H), 2.96 (t, J = 7.1 Hz, 2H), 2.72 (t, J = 7.1 Hz, 2H), 2.60–2.49 (m, 2H), 2.41 (s, 3H), 2.39–2.24 (m, 2H), 1.99 (m, 1H), 1.95 (s, 3H), 1.77–1.64 (m, 2H), 1.40 (td, J = 10.4, 3.3 Hz, 1H), 1.11–0.83 (m, 3H).13C NMR (100 MHz, methanol-d4, δ = ppm) δ 173.92, 171.14, 170.70, 150.63, 132.40, 129.19, 123.44, 118.60, 117.64, 89.69, 59.63, 46.70, 45.71, 42.20, 41.61, 36.19, 35.75, 35.43, 34.50, 27.95, 21.66, 21.36, 20.06, 19.97. HRMS-ESI (m/z): [M + H]+ calcd for C24H33N4O4S: 473.2223; found: 473.2229.
MST Measurements and Calculation of Dissociation Constants (Kd)
The binding affinities of 6-AmHap-mAb to 6-AmHap-Cy5 and morHap-Cy5 were determined at a 0.5 nM hapten-fluorophore tracer concentration using the conventional MST assay. The working solutions of mAbs and tracers were prepared in 1× DPBS with 0.05% BSA and 0.05% Tween 20.
The starting/highest concentration of 6-AmHap-mAb was 50 nM, which was serially diluted (1:1) in 200 μL PCR vials with a 1× DPBS/Tween/BSA solution to yield 16 different mAb concentrations. Each concentration/dilution was mixed with hapten-fluorophore tracer (e.g., 6-AmHap-Cy5, morHap-Cy5) in a 1:1 ratio and incubated in the dark for 20 min. The 6-AmHap-mAb:hapten-fluorophore mixtures were then loaded in capillary tubes for MST measurements. Kd values were determined by plotting the normalized fluorescence against [mAb] in MO. Affinity Analysis software,42 which provided a curve fitting that estimates the Kd values.
Experimental Methods for Determining the Antibody Binding-Site Concentration in Serum
The antibody binding-site concentration in serum was fitted from the ED of mAb or sera against the known concentration of a tracer. In the plot of fraction bound (FB) as a function of log[antibody] or serum dilution, the point at which the FB value started to deviate from 1.0 can be used to estimate the antibody binding-site concentration in serum, that is, [tracer] = [antibody binding-site]. The fraction bound (FB) was calculated using eq 1
| 1 |
where [sample] and [buffer] are the concentrations of the tracer in the sample and buffer chambers, respectively. The proof-of-concept was established from the equilibrium dialysis of mAb against the known concentration of ligand, using UPLC-MS/MS as a method of quantification.
ED-UPLC-MS/MS
Traditional Approach: Equilibrium Dialysis of mAb against Constant Tracer Concentrations
ED experiments of 6-AmHap-mAb against 5 and 50 nM 6-AmHap-acetamide tracers in equilibrium dialysis buffer (EDB) were performed according to previous reports.18,43
EDB was prepared by adding 125 μL of the BSA standard ampule to 500 mL of 1× DPBS. The solution was shaken thoroughly and stored at 4 °C when not in use. In the case of equilibrium dialysis of 6-AmHap-mAb against 5 nM 6-AmHap-acetamide, the highest/starting concentration of mAb was 40 nM, which was serially diluted with 5 nM 6-AmHap-acetamide in EDB in a 1:1 ratio to generate 24 different mAb concentrations. The equilibrium dialysis was performed in the RED plate, which has 48 pairs of buffer/sample chambers separated by a 12 kDa MWCO dialysis membrane. Each buffer chamber (left) was loaded with 300 μL EDB, while their corresponding sample chambers (right) contain 100 μL of each mAb concentration or dilution. There are two trials in every equilibrium dialysis set up. The RED plate was covered with an adhesive film and incubated at 4 °C and 300 rpm for 24 h in a thermomixer. After 24 h, 90 μL of solution from buffer and sample chambers was drawn to a 1 mL recovery vial for the UPLC-MS/MS quantification of 6-AmHap-acetamide tracer. The instrument settings and parameters are described in the next section.
The plot of FB 6-AmHap-acetamide vs log[mAb] was generated. The [antibody binding-site] was extrapolated from the binding curve using (i) linear regression at 0 < FB < 1 and (ii) second derivative of the 4 PL model (describe below). The calculated [antibody binding-site] was compared to a known [6-AmHap-acetamide]. The same procedure was followed in the case of the ED at 50 nM 6-AmHap-acetamide.
Alternative Approach: Equilibrium Dialysis of Constant mAb Concentration against Different Tracer Concentrations
The ED experiments of 5 and 50 nM 6-AmHap-mAb against 6-AmHap-acetamide tracer in EDB were accomplished according to the previous report with slight modifications.18 In the case of ED of 5 nM 6-AmHap-mAb against 6-AmHap-acetamide, the highest/starting concentration of 6-AmHap-acetamide was 40 nM, which was serially diluted with 5 nM 6-AmHap-mAb in EDB in a 1:1 ratio to generate 24 different 6-AmHap-acetamide concentrations. As described above, equilibrium dialysis was performed in the RED plate. Each buffer chamber (left) was seeded with 300 μL EDB while their corresponding sample chambers (right) were loaded with 100 μL of each 6-AmHap-acetamide concentration. The RED plate was covered with an adhesive film and incubated at 4 °C and 300 rpm for 24 h in a thermomixer. After 24 h, 90 μL of solution from the buffer and sample chambers was drawn to a 1 mL recovery vial for the UPLC-MS/MS quantification of 6-AmHap-acetamide. The instrument settings and parameters are described in the next section.
The plot of FB 6-AmHap-acetamide vs log[6-AmHap-acetamide] was generated. The [antibody binding-site] was extrapolated from the binding curve using (i) linear regression at 0 < FB < 1 and (ii) the second derivative of the 4 PL model (described below). The same procedure was followed for the equilibrium dialysis at 50 nM 6-AmHap-mAb.
UPLC-MS/MS Quantification
Quantification of 6-AmHap-acetamide was performed in a Thermo Scientific Vanquish UPLC coupled with a Q-Exactive Quadrupole-Orbitrap detector. The water’s HSS T3 column (2.1 mm × 100 mm, 1.8 μm particle size; Waters, Milford, MA) and the following mobile phases were used: A (water with 10 mM NH4COOH and 0.1% HCOOH) and B (MeOH with 0.1% HCOOH). The UPLC gradient used is described in Table S6. The column was maintained at 45 °C at a flow rate of 350 μL/min. The injection volume was 10 μL. All data were acquired using positive electrospray ionization (ESI) in a parallel reaction monitoring (PRM) mode. The electrospray and source settings were as follows: 3.5 kV (capillary voltage), 320 °C (capillary temperature), 25 AU (sheath gas flow rate), 10 AU (Aux gas flow rate), and 300 °C (Aux gas temperature). The analyte (6-AmHap-acetamide) was detected as [M + H]+ with the PRM transition of 473.2215 > 129.0004 at 5.73 min (chromatographic retention time). Quantification was performed using the external calibration method with a 1/X2 weighting scheme in TraceFinder 5.1 (Thermo Scientific, Waltham, MA).
ED-Fluorimetry
Equilibrium Dialysis of mAb against 6-AmHap-Cy5
The binding curve of FB vs log[antibody] or serum dilution from the equilibrium dialysis of mAb or serum against the known concentration of hapten-fluorophore tracer was constructed to fit the antibody binding-site concentration in mAb or serum. The applicability of this concept in ED-fluorimetry was established using ED experiments of 6-AmHap-mAb against 5 and 50 nM 6-AmHap-Cy5 in EDB.
EDB for ED-fluorimetry was prepared by adding 125 μL of the BSA standard ampule and 250 μL of Tween 20 to 500 mL of 1× DPBS. The solution was shaken carefully and stored at 4 °C when not in use. In the case of 6-AmHap-mAb against 5 nM 6-AmHap-Cy5, the highest/starting concentration of mAb was 40 nM, which was serially diluted with 5 nM 6-AmHap-Cy5 in EDB in a 1:1 ratio to generate 24 different mAb concentrations. The ED was done in a similar manner as described for ED-UPLC-MS/MS using the RED plate. Briefly, each buffer chamber (left) was loaded with 300 μL EDB while their corresponding sample chambers (right) contain 100 μL of each mAb concentration or dilution. The RED plate was covered with an adhesive film and incubated at 4 °C and 300 rpm for 24 h in a thermomixer. After 24 h, the 6-AmHap-mAb:6-AmHap-Cy5 mixtures were then loaded in capillary tubes for fluorescence measurements. The instrument settings and parameters are described in the next section.
The fraction bound tracer (FB) was calculated from fluorescence data using the following equation
| 2 |
where Fsample and Fbuffer are the fluorescence signals from the sample and buffer chambers, respectively.
The binding curve of FB 6-AmHap-Cy5 vs log[antibody] was generated. The binding-site concentration in mAb was estimated from this plot using (i) linear regression at 0 < FB < 1 and (ii) the second derivative of the 4 PL model (described below). The same procedure was followed for the equilibrium dialysis at 50 nM 6-AmHap-Cy5.
Effect of mAb Dilution with Preimmune Rat Sera
The possible matrix effect was investigated by diluting 6-AmHap-mAb with preimmune sera. Equilibrium dialysis of 6-AmHap-mAb/serum mixture against 6-AmHap-Cy5 was performed in a similar manner as described for mAb with slight modification. Briefly, 4 μM 6-AmHap-mAb in week 0 rat sera was prepared and served as a stock solution. The highest concentration, 40 nM 6-AmHap-mAb, was prepared and was serially diluted with 5 nM 6-AmHap-Cy5 in EDB in a 1:1 ratio to generate 16 different mAb concentrations. ED was performed in the RED plate, where each buffer chamber (left) was loaded with 300 μL EDB while their corresponding sample chambers (right) contain 100 μL of each mAb concentration or dilution. The RED plate was covered with an adhesive film and incubated at 4 °C and 300 rpm for 24 h in a thermomixer. After 24 h, 6-AmHap-mAb:6-AmHap-Cy5 samples were then loaded in capillary tubes for fluorescence measurements.
Equilibrium Dialysis of Rat Sera against 6-AmHap-Cy5
Equilibrium dialysis of sera against 6-AmHap-Cy5 tracer was done in a similar manner as described above for mAb. Briefly, a 5 nM 6-AmHap-Cy5 working solution was prepared in EDB. This was used to prepare different dilutions of a serum sample. The serum from TT-6-AmHap-immunized rats was diluted with 5 nM 6-AmHap-Cy5 in a 1:100 ratio. This diluted serum was then further serially diluted to generate 16 different serum dilutions. In the RED plate, each buffer chamber (left) was loaded with 300 μL of EDB while their corresponding sample chambers (right) contain 100 μL of each serum dilution. The RED plate was covered with an adhesive film and incubated at 4 °C and 300 rpm for 24 h in a thermomixer. After 24 h, the 6-AmHap-mAb:6-AmHap-Cy5 samples were then loaded in capillary tubes for fluorescence measurements. The antibody binding-site concentration was determined from the plot of FB as a function of serum dilutions, in a similar manner as described in the case of mAb.
Fluorescence Measurement
The fluorescence measurements were performed on a Monolith NT.115 instrument from NanoTemper Technology, GmbH. This instrument was equipped with an IR laser (wavelength, 1475 ± 15 nm; power 120 mW maximum) and a red fluorescence channel suitable for detecting red dyes such as Cy5 and Alexa Fluor 647.
Since we were dealing with different concentrations of tracers used in the dialysis experiment, variable % LED and % MST powers were utilized to avoid the saturation of the detector. In the case of equilibrium dialysis at 5 nM 6-AmHap-Cy5, fluorescence measurements were done at 20% LED/25% MST powers (high laser power). On the other hand, ED at 50 nM 6-AmHap-Cy5 utilized 2% LED/25% MST powers (low laser power).
Data Analysis
Statistical analyses and graphing of binding curves were performed in a GraphPad Prism 9.0. Correlation between ED-UPLC-MS/MS and ED-fluorimetry was established using the Pearson correlation analysis. Comparisons of % relative errors among methods and between concentrations were done using T-test. Differences among values are statistically significant if p ≤ 0.05.
Acknowledgments
This research work was supported by the National Institutes of Health (NIH; UG3DA048351 and 1DP1DA034787 to GRM). The work of GRM, OBT, EGA, RCB, CW, and TO was supported through a Cooperative Agreement Award (no. W81XWH-07-2-067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Army Medical Research and Medical Command (MRMC). The work of AS, JFA, AEJ, and KCR in the Drug Design and Synthesis Section, MTMDB, was supported by the Intramural Research Programs of the National Institute on Drug Abuse and the National Institute of Alcohol Abuse and Alcoholism. The authors would like to thank Dr. William Leister (NCATS, NIH) for his help with the preparative HPLC and Dr. John Lloyd (Mass Spectrometry Facility, NIDDK) for the mass spectral data. The authors would also like to thank Dr. Zoltan Beck, Dr. Christopher Karch, Alexander Anderson, David McCurdy, and Nadine Nehme for outstanding technical assistance.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c03237.
Details of the quantitation of antibody binding affinity (Kd) and binding sites in sera by ED-UPLC-MS/MS; 6-AmHap-mAb production, purification, and characterization; and complete raw data for the measured antibody binding-site by ED-fluorimetry and ED-UPLC-MS/MS (PDF)
Author Present Address
∥ BioFire Diagnostics, 515 Colorow Dr., Salt Lake City, Utah 84108, United States
Author Present Address
⊥ Merck Sharp & Dohme LLC., 126 E. Lincoln Ave., Rahway, New Jersey 07065, United States
Author Present Address
# Department of Obstetrics and Gynecology, Northwestern University, Evanston, Illinois 60208, United States
Author Present Address
∇ Braselmann Lab, Department of Chemistry, Georgetown University, Regents Hall 523 57th & O St. NW, Washington, DC 20057, United States
Author Present Address
○ Merck Sharp & Dohme LLC., 126 E. Lincoln Ave., Rahway, New Jersey 07065, United States
Author Contributions
The manuscript was written through contributions of all authors. Analytical and biological experiments were performed at the Laboratory of Adjuvant and Antigen Research, the Walter Reed Army Institute of Research, and chemical syntheses were accomplished in the Drug Design and Synthesis Section, MTMDB, at the National Institute on Drug Abuse, National Institutes of Health. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): G.R.M., K.C.R., A.E.J., and C.R.A. are co-inventors in a related U.S. patent owned by the U.S. Army and NIDA.
Notes
This material has been reviewed by the Walter Reed Army Institute of Research and the National Institute on Drug Abuse. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting true views of the Department of the Army, the Department of Defense, NIDA, NIH, or the U.S. government.
Supplementary Material
References
- Baehr C.; Pravetoni M. Vaccines to Treat Opioid Use Disorders and to Reduce Opioid Overdoses. Neuropsychopharmacology 2019, 44, 217–218. 10.1038/s41386-018-0197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick P. The Opioid Epidemic: Crisis and Solutions. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 143–159. 10.1146/annurev-pharmtox-010617-052534. [DOI] [PubMed] [Google Scholar]
- Bremer P. T.; Janda K. D. Conjugate Vaccine Immunotherapy for Substance Use Disorder. Pharmacol. Rev. 2017, 69, 298–315. 10.1124/pr.117.013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe G. N.; Vendruscolo L. F.; Edwards S.; Schlosburg J. E.; Misra K. K.; Schulteis G.; Mayorov A. V.; Zakhari J. S.; Koob G. F.; Janda K. D. A Vaccine Strategy to Induce Protective Immunity Against Heroin. J. Med. Chem. 2011, 54, 5195–5204. 10.1021/jm200461m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer L. Inner Workings: Using Vaccines to Harness the Immune System and Fight Drugs of Abuse. Proc. Natl. Acad. Sci. U.S.A. 2021, 118, 1–4. 10.1073/pnas.2121094118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. C.; Janda K. D. Development of Effective Therapeutics for Polysubstance Use Disorders. Curr. Opin. Chem. Biol. 2022, 66, 102105 10.1016/j.cbpa.2021.102105. [DOI] [PubMed] [Google Scholar]
- Kosten T. R.; Domingo C. B. Can You Vaccinate Against Substance of Abuse. Expert Opin. Biol. Ther. 2013, 13, 1093–1097. 10.1517/14712598.2013.791278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda K. D.; Treweek J. B. Vaccines Targeting Drugs of Abuse: Is the Glass Half-empty or Half-full?. Nat. Rev. Immunol. 2012, 12, 67–72. 10.1038/nri3130. [DOI] [PubMed] [Google Scholar]
- Landsteiner K.; Jacobs J. Studies on the Sensitization of Animals with Simple Compounds, II. J. Exp. Med. 1936, 64, 625–639. 10.1084/jem.64.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsteiner K.; Jacobs J. Studies of the Sensitization of Animals with Simple Chemical Compounds. J. Exp. Med. 1935, 61, 643–656. 10.1084/jem.61.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pravetoni M.; Comer S. D. Development of Vaccines to Treat Opioid Use Disorders and Reduce Incidence of Overdose. Neuropharmacology 2019, 158, 107662 10.1016/j.neuropharm.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone A. E.; Scheuemann S. E.; Haile C. N.; Cuny G. D.; Velasquez M. L.; Linhuber J. P.; Duddupudi A. L.; Vigliaturo J. R.; Pravetoni M.; Kosten T. A.; Kosten T. R.; Norton E. B. Fentanyl Conjugate Vaccine by Injected or Mucosal Delivery with dmLT or LTA1 Adjuvants Implicates IgA in Protection from Drug Challenge. npj Vaccines 2021, 6, 69 10.1038/s41541-021-00329-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C.; Gradinati V.; Hamid F.; Baehr C.; Crouse B.; Averick S.; Kovaliov M.; Harris D.; Runyon S.; Baruffaldi F.; LeSage M.; Comer S.; Pravetoni M. Therapeutic and Prophylactic Vaccines to Counteract Fentanyl Use Disorders and Toxicity. J. Med. Chem. 2020, 63, 14647–14667. 10.1021/acs.jmedchem.0c01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulima A.; Jalah R.; Antoline J. F.; Torres O. B.; Imler G.; Deschamps J. R.; Beck Z.; Alving C. R.; Jacobson A. E.; Rice K. C.; Matyas G. R. A Stable Heroin Analogue That Can Serve as a Vaccine Hapten to Induce Antibodies That Block the Effects of Heroin and Its Metabolites in Rodents and That Cross-React Immunologically with Related Drugs of Abuse. J. Med. Chem. 2018, 61, 329–343. 10.1021/acs.jmedchem.7b01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil Stowe G.; Schlosburg J. E.; Vendruscolo L. F.; Edwards S.; Misra K. K.; Schulteis G.; Zakhari J. S.; Koob G. F.; Janda K. Developing a Vaccine Against Multiple Psychoactive Targets: A Case Study of Heroin. CNS Neurol Disord: Drug Targets 2011, 10, 865–875. 10.2174/187152711799219316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogen I. L.; Boix F.; Nerem E.; Morland J.; Andersen J. M. A Monoclonal Antibody Specific for 6-monoacetylmorphine Reduces Acute Heroin Effects in Mice. J. Pharmacol. Exp. Ther. 2014, 349, 568–576. 10.1124/jpet.113.212035. [DOI] [PubMed] [Google Scholar]
- Torres O. B.; Duval A. J.; Sulima A.; Antoline J. F.; Jacobson A. E.; Rice K. C.; Alving C. R.; Matyas G. R. A Rapid Solution-based Method for Determining the Affinity of Heroin Hapten-induced Antibodies to Heroin, Its Metabolites, and Other Opioids. Anal. Bioanal. Chem. 2018, 410, 3885–3903. 10.1007/s00216-018-1060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres O. B.; Antoline J. F.; Li F.; Jalah R.; Jacobson A. E.; Rice K. C.; Matyas G. R. A Simple Nonradiative Method for the Determination of the Binding Affinities of Antibodies Induced by Hapten Bioconjugates for Drugs of Abuse. Anal. Bioanal. Chem. 2016, 408, 1191–1204. 10.1007/s00216-015-9223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orson F. M.; Kinsey B. M.; Singh R. A.; Wu Y.; Gardner T.; Kosten T. R. The Future of Vaccines in the Management of Addictive Disorders. Curr. Psychiatry Rep. 2007, 9, 381–387. 10.1007/s11920-007-0049-z. [DOI] [PubMed] [Google Scholar]
- Orson F. M.; Kinsey B. M.; Singh R. A.; Wu Y.; Gardner T.; Kosten T. R. Substance Abuse Vaccines. Ann. N. Y. Acad. Sci. 2008, 1141, 257–269. 10.1196/annals.1441.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresnais M.; Longuespee R.; Sauter M.; Schaller T.; Arndt M.; Krauss J.; Blank A.; Haefeli W. E.; Burhenne J. Development and Validation of an LC-MS-based Quantification Assay for New Therapeutic Antibodies: Application to a Novel Therapy Against Herpes Simplex Virus. ACS Omega 2020, 5, 24329–24339. 10.1021/acsomega.0c02547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damen C. W.; de Groot E.; Heij M.; Boss D.; Schellens J.; Rosing H.; Beijnen J.; Aarden L. Development and Validation of an Enzyme-linked Immunosorbent Assay for the Quantification of Trastuzumab in Human Serum and Plasma. Anal. Biochem. 2009, 391, 114–120. 10.1016/j.ab.2009.05.030. [DOI] [PubMed] [Google Scholar]
- Mulleman D.; Meric J-C.; Paintaud G.; Ducourau E.; Beuzelin C.; Valat J. P.; Goupille P. Infliximab Concentration Monitoring Improves the Control of Disease Activity in Rheumatoid Arthritis. Arthritis Res. Ther. 2009, 11, 1–6. 10.1186/ar2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri D.; Montigiani S.; Kirkham P. M. Biophysical Methods for the Determination of Antibody-antigen Affinities. Trends Biotechnol. 1996, 14, 465–470. 10.1016/S0167-7799(96)10067-6. [DOI] [PubMed] [Google Scholar]
- Butler J. E. Solid Supports in Enzyme-Linked Immunosorbent Assay and Other Solid-Phase Immunoassays. Methods 2000, 22, 4–23. 10.1006/meth.2000.1031. [DOI] [PubMed] [Google Scholar]
- Friguet B.; Chaffotte A. F.; Djavadi-Ohaniance L.; Goldberg M. E. Measurements of the True Affinity Constant in Solution of Antigen–antibody Complexes by Enzyme-linked Immunosorbent Assay. J. Immunol. Methods 1985, 77, 305–319. 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- Butler J. E.; Weber P.; Sinkora M.; Sun J.; Ford S. J.; Christenson R. K. Antibody Repertoire Development in Fetal and Neonatal Piglets. II. Characterization of Heavy Chain Complementarity-Determining Region 3 Diversity in the Developing Fetus. J. Immunol. 2000, 165, 6999–7010. 10.4049/jimmunol.165.12.6999. [DOI] [PubMed] [Google Scholar]
- Barrette R. W.; Urbonas J.; Silbart L. Quantifying Specific Antibody Concentrations by Enzyme-linked Immunosorbent Assay Using Slope Correction. Clin. Vaccine Immunol. 2006, 13, 802–805. 10.1128/CVI.00422-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson R.; Fagerstam L.; Nilshans H.; Persson B. Analysis of Active Antibody Concentration. Separation of Affinity and Concentration Parameters. J. Immunol. Methods 1993, 166, 75–84. 10.1016/0022-1759(93)90330-A. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y.; Uno S.; Nanami M.; Yoshimura Y.; Ida S.; Fukushima N.; Tsuchiya M. Determination of Concentration and Binding Affinity of Antibody Fragment by Use of Surface Plasmon Resonance. J. Biosci. Bioeng. 2005, 100, 311–317. 10.1263/jbb.100.311. [DOI] [PubMed] [Google Scholar]
- Lisi P. J.; Teipel J. W.; Goldstein G.; Schiffman M. Improved Radioimmunoassay Technique for Measuring Serum Thymopoietin. Clin. Chim. Acta 1980, 107, 111–119. 10.1016/0009-8981(80)90420-9. [DOI] [PubMed] [Google Scholar]
- Kolb A. J.; Kaplita P. V.; Hayes D. J.; Park Y-W.; Pernell C.; Major J. S.; Mathis G. Tyrosine Kinase Assays Adapted to Homogeneous Time-resolved Fluorescence. Drug Discovery Today 1998, 3, 333–342. 10.1016/S1359-6446(98)01204-5. [DOI] [Google Scholar]
- Turek T. C.; Small E. C.; Bryant R. W.; Hill W. A. Development and Validation of a Competitive AKT Serine/Threonine Kinase Fluorescence Polarization Assay Using a Product-specific Anti-phosphoserine Antibody. Anal. Biochem. 2001, 299, 45–53. 10.1006/abio.2001.5412. [DOI] [PubMed] [Google Scholar]
- Braunwalder A. F.; Yarwood D. R.; Sills M. A.; Lipson K. E. Measurement of the Protein Tyrosine Kinase Activity of C-SRC Using Time-Resolved Fluorometry of Europeum Chelates. Anal. Biochem. 1996, 238, 159–164. 10.1006/abio.1996.0269. [DOI] [PubMed] [Google Scholar]
- Chiu H.-H.; Liao H.; Shao Y.; Lu Y.; Lin C.; Tsai I.; Kuo C. Development of a General Method for Quantifying IgG-based Therapeutic Monoclonal Antibodies in Human Plasma Using Protein G Purification Coupled with a Two Internal Standard Calibration Strategy Using LC-MS/MS. Anal. Chim. Acta 2018, 1019, 93–102. 10.1016/j.aca.2018.02.040. [DOI] [PubMed] [Google Scholar]
- Li F.; Cheng K.; Antoline J. F.; Iyer M. R.; Matyas G. R.; Torres O. B.; Jalah R.; Beck Z.; Alving C. R.; Parrish D. A.; Deschamps J. R.; Jacobson A. E.; Rice K. C. Synthesis and Immunological Effects of Heroin Vaccines. Org. Biomol. Chem. 2014, 12, 7211–7232. 10.1039/C4OB01053A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerabek-Willemsen M.; Andre T.; Wanner R.; Roth H. M.; Duhr S.; Baaske P.; Breitsprecher D. MicroScale Thermophoresis: Interaction Analysis and Beyond. J. Mol. Struct. 2014, 1077, 101–103. 10.1016/j.molstruc.2014.03.009. [DOI] [Google Scholar]
- Matyas G. R.; Rice K. C.; Cheng K.; Li F.; Antoline J. F.; Iyer M. R.; Jacobson A. E.; Mayorov A. V.; Beck Z.; Torres O. B.; Alving C. R. Facial Recognition of Heroin Vaccine Opiates: Type 1 Cross-reactivities of Antibodies Induced by Hydrolytically Stable Haptenic Surrogates of Heroin, 6-acetylmorphine, and Morphine. Vaccine 2014, 32, 1473–1479. 10.1016/j.vaccine.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer N.; Steinicke F.; Lindigkeit R.; Ernst L.; Beuerle T. Determination of Cross-reactivity of Poly- and Monoclonal Antibodies for Synthetic Cannabinoids by Direct SPR and ELISA. Forensic Sci. Int. 2017, 280, 25–34. 10.1016/j.forsciint.2017.09.011. [DOI] [PubMed] [Google Scholar]
- Matyas G. R.; Wieczorek L.; Bansal D.; Chenine A-L.; Sanders-Buell E.; Tovanabutra S.; Kim J. H.; Polonis V.; Alving C. R. Inhibition of HIV-1 Infection of Peripheral Blood Mononuclear Cells by a Monoclonal Antibody that Binds to Phosphoinositides and Induces Secretion of b-chemokines. Biochem. Biophys. Res. Commun. 2010, 402, 808–812. 10.1016/j.bbrc.2010.10.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos R. C.; Bow E. W.; Whalen C.; Torres O. B.; Sulima A.; Beck Z.; Jacobson A. E.; Rice K. C.; Matyas G. R. Novel Vaccine That Blunts Fentanyl Effects and Sequesters Ultrapotent Fentanyl Analogues. Mol. Pharm. 2020, 17, 3447–3460. 10.1021/acs.molpharmaceut.0c00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NanoTemper Technologies GmbH: Floessergasse 4, 81369 Munich, Germany. [Google Scholar]
- Wu G.Assay Development: Fundamentals and Practices; Wiley: Hoboken, 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.