Abstract
Abnormal cerebral accumulation of amyloid-beta peptide (Aβ) is a major hallmark of Alzheimer’s disease. Non-invasive monitoring of Aβ deposits enables assessing the disease burden in patients and animal models mimicking aspects of the human disease as well as evaluating the efficacy of Aβ-modulating therapies. Previous in vivo assessments of plaque load have been predominantly based on macroscopic fluorescence reflectance imaging (FRI) and confocal or two-photon microscopy using Aβ-specific imaging agents. However, the former method lacks depth resolution, whereas the latter is restricted by the limited field of view preventing a full coverage of the large brain region. Here, we utilized a fluorescence molecular tomography (FMT)-magnetic resonance imaging (MRI) pipeline with the curcumin derivative fluorescent probe CRANAD-2 to achieve full 3D brain coverage for detecting Aβ accumulation in the arcAβ mouse model of cerebral amyloidosis. A homebuilt FMT system was used for data acquisition, whereas a customized software platform enabled the integration of MRI-derived anatomical information as prior information for FMT image reconstruction. The results obtained from the FMT-MRI study were compared to those from conventional planar FRI recorded under similar physiological conditions, yielding comparable time courses of the fluorescence intensity following intravenous injection of CRANAD-2 in a region-of-interest comprising the brain. In conclusion, we have demonstrated the feasibility of visualizing Aβ deposition in 3D using a multimodal FMT-MRI strategy. This hybrid imaging method provides complementary anatomical, physiological and molecular information, thereby enabling the detailed characterization of the disease status in arcAβ mouse models, which can also facilitate monitoring the efficacy of putative treatments targeting Aβ.
1. Introduction
The abnormal accumulation and spread of amyloid-beta (Aβ) deposits play a central role in the pathogenesis of Alzheimer’s disease (AD), leading to neurotoxicity, inflammation and synaptic dysfunction [1–3]. Aβ assembles into Aβ oligomers, Aβ fibrils and Aβ plaques [4] in a spontaneous manner. In preclinical research, in vivo Aβ detection and longitudinal monitoring in animal models of AD amyloidosis provided a handle for studying the effect of targeted molecular interventions on this molecular phenotype, thereby providing indirect insights into the mechanism underlying Aβ deposition and potential biomarkers for assessing the effects of Aβ-modulating therapeutics [5].
Detection of amyloid deposits has been made possible by using imaging tools covering different ranges of spatial resolution and fields-of-view (FOV) [6]. At a macroscopic level, positron emission tomography (PET) using various amyloid tracers, e.g., [11C]PiB [7], [18F]florbetaben [8] and [18F]florbetapir [9], has shown higher cortical fibrillar Aβ loads in patients with AD continuum compared to cognitive normal controls [10, 11]. In general, PET has been promoted as an important clinical tool for the early and differential diagnosis of AD [12]. Imaging of Aβ deposits at a mesoscopic resolution has also been demonstrated using 19F- and 1H-magnetic resonance imaging (MRI) with or without contrast agents [13,14] or using near-infrared fluorescence (NIRF) imaging in combination with fluorescence probes [15]. The development of Aβ-specific fluorescent probes such as AOI987 [16], CRANAD-2/-3 [17,18], and luminescent conjugated oligothiophenes [19–21] using methoxy-X04, BTA-1 and PiB [22–25] enabled the application of various optical imaging methods in preclinical studies. Optical detection of Aβ deposits by using ex vivo optical projection tomography [26], light-sheet microscopy [27] and various fluorescence microscopy methods, including confocal and multiphoton microscopy, enabled monitoring of Aβ at m spatial resolution [28]. However, these conventional imaging techniques are mostly hampered by a limited FOV due to strong light scattering in tissues. In addition, the cranial opening operation, which is commonly used in many microscopic technologies to expose the brain surface and reduce tissue scattering, may affect the physiological condition of the brain.
In contrast to the aforementioned optical imaging methods, fluorescence molecular tomography (FMT) is a truly non-invasive alternative that covers a large FOV comparable to traditional fluorescence reflectance imaging (FRI) and resolves the 3D fluorophore distribution in tissue [29]. More specifically, FMT belongs to a subclass of diffuse optical imaging modalities that adopt near-infrared light for exciting the fluorescence probe and utilize a model-based reconstruction algorithm to recover the 3D distribution of the fluorophore [30]. In the last decade, FMT has been applied widely in preclinical settings focusing predominantly on early tumor detection and monitoring [31,32]. In the study of AD, a multimodal imaging method that combines FMT and X-ray computed tomography (CT) has been applied to map the distribution of Aβ plaques employing the oxazine derivative AO1987 probe in a static manner [33]. The multimodal strategy accounts for the loss of structural information inherent to most molecular imaging techniques, including FMT and PET. Registration of molecular imaging data to anatomical reference image recorders under identical conditions is of value for predefined volume-of-interest (VOI)-based quantitative data analysis. Compared with FMT-CT, the combination of FMT and MRI has several advantages: MRI provides better image contrast on different soft tissue types, especially for the brain. In addition, both clinical and preclinical high-field MRI scanners can offer versatile physiological (e.g., regional cerebral blood flow, blood-brain barrier integrity, cerebral oxygenation), functional (e.g., functional neuronal networks) and structural information (e.g., white matter integrity, brain atrophy). Thus, the FMT-MRI combination allows the spatial linkage between functional/structural (MRI) and molecular information (FMT). FMT-MRI has been applied to study aspects of tumor vascularization in a mouse model [34] and longitudinal monitoring of ex vivo biological samples [35]. To the best of our knowledge, the hybrid approach has not yet been applied for studying cerebral pathologies such as cerebral amyloidosis.
In the present study, we demonstrated whole-brain mapping of Aβ deposits at mm resolution in arcAβ mouse models of cerebral amyloidosis, as observed in AD [36], using the Aβ fibril binding curcumin derivative CRANAD-2 as a fluorescent reporter. The schematic design of the multimodal FMT-MRI experimental pipeline is illustrated in Fig. 1(a). The in vivo imaging results were validated by immunofluorescence staining using CRANAD-2 with the anti-Aβ antibody 6E10 on mouse brain sections.
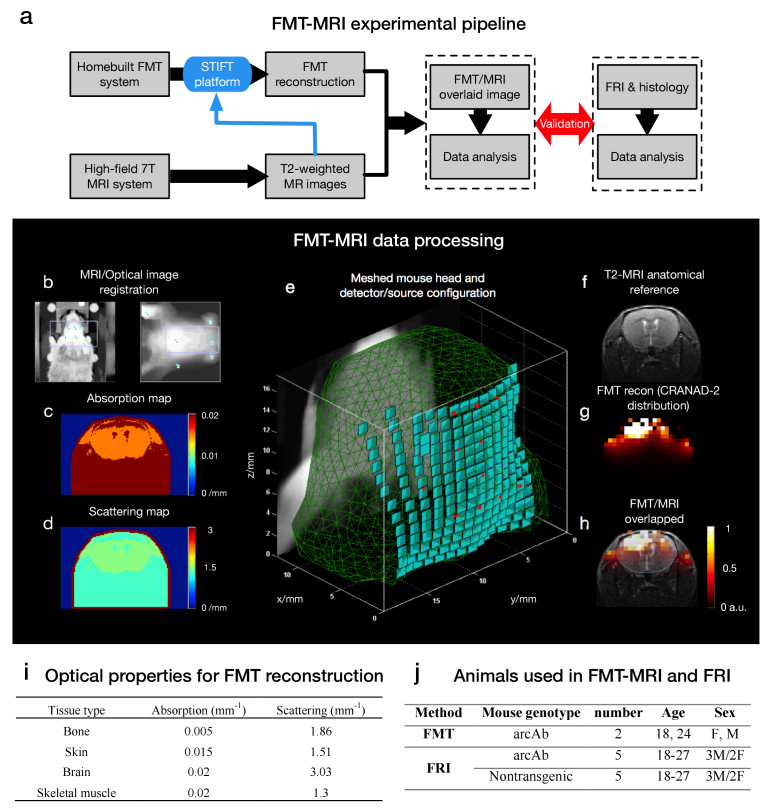
Fig. 1.
The multimodal FMT-MRI experimental pipeline and data processing framework are illustrated in the upper (a) and middle (b-h) panels, respectively. Regarding the framework of FMT-MRI data processing, the topological surface of anatomical MRI (b, right) was first registered with the optical image (b, left) with fiducial markers indicated on both images (blue dot). Once the two types of images were registered, both an absorption map (c) and a scattering map (d) were generated according to the intensity-based segmentation. (e) MRI-derived structural information was used to reconstruct the object in 3D, and adaptive meshing was performed with detector elements (green patches) and laser illumination points (red spots) assigned to the surface of the generated mesh. (f) T2-weighted MR image (f) provides an important anatomical reference that can significantly improve the localization of the FMT signal (g). (h) shows an overlaid FMT-MRI image containing both molecular and structural information within the same mouse. (i) The absorption and scattering coefficients of four typical tissue categories (bone, skin, brain and skeletal muscle) were used for FMT reconstruction. (j) Summary of animals used in FMT-MRI and FRI experiments.
2. Materials and methods
2.1. Animal models
Five transgenic arcAβ mice overexpressing the human APP695 transgene containing the Swedish (K670N/M671L) and Arctic (E693G) mutations under the control of the prion protein promoter and five age-matched non-transgenic littermates of both sexes were used in this study (18-27 months of age) [36–39]. Animals were housed in ventilated cages inside a temperature-controlled room and under a 12-hour dark/light cycle. Pelleted food (3437PXL15, CARGILL) and water were provided ad libitum. In total, 2 arcAβ mice were used for FMT (including one mouse died after 15 min after the experiment); arcAβ and 5 nontransgenic mice of both sexes were used for FRI (age: 18-27 month) (Fig. 1(j)). All experiments were performed in accordance with the Swiss Federal Act on Animal Protection and were approved by the Cantonal Veterinary Office Zurich (permit number: ZH018/14). All procedures fulfilled the ARRIVE guidelines on reporting animal experiments [40].
2.2. Preparation of animals and fluorescence probe
Briefly, mice were first anesthetized with an initial dose of 4% isoflurane (Abbott, Cham, Switzerland) in an oxygen/air mixture (200/800 mL/min), and anesthesia was maintained at 1.5% isoflurane in oxygen/air (100/400 mL/min). The fur over the head was then removed. The mice were placed in the prone position on a heating pad that maintained a constant body temperature. Regarding the fluorescent reporter, we used the Aβ-specific curcumin derivative CRANAD-2 (Sigma, Switzerland) [17] dissolved in a buffer of 20% dimethyl sulfoxide/polyethylene glycol (DMSO/PEG, Sigma, Switzerland) for in vivo imaging of amyloid aggregates in arcAβ mice. CRANAD-2 is a NIR Aβ beta-sheet binding fluorescent probe, with affinity (Kd) of 38 nM [17] and has been used to noninvasively detect Aβ deposits in in vivo NIRF imaging in transgenic mouse models of amyloidosis [41]. The spectrum of CRANAD-2 shows an absorption maximum at 640 nm and an emission maximum at 715 nm upon interacting with Aβ aggregates, undergoing a 90 nm blueshift from 805 nm in the absence of Aβ [17]. Moreover, the target interaction has been reported to result in a large increase in the quantum yield.
2.3. Magnetic resonance imaging
MRI scans were performed on a 7 T small animal MRI BioSpec scanner (Bruker Biospin GmbH, Ettlingen, Germany) with a magnet bore diameter of 16 cm and equipped with an actively shielded gradient capable of switching 760 mT/m with an 80-μs rise time and operated by the ParaVision 6.0 software platform (Bruker Biospin GmbH, Ettlingen, Germany). A circular polarized volume resonator was used for signal transmission, and an actively decoupled mouse brain quadrature surface coil with an integrated combiner and preamplifier was used for signal reception. To obtain the anatomical reference with high soft tissue contrast, in vivo T2-weighted MR images of the mouse brain/head were recorded using a spin echo sequence using the following parameters: FOV 20 × 20 mm2, number of slices 35, dimension of reconstructed matrix 256 × 256 × 35, slice thickness 0.5 mm, repetition time 4200 ms, echo time 33 ms, and flip angle α 90 degree within a scan time 12 min 36 s.
2.4. Fluorescence molecular tomography
A homebuilt non-contact FMT system was used for data acquisition [42]. The system is equipped with a 16-bit charge-coupled device (CCD) camera (ANDOR, Belfast, Northern Ireland), a galvanometric driven mirror system (ScanLab, Puchheim, Germany), a continuous-wave (CW) solid-state laser generator with 670 nm wavelength for illumination (B&W Tek, Newark, USA), and custom-made animal support. The camera was cooled to -79 degrees for noise reduction. Before brain imaging, the fur overlying the head was first trimmed with an electrical shaver and then removed using a depilation cream (Nair, NJ, USA). The tail vein was then cannulated with a 30-gauge needle for subsequent intravenous injection with 2 mg/kg CRANAD-2 solution in 20% DMSO/PEG buffer. For excitation of the fluorescent probe, the CW laser power was set to 20 mW. The irradiation spot has a diameter of 0.5 mm with an adjacent distance of 1 mm. The illumination pattern was designed as a 7 × 7 grid covering the mouse head. Bandpass filters with central frequencies of 660 ± 13 nm and 720 ± 13 nm for excitation and emission procedures, respectively, were used. Measurements were carried out prior to injection, and 5 min, 10 min, 20 min, and 30 min after injection of CRANAD-2. To ensure continuous recording of changes in the fluorescence signal, only one excitation image and one white-light image were taken prior to label injection, while emission images were taken at the four time points as indicated before. Each measurement of FMT took 4-5 minutes involving image acquisition at both excitation and fluorescence wavelengths. The FMT reconstruction was performed offline via STIFT toolkit [43] costing less than 3 minutes including forward modeling and inversion (MATLAB R2019b, MathWorks, US; Intel Core i7@ 2.60 GHz, RAM 8.00 GB).
2.5. FMT-MRI data processing
We used our customized reconstruction platform to perform FMT-MRI image reconstruction [43]. The schematics of FMT-MRI data processing are shown schematically in Fig. 1. First. The T2-weighted MR images were used as the input, and the white-light image was registered to the topological MRI map, which represents the mouse surface, using a landmark-based registration method (Fig. 1(b)). With common coordinates established, a self-adaptive mesh was generated based on the MRI anatomy [44]. The values of absorption and scattering coefficients were also generated according to the intensity-based segmentation of the MR images using published values of optical properties for different biological tissue types [45]. For simplicity, only four tissue categories were considered (Fig. 1(i)). The refractive index n in tissue is typically ranging from 1.33 to 1.5 depending on the content of water [45]. For simplicity, the refractive index was set uniformly to n = 1.4. The virtual detector plane consisting of individual elements with an area of 1×1 mm2 was assigned to the surface of the mesh. The diameter of each laser point was set to 0.5 mm. To merge the FMT-MRI images and analyze the results, a linear interpolation scheme was used to transform from the mesh coordinates used for reconstructing the dye distribution to Cartesian grid coordinates of the anatomical reference image. A Tikhonov regularization term was introduced to the reconstruction, and the inverse problem was solved using the conjugation gradient (CG) method [42,43]. The regularization parameter λ was set to 10−4. The maximum iterations for CG convergence was set to 200, with an error tolerance of 10−9. Quantitative analysis was carried out following FMT reconstruction. The registered MR images provided an anatomical reference for defining 3D VOIs using the Allen Brain Atlas as Ref. [46]. We analyzed the temporal variation in the cortical and subcortical regions. The average signal intensities within these two VOIs were calculated pre-injection and 5 min, 10 min, 20 min, and 30 min after injection. The uptake curves of CRANAD-2 in cortical and subcortical regions are depicted.
2.6. In vitro fluorescence reflectance imaging
FRI, also referring to near-infrared fluorescence imaging, was performed for comparison with the results obtained from FMT. We used the Maestro 500 multispectral imaging system (CRi, MA, USA) [47]. Briefly, the imager was equipped with a bandpass filter (615 to 665 nm) for excitation. The fluorescence signal was detected by a CCD camera mounted on the top of the imaging chamber. Aβ1-42 fibrils were formed as described previously by incubating a solution of 2 μM recombinant Aβ1-42 expressed in E.coli in phosphate buffer (20 mM sodium phosphate, 0.2 mM ethylenediaminetetra-acetic acid, pH 8.0) [41]. FRI of CRANAD-2 and Aβ1-42 fibril binding using 2 μM CRANAD-2 (50 μl), 2 μM CRANAD-2 (25 μl) + 2 μM Aβ1-42 fibril (25 μl) solutions (PBS buffer) was performed at 10 min after mixing. We used the aforementioned Maestro 500 multispectral imaging system with the same bandpass filter (615 to 665 nm) for excitation. Images were acquired by increasing the emission filter central wavelengths by 10-nm increments. The recorded Image cubes were spectrally unmixed using Maestro software (CRi, MA, USA).
2.7. Immunofluorescence staining and confocal microscopy
For staining and immunofluorescence investigations, mice were perfused with 1 × phosphate buffered saline (PBS, pH 7.4) under ketamine/xylazine/acepromazine maleate anesthesia (75/10/2 mg/kg body weight, bolus injection) and decapitated. Brains were removed from the skull afterwards and fixed in 4% paraformaldehyde in 1 × PBS (pH 7.4). Brain hemispheres were embedded in paraffin following routine procedures and cut into 5 μm horizontal sections. Immunofluorescence staining using donkey-anti-rat Alexa488 (Jackson, AB-2340686, 1:400), anti-Aβ1−16 antibody 6E10 (Signet Lab, SIG-39320, 1:5000), thioflavin S, and CRANAD-2 was performed following a protocol described earlier with nuclei counterstained by 4’ 6-diamidino-2-phenylindole (DAPI, Sigma, D9542, 1:1000). To further assess the co-localization of different channels, confocal images of arcAβ mice and non-transgenic littermates were further obtained at ×20 magnification in the cortex and hippocampal areas using a Leica SP8 confocal microscope (Leica Microsystems GmbH, Germany) at ScopeM ETH Zurich. Sequential images were obtained by using 405 nm, 488 nm, and 647 nm lines. Identical resolution settings were used for the Z stacks (n = 15). The Allen brain atlas was used for anatomical Ref. [46].
3. Results
3.1. FMT reconstruction results and VOI analysis
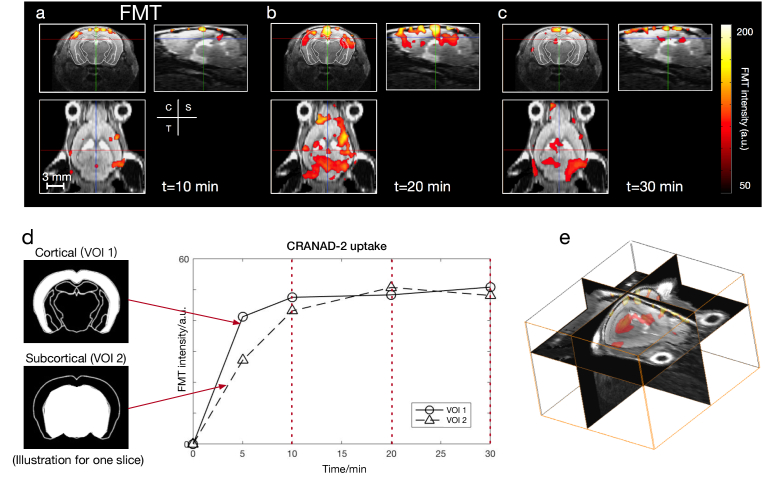
A mesh containing 7065 nodes, 42021 tetrahedral elements and 1910 surface elements was generated for forward modeling, leading to a recovered fluorescence distribution stored in a Cartesian grid of 20 × 31 × 28. Overlays of FMT and MRI data are shown for the three perpendicular planes (Fig. 2). The structural information is clearly depicted by the T2-weighted MR images, which helps localizing the fluorescence signal significantly. The displayed FMT signal indicates the progressive accumulation of the CRANAD-2 probe (by subtracting the baseline fluorescence intensity at t = 0 min) in the brain of the arcAβ mouse at t = 10, 20, and 30 min following the injection of the tracer (Fig. 2(a), (b), (c)). At all time points, the CRANAD-2 probe had a higher concentration in the cortex region (Fig. 2(d), upper left) than in the subcortical region (Fig. 2(d), lower left) in a diffuse manner. VOI analysis in the cortical region showed that the uptake of CRANAD-2 increased continuously for the first 20 minutes. For the subcortical region (dashed line, Fig. 2(d)), the concentration of CRANAD-2 dropped after 20 minutes (Fig. 2(d)).
Fig. 2.
FMT reconstruction for visualizing CRANAD-2 distribution within the brain of one representative arcAβ mouse. FMT reconstruction shows variations at time points at t = 10, 20, and 30 minutes (a, b, c). VOIs at cortical and subcortical regions are indicated by two images of different masks in (d). The temporal variation in CRANAD-2 in the two VOIs is shown in (d). For the cortex (solid line), the uptake of CRANAD-2 increased for the first 20 minutes. For the subcortical region (dashed line), the concentration of CRANAD-2 dropped after 20 minutes. (e) 3D visualization of FMT-MRI (t = 30 min).
3.2. Validation through dynamic FRI
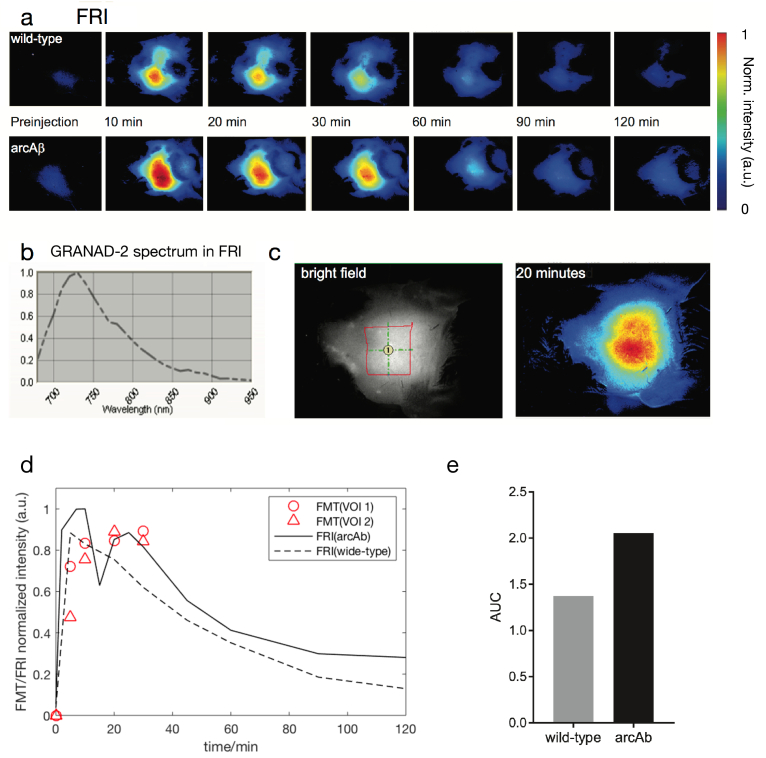
FMT results were compared to data from two-dimensional FRI performed on another mouse under identical physiological conditions. The protocol of CRANAD-2 probe administration was identical to that used in the FMT experiment. Planar fluorescence images of a wide-type mouse and an arcAβ mouse were recorded post- i.v. injection of CRANAD-2 at the same 4 time points as those in FMT measurement and subsequently every 10 min in a duration of 120 min. The region-of-interest (ROI) analysis of the dynamic CRANAD-2 uptake allowed differentiation of arcAβ mice from non-transgenic littermates (NTL) as shown in Fig. 3. The maximum FRI intensity was observed at t 10 min for both mice (Fig. 3(d)). However, FRI was not capable of disentangling the fluorescence distribution in the subcortical region due to the 2D nature of the imaging modality. The time course obtained from FRI showed a similar pattern as that detected by using FMT, except that the FMT result indicates a slower intensity variation during t = 10 to 20 min (Fig. 3(d)). Higher AUC was observed in arcAβ mice compared to NTL (Fig. 3(e)).
Fig. 3.
Time course analysis in fluorescence reflectance imaging (FRI) (a) Representative horizontal map of an NTL (upper row) and an arcAβ mouse (lower row) preinjection to 120 minutes after CRANAD-2 injection; (b) Spectra detected in the brains of arcAβ mice after injection; (c) White-light (left) and NIRF (right) images of one arcAβ mouse head with epilated skin over the head. Region of interest = 41 cm2; (d) Time course of NIRF signal in the brain of arcAβ and age-matched non-transgenic littermates; (e) Area under curve (AUC) of the cortical regions of arcAβ (n = 4) and NTL (n = 4) mice.
3.3. Ex vivo staining on mouse brain sections and in vitro FRI
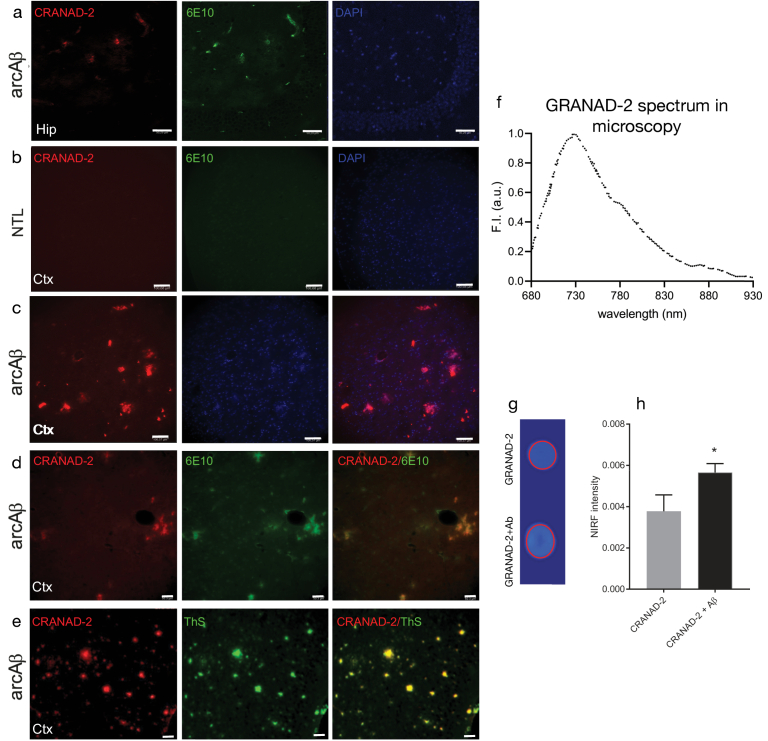
To confirm the specificity of CRANDA-2 binding to Aβ deposits in the mouse brain, horizontal brain tissue sections from arcAβ mice and non-transgenic littermates were stained with CRANAD-2 in addition to thioflavin S and anti-Aβ antibodies 6E10 [65] and were nuclear counterstained with DAPI (Fig. 4(a)-(d)). CRANAD-2 clearly co-stained with 6E10 and thioflavin S-stained parenchymal and vessel-associated Aβ deposits in the arcAβ mouse cortex and hippocampus.
Fig. 4.
Validation of CRANAD-2 binding to Aβ deposits. (a-e) CRANAD-2 staining in brain tissue sections from arcAβ mouse and non-transgenic mouse. Sections were stained with CRANAD-2 (red), DAPI (blue) and anti-Aβ1-16 antibody 6E10 (green) in the hippocampus and cortex of arcAβ mice at ×20 magnification. Scale bar = 100 m (b, c) and 50 m (a, d, e). No clear signal was detected in the cortex from non-transgenic littermate. (f) The emission spectrum of CRANAD-2 upon binding with Aβ1-42. (h) Comparison of NIRF signals with and without Aβ1-42 for ROI indicated on (g) NIRF imaging of probe on a slide wrapped with black tap, scale (a.u.) 0-1; Increased fluorescence when CRANAD-2 is incubated in the presence of Aβ fibrils. Hip, hippocampus; Ctx, cortex; ThS, Thioflavin S.
The binding and spectrum of the probe were further verified in vitro by assessing solutions of CRANAD-2 and Aβ1-42 fibrils with FRI (Fig. 4(f)-(h)). Enhanced signal was observed with FRI imaging in solutions of CRANAD-2 in the presence of Aβ42 fibril compared to CRANAD-2 only. Peak of the spectrum in the confocal microscopy is identified at 720 nm.
3. Discussion and conclusion
In this work, a sequential multimodal strategy combining FMT and MRI was applied to image the amyloid-fibril binding fluorescent probe in the brains of aged arcAβ transgenic mice, which displayed a high load of cerebral amyloid plaques. The NIRF probe CRANAD-2 shows high-affinity binding and a redshift for Aβ aggregates, and displays an increase in quantum yield upon binding with Aβ, thereby enhancing the overall signal-to-noise ratio (SNR). After data acquisition, we allocate the fluorescent signal to the registered high-resolution structural MRI reference. Furthermore, our FMT reconstruction incorporated a priori knowledge obtained by MRI to increase the accuracy of reconstruction [43]. In the end, quantitative VOI analysis of CRANAD-2 distribution was performed and guided by the MRI anatomical reference. Similar uptake curves were observed by using in vivo CRANAD-2 imaging with FMT and FRI, a routinely used planar imaging method.
Imaging Aβ plaque distribution in the mouse brain using FMT is a challenging problem, as we have to consider complex anatomical structures such as the scalp and skull during reconstruction. The problem is further complicated by the fact that we do not consider focal brain lesions, e.g., in a glioma model [29], but rather a plaque distribution that is diffuse across cortical and, to some extent, subcortical structures. Thus, a low target-to-background ratio for the distribution of fluorescent dye across large brain areas is expected. Time-dependent binding of the Aβ-specific fluorescent probe CRANAD-2 was monitored in both cortical and subcortical regions over 30 minutes pre- and post-injection. The average intensity in the cortical region increased continuously, while the intensity in subcortical structures decreased at approximately t = 20 minutes after probe administration. This might be explained by the fact that amyloid-targeted CRANAD-2 accumulated in the cortex, whereas it was gradually washed out from regions not displaying the target, i.e., aggregated beta-amyloid proteins. The differences in FMT and FRI uptake curves can be explained by the fact that FRI lacks depth information. Hence the fluorescence signals emitted from most cranial compartments including the skull and scalp are summed, leading to a heavily surface-weighted signal. In contrast, volumetric FMT imaging is capable of mapping the CRANAD-2 uptake in the MRI-guided anatomy more accurately in 3D. Therefore, FRI mainly reflects a mixture effect of wash-out signals from different types of tissues, but we believe FMT represents a more realistic cerebral kinetics. Notably, a similar slow wash-out curve of the same CRANAD-2 probe was also been observed in a optoacoustic tomography study where 3D rendering of probe kinetics was also implemented similar to FMT [41].
The combination of FMT and MRI appears more attractive than FMT-CT for brain applications, given the high versatility of MRI and its superior soft tissue contrast compared to CT [48]. An early study allowed the differentiation of fibroadenoma in the breast of female patients from normal breast tissue based on differences in local perfusion/vascular permeability using concurrent contrast-enhanced MRI with a gadolinium (Gd) contrast agent and diffuse optical tomography with administration indocyanine green [49]. Similarly, dual-tracer MRI-guided fluorescence tomography was used to study the uptake kinetics of fluorescence dyes, which allowed estimation of the epidermal growth factor receptor density [50]. We have previously developed a truly hybrid system which allows simultaneous FMT recording inside a 9.4T MRI scanner [51] and applied it for investigating tumor vascularity in vivo [34]. Nevertheless, In this work, we used separate imaging systems, i.e., a standalone FMT system and an MRI scanner. The main novelty in this work is to depict the fluorescence-labeled amyloid-beta deposits in brain in a long time duration. Current FMT technology is limited by its imaging speed. In our setup, 4-5 minutes are required to achieve a complete dataset that can be used as the input of 3D FMT reconstruction, which is not comparable with the imaging speed of DCE-MRI. In addition, the standalone FMT system is equipped with a -79 degree-cooled camera, providing high-SNR raw fluorescence images for reconstruction.
Registration between MRI and optical images, or more specifically, planar white-light images, serves as the first step in the FMT-MRI data processing protocol. The goal of this step is to align the MRI coordinates in the same way as optical images; thus, the MRI anatomical reference can be used in light propagation modeling and subsequent FMT image reconstruction. Herein, we used a landmark-based registration method by using identifiable anatomical structures, such as ear tips, nose tips, and eyes, that are clearly visible on both topological MRI and white-light images. To improve the registration accuracy, artificial landmarks with sufficient contrast in MRI and optics can also be introduced. In all these aforementioned studies, post-processing was rather time-consuming, as the whole procedure included registration of MR and optical images, sophisticated reconstruction using priors from MR structural images, and 3D visualization of the fluorophore distribution overlaid on the MRI images. In the present work, we successfully applied our customized software platform to efficiently tackle these steps. Our STIFT platform has the potential to further enhance the automation of FMT data acquisition by setting optimal parameters, such as the illumination pattern [52]. The traditional FMT requires the assumption of optical properties, i.e., absorption and scattering coefficients. As in vivo measurement of the optical properties in soft tissues remains challenging, adopting published values of optical properties for different tissue types is commonly used for FMT reconstruction, as these properties were systematically calculated and calibrated [45]. Time-domain diffuse optical tomography (td-DOT) using a Single-photon avalanche diode (SPAD) array [53] has been used to obtain quantitative optical properties (both absorption and scattering coefficients). A phantom study combining FMT and td-DOT has been recently reported [42].
There are still some limitations to the current work. First, the data acquisition procedure for a single FMT measurement is now 4-5 minutes considering the point-by-point laser scanning illumination, which limits the temporal resolution of FMT. The optimization of illumination patterns and usage of structured light illumination can significantly reduce the scanning time and improve the temporal resolution of FMT [43,52,54]. Second, in the current setup, FMT-MRI was implemented in a sequential mode, i.e., the mouse was measured by standalone FMT and an MRI scanner independently. In the early time frame (<10 minutes, perfusion phase), a major part of the signal came from non-labeled compounds in the circulation. Taking the time for animal preparation and transport into account, we therefore set the measurement time to 30 minutes where a stable retention phase was reached. To more accurately study the kinetics of the CRANAD-2 probe, a longer measurement time (>30 min) will be preferred. This can be implemented by employing a hybrid FMT-MRI imager [51], in which both MRI and optical images can be obtained simultaneously and the mouse can be measured for a longer time as no transport between different modalities is needed. Moreover, we calculated the AUC based on the total fluorescence signal. This is an intrinsic limitation of FMT at a single wavelength, as the signal source cannot be specifically unmixed. Notably, only one transgenic mouse was included in the FMT study, as shown in Fig. 2. The purpose of the current work is to explore the feasibility and establish analysis and registration pipeline of such a hybrid FMT-MRI method in the scenario of imaging amyloid-beta deposits in mice. We plan to increase the number of animals in the next study to enable group comparison. Last but not least, FMT-MRI can also be compared with other emerging hybrid imaging methods, such as optoacoustic tomography-MRI hybrid systems, that can record concurrent optoacoustic and MR images with high spatiotemporal resolution [55–57].
In conclusion, we demonstrated a non-invasive FMT-MRI multimodal strategy and a processing pipeline for in vivo imaging of cerebral Aβ deposits assisted with CRANAD-2 in AD amyloidosis mice. Such a pipeline may facilitate the understanding of the spread of amyloid deposits and the evaluation of Aβ clearing therapies.
Acknowledgments
The authors acknowledge support from Dr. Mark-Aurel Augath, Ms. Gloria Shi, Dr. Marie Rouault at Institute for Biomedical Engineering, ETH & University of Zurich; Dr. Alessia Villois at Department of Chemistry, ETH Zurich; Mr. Daniel Schuppli at Institute for Regenerative Medicine, University of Zurich.
Funding
Horizon 2020 Framework Programme10.13039/100010661 (801347-SENSITIVE); the Ministerio de Asuntos Económicos y Transformación Digital10.13039/501100010198 (FIS2020-115088RB-I00); Universität Zürich10.13039/501100006447 (MEDEF-20021); Helmut Horten Stiftung10.13039/501100013850; Stiftung Synapsis - Alzheimer Forschung Schweiz AFS10.13039/501100008947; Olga Mayenfisch Stiftung10.13039/501100012654; Vontobel-Stiftung10.13039/501100008494; Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung10.13039/501100001711 (320030_179277); National Natural Science Foundation of China10.13039/501100001809 (6210032228); ShanghaiTech University10.13039/501100012600.
Disclosures
The authors declare no conflicts of interest.
Data Availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
References
- 1.Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J., “Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo,” Nature 416(6880), 535–539 (2002). 10.1038/416535a [DOI] [PubMed] [Google Scholar]
- 2.Zott B., Simon M. M., Hong W., Unger F., Chen-Engerer H. J., Frosch M. P., Sakmann B., Walsh D. M., Konnerth A., “A vicious cycle of beta amyloid-dependent neuronal hyperactivation,” Science 365(6453), 559–565 (2019). 10.1126/science.aay0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Strooper B., Karran E., “The cellular phase of Alzheimer's disease,” Cell 164(4), 603–615 (2016). 10.1016/j.cell.2015.12.056 [DOI] [PubMed] [Google Scholar]
- 4.Cohen S. I., Linse S., Luheshi L. M., Hellstrand E., White D. A., Rajah L., Otzen D. E., Vendruscolo M., Dobson C. M., Knowles T. P., “Proliferation of amyloid-beta42 aggregates occurs through a secondary nucleation mechanism,” Proc. Natl. Acad. Sci. U. S. A. 110(24), 9758–9763 (2013). 10.1073/pnas.1218402110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni R., “Positron emission tomography in animal models of Alzheimer’s disease amyloidosis: translational implications,” Pharmaceuticals 14(11), 1179 (2021). 10.3390/ph14111179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Razansky D., Klohs J., Ni R., “Multi-scale optoacoustic molecular imaging of brain diseases,” Eur. J. Nucl. Med. Mol. Imaging 48(13), 4152–4170 (2021). 10.1007/s00259-021-05207-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klunk W. E., Lopresti B. J., Ikonomovic M. D., Lefterov I. M., Koldamova R. P., Abrahamson E. E., Debnath M. L., Holt D. P., Huang G. F., Shao L., DeKosky S. T., Price J. C., Mathis C. A., “Binding of the positron emission tomography tracer Pittsburgh compound-B reflects the amount of amyloid-beta in Alzheimer's disease brain but not in transgenic mouse brain,” J. Neurosci. 25(46), 10598–10606 (2005). 10.1523/JNEUROSCI.2990-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barthel H., Gertz H.-J., Dresel S., Peters O., Bartenstein P., Buerger K., Hiemeyer F., Wittemer-Rump S. M., Seibyl J., Reininger C., Sabri O., “Cerebral amyloid-β PET with florbetaben (18F) in patients with Alzheimer's disease and healthy controls: a multicentre phase 2 diagnostic study,” The Lancet Neurology 10(5), 424–435 (2011). 10.1016/S1474-4422(11)70077-1 [DOI] [PubMed] [Google Scholar]
- 9.Fleisher A. S., Chen K., Liu X., Roontiva A., Thiyyagura P., Ayutyanont N., Joshi A. D., Clark C. M., Mintun M. A., Pontecorvo M. J., Doraiswamy P. M., Johnson K. A., Skovronsky D. M., Reiman E. M., “Using positron emission tomography and florbetapir f 18 to image cortical amyloid in patients with mild cognitive impairment or dementia due to Alzheimer disease,” Arch. Neurol. 68(11), 1404–1411 (2011). 10.1001/archneurol.2011.150 [DOI] [PubMed] [Google Scholar]
- 10.Villemagne V. L., Dore V., Burnham S. C., Masters C. L., Rowe C. C., “Imaging tau and amyloid-beta proteinopathies in Alzheimer disease and other conditions,” Nat. Rev. Neurol. 14(4), 225–236 (2018). 10.1038/nrneurol.2018.9 [DOI] [PubMed] [Google Scholar]
- 11.Ni R., Gillberg P.-G., Bogdanovic N., Viitanen M., Myllykangas L., Nennesmo I., Långström B., Nordberg A., “Amyloid tracers binding sites in autosomal dominant and sporadic Alzheimer's disease,” Alzheimer's & Dementia 13(4), 419–430 (2017). 10.1016/j.jalz.2016.08.006 [DOI] [PubMed] [Google Scholar]
- 12.Jack C. R., Jr., Bennett D. A., Blennow K., Carrillo M. C., Dunn B., Haeberlein S. B., Holtzman D. M., Jagust W., Jessen F., Karlawish J., Liu E., Molinuevo J. L., Montine T., Phelps C., Rankin K. P., Rowe C. C., Scheltens P., Siemers E., Snyder H. M., Sperling R., “NIA-AA Research Framework: toward a biological definition of Alzheimer's disease,” Alzheimers Dement 14(4), 535–562 (2018). 10.1016/j.jalz.2018.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higuchi M., Iwata N., Matsuba Y., Sato K., Sasamoto K., Saido T. C., “19F and 1H MRI detection of amyloid beta plaques in vivo,” Nat Neurosci 8(4), 527–533 (2005). 10.1038/nn1422 [DOI] [PubMed] [Google Scholar]
- 14.Ni R., “Magnetic resonance imaging in animal models of Alzheimer’s disease amyloidosis,” Int. J. Mol. Sci. 22(23), 12768–533 (2021). 10.3390/ijms222312768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Xu D., Chan H. N., Poon C. Y., Ho S. L., Li H. W., Wong M. S., “Dual-modal NIR-fluorophore conjugated magnetic nanoparticle for imaging amyloid-beta species in vivo,” Small 14(28), 1800901 (2018). 10.1002/smll.201800901 [DOI] [PubMed] [Google Scholar]
- 16.Hintersteiner M., Enz A., Frey P., Jaton A. L., Kinzy W., Kneuer R., Neumann U., Rudin M., Staufenbiel M., Stoeckli M., Wiederhold K. H., Gremlich H. U., “In vivo detection of amyloid-beta deposits by near-infrared imaging using an oxazine-derivative probe,” Nat. Biotechnol. 23(5), 577–583 (2005). 10.1038/nbt1085 [DOI] [PubMed] [Google Scholar]
- 17.Ran C., Xu X., Raymond S. B., Ferrara B. J., Neal K., Bacskai B. J., Medarova Z., Moore A., “Design, synthesis, and testing of difluoroboron-derivatized curcumins as near-infrared probes for in vivo detection of amyloid-beta deposits,” J. Am. Chem. Soc. 131(42), 15257–15261 (2009). 10.1021/ja9047043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., Tian Y., Zhang C., Tian X., Ross A. W., Moir R. D., Sun H., Tanzi R. E., Moore A., Ran C., “Near-infrared fluorescence molecular imaging of amyloid beta species and monitoring therapy in animal models of Alzheimer’s disease,” Proc. Natl. Acad. Sci. 112(31), 9734–9739 (2015). 10.1073/pnas.1505420112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvo-Rodriguez M., Hou S. S., Snyder A. C., Dujardin S., Shirani H., Nilsson K. P. R., Bacskai B. J., “In vivo detection of tau fibrils and amyloid beta aggregates with luminescent conjugated oligothiophenes and multiphoton microscopy,” Acta Neuropathol Commun. 7(1), 171 (2019). 10.1186/s40478-019-0832-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirani H., Linares M., Sigurdson C. J., Lindgren M., Norman P., Nilsson K. P., “A palette of fluorescent thiophene-based ligands for the identification of protein aggregates,” Chem. Eur. J. 21(43), 15133–15137 (2015). 10.1002/chem.201502999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni R., Chen Z., Shi G., Villois A., Zhou Q., Arosio P., Nitsch R. M., Nilsson K. P. R., Klohs J., Razansky D., “Transcranial in vivo detection of amyloid-beta at single plaque resolution with large-field multifocal illumination fluorescence microscopy,” bioRxiv, 2020.2002.2001.929844 (2020).
- 22.Hefendehl J. K., Wegenast-Braun B. M., Liebig C., Eicke D., Milford D., Calhoun M. E., Kohsaka S., Eichner M., Jucker M., “Long-term in vivo imaging of beta-amyloid plaque appearance and growth in a mouse model of cerebral beta-amyloidosis,” J. Neurosci. 31(2), 624–629 (2011). 10.1523/JNEUROSCI.5147-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer-Luehmann M., Spires-Jones T. L., Prada C., Garcia-Alloza M., de Calignon A., Rozkalne A., Koenigsknecht-Talboo J., Holtzman D. M., Bacskai B. J., Hyman B. T., “Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer's disease,” Nature 451(7179), 720–724 (2008). 10.1038/nature06616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klunk W. E., Bacskai B. J., Mathis C. A., Kajdasz S. T., McLellan M. E., Frosch M. P., Debnath M. L., Holt D. P., Wang Y., Hyman B. T., “Imaging Aβ plaques in living transgenic mice with multiphoton microscopy and methoxy-X04, a systemically administered congo red derivative,” J. Neuropathol. Exp. Neurol. 61(9), 797–805 (2002). 10.1093/jnen/61.9.797 [DOI] [PubMed] [Google Scholar]
- 25.Bacskai B. J., Hickey G. A., Skoch J., Kajdasz S. T., Wang Y., Huang G.-F., Mathis C. A., Klunk W. E., Hyman B. T., “Four-dimensional multiphoton imaging of brain entry, amyloid binding, and clearance of an amyloid-β ligand in transgenic mice,” Proc. Natl. Acad. Sci. 100(21), 12462–12467 (2003). 10.1073/pnas.2034101100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen D., Uhlmann V., Planchette A. L., Marchand P. J., Van De Ville D., Lasser T., Radenovic A., “Supervised learning to quantify amyloidosis in whole brains of an Alzheimer's disease mouse model acquired with optical projection tomography,” Biomed. Opt. Express 10(6), 3041–3060 (2019). 10.1364/BOE.10.003041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni R., Dean-Ben X. L., Kirschenbaum D., Rudin M., Chen Z., Crimi A., Voigt F., Nilsson P. R., Helmchen F., Nitsch R. M., Aguzzi A., Razansky D., Klohs J., “Whole brain optoacoustic tomography reveals strain-specific regional beta-amyloid densities in Alzheimer`s disease amyloidosis models,” bioRxiv, DOI: 10.1101/2020.1102.1125.964064 (2020).
- 28.Pansieri J., Josserand V., Lee S.-J., Rongier A., Imbert D., Sallanon M. M., Kövari E., Dane T. G., Vendrely C., Chaix-Pluchery O., Guidetti M., Vollaire J., Fertin A., Usson Y., Rannou P., Coll J.-L., Marquette C., Forge V., “Ultraviolet–visible–near-infrared optical properties of amyloid fibrils shed light on amyloidogenesis,” Nat. Photonics 13(7), 473–479 (2019). 10.1038/s41566-019-0422-6 [DOI] [Google Scholar]
- 29.Ntziachristos V., Tung C. H., Bremer C., Weissleder R., “Fluorescence molecular tomography resolves protease activity in vivo,” Nat. Med. 8(7), 757–761 (2002). 10.1038/nm729 [DOI] [PubMed] [Google Scholar]
- 30.Arridge S. R., Schotland J. C., “Optical tomography: forward and inverse problems,” Inverse Probl. 25(12), 123010 (2009). 10.1088/0266-5611/25/12/123010 [DOI] [Google Scholar]
- 31.Darne C., Lu Y. J., Sevick-Muraca E. M., “Small animal fluorescence and bioluminescence tomography: a review of approaches, algorithms and technology update,” Phys. Med. Biol. 59(1), R1–R64 (2014). 10.1088/0031-9155/59/1/R1 [DOI] [PubMed] [Google Scholar]
- 32.Liu X., He X., Yan Z., Lu H., “4-D reconstruction of fluorescence molecular tomography using re-assembled measurement data,” Biomed Opt Express 6(6), 1963–1976 (2015). 10.1364/BOE.6.001963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyde D., de Kleine R., MacLaurin S. A., Miller E., Brooks D. H., Krucker T., Ntziachristos V., “Hybrid FMT-CT imaging of amyloid-beta plaques in a murine Alzheimer's disease model,” NeuroImage 44(4), 1304–1311 (2009). 10.1016/j.neuroimage.2008.10.038 [DOI] [PubMed] [Google Scholar]
- 34.Ren W., Elmer A., Buehlmann D., Augath M.-A., Vats D., Ripoll J., Rudin M., “Dynamic measurement of tumor vascular permeability and perfusion using a hybrid system for simultaneous magnetic resonance and fluorescence imaging,” Mol. Imaging Biol. 18(2), 191–200 (2016). 10.1007/s11307-015-0884-y [DOI] [PubMed] [Google Scholar]
- 35.Ren W., Cui S., Alini M., Grad S., Zhou Q., Li Z., Razansky D., “Noninvasive multimodal fluorescence and magnetic resonance imaging of whole-organ intervertebral discs,” Biomed. Opt. Express 12(6), 3214–3227 (2021). 10.1364/BOE.421205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merlini M., Meyer E. P., Ulmann-Schuler A., Nitsch R. M., “Vascular beta-amyloid and early astrocyte alterations impair cerebrovascular function and cerebral metabolism in transgenic arcAbeta mice,” Acta Neuropathol 122(3), 293–311 (2011). 10.1007/s00401-011-0834-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knobloch M., Konietzko U., Krebs D. C., Nitsch R. M., “Intracellular Abeta and cognitive deficits precede beta-amyloid deposition in transgenic arcAbeta mice,” Neurobiol Aging 28(9), 1297–1306 (2007). 10.1016/j.neurobiolaging.2006.06.019 [DOI] [PubMed] [Google Scholar]
- 38.Ni R., Kindler D. R., Waag R., Rouault M., Ravikumar P., Nitsch R., Rudin M., Camici G. G., Liberale L., Kulic L., Klohs J., “fMRI reveals mitigation of cerebrovascular dysfunction by Bradykinin receptors 1 and 2 inhibitor noscapine in a mouse model of cerebral amyloidosis,” Front. Aging Neurosci. 11, 27 (2019). 10.3389/fnagi.2019.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ni R., Rudin M., Klohs J., “Cortical hypoperfusion and reduced cerebral metabolic rate of oxygen in the arcAβ mouse model of Alzheimer’s disease,” Photoacoustics 10, 38–47 (2018). 10.1016/j.pacs.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Percie du Sert N., Hurst V., Ahluwalia A., Alam S., Avey M. T., Baker M., Browne W. J., Clark A., Cuthill I. C., Dirnagl U., Emerson M., Garner P., Holgate S. T., Howells D. W., Karp N. A., Lazic S. E., Lidster K., MacCallum C. J., Macleod M., Pearl E. J., Petersen O. H., Rawle F., Reynolds P., Rooney K., Sena E. S., Silberberg S. D., Steckler T., Würbel H., “The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research,” PLoS Biol 18(7), e3000410 (2020). 10.1371/journal.pbio.3000410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ni R., Villois A., Dean-Ben X. L., Chen Z., Vaas M., Stavrakis S., Shi G., deMello A., Ran C., Razansky D., Arosio P., Klohs J., “In-vitro and in-vivo characterization of CRANAD-2 for multi-spectral optoacoustic tomography and fluorescence imaging of amyloid-beta deposits in Alzheimer mice,” Photoacoustics 23, 100285 (2021). 10.1016/j.pacs.2021.100285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren W., Jiang J., Costanzo Mata A. D., Kalyanov A., Ripoll J., Lindner S., Charbon E., Zhang C., Rudin M., Wolf M., “Multimodal imaging combining time-domain near-infrared optical tomography and continuous-wave fluorescence molecular tomography,” Opt. Express 28(7), 9860–9874 (2020). 10.1364/OE.385392 [DOI] [PubMed] [Google Scholar]
- 43.Ren W., Isler H., Wolf M., Ripoll J., Rudin M., “Smart toolkit for fluorescence tomography: simulation, reconstruction, and validation,” IEEE Trans. Biomed. Eng. 67(1), 16–26 (2020). 10.1109/TBME.2019.2907460 [DOI] [PubMed] [Google Scholar]
- 44.Ren W., Elmer A., Augath M.-A., Rudin M., “FEM-based simulation of a fluorescence tomography experiment using anatomical MR images,” Proc. of SPIE 9788, 978828 (2016). 10.1117/12.2216454 [DOI] [Google Scholar]
- 45.Jacques S. L., “Optical properties of biological tissues: a review,” Phys. Med. Biol. 58(11), R37–R61 (2013). 10.1088/0031-9155/58/11/R37 [DOI] [PubMed] [Google Scholar]
- 46.Jones A. R., Overly C. C., Sunkin S. M., “The Allen Brain Atlas: 5 years and beyond,” Nat. Rev. Neurosci. 10(11), 821–828 (2009). 10.1038/nrn2722 [DOI] [PubMed] [Google Scholar]
- 47.Klohs J., Baeva N., Steinbrink J., Bourayou R., Boettcher C., Royl G., Megow D., Dirnagl U., Priller J., Wunder A., “In vivo near-infrared fluorescence imaging of matrix metalloproteinase activity after cerebral ischemia,” J. Cereb. Blood Flow Metab. 29(7), 1284–1292 (2009). 10.1038/jcbfm.2009.51 [DOI] [PubMed] [Google Scholar]
- 48.Ren W., Ji B., Guan Y., Cao L., Ni R., “Recent technical advances in accelerating the clinical translation of small animal brain imaging: hybrid imaging, deep learning, and transcriptomics,” Front. Med. 9, 771982 (2022). 10.3389/fmed.2022.771982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ntziachristos V., Yodh A. G., Schnall M., Chance B., “Concurrent MRI and diffuse optical tomography of breast after indocyanine green enhancement,” Proc. Natl. Acad. Sci. U. S. A. 97(6), 2767–2772 (2000). 10.1073/pnas.040570597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis S. C., Samkoe K. S., Tichauer K. M., Sexton K. J., Gunn J. R., Deharvengt S. J., Hasan T., Pogue B. W., “Dynamic dual-tracer MRI-guided fluorescence tomography to quantify receptor density in vivo,” Proc. Natl. Acad. Sci. U. S. A. 110(22), 9025–9030 (2013). 10.1073/pnas.1213490110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuker F., Baltes C., Dikaiou K., Vats D., Carrara L., Charbon E., Ripoll J., Rudin M., “Hybrid small animal imaging system combining magnetic resonance imaging with fluorescence tomography using single photon avalanche diode detectors,” IEEE Trans. Med. Imaging 30(6), 1265–1273 (2011). 10.1109/TMI.2011.2112669 [DOI] [PubMed] [Google Scholar]
- 52.Liu Y., Ren W., Ammari H., “Robust reconstruction of fluorescence molecular tomography with an optimized illumination pattern,” Inverse Probl. Imaging 14(3), 535–568 (2020). 10.3934/ipi.2020025 [DOI] [Google Scholar]
- 53.Bruschini C., Homulle H., Antolovic I. M., Burri S., Charbon E., “Single-photon avalanche diode imagers in biophotonics: review and outlook,” Light: Sci. Appl. 8(1), 87 (2019). 10.1038/s41377-019-0191-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Streeter S. S., Maloney B. W., McClatchy D. M., Jermyn M., Pogue B. W., Rizzo E. J., Wells W. A., Paulsen K. D., “Structured light imaging for breast-conserving surgery, part II: texture analysis and classification,” J. Biomed. Opt. 24(09), 1–12 (2019). 10.1117/1.JBO.24.9.096003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren W., Skulason H., Schlegel F., Rudin M., Klohs J., Ni R., “Automated registration of magnetic resonance imaging and optoacoustic tomography data for experimental studies,” Neurophotonics 6(2), 025001 (2019). 10.1117/1.NPh.6.2.025001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren W., Dean-Ben X. L., Augath M. A., Razansky D., “Development of concurrent magnetic resonance imaging and volumetric optoacoustic tomography: A phantom feasibility study,” J. Biophotonics 14(2), e202000293 (2021). 10.1002/jbio.202000293 [DOI] [PubMed] [Google Scholar]
- 57.Hu Y., Lafci B., Luzgin A., Wang H., Klohs J., Dean-Ben X. L., Ni R., Razansky D., Ren W., “Deep learning facilitates fully automated brain image registration of optoacoustic tomography and magnetic resonance imaging,” arXiv preprint, arXiv:2109.01880 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.