Abstract
Recent progress in the field of nanophotonics has opened up novel avenues for developing nanomaterial-based biosensing systems, which can detect various disease-specific biomarkers, including long noncoding RNAs (lncRNAs) known to circulate in biological fluids. Herein, we designed and developed a nanophotonic approach for rapid and specific capture of lncRNAs using oligonucleotide-conjugated graphene quantum-dot-nanoconjugates. The method offers accurate identification of the target lncRNAs with high selectivity, despite the presence of other molecules in the given sample. The observations also pointed toward the high feasibility and simplicity of the method in the selective determination of lncRNAs. Overall, the approach has the potential of assessing lncRNA expression as a function of disease initiation and progression.
Introduction
Nanophotonics permits the development of cutting-edge nanotechnologies, which are precise, sensitive, and rapid in disease diagnosis, prognosis, and therapeutic intervention. The use of nanoparticles (NPs) with exceptional properties such as stability, high quantum yield, and sensibility offers the manipulation of light even outside the diffraction limit, where photons can sturdily pair with either plasmons or phonons.1 This has also opened up the possibilities to develop novel nanoframework-based point-of-care assays that are capable of performing multiplexing and identifying the disease-specific biomarkers even at trace levels. In this regard, the flexible surface characteristics and optical merits of carbon-based quantum dots, particularly graphene quantum dots (GQDs), have proved to be superior. The GQDs are zero-dimensional in nature and comprise small sheets of graphene with lateral sizes of less than 10 nm.2,3 Thus, apart from the properties of graphene, these NPs also exhibit amazing physicochemical properties that include a quantum confinement effect, edge effects, and a nonzero band gap.4−6 In comparison to QDs and other metallic NPs, GQDs offer steady fluorescence properties, improved biocompatibility, and minimal toxicity and thus are considered superior for various biological applications.7,8 The other remarkable optoelectric features of GQDs, such as the excitation-dependent photoluminescence emission, transfer and redox of photo-induced electrons, and size-tunable optical characteristics, further allow flexibility in the selection of the desired wavelength with minimal spectral overlapping. This has also widened the use of these NPs for various multiplexed, highthroughput diagnostic applications.9−11
Moreover, recent advances in the field strongly indicate that combining these fluorescence-based nanophotonic approaches with transcriptomics may act as a breakthrough element in point-of-care diagnostics. This combinatorial approach can provide a detailed idea of the alterations in the ongoing biological processes, a major cause of the development and progression of various noncommunicable diseases. The circulating transcriptome serves as an important source of various circulating biomarkers, which precisely include different classes of RNA (both noncoding and coding).12 Among the myriad of circulating RNA molecules (ncRNAs) found in biological fluids, long noncoding RNA molecules (lncRNAs) has gathered significant attention as promising biomarkers. Owing to their interaction with other nucleic acids, these biomolecules serve as flexible molecular scaffolds for the recruitment of vital transcription factors and act as the crucial regulators of ongoing cellular processes. The disturbances in the given set of the lncRNA profile can reflect the possibilities of the onset of a disease.13,14 Therefore, in the present work, we designed and developed a simplified, pragmatic, and reliable GQD-based nanophotonic approach for rapid and specific characterization of circulating lncRNAs directly in intricate biological samples.
Materials and Methods
Isolation and Differential Expression Analysis of lncRNAs
The isolation of circulating cell-free RNAs (ccf-RNAs) was performed by using QIAamp circulating nucleic acid isolation kit (Qiagen, Hilden, Germany) as per the suggested manufacturer’s instructions. After quantification of the eluted ccf-RNAs, cDNA was prepared by using the PrimeScript 1st strand cDNA synthesis kit from Takara Bio Inc. (Shiga, Japan). Thus, prepared cDNAs were ligated with appropriate primers (a plex of 11 lncRNAs, as mentioned in Table 1) procured from Integrated DNA Technologies, Coralville, Iowa, USA, for expression analysis. The assay was performed by using the qPCR master mix (Luna universal qPCR master mix, New England Biolabs, Ipswich, Massachusetts, USA) and suitable thermal cycling conditions (30 °C for 10 min, 42 °C for 60 min, 70 °C for 15 min) in Insta Q96 Real-Time PCR (Himedia Laboratories, Mumbai, MH, India). The obtained Ct values were analyzed to assess the expression patterns.
Table 1. Primers and Uracil-Modified Oligonucleotide Sequences Used for Profiling of lncRNAs.
| oligonucleotides and primers used in the present work | sequence (5′→3′) |
|---|---|
| Primer Sequences | |
| DLX6-AS1-F | AGTTTCTCTCTAGATTGCCTT |
| DLX6-AS1-R | ATTGACATGTTAGTGCCCTT |
| UCA1-F | ACGCTAACTGGCACCTTGTT |
| UCA1-R | TGGGGATTACTGGGGTAGGG |
| PVT1-F | CCCATTACGATTTCATCTC |
| PVT1-R | GTTCGTACTCATCTTATTCAA |
| HPRT1-F | AGCCTAAGATGAGAGTTC |
| HPRT1-R | CACAGAACTAGAACATTGATA |
| HOTAIR-F | CAGTGGGGAACTCTGACTCG |
| HOTAIR-R | GTGCCTGGTGCTCTCTTACC |
| SOX2OT-F | CCTCGTGGCTTAGGAGATTG |
| SOX2OT-R | CTGGCAAAGCATGAGGAACT |
| GAS-5-F | TATGGTGCTGGGTGCGGAT |
| GAS-5-R | CCAATGGCTTGAGTTAGGCTT |
| H-19-F | ATCGGTGCCTCAGCGTTCGG |
| H-19-R | CTGTCCTCGCCGTCACACCG |
| PICART1-F | AGGCAGCTACTGTAATAAT |
| PICART1-R | GTACCCTGGGCCTTTCTTAC |
| CHRF-F | AGATTCACATGGTATCCTGAAC |
| CHRF-R | TAGTCTGGCCACATTTTGTCTC |
| SNHG1-F | AGGCTGAAGTTACAGGTC |
| SNHG1-R | TTGGCTCCCAGTGTCTTA |
| Uracil-Modified Oligonucleotide Sequence | |
| DLX6-AS1 | AGUUUCUCUCUAGAUUGCCUUCUUCAUUUU |
| GAS-5 | UAUGGUGCUGGGUGCGGAUGCAGUGUGGCU |
| HOTAIR | GUGCCUGGUGCUCUCUUACCCUGUGUUUUC |
| SNHG-1 | AGGCUGAAGUUACAGGUCUGAGCAAAUAAG |
Preparation and Evaluation of Biotinylated Oligonucleotide Probes
The target lncRNA complementary uracil-modified oligonucleotide probes (DLX6, HOT-AIR, GAS-5, and SNHG1) were purchased from Integrated DNA Technologies (Coralville, Iowa, USA). The probes were attached with a biotin molecule at the 3′-position using the Pierce 3′-end biotinylation kit purchased from Thermo Fisher Scientific (Waltham, MA, USA). Concisely, uracil-modified oligonucleotides probes (50 pmol) were incubated for 5 min at 85 °C. After incubation, the probes were immediately cooled and mixed with the recommended volumes of 10X RNA ligation buffer, biotinylated cytidine (Bis) phosphate, RNase inhibitor, T4 RNA Ligase enzyme, and 30% PEG. The mixture was incubated overnight at 4 °C. Subsequently, to remove RNA ligase, nuclease-free water (NFW) followed by the chloroform/isoamyl alcohol (in 24:1 ratio) was added, vortexed, and centrifuged at high-speed. The obtained aqueous layer, ice-cold 100% ethanol, glycogen, and 5 M NaCl were mixed and incubated at −20 °C (1 h) for precipitation. After centrifugation, the pellet was carefully separated and washed with 70% ethanol. The obtained pellet was allowed to air-dry, resuspended in NFW (20 μL), and stored at −80 °C. The biotin molecule attached at the 3′-end of the oligonucleotide probe was confirmed by using a gel electrophoretic mobility assay. Briefly, the nonbiotinylated RNA samples and biotinylated oligonucleotide probes were mixed with 6× gel loading dye (HiMedia Laboratories, Mumbai, MH, India), 2 μL of SYBR Safe (Thermo Fisher Scientific, Waltham, MA, USA), and MOPS buffer, followed by loading in 1.2% agarose gel. The gel was subjected to 70 V electrophoretic conditions for 20 min in MOPS buffer, and the mobility was analyzed using a Gel Doc XR+ molecular imager (Bio-Rad, Hercules, California, USA).
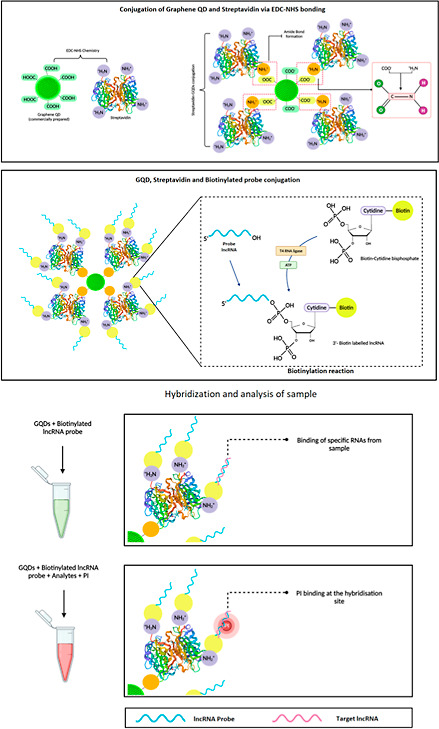
Preparation of the Streptavidin-Tethered GQD-Biotinylated Oligonucleotide Nanohybrid
The GQDs were purchased from Merck KGaA, Darmstadt, Germany. Prior to the nanohybrid preparation, GQDs were activated and attached with streptavidin using carbodiimide chemistry. The characterization of GQDs was performed by using Fourier transform infrared spectroscopy (Bruker Tensor-27 FTIR, Billerica, Massachusetts, US). For the activation, 50 μL of GQDs (0.5 M solution) were mixed with 50 μL of EDC (1 mg/mL solution) and 50 μL of NHS (1 mg/mL solution) in 250 μL 1× PBS. The mixture was incubated at room temperature (RT) for 25 min. This was subsequently followed by the addition of 0.1 μg of streptavidin and incubation for 15 min at RT. Following the incubation, a second aliquot of 50 μL of EDC and 50 μL of NHS was added to the tube containing the conjugate to increase the conjugation efficiency of the amine-carboxyl coupling reaction for 15 min at RT with stirring. The free GQDs were separated from streptavidin-coated GQDs by centrifugation at 6000 rpm for 15 min. After the supernatant removal, the pelleted streptavidin-coated GQD conjugates were stored at 4 °C until further use.
Following the attachment of streptavidin to the surface of GQDs, previously biotinylated oligonucleotides probes were conjugated using streptavidin–biotin coupling chemistry for the detection of target lncRNAs in each sample. For the conjugation reaction, streptavidin-tethered GQDs were added to 100 μL of 1× PBS (pH 7.4), vortexed, and mixed with the biotinylated probes in a ratio of 3:1, 1:1, and 1:3. The reaction mixture was then subjected to vortexing (1 h) at RT and incubated overnight at 4 °C. The biotinylated probes attached to the streptavidin moiety of GQDs would assist in the precise recognition of target lncRNAs in given samples. An electrophoretic mobility shift assay using was performed to confirm the conjugation between the oligonucleotide probes and GQDs. Briefly, the unconjugated GQDs and prepared nanohybrid (each 10 μL) were mixed with 1X TAE buffer and 2 μL of 6× gel loading dye and loaded in separate lanes of agarose gel (1%). The electrophoretic mobility was assessed at 70 V for 20 min in 1× TAE buffer, and the differential migration was analyzed by using Gel DocTM XR+ molecular imager (Bio-Rad, Hercules, California, USA). For the analysis of change in the fluorescence of GQD on attachment with streptavidin, 10 μL of unconjugated GQD and GQD-probe were analyzed by flow cytometry (Attune Nxt Flow Cytometer, Thermo Fisher, Massachusetts, USA) and fluorometry (Spark multimode microplate reader, TECAN, Seestrasse 103, Männedorf, Switzerland).
Targeted Detection of lncRNAs in Given Samples
The targeted detection of the lncRNAs by the developed nanohybrid structures was determined by flow cytometry (Attune Nxt Flow Cytometer, Thermo Fisher, Massachusetts, USA), followed by fluorometric confirmation using a Spark multimode microplate reader (TECAN, Seestrasse 103, Männedorf, Switzerland). The flow cytometric examination was performed for the assessment of the capability of detection of lncRNAs of interest by the developed nanohybrid structures in plasma samples. For the analysis, two sets of plasma samples (250 μL), one subjected to high-speed centrifugation (samples S1 and S2) and one set of samples (S3 and S4) subjected to filtration with 0.45μ filters, were added to 30 μL of GQD-biotin probe nanohybrid solution and 20 μL of propidium iodide (PI). The mixture was incubated for 3–5 min in ice-cold conditions, and evaluation was done in the BL2-H channel. Furthermore, we assessed the prepared nanoconjugates for the accurate determination of target lncRNAs (PICART, DLX6, SNHG, Gas5, H19, and SOX2OT) in isolated samples using fluorometry. For the analysis, 10 μL of nanoconjugates with PI was added to lncRNA samples, and readings were recorded. All the measurements were recorded at different concentrations (1X and 5X) in triplicate sets.
Specificity Assessment of the Nanoconjugates
The prepared conjugates were tested for their specificity to precisely identify the target lncRNA in a pool of mixed lncRNA species amplicons. For the specificity investigation, different lncRNA amplicons were pooled together. This pooled sample (1 μL) was mixed with 10 μL of nanoconjugates and analyzed by flow cytometry.
Applicability of the Method Using Biological Samples
The developed assay system was further evaluated to determine its applicability as a routine analytical procedure in intricate biological samples such as native plasma. This is essential to demonstrate the adaptability and practicality of the assay system for real-time and rapid diagnostic applications. The recognition of the lncRNAs in isolated plasma samples was quantitatively confirmed using fluorometry. For the investigation, 10 μL of GQD-probe with plasma samples and a nucleobase intercalating dye PI (10 μL) that result in the generation of a GQD-probe: ccf-RNAs: PI nanoarchitecture was evaluated with respect to the GQD-probe in NFW for the determination of fluorescence intensity measurements.
Results and Discussion
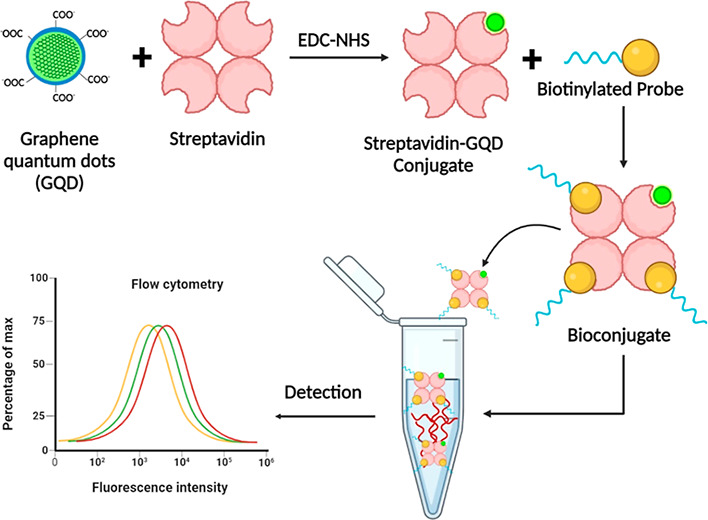
With the advent of the nanophotonic-based fluorescence methodologies, combinatorial approaches that involve fluorescently tagged oligonucleotide probes to target circulating biomolecules have attracted great attention.15−18 In the present study, we designed a system to detect circulating lncRNAs directly in the biological fluids using fluorescent semiconducting NPs. The method is simple to use and omits the requirement of additional sample processing and RNA isolation procedures. It mainly involves the conjugation of biotinylated probes with streptavidin-labeled GQDs that exhibit specificity toward the target molecule of interest, irrespective of the presence of other molecules. The combination of the high selectivity of biotinylated oligonucleotides probes with the optical attributes of the streptavidin-conjugated GQDs offers a precise nanohybrid-based approach for the detection of lncRNAs (Figure 1).
Figure 1.
Diagrammatic representation of the developed GQD-based assay for the selective detection of circulating lncRNAs in a given sample.
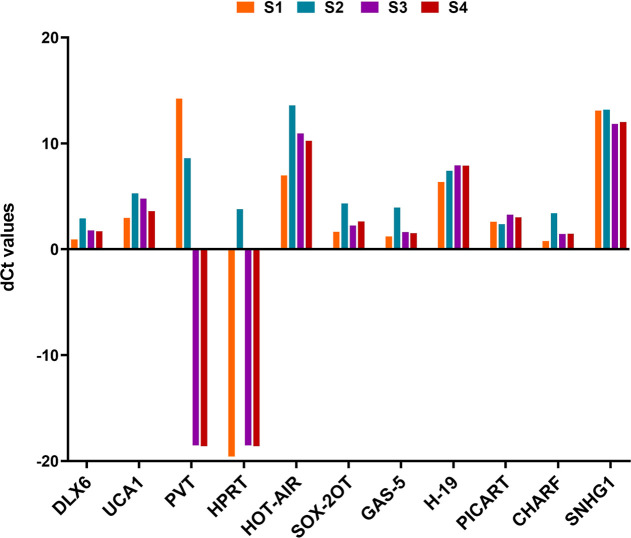
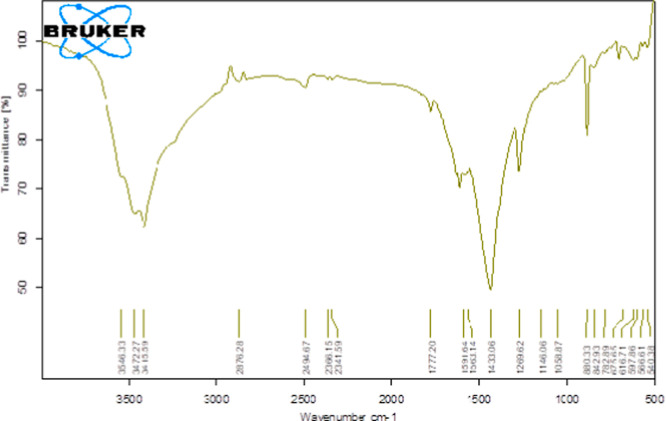
The lncRNAs are important regulatory RNA transcripts, known to circulate in different bodily fluids at differential levels in diseased and normal conditions.19−21 As lncRNAs can potentially serve as a diagnostic biomarker, rapid screening of these circulating biomolecules has a remarkable clinical importance. Owing to this, we developed a simplified nanophotonic approach for the rapid, sensitive, and specific detection of lncRNAs. Initially, we screened a panel of lncRNAs that have a profound role in several developmental and pathological processes (Figures 2, S1). The graph depicts the differential expression of panel of 11 lncRNAs, among which the highly expressive lncRNAs (such as HOTAIR and SNHG1) and lncRNAs with low expression patterns (DLX6 and GAS-5) were selected for the study. Following the selection of target molecules, a nanohybrid comprising streptavidin-coated GQDs and biotinylated oligonucleotide probes possessing sequence complementarity to the molecules of interest was prepared. The GQDs were used as they exhibit excellent optical properties, high quantum yield, and stability, which make them ideal candidates for such applications. Owing to the quantum confinement and edge effect among GQDs, these NPs possess rapid electron transport, which further boosts their use as a sensing material. In the present study, the FTIR spectrum of GQDs revealed −OH stretching vibrations at 3415 cm–1, C=C alkene stretching at 1591 cm–1, CH2 and CH3 bending at 1433 cm–1, and C–O of carboxylic group stretching at 1269 cm–1 (Figure 3).
Figure 2.
Graph depicting the aberrant expression patterns of a panel of 11 lncRNAs (n = 3) analyzed through quantitative real-time PCR.
Figure 3.
Figure showing GQD characterization performed through FTIR spectroscopy.
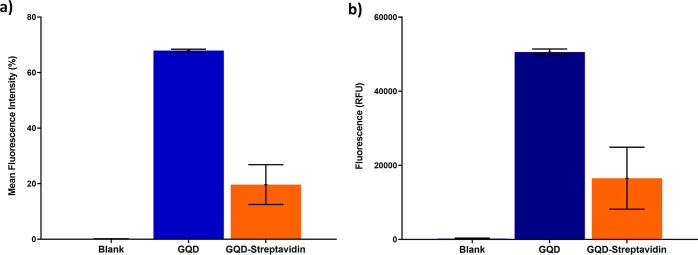
Furthermore, prior to the fabrication of the nanohybrid, the uracil-modified oligonucleotide probes were attached with biotin molecules at the 3′-end that provided a strong binding affinity for the streptavidin molecule. The biotinylation was confirmed by using the electrophoretic mobility shift assay, which showed constrained mobility of the biotinylated probes in comparison to the nonbiotinylated probes (Figure 4). This restriction in mobility is due to an increase in the size and protein content upon attachment of a biotin molecule at the 3′-end of the oligonucleotide probes. The biotinylated probes were then attached to the streptavidin-conjugated GQDs using the streptavidin–biotin coupling chemistry.22,23 Upon evaluation, we observed that the hybridization of the biotinylated probes with the streptavidin-conjugated GQDs results in a decrease in the fluorescence intensity of the GQDs (Figure 5a,b). This decrease in the fluorescence intensity is due to the phenomena of “blueshift” in the fluorescence of QDs. Since GQDs exhibit a quantum confinement effect, the attachment of streptavidin, a protein, has a potential influence on the size or the inner effectual coupling length or the sp2 domain size in the carbon cores, leading to a dramatic alteration in the confined wavelength and emission wavelength. Alteration in the electronic quantum-confined effects caused due to reduction of the effective GQD size upon conjugation with streptavidin, owing to energy gap alteration in the π-electron system of the carbon cores, might also result in the “blue” shift of fluorescence. The conjugation of proteins to the external GQD surface significantly changes the energy balance, which results in the generation of van der Waals forces. The additional charge forms a blocking electric field, leading to compression of the effective volume of QDs, which further increases upon attachment of biotinylated probes, as a result of which a variation in the emission light is observed.24−26
Figure 4.
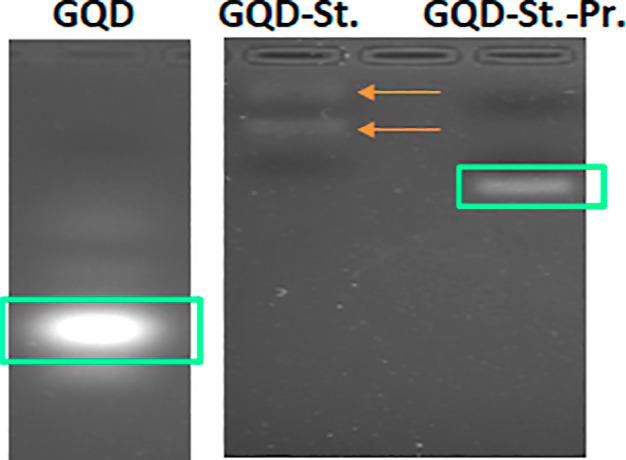
Gel electrophoretic mobility assay of the unconjugated GQDs, GQDs conjugated with streptavidin (GQD-St), and GQD-streptavidin-biotinylated probe nanohybrid (GQD-St.-Pr.), as observed in the gel imager.
Figure 5.
Graphs illustrating the comparison of the fluorescent measurements of GQD and GQD-streptavidin by (a) flow cytometry and (b) fluorometry. The values are shown as mean ± SE.
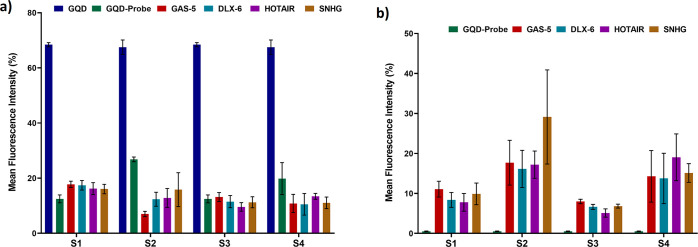
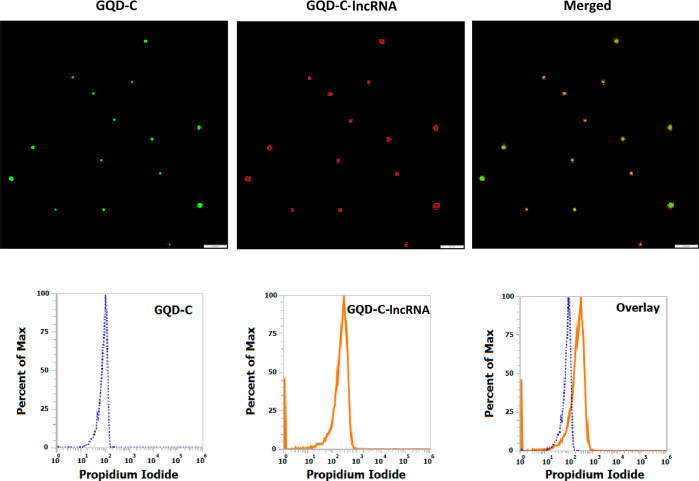
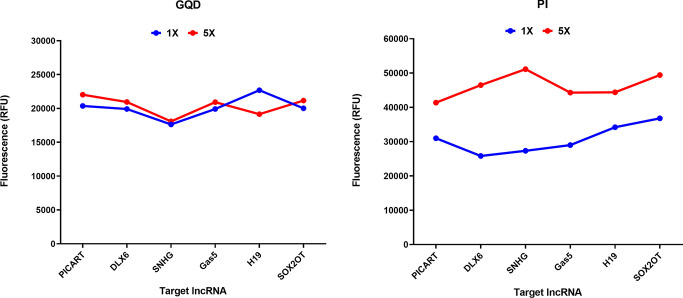
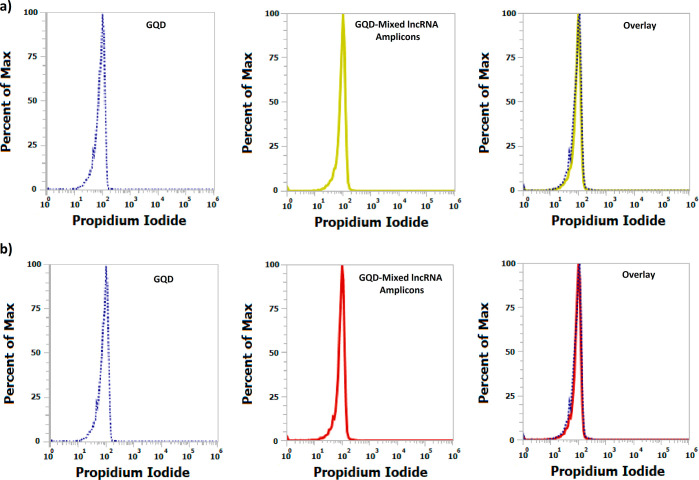
The system was further subjected to the assessment of the capability of selective detection of the molecules of interest in intricate plasma samples using flow cytometry. For the analysis, the nanohybrid solution comprising the GQD-biotinylated probes nanohybrid was incubated with plasma samples. The two sets of samples were prepared to obtain plasma devoid of any large extracellular vesicles, such as microvesicles, apoptotic, and necrotic bodies, that might influence the analysis procedure. The oligonucleotide probes in the nanohybrid framework possess sequence complementary to the target lncRNA molecules, allowing selective and precise binding with high specificity. In comparison to the unbound nanohybrids, the precise hybridization of the target lncRNAs resulted in a decrease in the fluorescence intensity of the lncRNA-bound nanohybrids (Figure 6a). However, as depicted in Figures 6b and 7, an increase in the fluorescence intensity of PI was observed, which can be correlated with the expression patterns or the circulating levels of lncRNAs. Moreover, an increase in the circulating levels should result in the increased formation of the nanohybrid:lncRNA complex and detection with the nanohybrid system. To assess this, the prepared nanoconjugates were added with two different concentrations of the lncRNA sample from 1× to 5×. The attachment resulting in the formation of the nanohybrid:lncRNA complex was determined as a function of the increase in fluorescence at higher concentrations using fluorometry. The green-red dual-fluorescent characteristics owing to PI and the GQDs were separately determined at two different wavelengths. An increase in the fluorescence was observed at a 5× concentration similar to that of PI, indicating the selectivity and sensitivity of the approach (Figure 8). In the specificity assay, upon addition of the mixed lncRNA amplicon pool (lacking the target lncRNA amplicon), no signals were observed. This absence of cross-reactivity among the probe and other lncRNAs confirmed the specificity of the developed nanohybrid toward its target lncRNA (Figure 9).
Figure 6.
Graph showing the change in the fluorescence intensity of the GQD—probe nanohybrids upon the precise hybridization of the target lncRNAs. (a) GQD fluorescence (b) PI. The values are observed by flow cytometry and expressed in terms of mean ± SE (n = 3) for GQD alone, GQD-probe nanoconjugate, and GQD-probe nanoconjugate with different lncRNAs (GAS5, DLX, HOTAIR, and SNHG).
Figure 7.
Representative image demonstrating the detection of lncRNAs using GQD—streptavidin-biotinylated probe nanohybrid and PI observed through fluorescent microscopy (upper panel) and flow cytometry (lower panel). The GQD-C: GQD-probe nanoconjugate and GQD-C: GQD-probe nanoconjugate with lncRNAs along with overlay images are shown.
Figure 8.
Graphs demonstrating the capability of the GQD—streptavidin-biotinylated probe nanohybrid in the detection of lncRNAs of interest at two different concentrations (1× and 5×). The values observed by fluorometry are shown in terms of relative fluorescence unit (RFU) for both GQD and PI.
Figure 9.
Representative image showing the detection of lncRNAs using GQD-streptavidin-biotinylated probe nanohybrid. Histogram analysis of (a) GQD: GQD-probe nanoconjugate and GQD: GQD-probe nanoconjugate (GAS5) with mixed lncRNA amplicons (DLX, HOTAIR, and SNHG) and (b) GQD: GQD-probe nanoconjugate and GQD: GQD-probe nanoconjugate (DLX) with mixed lncRNA amplicons (GAS5, HOTAIR, and SNHG) is shown.
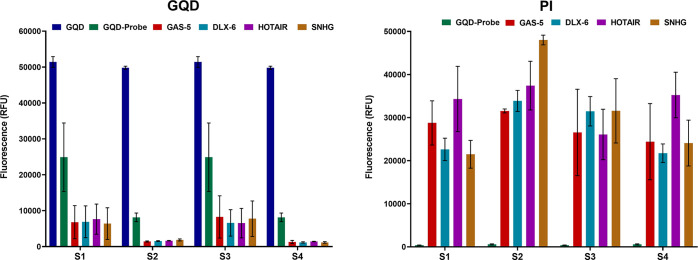
We further assessed the capability of the method to identify the target lncRNAs in the given biological samples. As depicted in Figure 10, a decrease in the fluorescence intensity of the nanohybrid was observed upon hybridization of the lncRNAs with the respective oligonucleotides in the nanohybrid structure. While a significant increment in the fluorescence intensity of PI was reflected upon attachment of the lncRNAs with the nanohybrid, which allowed efficient intercalation of PI in the double helical structure of the oligonucleotide probe: lncRNA in the nanoframework. The fluorometric measurements displayed a similar pattern of alteration in the RFU of both the nanoconjugates and PI, as demonstrated in the cytometric analysis, conferring the applicability of the approach as a routine analytical method for the detection of the lncRNAs in control and experimental settings.
Figure 10.
Graphs demonstrating the comparison of the ability of the GQD-streptavidin-biotinylated probe nanohybrid in the detection of lncRNAs of interest with respect to unconjugated GQDs and GQD-probe nanohybrids. The values are observed by fluorometry and expressed as mean ± SE (n = 3) for both GQD and PI.
Conclusions
The present research utilizes the exceptional photonic attributes of GQDs for the development of a simplified and rapid fluorescence-based approach for the precise recognition of circulating lncRNAs directly in a given clinical sample. The method efficiently performs the detection of target lncRNAs by sequence-specific biotinylated oligonucleotide probes conjugated to streptavidin-labeled GQDs. Overall, our results demonstrated that the developed assay system offers a rapid and feasible approach for the real-time analysis of lncRNAs in biological samples.
Translational Perspectives
In the modern scientific era of innovations, medical science is rapidly gravitating toward the nanoscale for making large-scale breakthroughs in disease management. Continuous attempts in this direction are being made to improve clinical precision, which includes the development of novel point-of-care tests for well-timed medical decisions in various life-threatening diseases. With exceptional features, the novel nanoframework systems can allow fast, precise, responsive, consistent, and cost-effective point-of-care detection of target bioanalytes, including circulating lncRNAs. As these biomolecules can act as a biomarker for various noncommunicable diseases, the development of a rapid and sensitive POC approach can assist in early disease diagnosis and therapeutic intervention. The GQDs, with their attributes superior to typical conventional fluorophores, have emerged as new players in high-performance multiplexed diagnostic applications. The GQD-based nanophotonic approach discussed herein is a feasible approach for the real-time characterization of lncRNAs in clinical settings and has a high translational value for the identification of altered biological processes. It is capable of performing rapid lncRNA detection that can help in taking timely decisions related to therapeutic intervention and patient monitoring. The approach has the potential to fulfill the features based on affordability, usability, sensitivity and selectivity, fast reaction time and robustness, and equipment-free and patient-specific delivery (ASSURED) and may contribute to patient-centered healthcare. Of course, after appropriate validation in the clinical setting, such POCT-based strategies are likely to expand significantly in the next decade.
Acknowledgments
The authors are thankful to the India Cancer Research Consortium (ICRC Task Force Project ID: 5/13/1/PKM/ICRC/2020/NCD-III), Department of Health Research (DHR), Ministry of Health and Family Welfare (MoHFW), Government of India, New Delhi, for the project funding support. A.B. and R.K. are recipients of the Post-Doctoral Research Fellowship and Senior Research Fellowship, respectively, from the Indian Council of Medical Research (ICMR), Government of India, New Delhi.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c02802.
Expression profiling of circulating lncRNAs (PICART, DLX, H19, SOX2OT, GAS-5, and SNHG) (PDF)
Author Contributions
R.S. and A.B. have contributed equally. P.K.M.: conceptualization, supervision, project administration, funding acquisition, writing-original draft preparation, final approval; R.S.: investigation, writing-original draft preparation, final approval; A.B.: investigation, writing-original draft preparation, final approval; P.R.: investigation, visualization, final approval; R.K.: investigation, visualization, final approval; R.T.: editing, final approval; P.C.: visualization, final approval.
The authors declare no competing financial interest.
Supplementary Material
References
- Mishra P. K.; Shandilya R. Nanophotonic biosensors as point-of-care tools for preventive health interventions. Nanomedicine 2020, 15, 1541–1544. 10.2217/nnm-2020-0162. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Hu T.; Liang R.; Wei M. Application of zero-dimensional nanomaterials in biosensing. Front. Chem. 2020, 8, 320. 10.3389/fchem.2020.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S.; Jha A.; Ahmad H.; Islam S. S. Avenue to Large-Scale Production of Graphene Quantum Dots from High-Purity Graphene Sheets Using Laboratory-Grade Graphite Electrodes. ACS Omega 2020, 5, 18831–18841. 10.1021/acsomega.0c01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P.; Tang L.; Teng K. S.; Lau S. P. Graphene quantum dots from chemistry to applications. Mater. Today Chem. 2018, 10, 221–258. 10.1016/j.mtchem.2018.09.007. [DOI] [Google Scholar]
- Lee S. Y.; Kwon M.; Raja I. S.; Molkenova A.; Han D. W.; Kim K. S. Graphene-Based Nanomaterials for Biomedical Imaging. Adv. Exp. Med. Biol. 2022, 1351, 125–148. 10.1007/978-981-16-4923-3_7. [DOI] [PubMed] [Google Scholar]
- Abbas A.; Tabish T. A.; Bull S. J.; Lim T. M.; Phan A. N. High yield synthesis of graphene quantum dots from biomass waste as a highly selective probe for Fe3+ sensing. Sci. Rep. 2020, 10, 21262. 10.1038/s41598-020-78070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffarkhah A.; Hosseini E.; Kamkar M.; Sehat A. A.; Dordanihaghighi S.; Allahbakhsh A.; van der Kuur C.; Arjmand M. Synthesis, Applications, and Prospects of Graphene Quantum Dots: A Comprehensive Review. Small 2022, 18, e2102683 10.1002/smll.202102683. [DOI] [PubMed] [Google Scholar]
- Huang H.; Su S.; Wu N.; Wan H.; Wan S.; Bi H.; Sun L. Graphene-Based Sensors for Human Health Monitoring. Front. Chem. 2019, 7, 399. 10.3389/fchem.2019.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldoni R.; Farronato M.; Connelly S. T.; Tartaglia G. M.; Yeo W. H. Recent advances in graphene-based nanobiosensors for salivary biomarker detection. Biosens. Bioelectron. 2021, 171, 112723. 10.1016/j.bios.2020.112723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Loh K. J.; Chiang W. H.; Manna K. Micro-patterned graphene-based sensing skins for human physiological monitoring. Nanotechnology 2018, 29, 105503. 10.1088/1361-6528/aaa709. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Huang X.; Song W. Physical and Chemical Sensors on the Basis of Laser-Induced Graphene: Mechanisms, Applications, and Perspectives. ACS Nano 2021, 15, 18708–18741. 10.1021/acsnano.1c05806. [DOI] [PubMed] [Google Scholar]
- Chauhan P.; Bhargava A.; Kumari R.; Ratre P.; Tiwari R.; Kumar Srivastava R. K.; Yu Goryacheva I.; Kumar Mishra P. K. Surface-enhanced Raman scattering biosensors for detection of oncomiRs in breast cancer. Drug Discov. Today 2022, 27, 2121–2136. 10.1016/j.drudis.2022.04.016. [DOI] [PubMed] [Google Scholar]
- Qian Y.; Shi L.; Luo Z. Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. 10.3389/fmed.2020.612393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia C. C. M.; Rodrigues L. F.; de Avila Pelozin B. R.; Oliveira E. M.; Fernandes T. Long Non-Coding RNAs in Cardiovascular Diseases: Potential Function as Biomarkers and Therapeutic Targets of Exercise Training. Noncoding RNAs 2021, 7, 65. 10.3390/ncrna7040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan S.; Jain S.; Bhargava A.; Shandilya R.; Srivastava R. K.; Mishra P. K. Lateral flow assay-based detection of long non-coding RNAs: A point-of-care platform for cancer diagnosis. J. Pharm. Biomed. Anal. 2021, 204, 114285. 10.1016/j.jpba.2021.114285. [DOI] [PubMed] [Google Scholar]
- Shandilya R.; Ranjan S.; Khare S.; Bhargava A.; Goryacheva I. Y.; Mishra P. K. Point-of-care diagnostics approaches for detection of lung cancer-associated circulating miRNAs. Drug Discov. Today 2021, 26, 1501–1509. 10.1016/j.drudis.2021.02.023. [DOI] [PubMed] [Google Scholar]
- Tabish T. A.; Scotton C. J.; J Ferguson D. C. J.; Lin L.; der Veen A. V.; Lowry S.; Ali M.; Jabeen F.; Ali M.; Winyard P. G.; Zhang S. Biocompatibility and toxicity of graphene quantum dots for potential application in photodynamic therapy. Nanomedicine 2018, 13, 1923–1937. 10.2217/nnm-2018-0018. [DOI] [PubMed] [Google Scholar]
- Shandilya R.; Bunkar N.; Kumari R.; Bhargava A.; Chaudhury K.; Goryacheva I. Y.; Mishra P. K. Immuno-cytometric detection of circulating cell free methylated DNA, post-translationally modified histones and micro RNAs using semi-conducting nanocrystals. Talanta 2021, 222, 121516. 10.1016/j.talanta.2020.121516. [DOI] [PubMed] [Google Scholar]
- Chen S.; Fang Y.; Sun L.; He R.; He B.; Zhang S. Long Non-Coding RNA: A Potential Strategy for the Diagnosis and Treatment of Colorectal Cancer. Front. Oncol. 2021, 11, 762752. 10.3389/fonc.2021.762752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z.; Jia J.; Yao L.; Li Z. Crosstalk of Exosomal Non-Coding RNAs in The Tumor Microenvironment: Novel Frontiers. Front. Immunol. 2022, 13, 900155. 10.3389/fimmu.2022.900155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandilya R.; Kumari R.; Singh R. D.; Chouksey A.; Bhargava A.; Goryacheva I. Y.; Mishra P. K. Gold based nano-photonic approach for point-of-care detection of circulating long non-coding RNAs. Nanomedicine 2021, 36, 102413. 10.1016/j.nano.2021.102413. [DOI] [PubMed] [Google Scholar]
- Avraham O.; Bayer E. A.; Livnah O. Crystal structure of afifavidin reveals common features of molecular assemblage in the bacterial dimeric avidins. FEBS J. 2018, 285, 4617–4630. 10.1111/febs.14685. [DOI] [PubMed] [Google Scholar]
- Gruber S.; Löf A.; Sedlak S. M.; Benoit M.; Gaub H. E.; Lipfert J. Designed anchoring geometries determine lifetimes of biotin-streptavidin bonds under constant load and enable ultra-stable coupling. Nanoscale 2020, 12, 21131–21137. 10.1039/d0nr03665j. [DOI] [PubMed] [Google Scholar]
- Kim S.; Hwang S. W.; Kim M. K.; Shin D. Y.; Shin D. H.; Kim C. O.; Yang S. B.; Park J. H.; Hwang E.; Choi S. H.; Ko G.; Sim S.; Sone C.; Choi H. J.; Bae S.; Hong B. H. Anomalous behaviors of visible luminescence from graphene quantum dots: interplay between size and shape. ACS Nano 2012, 6, 8203–8208. 10.1021/nn302878r. [DOI] [PubMed] [Google Scholar]
- Jeong S.; Pinals R. L.; Dharmadhikari B.; Song H.; Kalluri A.; Debnath D.; Wu Q.; Ham M. H.; Patra P.; Landry M. P. Graphene Quantum Dot Oxidation Governs Noncovalent Biopolymer Adsorption. Sci. Rep. 2020, 10, 7074. 10.1038/s41598-020-63769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H.; Li X. H.; Chen X. B.; Wei J. S.; Li X. B.; Xiong H. M. Surface states of carbon dots and their influences on luminescence. J. Appl. Phys. 2020, 127, 231101. 10.1063/1.5143819. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.