Abstract
The genus Glycyrrhiza, comprising approximately 36 spp., possesses complex structural diversity and is documented to possess a wide spectrum of biological activities. Understanding and finding the mechanisms of efficacy or safety for a plant-based therapy is very challenging, yet it is crucial and necessary to understand the polypharmacology of traditional medicines. Licorice extract was shown to modulate the xenobiotic receptors, which might manifest as a potential route for natural product-induced drug interactions. However, different mechanisms could be involved in this phenomenon. Since the induced herb–drug interaction of licorice supplements via Pregnane X receptor (PXR) is understudied, we ventured out to analyze the potential modulators of PXR in complex mixtures such as whole extracts by applying computational mining tools. A total of 518 structures from five species of Glycyrrhiza: 183 (G. glabra), 180 (G. uralensis), 100 (G. inflata), 33 (G. echinata), and 22 (G. lepidota) were collected and post-processed to yield 387 unique compounds. Visual inspection of top candidates with favorable ligand–PXR interactions and the highest docking scores were identified. The in vitro testing revealed that glabridin (GG-14) is the most potent PXR activator among the tested compounds, followed by licoisoflavone A, licoisoflavanone, and glycycoumarin. A 200 ns molecular dynamics study with glabridin confirmed the stability of the glabridin-PXR complex, highlighting the importance of computational methods for rapid dereplication of potential xenobiotic modulators in a complex mixture instead of undertaking time-consuming classical biological testing of all compounds in a given botanical.
1. Introduction
Complementary and alternative medicine is an integral part of various traditional practices and continues to gain popularity in the US and elsewhere. Approximately 80% of the worldwide population uses herbal medicines in their daily life.1−3 Traditional Chinese Medicine (TCM) including herbal supplements in a country’s medical system has gained increased importance compared to western medicine.4 The discovery of artemisinin, a natural antimalarial drug isolated from the plant Artemisia annua, led to the award of the 2015 Nobel Prize in Medicine to Chinese medical scientist Youyou Tu, perhaps further facilitating the acceptance of TCM into China’s healthcare system.5 There is a common popular belief that all the nature-derived products are harmless and safe to consume. The ongoing pandemic situation with coronavirus disease 2019 (COVID-19) has further boosted the quest for untested botanicals as antiviral remedies. However, the pertinent safety data on these natural products are limited and warrant scientific rigor before these products are introduced to the consumer.1,6
Licorice is among the most popular medicinal plants marketed in the US to alleviate multiple ailments, including cough, asthma, and menopausal complaints, among others. In addition, it is recognized as one of the most studied herbs in contemporary alternative medicine.7−11 However, there is no conclusive explanation or even an appropriate recommendation of either its efficacy or safety. This includes its use as an alternative medicine for hormone replacement therapy, a chemopreventive agent, adjuvant therapy in cancer treatments,8,12 or antiviral remedy to treat COVID-19.13
In addition, consumed “licorice” supplements can be prepared from closely related species of Glycyrrhiza.14,15 Often, in terms of chemistry, biologically active secondary metabolites are species-specific, and hence they are believed to possess a different spectrum of end biological effects.16−18 This situation warrants the use of more sophisticated methods to empower and delineate activity endpoint detection in clinical trials or to detect pharmacokinetic liabilities before these botanicals are accepted as drugs.15,19
The Glycyrrhiza genus belongs to the family Fabaceae and consists of approximately 36 species. However, three clinically relevant species are described in the pharmacopoeias: G. glabra L., G. uralensis Fisch, and G. inflata Batal.16Glycyrrhiza has been prioritized by the Center of Excellence for Natural Product Drug Interaction Research as one of the high-risk herbal constituents for inducing adverse effects such as herb–drug interactions.12 In the realm of drug metabolism and excretion, regnane X receptor (PXR) has a major effect on the expression of drug-metabolizing cytochrome P450 enzymes (CYPs) and transporters. PXR has a paramount role as a xenosensor by controlling the expression of many metabolic enzymes and transporters responsible for drug or “xenobiotics” disposition. Furthermore, PXR has viable but less studied roles in endobiotic synthesis, metabolism, and homeostasis of bile acids, lipids, glucose, bilirubin, vitamins, and steroidal hormones. The broad range of test substrates highlights the diverse and crucial role of PXR in drug metabolism (efficacy, toxicity, drug interactions, and drug resistance) and its associated diseases (metabolic syndrome, cancer, and inflammation).20−23 Integration of recent advancements and developments of computational approaches for identifying drug targets of natural products has gained popularity in drug discovery programs.24 Therefore, the primary objective of this study is to rapidly dereplicate potential PXR modulators from the chemical reservoirs of five Glycyrrhiza species with the help of computational tools before conducting time-consuming, expensive in vitro and in vivo methods to gauge the deleterious effects of potential candidates. Further computational validation studies involving putative binding and interaction profiles of the most promising candidates and ultimately experimental confirmation would rapidly identify xenobiotics in complex mixtures such as botanical extracts, including closely related species within the same genus.
2. Results and Discussion
The native or self-docking method was considered as a measure of the accuracy of the docking procedure.25 The virtual screening workflow (VSW)26 docking protocol was reliable enough to reproduce the poses of the co-crystallized ligands with the lower root mean-square deviation (RMSD) values (<1.1 Å). The ensemble docking approach correctly mimics the pose of co-crystallized ligands bound with the parent protein by fulfilling the necessary ligand residue interactions for each native ligand, as shown in Table 1. Several computational and site-directed mutagenesis studies have deduced global features as the primary requirements needed for PXR activation.27−30 For instance, large hydrophobic molecules have a better chance of interacting with PXR; this includes a molecular weight larger than 300 Daltons and hydrophobic features that allow multiple hydrophobic interactions and π–π interactions with a list of recognized hydrophobic amino acids, mainly those occupying the aromatic sub-pocket surrounded by Phe288, Trp299, and Tyr306. Further studies recognized Ser247, Gln285, and His407 as indispensable residues for PXR induction. Thus, at least one hydrogen bond interaction with one of the key amino acids is considered an essential requirement for activation.27−31
Table 1. Positive Control Used in the Docking Study.
The X-ray crystal structure of the human PXR ligand-binding domain (LBD) in complex with SR12813 (PDB ID: 1NRL)29 showed H-bonding interactions with Ser247 and His407 and had hydrophobic interactions with Leu209, Met243, Phe288, and Leu411. Similarly, hyperforin (PDB ID: 1M13),32 the psychoactive agent in St. John’s wort, complexed with the LBD of the human PXR X-ray crystal structure, had the interactions identical to SR12813, and showed one additional H-bonding with Gln285. The experimentally used positive control, rifampicin (PDB ID: 1SKX),33 forms hydrogen bonds with Ser247, Gln285, and His407. Rifampicin interacts with several residues, including Val211, Leu239, Leu308, and Arg410, which were not close to ligands in the previous PXR complexes involving SR12813 or hyperforin.
Recent studies have raised concerns about licorice-induced PXR activity and indicated glycyrrhizin as the principal source for PXR modulation. However, only weak PXR activation has been detected with the in vitro reporter gene assays for glycyrrhizin compared to the positive control, rifampicin.12,34,35 In the current study, we aimed to identify the Glycyrrhiza metabolites that might modulate PXR activity, based on the notion that the correlation of the PXR activity to only a few of the compounds is an oversimplification of the multitude of the constituents present in Glycyrrhiza. Hence, a library of 387 unique compounds reported from the five species of Glycyrrhiza (glabra, uralensis, inflata, echinata, and lepidota) was docked into the two PXR crystal structures, 1NRL and 1M13. The ligand preparations of 387 unique compounds yielded 456 structures, including possible isoforms of some of the compounds with undefined absolute configurations. A total of 323 out of 456 structures were docked successfully with PXR crystal structures and had binding free-energies ranging from −2.0 to −73.39 kcal/mol. Since the prime molecular mechanics/generalized born surface area (prime/MM-GBSA) binding free energies (ΔG) of positive controls were computed as approximately −64 kcal/mol (Table 1), therefore, the best complexes were selected based on prime MM-GBSA binding free energies using ≤−50 kcal/mol as an energy cutoff value.
According to our predefined threshold for potential PXR modulation, a total of 55 compounds including 38 species-specific metabolites from G. glabra, 41 compounds including 23 unique metabolites from G. uralensis, 37 compounds including 21 unique metabolites from G. inflata, five compounds with three species-specific compounds from G. lepidota, and four compounds with only one unique metabolite from G. echinata have scored below −50 kcal/mol (Table S1, Supporting Information). Our docking analysis revealed that the binding pocket of PXR exhibits a “one-size-fits-all” type of structure that supports previously reported studies.
In the next step, the selected Glycyrrhiza compounds were clustered according to the Tanimoto index,36 one of the most commonly used similarity indexes using the hierarchical-clustering (HC)36,37 method. After the HC, all the output clusters were manually visualized to assure the accuracy of similarity indexes. Overall, 9 clusters belonging to G. glabra, 10 clusters for G. uralensis, and 8 for G. inflata (Table S1, Supporting Information) were established.
To facilitate results analysis and discussion, the clusters were classified into the following structural scaffolds: flavonols, flavones, flavanones, isoflavones, isoflavanones, isoflavenes, isoflavans, pterocarpans, chalcones, dihydrostilbenes, dihydrophenanthrenes, arylcoumarins, and phenylbenzofurans as shown in Figure 1.
Figure 1.
Representation of the main structural scaffolds identified in Glycyrrhiza species.
Furthermore, the average binding free-energies of the species-specific secondary metabolites present in each cluster were calculated, averaged, and plotted against each scaffold. This graph showed that the isoflavan and flavanone marker constituents of G. glabra appear to possess better binding affinities toward PXR than the other species of Glycyrrhiza, except G. inflata (Figure 2). Based on a thorough literature search and Dr. Duke’s database (https://phytochem.nal.usda.gov/phytochem/search) on phytochemicals, compounds such as glabridin (isoflavan) and glabrol (flavanone) are typically found in considerable concentrations in G. glabra species14,38 (Table 2 and Table S1, Supporting Information). Similarly, gancaonin P, gancaonin O, the arylcoumarin scaffold (glycycoumarin), and kanzonol series (G, I, and J) present in G. uralensis(38) showed promising affinity toward PXR activity with favorable ligand–receptor interactions (Table 3 and Table S1, Supporting Information). The visual inspection of the docking poses of various metabolites specifically belonging to G. glabra and G. uralensis were identified with vital hydrophobic and π–π interactions with Phe288, Trp299, and Trp306, and H-bond interactions with the key polar residues Ser247, Gln285, and His407.27−32 All other metabolites were found to possess partial interactions either with one or two key residues along with many hydrophobic interactions. Details on associated binding free energies with these specific interactions are delineated in Figure 2 for each scaffold.
Figure 2.
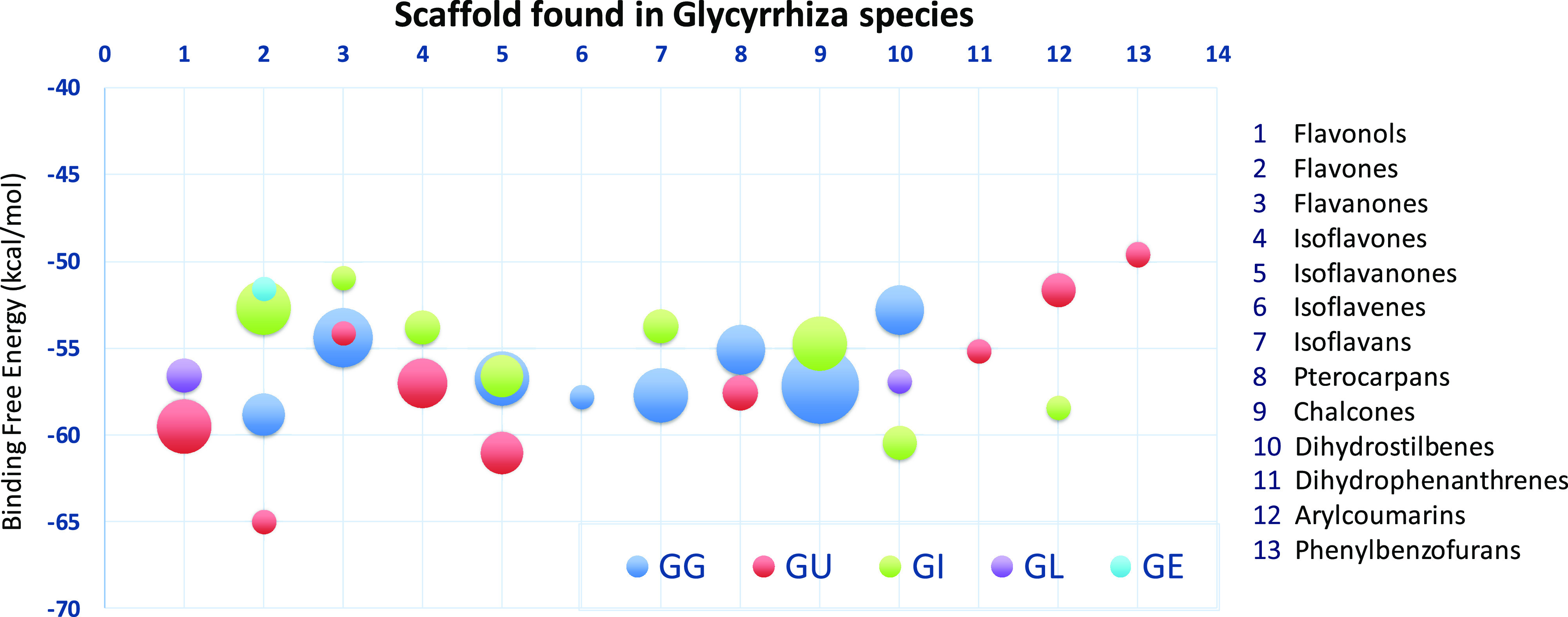
Bubble graph showing the PXR affinity of the compounds from Glycyrrhiza species.
Table 2. Best Ranking Compounds of G. glabra Docked with the PXR Protein.
| code no. | cluster no. | CAS no. | common namea | total | common | key interactions (H-bonds and π–π stacking) | BFEb |
|---|---|---|---|---|---|---|---|
| GG-105 | 1 | 91433-17-9 | licoflavone B | 5 | GI-29 | Gln285, His407 | –57.25 |
| GG-120 | 153-18-4 | rutin | unique | Leu209, Lys210, Leu239, Ser247 | –61.31 | ||
| GG-121 | 480-10-4 | astragalin | GL-19 (16 ppm) | Gln285 | –57.84 | ||
| GG-128 | 482-35-9/21637-25-2 | isoquercitrin | unique | Gln285, His407 | –54.54 | ||
| GG-131 | 54542-51-7 | quercetin 3-O-(6-acetyl glucoside) | unique | Leu209, Leu239, Trp299, His407 | –60.70 | ||
| GG-13 | 2 | 59870-65-4 | glabrol (440) | 11 | GI-30 | His407, Leu239, Phe281 | –57.17 |
| GG-26 | 82345-36-6 | xambioona | unique | Phe288, Trp306 | –51.88 | ||
| GG-29 | 125140-20-7 | euchrenone a5 | GI-56 | His407, Phe288, Trp306 | –50.0 | ||
| GG-32 | 157414-03-4 | shinflavanone | unique | His407, Phe288 | –50.29 | ||
| GG-45 | 220860-37-7 | kanzonol Z (10) | unique | His407, Leu239 | –52.21 | ||
| GG-49 | 2083619-55-8 | prenylflavan-3-ol | unique | Leu209, Ser247, Gln285 | –60.56 | ||
| GG-89 | 1217305-78-6 | prenylflavanone-3-ol | unique | Leu209, Ser247, Phe288, His407 | –50.82 | ||
| GG-98 | 551-15-5 | liquiritin (2300) | GU01 (300000), GE12 | Leu209, Lys210 | –49.95 | ||
| GG-104 | 74639-14-8 | liquiritin apioside | GU-24, GE-11 | Val211, Leu239, Gln285, His407 | –49.61 | ||
| GG-133 | 75829-43-5 | pinocembroside | unique | Asp205, Phe288, Trp299, His407 | –60.57 | ||
| GG-165 | 202657-63-4 | licorice glycoside D1 | GU-169, GE-17 | Lys210, Leu239, Ser247, Trp306 | –53.89 | ||
| GG-97 | 3 | 486-62-4 | formononetin 7-O-glucoside (300) | 4 | GU-30 (6000) | Lys210, Gln285 | –61.28 |
| GG-156 | 125310-04-5 | glycyroside | GU-145 | Lys210, Ser247, Gln285, His407 | –68.59 | ||
| GG-173 | 552-66-9 | daidzin | GU-110 | Lys210, Leu239, Ser247, Gln285 | –49.91 | ||
| GG-28 | 104691-86-3 | 8-prenylgenistein | GU-139 | Ile236, His407 | –50.37 | ||
| GG-27 | 4 | 94388-78-0 | prenylisoflavanone | 5 | all unique | Leu209, Trp299 | –50.29 |
| GG-53 | 2137446-32-1 | glycybridin J | Gln285, Phe281 | –51.98 | |||
| GG-82S | 905708-41-0 | pyranoisoflavanone-3-ol | His407, Phe281 | –60.44 | |||
| GG-83R | 905708-41-0 | pyranoisoflavanone-3-ol | Ser247, His407, Phe281, Gln285, Phe288 | –52.77 | |||
| GG-182 | 1201428-07-0 | glabra isoflavanone B | Trp299, His407, Trp299 | –68.1 | |||
| GG-56 | 5 | 2138843-57-7 | glycybridin D | 1 | unique | Leu209, Phe281 | –57.79 |
| GG-19 | 6 | 68978-03-0 | hispaglabridin A (127) | 5 | all unique | His407, Phe281, Gln285, Phe288, Trp299 | –53.23 |
| GG-37 | 175554-12-8 | 8-prenylphaseollinisoflavan | His407, Phe281, Gln285, Phe288, Trp299 | –52.03 | |||
| GG-66 | 156250-73-6 | kanzonol R (10) | His407, Phe281, Gln285 | –63.55 | |||
| GG-86 | 938190-33-1 | 4″-hydroxyglabridin | Leu209, Lys210, Leu239, Ser247, Gln285 | –56.46 | |||
| GG-14 | 59870-68-7 | glabridin (5700) | Phe281,Trp299, Phe429, His407 | –63.15 | |||
| GG-15 | 7 | 59901-97-2 | dehydroglyceollin I | 4 | all unique | His407, Phe288, Trp299 | –50.03 |
| GG-43 | 202815-29-0 | licoagrocarpin | His407, Trp299 | –55.28 | |||
| GG-78 | 157479-38-4 | O-methylshinpterocarpin | His407 | –51.46 | |||
| GG-96 | 66446-92-2 | hemileiocarpin | His407, Phe288, Trp299 | –63.53 | |||
| GG-12 | 8 | 58749-22-7 | licochalcone A | 16 | GU-17, GI-26 | Trp299, His407 | –56.55 |
| GG-30 | 151135-82-9 | kanzonol C | GI-58 | Ser247, His407 | –58.35 | ||
| GG-34 | 161099-57-6 | paratocarpin B | GI-96 | Phe288, Trp299 | –57.36 | ||
| GG-41 | 184584-87-0 | kanzonol Y | unique | Ser247, Phe281, Tyr306, His407 | –66.66 | ||
| GG-50 | 2083623-32-7 | unique | Gln285, His407 | –49.69 | |||
| GG-52 | 2088505-67-1 | glycybridin C | unique | Met243, Phe288, Tyr306, His407 | –67.93 | ||
| GG-54 | 2138843-55-5 | glycybridin A | unique | Ser247, Gln285, Tyr306, His407 | –53.41 | ||
| GG-55 | 2138843-56-6 | glycybridin B | unique | Ser247, Gln285, Tyr306, His407 | –55.73 | ||
| GG-77 | 151135-83-0 | DMDBP | unique | Ile236, Trp299 | –56.10 | ||
| GG-81 | 905708-40-9 | prenylchalcone | unique | Leu209, Phe281, Gln285, His407 | –56.75 | ||
| GG-92 | 944257-60-7 | dipyranochalcone | unique | only hydrophobic interactions | –55.42 | ||
| GG-87 | 938190-35-3 | prenyldihydrochalcone | unique | Leu209, Ser247, His407 | –56.77 | ||
| GG-101 | 29913-71-1 | licuracide | GU-31 | Lys210, Ser247, Gln285, His407 | –57.80 | ||
| GG-103 | 59122-93-9 | neoisoliquiritigenin | GU-38 | Ser247, His407 | –63.99 | ||
| GG-168 | 144506-14-9 | licochalcone C | GI-32 | only hydrophobic interactions | –50.14 | ||
| GG-170 | 2129164-89-0 | glycyglabrone | unique | Val211, Phe288, Trp299 | –53.24 | ||
| GG-134 | 9 | 266997-59-5 | stilbenoidglycoside | 4 | all unique | Ser247, Gln285, Trp299, His407 | –56.06 |
| GG-135 | 525585-29-9 | diprenylstilbenoid | Phe288, Trp299, His327 | –49.88 | |||
| GG-137 | 525585-31-3 | prenylstilbenoid | Ser247, Phe288, Trp299, His327 | –51.17 | |||
| GG-139 | 525585-33-5 | prenylstilbenoid | Val211, Ile236, Leu239, His407 | –54.08 |
Adopted from Dr. Duke’s Phytochemical and Ethnobotanical Database. The abundance in ppm is indicated in parentheses after the compound name.
Binding free energy in kcal/mol. Unique: species-specific.
Table 3. Best Ranking Compounds of G. uralensis Docked with PXR Proteins.
| code no. | cluster no. | CAS no. | common namea | total | common | key interactions (H-bonds and π–π stacking) | BFEb |
|---|---|---|---|---|---|---|---|
| GU-59 | 1 | 129145-54-6 | gancaonin P (6) | 6 | unique | Leu209, Val211, Ile236, His407 | –67.76 |
| GU-66 | 94805-83-1 | isolicoflavonol | GL-14 | Leu209, Leu239, Trp299 | –52.77 | ||
| GU-71 | 60197-60-6 | licoflavonol | unique | Ser247, Tyr306, His407 | –58.18 | ||
| GU-140 | 109605-79-0 | topazolin | unique | Lys210 | –52.45 | ||
| GU-147 | 139163-15-8 | uralenol | unique | Ser247, Phe281, Gln285, His407 | –60.14 | ||
| GU-157 | 1307578-72-8 | prenylflavone-3-ol | unique | Ser247, Phe281, Gln285, Trp299, His407 | –58.86 | ||
| GU-34 | 2 | 72357-31-4 | licoflavone C | 3 | GI-25 | Phe288, His407 | –59.95 |
| GU-58 | 129145-53-5 | gancaonin O (5) | unique | Val211, His407 | –65.01 | ||
| GU-174 | 134958-52-4 | gancaonin Q | GI-61 | Gln285, His407 | –55.63 | ||
| GU-35 | 3 | 833488-05-4 | 6″-O-α-OH-propionylliquiritin | 1 | unique | Lys210, Leu239, Ser247, Gln285 | –54.13 |
| GU-18 | 4 | 122290-50-0 | 7-O-methylluteone | 10 | GI-74 | Phe281, Gln285, His407, Phe429 | –57.53 |
| GU-56 | 129145-51-3 | gancaonin M (6) | unique | Leu209, Phe281 | –56.16 | ||
| GU-104 | 1879910-27-6 | glycyuralin F | unique | Leu209, Lys210, Leu239, Gln285 | –57.80 | ||
| GU-123 | 51225-28-6 | 6,8-diprenylgenistein | GI-10, 51 | Ser247, Phe281, His407 | –53.20 | ||
| GU-124 | 51225-30-0 | erythrinin B | GI-67 | Leu209, Phe281, Gln285, Phe429 | –55.17 | ||
| GU-128 | 66056-19-7 | licoisoflavone A | GI-65 | Ser247, Phe281, Gln285, His407 | –55.61 | ||
| GU-129 | 66056-30-2 | licoisoflavone B | GI-11 | Phe281, Tyr306, His407, Phe429 | –52.45 | ||
| GU-143 | 121747-89-5 | isoderrone | GI-09 | Phe281, Tyr306, His407, Phe429 | –54.95 | ||
| GU-144 | 121747-94-2 | 2′-hydroxylisolupalbigenin | unique | Ser247, His407 | –54.91 | ||
| GU-150 | 162616-70-8 | isolupalbigenin | unique | Tyr306 | –59.20 | ||
| GU-51 | 5 | 152511-45-0 | kanzonol G (4) | 3 | all unique | Phe281, Gln285, His407 | –51.82 |
| GU-97 | 142488-54-8 | glyasperin B | Ser247, Gln285, Phe429 | –57.82 | |||
| GU-130 | 66067-26-3 | licoisoflavanone | Ser247, Tyr306, His407 | –73.39 | |||
| GU-46 | 6 | 30508-27-1 | licoricidin (11) or licorisoflavan B | 3 | species-specific | Gln285 | –52.51 |
| GU-53 | 152511-47-2 | kanzonol J (2) | Phe281, Gln285, His407 | –49.93 | |||
| GU-54 | 152546-94-6 | kanzonol I (4) | Gln285 | –57.92 | |||
| GU-32 | 7 | 52766-70-8 | 3-glucopyranosyloxymedicarpin | 2 | unique | Ser247, Trp299, His407 | –59.52 |
| GU-102 | 1879910-25-4 | glycyuralin D | Gln285, Phe288, His407 | –55.61 | |||
| GU-180 | 8 | 134958-56-8 | gancaonin U (12) | 1 | unique | Lys210, Val211 | –55.2 |
| GU-03 | 9 | 94805-82-0 | glycycoumarin (1750) | 3 | GG-159 (710) | Leu209, Ser247, Gln285 | –50.77 |
| GU-72 | 66056-18-6 | glycyrin (400) | unique | Ser247, Gln285 | –53.67 | ||
| GU-146 | 125709-31-1 | licoarylcoumarin | unique | Ser247, Phe281, Trp299, His407 | –49.61 | ||
| GU-05 | 10 | 1253641-15-4 | glycybenzofuran | 1 | unique | Gln285,Trp299, Tyr306, His407 | –49.61 |
Adopted from Dr. Duke’s Phytochemical and Ethnobotanical Database. The abundance in ppm is indicated in parentheses after the compound name.
Binding free energy in kcal/mol. Unique: species-specific.
2.1. Flavonols
The metabolites representing the flavonol scaffold (cluster 1), found in the G. uralensis and G. lepidota (Table S1, Supporting Information), exhibited strong interactions with the key residues Ser247 and His407 and showed good binding free energies [gancaonin P (GU-59); ΔG = −67.76 kcal/mol and glepidotin A (GL-08); −53.35 kcal/mol] with PXR. Gancaonin P formed additional H-bonding with Leu209, Val211, and Ile236. Glepidotin A, which is found in G. lepidota, formed strong H-bonding with Ser247 and His407 (OH distance at C-7 = 2.87 Å). In addition, it exhibited strong π–π interactions with Phe288, Trp299, and Tyr306. The 2D interactions diagram of representing flavonols [gancaonin P (GU-59) and glepidotin A (GL-08)] are shown in Figure S1 (Supporting Information).
2.2. Flavones and Isoflavones
Some species-specific metabolites belonging to the flavone and isoflavone scaffolds were reported to be constituents of G. glabra, G. uralensis, G. inflata, and G. lepidota (Table S1, Supporting Information). In general, the metabolites originating from G. glabra showed favorable ligand–PXR interactions (strong H-bonding with Ser247 and His407) compared to metabolites reported from the two other species, G. uralensis and G. inflata. The 2D interaction diagrams of the representative flavone, rutin (GG-120), and isoflavone, glycyroside (GG-156), scaffolds are shown in Figure S2 (Supporting Information).
2.3. Flavanones and Isoflavanones
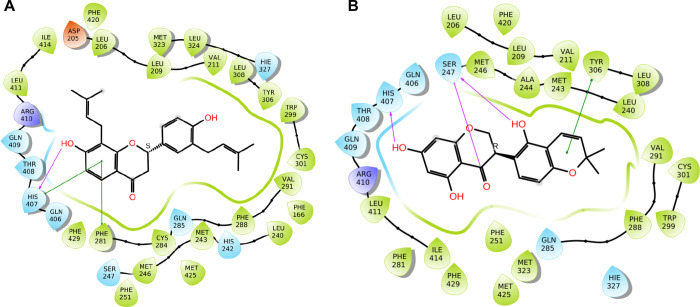
The categorical representation of the metabolites belonging to the flavanone scaffold was found in G. glabra, G. uralensis, G. inflata, and G. echinata. These metabolites were found to exhibit strong H-bond interaction with His407, a critical residue for PXR activity. The 2D interaction profile of the representative flavanone, glabrol (GG-13), found in G. glabra showed that the C-7 hydroxy formed strong H-bond interaction with His407 of the PXR protein. In addition, strong π–π interactions were observed between Phe281, His407, and ring A of glabrol (Figure 3A). Similarly, the C-7 hydroxy group of licoisoflavanone (GU-130) found in G. uralensis formed strong H-bonding with His407 and the C-4 carbonyl group (ring C), while the C-2′ hydroxy group (ring B) formed H-bonding with Ser247 (Figure 3B).
Figure 3.
2D interaction diagram of (A) glabrol and (B) licoisoflavanone with PXR X-ray crystal structure (1NRL).
2.4. Isoflavans
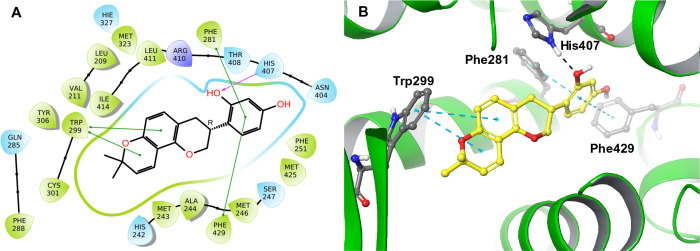
Some species-specific metabolites of the isoflavan-type were reported to be constituents of G. glabra and G. inflata. Isoflavan glabridin (GG-14) (Table 2 and Table S1, Supporting Information), identified in G. glabra, showed strong H-bonding interaction with His407 and had π–π interactions with Phe281, Trp299, and Phe429. In addition, glabridin forms a network of hydrophobic interactions with PXR residues Val211, Met243, Phe251, Phe288, Trp299, Cys301, Tyr306, Leu411, Met425, and Met426 (Figure 4), suggesting that candidates with an isoflavan scaffold are promising PXR modulators. Based on chemical fingerprinting and differentiation of the five studied species, glabridin (acquisition time = 30.7 min) was identified as a principal isoflavan in all G. glabra samples and was not detected in other analyzed Glycyrrhiza species (Figures S3 and S4, Supporting Information).39
Figure 4.
2D (A) and 3D (B) interaction diagrams of glabridin (GG-14) with 1NRL PXR X-ray crystal structure.
2.5. Isoflavenes
The only isoflavene-like structure, glycybridin D (GG-56), was found in cluster 5 of G. glabra (Table 2 and Table S1, Supporting Information). This secondary metabolite showed hydrogen bonding interaction with the carbonyl backbone of Leu209 and exhibited π–π interaction with Phe281. The schematic 2D interactions of glycybridin D with the PXR X-ray crystal are shown in Figure 5.
Figure 5.
2D interactions diagram of glycybridin D with 1NRL X-ray crystal structure.
2.6. Pterocarpans
Pterocarpans are derivatives of isoflavonoids and are present in the Fabaceae family. Cluster 7 of G. glabra contains compounds with a ptereocarpan scaffold in their structure. The ptereocarpan derivatives that showed good binding free-energies within G. glabra species are dehydroglyceollin I, licoagrocarpin, O-methylshinpterocarpin, and hemileiocarpin. Among these four compounds, hemileiocarpin (GG-96) showed better predicted binding affinity (ΔG = −63.53 kcal/mol) with the PXR receptor primarily due to strong H-bonding with His407 and π–π interactions with Phe288, Trp299, and His407 as well as other hydrophobic interactions with Leu209, Val211, Leu240, Met243, Met246, and Met250 (Figure 6A).
Figure 6.
2D interactions diagrams of (A) hemileiocarpin, a component of G. glabra, and (B) 3-glucopyranosyloxymedicarpin, a component of G. uralensis with 1NRL X-ray crystal structure.
On the other hand, cluster 7 of G. uralensis possesses only two pterocarpan-like metabolites: 3-glucopyranosyloxy-medicarpin (GU-32) and glycyuralin D. 3-Glucopyranosyloxy-medicarpin showed strong interactions with PXR compared to glycyuralin D. 3-Glucopyranosyloxy-medicarpin exhibited strong H-bonding with Ser247 and His407 and had π–π interactions with Trp299. The whole structure was surrounded by an array of hydrophobic residues such as Leu209, Val211, Leu240, Met243, Phe251, Phe288, Cys301, Tyr306, Leu411, and Met425 (Figure 6B). However, the aglycone medicarpin showed weaker ligand–PXR interactions than the prenylated analogue, hemileiocarpin, which is present in multiple samples of G. glabra, but at least one log unit less than glabridin.39
2.7. Chalcones
Chalcone-like compounds were found in G. glabra, G. inflata, and G. uralensis species. Cluster 8 of G. glabra consists of 16 compounds, including 10 species-specific markers. The main interacting residues for these compounds were Ser247, Gln285, Trp299, and His407. Most of the compounds formed at least one H-bond with the key residues Ser285 or His407. Among the 16 compounds, only two compounds had only hydrophobic interactions with the other key residues. This cluster represents the better binding free energy and has the highest number of unique compounds among all the clusters of the G. glabra species. The 2D interaction diagram representing glycybridin C (GG-52), which showed the highest negative binding free (ΔG = −67.93 kcal/mol), is reported in Figure S5 (Supporting Information).
On the other hand, cluster 6 of G. inflata has five unique chalcone-type compounds (Table 4 and Table S1) in which licochalcone K (GI-82) showed a better predicted binding free energy (ΔG = −59.69 kcal/mol) with strong favorable interactions with Ser247, Trp299, and His407 residues of PXR (Figure S6, Supporting Information). These strong interactions indicate that this compound and structurally similar mimics viz., licochalcones, G, D, and I could act as promising PXR modulators. Indeed, these metabolites indicate the unique yellow color associated with G. inflata extracts and are present across multiple samples of G. inflata.39
Table 4. Best-Ranking Compounds of G. inflata Docked within PXR Crystal Structures.
| code no. | cluster no. | CAS no. | common namea | total | common | key interactions (H-bonds and π–π stacking) | BFEb |
|---|---|---|---|---|---|---|---|
| GI-60 | 1 | 70872-32-1 | 6,8-diprenylapigenin | 5 | all unique | Leu209 | –51.74 |
| GI-78 | 2139264-68-7 | licoflavone D | Lys210, Gln285 | –54.42 | |||
| GI-85 | 155233-21-9 | kanzonol E | only hydrophobic interactions | –55.74 | |||
| GI-86 | kanzonol D | kanzonol D | Ser247, Phe281, Trp299, His407 | –50.24 | |||
| GI-93 | 106593-04-8 | retusin/3,4,3′,4′-tetrahydroxychalcone | Trp299, His407 | –51.25 | |||
| GI-53 | 2 | 80510-05-0 | euchestraflavanone A | 1 | unique | Leu239, His407 | –50.94 |
| GI-69 | 3 | 199331-53-8 | glyurallin B | 2 | all unique | Leu239, Gln285 | –56.27 |
| GI-76 | 2139264-65-4 | licoisoflavone D | Gln285, His407 | –51.35 | |||
| GI-62 | 4 | 21554-71-2 | dihydrogenistein | 3 | all unique | Leu209, Gln285, Trp299, Tyr306 | –57.06 |
| GI-81 | 2139264-71-2 | licoisoflavanone C | Ser247, Gln285, Trp299, His407 | –56.50 | |||
| GI-100 | 2137884-97-8 | prenylisoflavanone | Trp299, His407 | –56.30 | |||
| GI-06 | 5 | 164123-55-1 | glyinflanin I | 2 | all unique | Leu239, His327 | –54.67 |
| GI-12 | 160825-67-2 | gancaonin Z | Ser247, Gln285, Phe288, Tyr306 | –52.82 | |||
| GI-23 | 6 | 144506-15-0 | licochalcone D | 5 | all unique | Lys210 | –53.19 |
| GI-35 | 1083200-74-1 | chalcone derivative | Leu209, Lys210, Leu239, His407 | –49.94 | |||
| GI-41 | 151410-32-1 | xinjiachalcone A | His407 | –51.99 | |||
| GI-82 | 2139264-64-3 | licochalcone K | Ser247, Trp299, His407 | –59.69 | |||
| GI-88 | 775351-90-1 | corylifol B | Leu209, Val211, Gln285 | –58.91 | |||
| GI-39 | 7 | 1174167-73-7 | xinjiastilbene A | 2 | all unique | Ile236, His407 | –68.2 |
| GI-40 | 1174167-74-8 | xinjiastilbene B | Leu209, Phe288, Trp299, Tyr306 | –52.7 | |||
| GI-42 | 8 | 158446-33-4 | inflacoumarin A | 1 | unique | Phe288, Trp299, His407 | –58.45 |
Adopted from Dr. Duke’s Phytochemical and Ethnobotanical Database. The abundance in ppm is indicated in parentheses after the compound name.
Binding free energy in kcal/mol. Unique: species-specific.
2.8. Dihydrostilbenes
The secondary metabolites comprising the dihydrostilbene scaffold were found in G. glabra, G. inflata species, and G. lepidota. A total of four metabolites representing a dihydrostilbene scaffold (cluster 9) were identified as components of G. glabra and appeared to be species-specific markers. Two secondary metabolites, xinjiastilbenes A and B (cluster 7), were identified as G. inflata-specific secondary metabolites. The latter compound showed superior ligand–protein interactions resulting in a binding free energy of ΔG = −68.2 kcal/mol (Table 4 and Figure S7). Glepidotin D (Table 5) is the only metabolite identified as a component of G. lepidota and appeared to be species-specific markers.
Table 5. Best-Ranking Compounds of G. echinata and G. lepidota Docked within PXR Crystal Structures.
| code no. | cluster no. | CAS no. | common namea | total | common | key interactions (H-bonds and π–π stacking) | BFEb |
|---|---|---|---|---|---|---|---|
| GE-04 | 1 | 76690-67-0 | 7,4′-dihydroxy-8-prenylflavone | 1 | unique | Ile236, His407 | –52 |
| GL-08 | 1 | 42193-83-9 | glepidotin A | 2 | all unique | Ser247, Phe288, Trp299, Tyr306 | –53 |
| GL-20 | 17650-84-9 | nicotiflorin | Ser208, Ser247, Phe288, Trp299 | –60 | |||
| GL-11 | 2 | 374750-10-4 | glepidotin D | 1 | unique | Ile236, Leu239, His407 | –57 |
Adopted from Dr. Duke’s Phytochemical and Ethnobotanical Database.
Binding free energy in kcal/mol. Unique: species-specific.
2.9. Dihydrophenanthrenes and Phenylbenzofurans
The species-specific secondary metabolites belonging to the dihydrophenanthrene and phenylbenzofuran scaffolds (Table 3, clusters 8 and 10) were only found in G. uralensis and are represented by gancaonin U and glycybenzofuran, respectively. In the PXR ligand-binding domain, gancaonin U was surrounded by essential interactions with hydrophobic residues and devoid of typical hydrogen-bond interactions with any of the key residues (Ser247, Gln285, and His407); instead, strong H-bonding with Lys210 and Val211 (carbonyl backbone) was observed (Figure 7A). The glycybenzofuran compound representing the phenylbenzofuran scaffold exhibited typical strong H-bonding with Gln285 and His407, known for PXR induction. In addition, this compound also showed π–π interactions with Trp299 and Tyr306. Most parts of the molecule were surrounded by an array of hydrophobic residues shown in Figure 7B. Avula et al. did not find these compounds in their extracts in the other four Glycyrrhiza species.39
Figure 7.
2D interactions diagram of G. uralensis species-specific secondary metabolites, (A) gancaonin U and (B) glycybenzofuran within the 1M13 X-ray crystal structure.
2.10. Arylcoumarins
The compounds based on the coumarin aryl scaffold were found in G. glabra, G. uralensis, and G. inflata species. However, only a few members belong to these clusters. Glycyrin, glycycoumarin, and licoarylcoumarin are the only three metabolites reported for G. uralensis in which glycyrin (GU-72) and glycycoumarin (GU-03) were reported at the 400 and 1750 ppm levels, respectively. The major structural difference between glycycoumarin and glycyrin involves C-7 carrying hydroxy and methoxy moieties, respectively. These compounds showed strong interactions with the key residues Ser247 and Gln285 and fit well within the PXR active site (Figure 8A). The glycycoumarin with a C-7 hydroxy group showed additional hydrogen bonding with Leu209 (Figure S8, Supporting Information). In contrast, cluster 8 of G. inflata contains only one compound, inflacoumarin A (GI-42), which is reported to be the species-specific marker. It exhibited H-bond interaction with His407 and π–π stacking with Phe288 and Trp299 as well as the whole molecule being surrounded by an array of hydrophobic residues (Figure 8B). Based on LC–MS chemical fingerprinting of multiple samples of Glycyrrhiza species, inflacoumarin A in G. inflata and glycyrin in G. uralensis were tentatively identified.39
Figure 8.
2D interactions diagram of (A) glycyrin, a component of G. uralensis, and (B) inflacoumarin A, the component of G. inflata with 1NRL X-ray crystal structure.
2.11. Pregnane X Receptor (PXR) Activation
A luciferase reporter gene assay40 was employed to determine the effect of potential hits on the transcriptional activity of PXR. We tested six compounds, namely, (3R)-glabridin (GG-14), licoisoflavone A (GU-128), (2S)-liquiritin (GG-98), glycycoumarin (GU-03), (3R)-isoliquiritigenin (GG-19), and (3R)-licoisoflavanone (GU-130), from the list of potential hits identified, which were available in our in-house library (Table 6). The in vitro assay results showed that among the selected six compounds, glabridin, licoisoflavone A, glycycoumarin, and licoisoflavanone exhibited concentration-dependent PXR agonistic effects. The highest activity was found for glabridin (6.53-fold increase) followed by licoisoflavone A (3.88-fold increase), licoisoflavanone (3.29-fold increase), and glycycoumarin (2.46-fold increase) at 30 μM concentrations. Glabridin, licoisoflavone A, and licoisoflavanone retained high activity even at a lower concentration of 10 μM with 3.29, 2.53, and 2.40-fold increases, respectively. Other promising hits such as liquiritin and isoliquiritigenin showed minimal activity (<1.5-fold increase). In general, an increase in fold-activation of PXR correlates with the induction of certain CYP450 isozymes. No significant increase in 3A4 enzymatic levels was noticed, even though at least 6-fold PXR activation was observed with glabridin.41 On the contrary, 2,4-dihydroxyphenyl group on the glabridin scaffold were critical for CYP450 interactions and its antioxidant properties.42,43 Moreover, glabridin was identified to inactivate the enzymatic activities of 3A4, 2B6,44 and other isozymes and transporters using a UHPLC–MS/MS cocktail assay45 and serve as a competitive inhibitor of CYP2E1 isozyme.46 Nevertheless, further in-depth translational studies are warranted to establish the clinical relevancy of potential herb–drug interactions associated with glabridin. Analysis of the interaction profile of the docked pose of liquiritin with PXR showed H-bonding of liquirtin with Leu209 and π–cation interactions with Lys210, instead of typical key interactions with critical residues Ser285, His407, or Gln285 and hence displayed weaker PXR activation results. In order to gauge the overall modulatory effects on PXR, aq. ethanol extracts and pure compounds of five Glycyrrhiza species were evaluated for their effects on major CYP isoforms, which are regulated by PXR (3A4 and 1A2).41 The study revealed that glycycoumarin showed the highest induction (3-fold) of CYP3A4 enzymatic activity at 30 μM followed by licoisoflavone A with >2-fold induction. It is worth noting that glabridin did not induce CYP3A4 enzymatic activity despite its high PXR activation. Furthermore, glycycoumarin showed a ∼2-fold increase in CYP1A2 enzymatic activities at 30 and 10 μM, with no effect at the lower concentrations. Glabridin showed 2-fold induction of CYP1A2 activity at 10 μM. In contrast, licoisoflavone A did not induce CYP1A2 enzymatic activity at any tested concentration. Furthermore, selected extracts and marker compounds including glycycoumarin were also evaluated for their effects on the metabolism of two antiretroviral drugs, rilpivirine and dolutegravir, in hepatocytes.41
Table 6. PXR Activation by Compounds from Glycyrrhiza Species.
| fold increase in PXR activity at |
||||
|---|---|---|---|---|
| code | compound name | 30 μM ± SD | 10 μM ± SD | 3 μM ± SD |
| GG-14 | (3R)-glabridin | 6.53 ± 0.38 | 3.29 ± 0.09 | 1.99 ± 0.30 |
| GU-128 | licoisoflavone A | 3.88 ± 0.41 | 2.53 ± 0.17 | 1.87 ± 0.19 |
| GG-98 | liquiritin | 1.28 ± 0.07 | 1.07 ± 0.22 | 0.82 ± 0.15 |
| GU-03 | glycycoumarin | 2.46 ± 0.22 | 1.44 ± 0.05 | 1.19 ± 0.14 |
| GI-19 | isoliquiritigenina | 1.34 ± 0.03 | 1.20 ± 0.16 | 0.90 ± 0.02 |
| GU-130 | licoisoflavanone | 3.29 ± 0.31 | 2.40 ± 0.31 | 1.90 ± 0.31 |
| positive control | rifampicin | 3.51 ± 0.42 | 3.20 ± 0.80 | 2.70 ± 0.50 |
Close mimic for neoisoliquiritigenin (GG-103).
2.12. Molecular Dynamics
To explore the structural dynamics and stability of protein–ligand complexes, the RMSD analysis of the α-atoms of 1NRL-glabridin (GG-14) and 1NRL-rifampicin were calculated based on the starting frame at a 0 ns time interval and are plotted in Figure 9A. The RMSD plots indicate that the conformation of the protein of 1NRL-glabridin and 1NRL-rifampicin complexes were significantly stable compared to 1NRL protein only. It indicates that when the ligand binds with the PXR 1NRL protein and formed stable and strong interactions with the protein, it prevents high mobility or fluctuation during the MD simulations (Figure 9B). Similarly, the conformations of the ligands, rifampicin and glabridin, were quite stable during the 200 ns simulations except for 70–90 ns for the glabridin complex. During this time interval, the RMSD varies between ∼0.5 and 2.0 Å. After 90 ns, the conformational changes of both ligands were highly stable at the binding site for the remainder of the simulation.
Figure 9.
Root mean square deviation (RMSD) analysis of the molecular dynamic (MD) simulation trajectory. RMSD plot obtained for (A) C-α atoms of the protein PXR (PDB ID: 1NRL) with glabridin (GG-14) and rifampicin complexes; and (B) ligand-heavy atoms for glabridin- and rifampicin-PXR complexes (PDB ID: 1NRL), for the reference frame at time 0 ns.
The RMSF plots of 1NRL-glabridin and 1NRL-rifampicin complexes were further assessed to comprehend the individual residue flexibility or how much a particular residue fluctuates during an MD simulation. The RMSF plots revealed that the proteins did not show any significant fluctuations upon binding of glabridin to the protein residues (Figure 10).
Figure 10.
The root mean square fluctuation (RMSF) plot is based on C-α atoms of PXR protein (PDB ID: 1NRL) with (A) rifampicin and (B) glabridin. Protein residues that interact with the respective ligands are marked with green vertical bars.
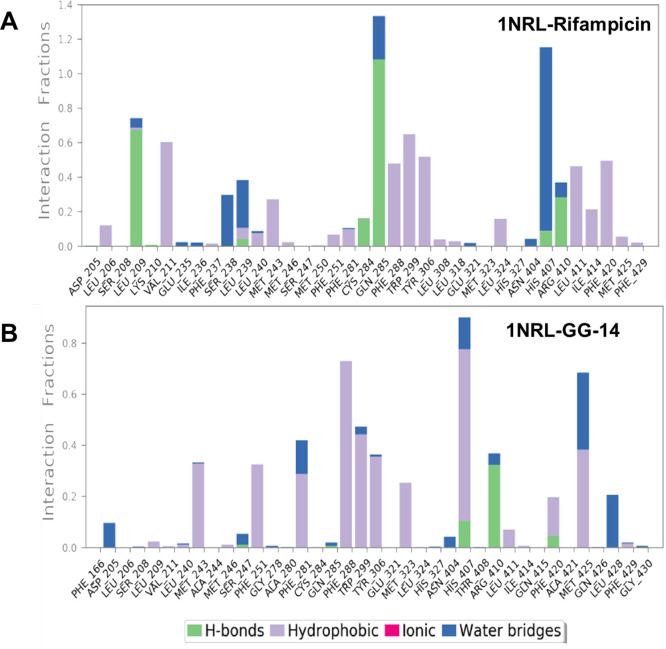
The interaction histogram of 1NRL-glabridin and 1NRL-rifampicin complexes revealed that both compounds formed major interactions with amino acids Met243, Phe288, Trp299, Trp306, His407, and Arg410 (Figure 11).
Figure 11.
Analysis of molecular interactions for (A) rifampicin and (B) glabridin with protein PXR (PDB ID: 1NRL) after MD simulation.
The 1NRL-rifampicin complex showed additional interactions with Leu209 (H-bonding), Val211 (hydrophobic), Ser238 (water-mediated), and Gln285 (H-bonding) (Figure 12A). These additional interactions resulted due to its extended molecular architecture. In contrast, glabridin showed strong π–π interactions with Phe251, Phe288, Trp299, Trp306, and His407 and H-bonding with Arg410 (Figure 12B). Interestingly, glabridin showed a hydrogen bond interaction between a phenolic OH (at C-6′) and His407 during docking; however, the same OH (at C-6′) is hydrogen bonded to Arg410 after MD simulation. The His407 exhibited strong π–π interactions with ring B. Overall, glabridin exhibited strong and stable interactions with the key residues of PXR, being known for its activity.
Figure 12.
Analysis of the type of contacts (2D interaction contour map with the key protein residues) for (A) rifampicin and (B) glabridin with protein PXR (PDB ID: 1NRL) after MD simulation.
3. Conclusions
Glycyrrhiza is considered as one of the alarmingly high-risk herbal constituents for inducing adverse effects; therefore, we conducted an exhaustive in silico screening of the known components of different yet closely related Glycyrrhiza species for potential PXR modulation by using docking and binding free-energy calculations. The results showed that compounds from G. glabra have diverse and unique scaffolds and exhibit better predicted binding affinities toward PXR than other Glycyrrhiza species. In addition, with their unique compounds, G. uralensis and G. inflata were also predicted to have a better binding affinity for PXR. A few compounds from the list of potential PXR modulators were tested through an assay for PXR activation based on their ready availability. Among the six tested, four compounds showed strong PXR activation effects, which were concentration-dependent; among them, (3R)-glabridin (GG-14) was found to be the most active. The MD results revealed that glabridin formed strong and stable interactions with the key residues of PXR and remained stable during the entire 200 ns simulation. In the future, the remaining highly alarming components from the studied Glycyrrhiza species based on their binding free-energy data and 3D interactions with the key residues of PXR will be further investigated by the application of cell-based in vitro assays for activation of the potential nuclear receptors and their modulatory effects on the susceptible target genes. A limitation of the current computational approach necessitates prior structural knowledge of compounds present in a complex natural product mixture. Before undertaking an in silico approach, an extensive literature search to collect all known phytochemicals is mandatory. In some cases, the exact stereo structures are not defined in the literature, and hence, all plausible stereostructures need to be considered in computational data mining. Therefore, natural products have to be isolated at some point to determine their absolute stereostructure and thus their identity and purity. Nevertheless, if not readily available, one must undertake pharmacognostic investigations to isolate target compounds and test their modulatory effects on desired biological targets to validate and confirm the predictive attributes. The utility of computer-aided tools when prior structural knowledge of compounds is available can be exploited to rapidly identify the potential nuclear receptor modulators in complex botanical extracts with demonstrable success and avoidance of time-consuming isolation efforts. Implementation of such tools would expedite and complement traditional pharmacognostic investigations and provide a platform to understand the safety and polypharmacology of the traditional herbs.
4. Experimental Section
4.1. Database Preparation
Literature reports16,38,47−106 including chemical databases such as Dictionary of Natural Products107 and SciFinder108 were searched for all known secondary metabolites of Glycyrrhiza species. Further, the unique CAS number associated with each specific compound isolated from the five species of Glycyrrhiza (glabra, uralensis, inflata, echinata, and lepidota) were collected. In the next step, the 2D structures (SMILES notation) of all the isolated compounds were retrieved via CAS numbers using the PubMed search. A total of 518 compounds from various species of Glycyrrhiza, 183 (GG, G. glabra), 180 (GU, G. uralensis), 100 (GI, G. inflata), 33 (GE, G. echinata), and 22 (GL, G. lepidota), were retrieved. Using CAS numbers, duplicate structures were removed to furnish 387 unique compounds. The ligand preparations of 387 unique compounds yielded 456 structures, which included possible isoforms of some of the compounds where stereogenic centers were undefined. The simplified molecular input line entry system (SMILES) notations of these compounds were converted into 2D structures using Canvas software. The 2D structures of 387 compounds were further converted into 3D-energy minimized structures using the LigPrep109 module implemented in the Schrödinger release 2020-4 suite at physiological pH 7.4. Compounds that had no defined absolute configuration were converted into all the possible stereoisomers. This step led to a total of 456 unique metabolites of Glycyrrhiza.
4.2. Protein Preparation
The 3D XYZ coordinates of X-ray crystal structures of PXR (PDB IDs: 1NRL(29) and 1M13(32) were downloaded from the protein data bank (https://www.rcsb.org/). The proteins were prepared using the Protein Prep Wizard module110,111 implemented in the Schrödinger software by adding missing hydrogen, correcting bond orders, proper ionization at pH 7.4, adding missing side chains, removing water molecules 5 Å away from the co-crystalized ligand, and having <2 hydrogen bonds with non-water molecules. The preprocessed proteins were further optimized for the H-bond assignment, and at the last hydrogen, only restraint minimization was performed.
4.3. Grid Generation
The grids for the active site of PXR proteins were prepared considering the centroid of the co-crystalized ligands in their corresponding proteins. No additional constraints were included during grid generation.
4.4. Native Docking
Before docking the dataset of compounds collected from Glycyrrhiza species, re-docking the cognate ligands into their corresponding proteins was performed. Cognate ligand docking effectively establishes a benchmark for a virtual screen of compounds with unknown binding activity against a given target. The docked pose of cognate ligands was compared with co-crystallized ligands in their corresponding proteins. The lower RMSD (<0.5 Å) of the docked pose with co-crystallized ligand pose indicates the suitability of the docking protocol.
4.5. Virtual Screening Workflow
Two X-ray crystal structures of PXR (PDB IDs: 1NRL(29) and 1M13(32)) were used for the ensemble docking via a virtual screening workflow (VSW)25 module implemented in the Schrödinger software. The VSW consists of docking Glycyrrhiza compounds collected from various species of Glycyrrhiza using Glide standard precision (SP) followed by Glide extra precision (XP).112,113 The final outputs of XP docking were further subjected to prime114,115 molecular mechanics/generalized born surface area (MM-GBSA) binding free-energy calculations. The flexible ligand sampling method was used for both docking methods. Three poses per ligand were allowed to store in the VSW method.
4.6. 2D Fingerprint-Based Clustering
After docking, the best complexes were selected based on prime MM-GBSA binding free energies using ≤−50 kcal/mol as a cutoff value. This criterion led to a total of 55 compounds from G. glabra, 41 compounds from G. uralensis, 37 compounds from G. inflata, five compounds from G. lepidota, and four compounds from G. echinata (Table S1, Supporting Information, and Figures 13–16). Among them, species-specific metabolites are 38, 23, 21, 3, and 1 for G. glabra, G. uralensis, G. inflata, G. lepidota, and G. echinata, respectively. In the next step, the 2D fingerprint of these compounds was calculated using the hashed linear fingerprint method116 implemented in the Canvas software.117,118 Fingerprints were generated using the default atom-typing scheme (daylight invariant atom types) and no bit scaling. Hashed fingerprints were mapped into a 32-bit address space. Compounds were clustered based on the Tanimoto index,36 one of the most commonly used similarity indexes in the literature using the hierarchical-clustering (HC) method.36,37 The clustering was performed using the average cluster linkage method, and Kelly criteria were used to identify the total number of clusters for HC.
Figure 13.

Structures of secondary metabolites found in G. glabra clustered according to the main scaffold.
Figure 16.
Structures of all the clusters of secondary metabolites found in G. lepidota (GL) andG. echinata (GE).
Figure 14.

Structures of all the clusters of secondary metabolites found in Glycyrrhiza uralensis.
Figure 15.
Structures of all the clusters of secondary metabolites found in G. inflata.
4.7. Isolation and Purification of Licorice Root Components
The preparation of licorice root extracts were achieved as described by Haron et al.41 The pure compounds, glabridin, isoliquiritigenin, licoisoflavone A, liquiritin, licoisoflavanone, and glycycoumarin were obtained from the methanol extract of botanically verified licorice by column chromatography over normal silica gel, reversed-phase silica (RP-18), and Sephadex LH-20, as well as preparative thin-layer chromatography (PTLC).119 Product integrity and purity of all six flavonoids were characterized using NMR, high-resolution mass spectrometry, and HPLC.41,119 Isoliquiritigenin, glabridin, and glycycoumarin were identified with >95% purity. Whereas, liquritin, licoisoflavonone, and licoisoflavone A were identified with >90% purity.
4.8. Biological Assay
4.8.1. PXR Activation Using Reporter Gene Assay
To further validate the computational results, some pure compounds from licorice available in-house with promising in silico results and some control compounds with proven ligand–PXR interactions were tested using a reporter gene assay described earlier.40 In brief, the cells were transiently transfected with 25 μg of pSG5-hPXR and 25 μg of PCR5 plasmid DNA by electroporation and seeded in 96-well plates (50,000 cells per well). After incubating for 24 h, the test compounds and the positive control were added at various concentrations (30, 10, and 3 μM). After an incubation of 24 h with the test samples, media was aspirated, and 40 μL of luciferase reagent (Promega Corporation, Madison, WI, USA) was added to each well. Luminescence was measured on a Spectramax M5 plate reader (Molecular Devices, Sunnyvale, CA, USA). The increase in luciferase activity of the sample-treated cells was calculated in comparison with vehicle-treated cells. Rifampicin was used as a positive control. Each compound was tested in triplicate unless stated otherwise.
4.9. Molecular Dynamics Simulation
(3R)-Glabridin, which showed the highest PXR activation among all the tested compounds, was subjected to all-atom molecular dynamics (200 ns) simulation along with the positive control, rifampicin, using Desmond software, (Schrödinger).120 The selected ligand–protein complexes were first solvated with a TIP3P explicit water model. The whole system was neutralized using NaCl and set to an ionic strength of 0.15 M. An orthorhombic box was used with a buffer distance of 10 Å for each dimension by the Desmond software. A modified relaxation protocol was used as described in our previous publication.121 In brief, the first step involved in the equilibrium stage was Brownian NVT dynamics (constant volume and temperature) with T = 10 K and restraints on solute heavy atoms for 1 ns; the second step includes T = 100 K, H2O barrier, Brownian NPT (constant pressure and temperature), membrane restrained in the Z direction, and protein restrained for 100 ps; the third step consists of NPγT, heating from 100 → 300 K, H2O barrier, and gradual release of restraints; the fourth step includes NVT production, T = 300 K and no restraints for 500 ps; and the final step involves NPT production, T = 300 K and no restraints for 5 ns. After the equilibration stage, the production run was performed in the NPT ensemble using a Langevin thermostat at a temperature of 300 K and 1.63 bar pressure over 200 ns (time-step 2 fs) with recording intervals of 1.2 ps for energy and 200 ps for trajectory. Simulations were run with the OPLS3e force field. Plots and figures were generated with the Desmond simulation interaction diagram (SID) implemented in the Desmond software.
Acknowledgments
This research was supported by the “Holistic Approach for Potential Drug Interactions with Botanical Drugs – Importance of Chemical Fingerprinting and Biosimilarity” funded by the US Food and Drug Administration grant #HHSF223201810175. Excellent technical support in the bioassays by Ms. Olivia Dale and Ms. Katherine Martin is also acknowledged.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c03240.
2D interactions diagram of representative compounds from the different scaffolds of Glycyrrhiza species and comparative chromatograms of various licorice species (PDF)
Author Present Address
∥ Faculty of Pharmacy, Philadelphia University, Amman, Jordan
Author Contributions
$ M.A. and P.P. contributed equally to this work.
This publication reflects the views of the authors and should not be construed to represent US FDA’s views or policies.
The authors declare no competing financial interest.
Supplementary Material
References
- Grimstein M.; Huang S. M. A regulatory science viewpoint on botanical-drug interactions. J. Food Drug Anal. 2018, 26, S12–S25. 10.1016/j.jfda.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization WHO global report on traditional and complementary medicine, 2019; World Health Organization: Geneva, Switzerland, 2019; p 226. [Google Scholar]
- World Health Organization WHO traditional medicine strategy. 2002–2005; World Health Organization: Geneva, 2002, p 61. [Google Scholar]
- World Health Organization WHO traditional medicine strategy. 2014–2023; World Health Organization: Geneva, 2013, p 76. [Google Scholar]
- Newman D. J. Modern traditional Chinese medicine: Identifying, defining and usage of TCM components. Adv. Pharmacol. 2020, 87, 113–158. 10.1016/bs.apha.2019.07.001. [DOI] [PubMed] [Google Scholar]
- Gaston T. E.; Mendrick D. L.; Paine M. F.; Roe A. L.; Yeung C. K. “Natural” is not synonymous with “Safe:” Toxicity of natural products alone and in combination with pharmaceutical agents. Regul. Toxicol. Pharmacol. 2020, 113, 104642 10.1016/j.yrtph.2020.104642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinzadeh H.; Nassiri-Asl M. Pharmacological effects of Glycyrrhiza spp. and its bioactive constituents: Update and review. Phytother. Res. 2015, 29, 1868–1886. 10.1002/ptr.5487. [DOI] [PubMed] [Google Scholar]
- Dietz B. M.; Hajirahimkhan A.; Dunlap T. L.; Bolton J. L. Botanicals and their bioactive phytochemicals for women’s health. Pharmacol. Rev. 2016, 68, 1026–1073. 10.1124/pr.115.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino G.; Cornara L.; Soares S.; Rodrigues F.; Oliveira M. B. P. P. Liquorice (Glycyrrhiza glabra): A phytochemical and pharmacological review. Phytother. Res. 2018, 32, 2323–2339. 10.1002/ptr.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikkeyan G.; Najar M. A.; Pervaje R.; Pervaje S. K.; Modi P. K.; Prasad T. S. K. Identification of molecular network associated with neuroprotective effects of yashtimadhu (Glycyrrhiza glabra L.) by quantitative proteomics of rotenone-induced Parkinson’s disease model. ACS Omega 2020, 5, 26611–26625. 10.1021/acsomega.0c03420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asl M. N.; Hosseinzadeh H. Review of pharmacological effects of Glycyrrhiza sp. and its bioactive compounds. Phytother. Res. 2008, 22, 709–724. 10.1002/ptr.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. J.; González-Peréz V.; Tian D.-D.; Lin Y. S.; Unadkat J. D.; Rettie A. E.; Shen D. D.; McCune J. S.; Paine M. F. Selection of priority natural products for evaluation as potential precipitants of natural product-drug interactions: A NaPDI center recommended approach. Drug Metab. Dispos. 2018, 46, 1046–1052. 10.1124/dmd.118.081273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C.; Vergoten G. Glycyrrhizin: An alternative drug for the treatment of COVID-19 infection and the associated respiratory syndrome?. Pharmacol. Ther. 2020, 214, 107618 10.1016/j.pharmthera.2020.107618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.; Nikolic D.; van Breemen R. B. Identification and chemical standardization of licorice raw materials and dietary supplements using UHPLC-MS/MS. J. Agric. Food Chem. 2016, 64, 8062–8070. 10.1021/acs.jafc.6b02954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W. C.; Lin Y.-H.; Chang T.-M.; Huang W.-Y. Identification of two licorice species, Glycyrrhiza uralensis and Glycyrrhiza glabra, based on separation and identification of their bioactive components. Food Chem. 2012, 132, 2188–2193. 10.1016/j.foodchem.2011.12.051. [DOI] [Google Scholar]
- Wong Y. F.; Cacciola F.; Fermas S.; Riga S.; James D.; Manzin V.; Bonnet B.; Marriott P. J.; Dugo P.; Mondello L. Untargeted profiling of Glycyrrhiza glabra extract with comprehensive two-dimensional liquid chromatography-mass spectrometry using multi-segmented shift gradients in the second dimension: Expanding the metabolic coverage. Electrophoresis 2018, 1993. 10.1002/elps.201700469. [DOI] [PubMed] [Google Scholar]
- Hajirahimkhan A.; Simmler C.; Yuan Y.; Anderson J. R.; Chen S.-N.; Nikolić D.; Dietz B. M.; Pauli G. F.; van Breemen R. B.; Bolton J. L. Evaluation of estrogenic activity of licorice species in comparison with hops used in botanicals for menopausal symptoms. PLoS One 2013, 8, e67947 10.1371/journal.pone.0067947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajirahimkhan A.; Simmler C.; Dong H.; Lantvit D. D.; Li G.; Chen S.-N.; Nikolić D.; Pauli G. F.; van Breemen R. B.; Dietz B. M.; Bolton J. L. Induction of NAD(P)H:quinone oxidoreductase 1 (NQO1) by Glycyrrhiza species used for women’s health: differential effects of the Michael acceptors isoliquiritigenin and licochalcone A. Chem. Res. Toxicol. 2015, 28, 2130–2141. 10.1021/acs.chemrestox.5b00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.; Huang K.; Nikolic D.; van Breemen R. B. High-throughput Cytochrome P450 cocktail inhibition assay for assessing drug-drug and drug-botanical interactions. Drug Metab. Dispos. 2015, 43, 1670–1678. 10.1124/dmd.115.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J.; Xie W. A brief history of the discovery of PXR and CAR as xenobiotic receptors. Acta Pharm. Sin. B 2016, 6, 450–452. 10.1016/j.apsb.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C. Novel functions of PXR in cardiometabolic disease. Biochim. Biophys. Acta 2016, 1859, 1112–1120. 10.1016/j.bbagrm.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M.; Robbins D.; Chen T. Targeting xenobiotic receptors PXR and CAR in human diseases. Drug Discovery Today 2015, 20, 618–628. 10.1016/j.drudis.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladimeji P. O.; Chen T. PXR: More than just a master xenobiotic receptor. Mol. Pharmacol. 2018, 93, 119–127. 10.1124/mol.117.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollinger J. M.; Langer T.; Stuppner H. Integrated in Silico Tools for Exploiting the Natural Products’ Bioactivity. Planta Med. 2006, 72, 671–678. 10.1055/s-2006-941506. [DOI] [PubMed] [Google Scholar]
- Ramírez D.; Caballero J. Is It Reliable to Take the Molecular Docking Top Scoring Position as the Best Solution without Considering Available Structural Data?. Molecules 2018, 23, 1038. 10.3390/molecules23051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Release 2020–4: Glide; Schrödinger, LLC: New York, NY, 2020. [Google Scholar]
- Honkakoski P.; Sueyoshi T.; Negishi M. Drug-activated nuclear receptors CAR and PXR. Ann. Med. 2003, 35, 172–182. 10.1080/07853890310008224. [DOI] [PubMed] [Google Scholar]
- Banerjee M.; Chai S. C.; Wu J.; Robbins D.; Chen T. Tryptophan 299 is a conserved residue of human pregnane X receptor critical for the functional consequence of ligand binding. Biochem. Pharmacol. 2016, 104, 131–138. 10.1016/j.bcp.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins R. E.; Davis-Searles P. R.; Lambert M. H.; Redinbo M. R. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J. Mol. Biol. 2003, 331, 815–828. 10.1016/S0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- Östberg T.; Bertilsson G.; Jendeberg L.; Berkenstam A.; Uppenberg J. Identification of residues in the PXR ligand binding domain critical for species specific and constitutive activation. Eur. J. Biochem. 2002, 269, 4896–4904. 10.1046/j.1432-1033.2002.03207.x. [DOI] [PubMed] [Google Scholar]
- Ekins S.; Chang C.; Mani S.; Krasowski M. D.; Reschly E. J.; Iyer M.; Kholodovych V.; Ai N.; Welsh W. J.; Sinz M.; Swaan P. W.; Patel R.; Bachmann K. Human pregnane X receptor antagonists and agonists define molecular requirements for different binding sites. Mol. Pharmacol. 2007, 72, 592–603. 10.1124/mol.107.038398. [DOI] [PubMed] [Google Scholar]
- Watkins R. E.; Maglich J. M.; Moore L. B.; Wisely G. B.; Noble S. M.; Davis-Searles P. R.; Lambert M. H.; Kliewer S. A.; Redinbo M. R. 2.1 Å crystal structure of human PXR in complex with the St. John’s wort compound hyperforin. Biochemistry 2003, 42, 1430–1438. 10.1021/bi0268753. [DOI] [PubMed] [Google Scholar]
- Chrencik J. E.; Orans J.; Moore L. B.; Xue Y.; Peng L.; Collins J. L.; Wisely G. B.; Lambert M. H.; Kliewer S. A.; Redinbo M. R. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol. Endocrinol. 2005, 19, 1125–1134. 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- Feng X.; Ding L.; Qiu F. Potential drug interactions associated with glycyrrhizin and glycyrrhetinic acid. Drug Metab. Rev. 2015, 47, 229–238. 10.3109/03602532.2015.1029634. [DOI] [PubMed] [Google Scholar]
- Wang Y.-G.; Zhou J.-M.; Ma Z.-C.; Li H.; Liang Q.-D.; Tan H.-L.; Xiao C.-R.; Zhang B.-L.; Gao Y. Pregnane X receptor mediated-transcription regulation of CYP3A by glycyrrhizin: A possible mechanism for its hepatoprotective property against lithocholic acid-induced injury. Chem.-Biol. Interact. 2012, 200, 11–20. 10.1016/j.cbi.2012.08.023. [DOI] [PubMed] [Google Scholar]
- Tanimoto T. T.Elementary mathematical theory of classification and prediction; IBM Corp., 1958. [Google Scholar]
- Bajusz D.; Rácz A.; Héberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations?. Aust. J. Chem. 2015, 7, 20. 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.; Qiao X.; Chen K.; Wang Y.; Ji S.; Feng J.; Li K.; Lin Y.; Ye M. Biosynthesis-based quantitative analysis of 151 secondary metabolites of licorice to differentiate medicinal Glycyrrhiza species and their hybrids. Anal. Chem. 2017, 89, 3146–3153. 10.1021/acs.analchem.6b04919. [DOI] [PubMed] [Google Scholar]
- Avula B.; Bae J. Y.; Chittiboyina A. G.; Wang Y. H.; Wang M.; Zhao J.; Ali Z.; Brinckmann J. A.; Li J.; Wu C.; Khan I. A. Chemometric Analysis and Chemical Characterization for the Botanical Identification of Glycyrrhiza species (G. glabra, G. uralensis, G. inflata, G. echinata and G. lepidota) using liquid chromatography-quadrupole time of flight mass spectrometry (LC-QToF). J. Food Compos. Anal. 2022, 112, 104679 10.1016/j.jfca.2022.104679. [DOI] [Google Scholar]
- Manda V. K.; Avula B.; Dale O. R.; Ali Z.; Khan I. A.; Walker L. A.; Khan S. I. PXR mediated induction of CYP3A4, CYP1A2, and P-gp by Mitragyna speciosa and its alkaloids. Phytother. Res. 2017, 31, 1935–1945. 10.1002/ptr.5942. [DOI] [PubMed] [Google Scholar]
- Haron M. H.; Avula B.; Ali Z.; Chittiboyina A. G.; Khan I. A.; Li J.; Wang V.; Wu C.; Khan S. I. Assessment of Herb-Drug Interaction Potential of Five Common Species of Licorice and Their Phytochemical Constituents. J. Diet. Suppl. 2022, 1–20. 10.1080/19390211.2022.2050875. [DOI] [PubMed] [Google Scholar]
- Vaya J.; Somjen D.; Tamir S.. The role of isoflavan’s 2′-hydroxyl in diverse biological activities; XI Biennial Meeting of the Society for Free Radical Research International, 2002. 567–574. [Google Scholar]
- Simmler C.; Pauli G. F.; Chen S. N. Phytochemistry and biological properties of glabridin. Fitoterapia 2013, 90, 160–184. 10.1016/j.fitote.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent U. M.; Aviram M.; Rosenblat M.; Hollenberg P. F. The licorice root derived isoflavan glabridin inhibits the activities of human cytochrome P450S 3A4, 2B6, and 2C9. Drug Metab. Dispos. 2002, 30, 709–715. 10.1124/dmd.30.6.709. [DOI] [PubMed] [Google Scholar]
- Li G.; Simmler C.; Chen L.; Nikolic D.; Chen S. N.; Pauli G. F.; van Breemen R. B. Cytochrome P450 inhibition by three licorice species and fourteen licorice constituents. Eur. J. Pharm. Sci. 2017, 109, 182–190. 10.1016/j.ejps.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S.; Kumar V.; Dogra A.; Ojha P. K.; Wazir P.; Sangwan P. L.; Singh G.; Nandi U. Amalgamation of in-silico, in-vitro and in-vivo approach to establish glabridin as a potential CYP2E1 inhibitor. Xenobiotica 2021, 51, 625–635. 10.1080/00498254.2021.1883769. [DOI] [PubMed] [Google Scholar]
- Bhardwaj D. K.; Murari R.; Seshadri T. R.; Singh R. Liqcoumarin, a novel coumarin from Glycyrrhiza glabra. Phytochemistry 1976, 15, 1182. 10.1016/0031-9422(76)85130-8. [DOI] [Google Scholar]
- Kinoshita T.; Tamura Y.; Mizutani K. The isolation and structure elucidation of minor isoflavonoids from licorice of Glycyrrhiza glabra origin. Chem. Pharm. Bull. 2005, 53, 847–849. 10.1248/cpb.53.847. [DOI] [PubMed] [Google Scholar]
- Li K.; Ji S.; Song W.; Kuang Y.; Lin Y.; Tang S.; Cui Z.; Qiao X.; Yu S.; Ye M. Glycybridins A-K, bioactive phenolic compounds from Glycyrrhiza glabra. J. Nat. Prod. 2017, 80, 334–346. 10.1021/acs.jnatprod.6b00783. [DOI] [PubMed] [Google Scholar]
- Fukai T.; Cai B.; Maruno K.; Miyakawa Y.; Konishi M.; Nomura T. An isoprenylated flavanone from Glycyrrhiza glabra and rec-assay of licorice phenols. Phytochemistry 1998, 49, 2005. 10.1016/S0031-9422(98)00389-6. [DOI] [Google Scholar]
- Kuroda M.; Mimaki Y.; Honda S.; Tanaka H.; Yokota S.; Mae T. Phenolics from Glycyrrhiza glabra roots and their PPAR-gamma ligand-binding activity. Bioorg. Med. Chem. 2010, 18, 962–970. 10.1016/j.bmc.2009.11.027. [DOI] [PubMed] [Google Scholar]
- Fukui H.; Goto K.; Tabata M. Two antimicrobial flavanones from the leaves of Glycyrrhiza glabra. Chem. Pharm. Bull. 1988, 36, 4174–4176. 10.1248/cpb.36.4174. [DOI] [PubMed] [Google Scholar]
- Kitagawa I.; Chen W. Z.; Hori K.; Harada E.; Yasuda N.; Yoshikawa M.; Ren J. Chemical studies of Chinese licorice-roots. I. Elucidation of five new flavonoid constituents from the roots of Glycyrrhiza glabra L. collected in Xinjiang. Chem. Pharm. Bull. 1994, 42, 1056–1062. 10.1248/cpb.42.1056. [DOI] [PubMed] [Google Scholar]
- Haraguchi H.; Yoshida N.; Ishikawa H.; Tamura Y.; Mizutani K.; Kinoshita T. Protection of mitochondrial functions against oxidative stresses by isoflavans from Glycyrrhiza glabra. J. Pharm. Pharmacol. 2000, 52, 219–223. 10.1211/0022357001773724. [DOI] [PubMed] [Google Scholar]
- Mitscher L. A.; Park Y. H.; Clark D.; Beal J. L. Antimicrobial agents from higher plants. Antimicrobial isoflavanoids and related substances from Glycyrrhiza glabra L. var. typica. J. Nat. Prod. 1980, 43, 259–269. 10.1021/np50008a004. [DOI] [PubMed] [Google Scholar]
- Siracusa L.; Saija A.; Cristani M.; Cimino F.; D’Arrigo M.; Trombetta D.; Rao F.; Ruberto G. Phytocomplexes from liquorice (Glycyrrhiza glabra L.) leaves--chemical characterization and evaluation of their antioxidant, anti-genotoxic and anti-inflammatory activity. Fitoterapia 2011, 82, 546–556. 10.1016/j.fitote.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Schmid C.; Dawid C.; Peters V.; Hofmann T. Saponins from European licorice roots (Glycyrrhiza glabra). J. Nat. Prod. 2018, 81, 1734–1744. 10.1021/acs.jnatprod.8b00022. [DOI] [PubMed] [Google Scholar]
- Chen J.-J.; Cheng M.-J.; Shu C.-W.; Sung P.-J.; Lim Y.-P.; Cheng L.-Y.; Wang S.-L.; Chen L.-C. A new chalcone and antioxidant constituents of Glycyrrhiza glabra. Chem. Nat. Compd. 2017, 53, 632–634. 10.1007/s10600-017-2077-1. [DOI] [Google Scholar]
- Elgamal M. H. A.; El-Tawil B. A. H. Constituents of local plants. XVIII. 28-hydroxyglycyrrhetic acid, a new triterpenoid isolated from the roots of Glycyrrhiza glabra. Planta Med. 1975, 27, 159–163. 10.1055/s-0028-1097778. [DOI] [PubMed] [Google Scholar]
- Khalaf I.; Vlase L.; Ivanescu B.; Lazǎr D.; Corciovǎ A. HPLC analysis of polyphenolic compounds, phytoestrogens and sterols from Glycyrrhiza glabra L. tincture. Stud. Univ. Babes-Bolyai, Chem. 2012, 57, 113–118. [Google Scholar]
- Li J. R.; Wang Y. Q.; Deng Z. Z. Two new compounds from Glycyrrhiza glabra. J. Asian Nat. Prod. Res. 2005, 7, 677–680. 10.1080/10286020310001625067. [DOI] [PubMed] [Google Scholar]
- Suman A.; Ali M.; Alam P. New prenylated isoflavanones from the roots of Glycyrrhiza glabra. Chem. Nat. Compd. 2009, 45, 487. 10.1007/s10600-009-9403-1. [DOI] [Google Scholar]
- Li S.; Li W.; Wang Y.; Asada Y.; Koike K. Prenylflavonoids from Glycyrrhiza uralensis and their protein tyrosine phosphatase-1B inhibitory activities. Bioorg. Med. Chem. Lett. 2010, 20, 5398–5401. 10.1016/j.bmcl.2010.07.110. [DOI] [PubMed] [Google Scholar]
- Lee Y. S.; Kim S. H.; Jung S. H.; Kim J. K.; Pan C.-H.; Lim S. S. Aldose reductase inhibitory compounds from Glycyrrhiza uralensis. Biol. Pharm. Bull. 2010, 33, 917–921. 10.1248/bpb.33.917. [DOI] [PubMed] [Google Scholar]
- Meng X.; Li H.; Song F.; Liu C.; Liu Z.; Liu S. Studies on triterpenoids and flavones in Glycyrrhiza uralensis Fisch. by HPLC-ESI-MSn and FT-ICR-MSn. Chin. J. Chem. 2009, 27, 299–305. 10.1002/cjoc.200990048. [DOI] [Google Scholar]
- Liu Y.-c.; Chen Y.-g.; Wang D.; Gao X.-y.; Guo H.-z.; Fu X.-t.; Wang W.-q. Studies on chemical constituents on roots of Glycyrrhiza uralensis. Chin. J. Pharm. Anal. 2011, 31, 1251–1255. [Google Scholar]
- Liu H. X.; Lin W. H.; Yang J. S. A new dihydroflavone glycoside from Glycyrrhiza uralensis. Chin. Chem. Lett. 2004, 15, 925–926. [Google Scholar]
- Pan Y. Isolation and identification of saxifragin from Glycyrrhiza uralensis Fisch. Zhongguo Zhongyao Zazhi 1999, 24, 295–319. [PubMed] [Google Scholar]
- Kitagawa I.; Hori K.; Uchida E.; Chen W. Z.; Yoshikawa M.; Ren J. Saponin and sapogenol. L. On the constituents of the roots of Glycyrrhiza uralensis Fischer from Xinjiang, China. Chemical structures of licorice-saponin L3 and isoliquiritin apioside. Chem. Pharm. Bull. 1993, 41, 1567–1572. 10.1248/cpb.41.1567. [DOI] [PubMed] [Google Scholar]
- Fukai T.; Nishizawa J.; Yokoyama M.; Nomura T. Five new isoprenoid-substituted flavonoids, kanzonols F-J, from Glycyrrhiza uralensis. Heterocycles 1993, 36, 2565–2576. 10.3987/COM-93-6486. [DOI] [Google Scholar]
- Fukai T.; Wang Q.-H.; Takayama M.; Nomura T. Structures of five new prenylated flavonoids, gancaonins L, M, N, O, and P from aerial parts of Glycyrrhiza uralensis. Heterocycles 1990, 31, 373–382. 10.3987/COM-89-5260. [DOI] [Google Scholar]
- Nomura T.; Fukai T.. Phenolic constituents of licorice (Glycyrrhiza Species). In Fortschritte der Chemie organischer Naturstoffe / Progress in the Chemistry of Organic Natural Products; 1998, 1–140, 10.1007/978-3-7091-6480-8_1. [DOI] [PubMed] [Google Scholar]
- Zheng Y.-F.; Qi L.-W.; Zhou J.-L.; Li P. Structural characterization and identification of oleanane-type triterpene saponins in Glycyrrhiza uralensis Fischer by rapid-resolution liquid chromatography coupled with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 3261–3270. 10.1002/rcm.4768. [DOI] [PubMed] [Google Scholar]
- Zhu D.-Y.; Song G.-Q.; Jian F. X.; Chang X.-R.; Guo Y. Studies on chemical constituent of Glycyrrhiza uralensis Fisch: the structures of isolicoflavonol and glycycomarin. Acta Chim. Sin. 1984, 42, 1080. [Google Scholar]
- Qiao X.; Liu C.-F.; Ji S.; Lin X.-H.; Guo D.-A.; Ye M. Simultaneous determination of five minor coumarins and flavonoids in Glycyrrhiza uralensis by solid-phase extraction and high-performance liquid chromatography/electrospray ionization tandem mass spectrometry. Planta Med. 2014, 80, 237–242. 10.1055/s-0033-1360272. [DOI] [PubMed] [Google Scholar]
- Ji S.; Li Z.; Song W.; Wang Y.; Liang W.; Li K.; Tang S.; Wang Q.; Qiao X.; Zhou D.; Yu S.; Ye M. Bioactive constituents of Glycyrrhiza uralensis (licorice): discovery of the effective components of a traditional herbal medicine. J. Nat. Prod. 2016, 79, 281–292. 10.1021/acs.jnatprod.5b00877. [DOI] [PubMed] [Google Scholar]
- Fukai T.; Nomura T. Isoprenoid-substituted flavonoids from roots of Glycyrrhiza inflata. Phytochemistry 1995, 38, 759–765. 10.1016/0031-9422(94)00674-I. [DOI] [Google Scholar]
- Fang S.; Qu Q.; Zheng Y.; Zhong H.; Shan C.; Wang F.; Li C.; Peng G. Structural characterization and identification of flavonoid aglycones in three Glycyrrhiza species by liquid chromatography with photodiode array detection and quadrupole time-of-flight mass spectrometry. J. Sep. Sci. 2016, 39, 2068–2078. 10.1002/jssc.201600073. [DOI] [PubMed] [Google Scholar]
- Yoon G.; Do Jung Y.; Cheon S. H. Cytotoxic allyl retrochalcone from the roots of Glycyrrhiza inflata. Chem. Pharm. Bull. 2005, 53, 694–695. 10.1248/cpb.53.694. [DOI] [PubMed] [Google Scholar]
- Kajiyama K.; Demizu S.; Hiraga Y.; Kinoshita K.; Koyama K.; Takahashi K.; Tamura Y.; Okada K.; Kinoshita T. Two prenylated retrochalcones from Glycyrrhiza inflata. Phytochemistry 1992, 31, 3229–3232. 10.1016/0031-9422(92)83481-D. [DOI] [Google Scholar]
- Kajiyana K.; Demizu S.; Hiraga Y.; Kinoshita K.; Koyama K.; Takahashi K.; Tamura Y.; Okada K.; Kinoshita T. New prenylflavones and dibenzoylmethane from Glycyrrhiza inflata. J. Nat. Prod. 1992, 55, 1197–1203. 10.1021/np50087a005. [DOI] [Google Scholar]
- Fu Y.; Chen J.; Li Y.-J.; Zheng Y.-F.; Li P. Antioxidant and anti-inflammatory activities of six flavonoids separated from licorice. Food Chem. 2013, 141, 1063–1071. 10.1016/j.foodchem.2013.03.089. [DOI] [PubMed] [Google Scholar]
- Simmler C.; Lankin D. C.; Nikolić D.; Van Breemen R. B.; Pauli G. F. Isolation and structural characterization of dihydrobenzofuran congeners of licochalcone A. Fitoterapia 2017, 121, 6–15. 10.1016/j.fitote.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. K.; Oh J. S.; Lee J.-K. Antinociceptive effect of glyasperin F isolated from Glycyrrhiza inflata in mice. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 541–545. 10.1007/s13765-013-3161-2. [DOI] [Google Scholar]
- Dao T. T.; Nguyen P. H.; Lee H. S.; Kim E.; Park J.; Lim S. I.; Oh W. K. Chalcones as novel influenza A (H1N1) neuraminidase inhibitors from Glycyrrhiza inflata. Bioorg. Med. Chem. Lett. 2011, 21, 294–298. 10.1016/j.bmcl.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Iwasaki N.; Baba M.; Aishan H. Studies of traditional folk medicines in Xinjiang Uighur Autonomous (2) Research for chemical constituents of Xinjiang licorice. Heterocycles 2009, 78, 1581–1587. 10.3987/COM-09-11673. [DOI] [Google Scholar]
- Wang Q.-E.; Lee F. S.-C.; Wang X. Isolation and purification of inflacoumarin A and licochalcone A from licorice by high-speed counter-current chromatography. J. Chromatogr. A 2004, 1048, 51–57. 10.1016/S0021-9673(04)01086-6. [DOI] [PubMed] [Google Scholar]
- Lin Y.; Kuang Y.; Li K.; Wang S.; Ji S.; Chen K.; Song W.; Qiao X.; Ye M. Nrf2 activators from Glycyrrhiza inflata and their hepatoprotective activities against CCl4-induced liver injury in mice. Bioorg. Med. Chem. Lett. 2017, 25, 5522–5530. 10.1016/j.bmc.2017.08.018. [DOI] [PubMed] [Google Scholar]
- Wang B.; Zou K.; Yang X.; He W.; Zhao Y.; Zhang R. Two new flavanone glycosides from Glycyrrhizia inflata. Acta Pharm. Sin. 1997, 32, 199–202. [PubMed] [Google Scholar]
- Zou K.; Zhao Y.-Y.; Zhang R.-Y. Structure Determination of Inflasaponin II and VI from Glycyrrhiza inflata Root. J. Chin. Pharm. Sci. 1995, 4, 113–117. [Google Scholar]
- Zou K.; Cai L.-N.; Zhang R.-Y. Structure identification of inflasaponin III and inflasaponin V. Chem. Res. Chin. Univ. 1994, 15, 845–848. [Google Scholar]
- Mirhom Y. W.; Hanna A. G.; Elgamal M. H. A.; Szendrei K.; Reisch J. Notizen: A novel triterpenoid isolated from the roots of Glycyrrhiza echinata L.. Z. Naturforsch., B: J. Chem. Sci. 1990, 45, 1111–1112. 10.1515/znb-1990-0738. [DOI] [Google Scholar]
- Ayabe S.-I.; Kobayashi M.; Hikichi M.; Matsumoto K.; Furuya T. Flavonoids from the cultured cells of Glycyrrhiza echinata. Phytochemistry 1980, 19, 2179–2183. 10.1016/S0031-9422(00)82219-0. [DOI] [Google Scholar]
- Mamedov N. A.; Egamberdieva D., Phytochemical constituents and pharmacological effects of licorice: A review. In Plant and Human Health; Ozturk M.; Hakeem K., Eds.; Springer, Cham, 2019; Vol. 3, pp. 1–21. [Google Scholar]
- Shibata S.; Saitoh T. Flavonoid compounds in licorice root. J. Indian Chem. Soc., Ind. News Ed. 1978, 55, 1184–1191. [Google Scholar]
- Kir’yalov N. P.; Bogatkina V. F. Isomacedonic acid from the roots of Glycyrrhiza echinata. Chem. Nat. Compd. 1971, 7, 117–117. 10.1007/BF01032052. [DOI] [Google Scholar]
- Murav’ev I. A.; Semenchenko V. F. Structure of triterpene saponins from the roots of Glycyrrhiza echinata. Chem. Nat. Compd. 1969, 5, 13–14. 10.1007/BF00564910. [DOI] [Google Scholar]
- Farag M. A.; Porzel A.; Wessjohann L. A. Comparative metabolite profiling and fingerprinting of medicinal licorice roots using a multiplex approach of GC–MS, LC–MS and 1D NMR techniques. Phytochemistry 2012, 76, 60–72. 10.1016/j.phytochem.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Ryu Y. B.; Kim J. H.; Park S.-J.; Chang J. S.; Rho M.-C.; Bae K.-H.; Park K. H.; Lee W. S. Inhibition of neuraminidase activity by polyphenol compounds isolated from the roots of Glycyrrhiza uralensis. Bioorg. Med. Chem. Lett. 2010, 20, 971–974. 10.1016/j.bmcl.2009.12.106. [DOI] [PubMed] [Google Scholar]
- Song W.; Si L.; Ji S.; Wang H.; Fang X.-m.; Yu L.-y.; Li R.-y.; Liang L.-n.; Zhou D.; Ye M. Uralsaponins M–Y, antiviral triterpenoid saponins from the roots of Glycyrrhiza uralensis. J. Nat. Prod. 2014, 77, 1632–1643. 10.1021/np500253m. [DOI] [PubMed] [Google Scholar]
- Kuroda M.; Mimaki Y.; Sashida Y.; Mae T.; Kishida H.; Nishiyama T.; Tsukagawa M.; Konishi E.; Takahashi K.; Kawada T.; Nakagawa K.; Kitahara M. Phenolics with PPAR-γ ligand-binding activity obtained from licorice (Glycyrrhiza uralensis Roots) and ameliorative effects of glycyrin on genetically diabetic KK-Ay mice. Bioorg. Med. Chem. Lett. 2003, 13, 4267–4272. 10.1016/j.bmcl.2003.09.052. [DOI] [PubMed] [Google Scholar]
- Ali Z.; Srivedavyasasri R.; Zhao J.; Avula B.; Chittiboyina A. G.; Khan I. A. Oleanane-type triterpenoid glucuronosides from Glycyrrhiza echinata L. root. Biochem. Syst. Ecol. 2020, 92, 104088 10.1016/j.bse.2020.104088. [DOI] [Google Scholar]
- Ali Z.; Hawwal M.; Ahmed M. M. A.; Avula B.; Chittiboyina A. G.; Li J.; Wu C.; Taylor C.; Chan Y.-M.; Khan I. A. Licochalcone L, an undescribed retrochalcone from Glycyrrhiza inflata roots. Nat. Prod. Res. 2021, 200–206. 10.1080/14786419.2022.2062755. [DOI] [PubMed] [Google Scholar]
- Ali Z.; Hawwal M.; Avula B.; Chittiboyina A. G.; Li J.; Wu C.; Khan I. A. Phenoxychromone and 4-hydroxyisoflavans from the roots of Glycyrrhiza uralensis. Nat. Prod. Res. 2021, 1–8. 10.1080/14786419.2021.1892668. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Li Y.; Ma X.; Ren H.; Fan W.; Leng F.; Yang M.; Wang X. Extraction, purification, and bioactivities analyses of polysaccharides from Glycyrrhiza uralensis. Ind. Crops Prod. 2018, 122, 596–608. 10.1016/j.indcrop.2018.06.011. [DOI] [Google Scholar]
- Chin Y. W.; Jung H. A.; Liu Y.; Su B. N.; Castoro J. A.; Keller W. J.; Pereira M. A.; Kinghorn A. D. Anti-oxidant constituents of the roots and stolons of licorice (Glycyrrhiza glabra). J. Agric. Food Chem. 2007, 55, 4691–4697. 10.1021/jf0703553. [DOI] [PubMed] [Google Scholar]
- Chapman H.Dictionary of natural products on DVD (23: 1); CRC Press: Taylor and Francis Group, 2014. [Google Scholar]
- American Chemical Society Chemical Abstracts , S. SciFinder; American Chemical Society: Columbus, Ohio, 2007. [Google Scholar]
- Schrödinger Release 2020–4: LigPrep; Schrödinger, LLC: New York, NY, 2020. [Google Scholar]
- Madhavi Sastry G.; Adzhigirey M.; Day T.; Annabhimoju R.; Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput.-Aided Mol. Des. 2013, 27, 221–234. 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Schrödinger Release 2020–4: Protein Preparation Wizard; Epik, Schrödinger, LLC: New York, NY, 2020. [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; Shaw D. E.; Francis P.; Shenkin P. S. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Halgren T. A.; Murphy R. B.; Friesner R. A.; Beard H. S.; Frye L. L.; Pollard W. T.; Banks J. L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Schrödinger Release 2020–4: Prime; Schrödinger, LLC: New York, NY, 2020. [Google Scholar]
- Jacobson M. P.; Pincus D. L.; Rapp C. S.; Day T. J. F.; Honig B.; Shaw D. E.; Friesner R. A. A hierarchical approach to all-atom protein loop prediction. Proteins 2004, 55, 351–367. 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Duan J.; Dixon S. L.; Lowrie J. F.; Sherman W. Analysis and comparison of 2D fingerprints: Insights into database screening performance using eight fingerprint methods. J. Mol. Graphics Modell. 2010, 29, 157–170. 10.1016/j.jmgm.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Schrödinger Release 2020–4: Canvas; Schrödinger, LLC: New York, NY, 2020. [Google Scholar]
- Sastry M.; Lowrie J. F.; Dixon S. L.; Sherman W. Large-scale systematic analysis of 2D fingerprint methods and parameters to improve virtual screening enrichments. J. Chem. Inf. Model. 2010, 50, 771–784. 10.1021/ci100062n. [DOI] [PubMed] [Google Scholar]
- Boonmuen N.; Gong P.; Ali Z.; Chittiboyina A. G.; Khan I.; Doerge D. R.; Helferich W. G.; Carlson K. E.; Martin T.; Piyachaturawat P.; Katzenellenbogen J. A.; Katzenellenbogen B. S. Licorice root components in dietary supplements are selective estrogen receptor modulators with a spectrum of estrogenic and anti-estrogenic activities. Steroids 2016, 105, 42–49. 10.1016/j.steroids.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Release 2020–4: Desmond Molecular Dynamics System, D. E.;Shaw Research: New York, NY, 2020. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2020. [Google Scholar]
- Pandey P.; Chatterjee S.; Berida T.; Doerksen R. J.; Roy S. Identification of potential non-nucleoside MraY inhibitors for tuberculosis chemotherapy using structure-based virtual screening. J. Biomol. Struct. Dyn. 2022, 4832. 10.1080/07391102.2020.1862705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.