Issues of thiol-disulfide redox chemistry were nowhere in our consciousness when we began to study the export of Escherichia coli alkaline phosphatase from the cytoplasm. We chose this periplasmic enzyme because it seemed an excellent tool for characterizing the mechanism of protein translocation across the cytoplasmic membrane. However, our selection and analysis of signal sequence mutants led us to the surprising discovery that alkaline phosphatase was enzymatically inactive when it was localized to the bacterial cytoplasm (32). Since the cytoplasm was reputedly a much more reducing environment than the periplasm, we reasoned that the lack of cytoplasmic alkaline phosphatase activity was due to the failure of the protein to form its two essential disulfide bonds. We later established that the cytoplasmic form of the enzyme did indeed lack disulfide bonds (14).
These results presented us with questions we had not considered before. Were there specific proteins in the cytoplasm that were responsible for keeping unwanted disulfide bonds from forming? Were there specific proteins in the periplasm that catalyzed formation of these bonds? Could we approach these questions genetically? From posing these questions, we moved on to develop genetic selections that might help us identify proteins involved in determining the oxidation state of cysteines in proteins of the two compartments. The results of these efforts along with those of a number of other labs have opened our eyes to an impressive array of E. coli proteins that are part of the thioredoxin superfamily. Furthermore, pushing ever deeper into the functions of members of this family which exhibit very similar structures raises a number of issues about the directions necessary for the success of studies in functional genomics.
FUNCTIONAL GENOMICS
The availability of microbial genome sequences holds the promise of advancing our understanding of many aspects of bacterial physiology. The depth of this understanding depends on the ability to deduce the functions of the thousands of individual open reading frames (ORFs) in each organism. At present, putative functions can be assigned to approximately 70% of all microbial genome ORFs based on sequence homology. Thus, a major endeavor for functional genomics is the exploration of the remaining ORFs, which have no homologues with known activities. Nevertheless, even the putative functional assignments based on sequence similarities usually allow only classification of a predicted protein as a member of a protein family with common activities. In many cases, such assignments do not allow the definition of specific physiological roles for such proteins. Furthermore, there are often many paralogues of ORFs within an organism that may reflect a history of gene duplication and domain shuffling in that organism. Multiple copies of members of such gene families provide for apparent functional redundancy of particular protein activities and are sometimes explained by a need of the cell for “backup systems.” However, the use of functional redundancy to account for the persistence of these multiple ORFs in a genome may simply be a way of covering up our lack of detailed knowledge of the physiology and ecology of the organism.
While exploring the functions of the ORFs with no homologues is an obvious challenge, we argue that an equally important effort is to try to understand the specific physiological roles of genes which, at first glance, seem to be redundant. We will call these sets of genes the gray areas of the genome. We will illustrate the issues involved by describing what has been learned about the specific roles of members of the thioredoxin superfamily in E. coli. We have chosen this family because of its high redundancy in most organisms and also because several aspects of the roles of these oxidoreductases cannot be predicted based on sequence similarities. We will conclude by discussing which approaches to the paralogue problem seem relevant to analysis of such ORF families.
THIOL-DISULFIDE OXIDOREDUCTASES
Members of the thioredoxin superfamily share two features in common: they contain a short sequence motif that includes a Cys-X1-X2-Cys sequence (the active site) and an overall structure containing this motif that corresponds to what is called a thioredoxin-like fold (29). The latter structural features have been determined directly by X-ray crystallography for some members of the family and by structural modelling in others (29). The actual role of each of these proteins in the cell is partly determined by the redox potential of the protein and partly by the direction of the electron transport pathway it participates in, showing the importance of both kinetic and thermodynamic factors. Thioredoxin 1 of E. coli, with a redox potential of −270 mV, is a major reductant in the cytoplasm; it is important for the reduction of such cytoplasmic enzymes as ribonucleotide reductase (26). DsbA, a periplasmic protein with a redox potential of −122 mV, is highly oxidizing; it is required for disulfide bond formation in the cell envelope (6).
While much knowledge of the specific roles of these two proteins of the thioredoxin family has been accumulated, genetic and biochemical studies plus sequence analysis have led to the discovery of nine other genes in E. coli that code for thioredoxin-like proteins or proteins that include a thioredoxin domain. For many of these proteins, specific roles cannot be assigned. When multiple genes are discovered for proteins with apparently identical functions, the genes are often described as redundant (e.g., 10). Sometimes, genetic and physiological studies support this characterization. Despite this evidence, there is increasing reason to believe that each of these proteins may have a specialized role to play, at least under some environmental conditions.
SPECIFICITY AND REDUNDANCY OF CYTOPLASMIC THIOL-DISULFIDE OXIDOREDUCTASES
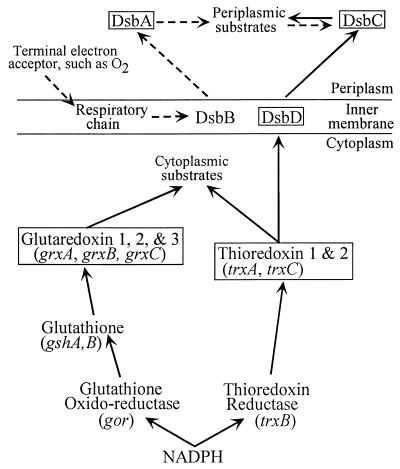
A number of cytoplasmic reductive enzymes become oxidized as part of their catalytic cycle and must be subsequently reduced for their continued function. These enzymes, such as ribonucleotide reductase, methionine sulfoxide reductase, and 3′-phosphoadenosine 5′-phosphosulfate reductase, can utilize thioredoxins or thioredoxin-like proteins to be continuously regenerated (49). The best-studied members of the cytoplasmic thioredoxin superfamily are thioredoxins 1 and 2 (TrxA and TrxC, respectively) (33, 49) and glutaredoxins 1, 2, and 3 (GrxA, GrxB, and GrxC, respectively) (Fig. 1) (4). The thioredoxins and glutaredoxins exhibit features common to the thioredoxin superfamily—the Cys-X1-X2-Cys active site and the thioredoxin fold. Despite sharing these structural properties, the thioredoxin and glutaredoxin subfamilies do not share sequence similarities. Furthermore, even though both sets of proteins ultimately derive their reducing power from NADPH, maintenance of the thioredoxins in the reduced state is accomplished by thioredoxin reductase (TrxB), while the glutaredoxins are reduced via glutathione. Finally, each member of this family exhibits its own characteristic redox potential. While most thioredoxin homologues are potent disulfide bond reductants, of the E. coli glutaredoxins, only glutaredoxin 1 is efficient in this respect both in vivo and in vitro (3, 49). The activity of glutaredoxins 2 and 3 may be limited to reduction of mixed disulfides containing glutathione (4).
FIG. 1.
Members of the thioredoxin superfamily in E. coli. Members of the family are boxed. Reduction (black arrow), oxidation (broken arrow), and disulfide bond isomerization (two arrows) are indicated. Three members of this family (NrdH, DsbG, and DsbE) are not shown, as their roles in these pathways are not clear.
Some evidence can be adduced to support the proposition that cytoplasmic thioredoxins and glutaredoxins are simply redundant and provide backup for each other. Null mutants for genes coding for each of these proteins are viable under usual laboratory growth conditions (41, 46). In fact, only by eliminating the three most effective reductants, thioredoxins 1 and 2 and glutaredoxin 1, is a lethal phenotype created (49). Apparently, any one of these three proteins can suffice to supply the disulfide reductase needs of the cell.
However, indications are beginning to accumulate that, while these proteins can substitute for one another under some conditions, there are certain pathways or environmental conditions that call largely on the utilization of only one of these proteins. One such example involves the enzyme methioninesulfoxide reductase. A mutant missing thioredoxin 1 is unable to utilize methionine sulfoxide as a source of methionine, indicating that thioredoxin 1 is the main source of reducing power for this enzyme (49). Overexpression of glutaredoxin 1 can partially substitute for the missing thioredoxin 1, whereas overexpression of thioredoxin 2 does not (49).
Another striking example of a specific role for a member of the thioredoxin family occurs under a stress response condition. When E. coli is challenged with hydrogen peroxide, the OxyR regulatory protein becomes active and induces the expression of a set of genes that destroy hydrogen peroxide and repair the damage done to the cell. The activation of OxyR occurs by the formation of a disulfide bond in the protein. One of the genes whose expression is increased by activated OxyR is grxA; the high levels of glutaredoxin 1 are then able to reduce the disulfide bond in OxyR, rendering the regulatory protein inactive. Thus, once the stress response has been sufficiently activated, this feedback mechanism returns the cell to its original state. Mutants lacking glutaredoxin 1 show a delay in the reduction of OxyR (53).
Finally, disulfide bond isomerization in the periplasm is accomplished by channeling of electrons from thioredoxin 1 across the cytoplasmic membrane (43, 44). High-level expression of thioredoxin 2 can only partially substitute for thioredoxin 1, and glutaredoxin 1 is completely inactive in this pathway (42).
Thus, each of these very similar proteins may fulfill specific and important roles in the cell in addition to their ability to backup one another under some conditions. The differing specificities may be due to the particular redox potential of each protein, to features of their amino acid sequences that permit interactions with particular substrate proteins, or to other yet-to-be understood properties of these proteins. We point out that at least one member of this family of proteins, thioredoxin 1, utilizes other features of its amino acid sequence for its role as a cofactor in the development of certain bacteriophages (16, 47). These latter activities do not involve the redox center of the protein. For instance, thioredoxin 1 forms a tight complex with the bacteriophage T7 DNA polymerase, contributing processivity to the functioning of this enzyme (7). Even when overexpressed, thioredoxin 2 cannot substitute for thioredoxin 1 in T7 development (50). The features of thioredoxin 1 structure that are responsible for its interaction with T7 DNA polymerase may also contribute to its unique role in other pathways. Parenthetically, it is interesting to note that the genome of bacteriophage T4 includes two genes encoding glutaredoxins (20). One of these two bacteriophage T4 proteins, identified as a glutaredoxin by its sequence, can use either thioredoxin reductase or glutathione as a source of electrons (38).
In addition to the thioredoxins and glutaredoxins referred to above, a gene encoding an additional cytoplasmic member of this family, NrdH (24), has been detected by analysis of E. coli genome sequences. There is little genetic or physiological evidence indicating the in vivo role of NrdH. Interestingly, this protein, which shows significant homology with glutaredoxins, does not interact with glutathione, but with thioredoxin reductase—a finding which would not have been predicted from sequence gazing (24).
SPECIFICITY AND REDUNDANCY OF THIOL-DISULFIDE OXIDOREDUCTASES OF THE CELL ENVELOPE
Two major roles for cell envelope thiol-disulfide oxidoreductases have been identified. The first role is the formation and isomerization of disulfide bonds in proteins localized to the bacterial cell envelope. Despite some suggestions of overlap of function among these proteins (1, 36), the evidence indicates that most of the well-characterized members of this family fulfill a specific well-defined function (43, 44). DsbA is required for the oxidation of pairs of cysteines to generate disulfide bonds in certain cell envelope proteins (6). The consequently reduced DsbA transfers its electrons to DsbB, thus becoming reoxidized (5, 8, 12, 35). DsbC is required for the isomerization of incorrectly formed disulfide bonds, an activity that depends on its Cys-X1-X2-Cys motif being maintained in the reduced state (36, 43, 48, 52). The reduction of DsbC is achieved by the membrane-bound DsbD (DipZ) protein (43, 44). Although a protein with strong homology to DsbC, DsbG, exists in the E. coli genome sequence (1, 51), no persuasive evidence indicating its physiological role has yet been obtained (9).
Certain virulent strains of Salmonella typhimurium contain two dsbA homologues—one on the chromosome and one on a virulence plasmid (30, 45). The latter gene product may be there to enhance the oxidizing potential in the periplasm or may provide substrate specificity, e.g., being more active as an oxidant on proteins encoded by the plasmid.
The second role for these cell envelope proteins is in the assembly of c-type cytochromes. At least two membrane proteins, DsbD (DipZ) (11) and CcmG (DsbE), (31) appear to be essential for maintaining the cysteines of cell envelope apocytochromes in the reduced state. The reduced cysteines are needed for the assembly of the heme moiety into the active holoenzyme.
All of the proteins identified, except for DsbB, contain domains that place them in the thioredoxin superfamily.
HOW THE ROLES OF FAMILY MEMBERS HAVE BEEN DEDUCED
One can readily pick out nearly all the genes encoding thiol-disulfide oxidoreductases by using homology programs for analysis of the E. coli genome sequence (18). This is possible because all but DsbB share sequence features with the prototypes, thioredoxin 1 or glutaredoxin 1. However, little can be said about the most interesting aspects of the roles of these proteins simply from examination of the sequence. For instance, the effectiveness in the process of disulfide bond reduction is largely dependent on the redox potential of these proteins which is not deducible simply by comparing protein sequences. Furthermore, the overall function of these proteins as oxidants or reductants is dependent on the redox status of the subcellular environment in which they are found. As far as is known, each of these proteins has the potential to act relatively efficiently both as an oxidase or reductase. For example, the first discovered thioredoxin (thioredoxin 1) of E. coli is the most efficient at reducing disulfide bonds. Nevertheless, when thioredoxin is oxidized—when its two cysteines of the Cys-X1-X2-Cys are joined in a disulfide bond—it is able to donate that bond to substrate proteins, an oxidative reaction (13, 28, 40, 49). This point is not purely academic, since many genomes contain thioredoxin variants with a signal sequence. Whether these variants act as oxidants or reductants in vivo apparently cannot be decided without experimental evidence.
Thus, it is instructive to recount how we have come to understand the functions of certain of these proteins in the life of the bacteria. This knowledge has accrued as a result of a variety of approaches, including genetic, physiological, and biochemical studies and sequence analysis. What is learned from an analysis of the thioredoxin superfamily is that sequence gazing in the absence of these other endeavors yields little insight into the physiological activity of each member. Furthermore, information on the role of a particular thiol-disulfide oxidoreductase in the bacteria has often arisen not from a linear process designed to elucidate function but rather has resulted fortuitously from studies designed to study some other problem.
Thioredoxin 1 was purified from E. coli as a reductant of disulfides in the active site of ribonucleotide reductase (26). The same assay was used to detect glutaredoxin 1 in extracts of cells missing thioredoxin 1 (23). The two additional glutaredoxins were isolated from extracts of mutants lacking thioredoxin 1 and glutaredoxin 1 (4). Although glutaredoxins 2 and 3 are more abundant than glutaredoxin 1, they show poor ability to act as disulfide bond reductants in vitro, and a null mutant of glutaredoxin 3 displays no clear phenotypes under the conditions tested (41). In these cases, a recognition that a reductant was needed for ribonucleotide reductase led to the search for such molecules; the physiological problem was understood and led to the discovery of candidate molecules.
Thioredoxin 2 was postulated based on the study of E. coli mutants that were able to introduce disulfide bonds into cytoplasmic proteins. The finding that the increased cytoplasmic oxidation potential in mutants lacking thioredoxin reductase (trxB) could not be explained solely by the altered state of thioredoxin 1 led to the suggestion that the bacteria expressed a second thioredoxin (15, 41). We found that the accumulation of oxidized forms of both thioredoxins 1 and 2 accounted for the high level of disulfide bond formation in the trxB mutants (49).
Despite the fact that most members of the cytoplasmic thioredoxin superfamily are reductants of ribonucleotide reductase, there is still no direct evidence for which one or what combination of these thiol-disulfide oxidoreductases performs this essential reductive step in vivo. Nevertheless, an indication that thioredoxin 1 and glutaredoxin 1 may play overlapping roles in this reaction in vivo is the finding that a trxA grxA double mutant shows a 25-fold induction of ribonucleotide reductase, while single mutants of these genes show only a very modest induction (34).
Overall, the role or roles of each individual thioredoxin or glutaredoxin in cell physiology remain an open question.
Several different genetic approaches led unexpectedly to the isolation of mutants of the dsb pathway. These included genetic selections or screening procedures for detecting mutants affecting (i) membrane protein assembly (6), (ii) bacterial virulence (39), (iii) bacterial motility (12), (iv) protein folding in the periplasm (8, 25), and (v) increased resistance or sensitivity to the reductant dithiothreitol (35). In no case were the genetic approaches designed to find genes promoting disulfide bond formation. The absence of efforts to purposely select for mutants affecting this function derives, in part, from the impression that protein disulfide bond formation occurred spontaneously and did not require a catalyst (2). It was only in-depth study of these fortuitously isolated mutants that led to the discovery of the in vivo roles of the proteins DsbA and DsbB. The dsbC gene was first identified in E. coli as a multicopy suppressor of a dsbA mutant (48). (Note that DsbC is ordinarily in the reduced state and unable to promote disulfide bond formation. However, when overexpressed, some oxidized DsbC presumably accumulates and acts as an oxidant.) The identification of the involvement of DsbC and DsbD in a pathway for disulfide bond isomerization came from a combination of biochemical and genetic studies (43, 44, 52). (The dsbD gene had previously been found and named dipZ via its role in cytochrome c biogenesis [11].)
WHITHER POSTGENOMICS OF THE PARALOGUES?
So, we are left with a problem. A large number of members of the thioredoxin family have been identified in E. coli. For some of these, a relatively specific role has been outlined, but for others, such specificity has not been achieved. We could finesse this problem by suggesting that many of these molecules are redundant, providing backup for each other. This is not a very satisfying explanation, particularly if one accepts a strictly evolutionary adaptationist explanation for the presence of genes in an organism. However, more importantly, evidence is beginning to accumulate for very specialized roles for certain of these proteins in addition to their ability to act as backup molecules.
This discussion leads to the question for the thioredoxin superfamily of how we go about determining what specialized roles these molecules might fulfill. More generally, for any family of proteins, members of which share common sequence features or even active sites, how do we go from their identification via genome analysis to an understanding of their function? Obviously, some of the functional genomic approaches already being taken will yield information. The availability of oligonucleotide microarray chips and other genome array techniques allows determination of the transcription patterns of all genes in an organism. If such data are collected under numerous environmental conditions, it seems probable that many ORFs that appear redundant will show rather specific expression patterns. In addition, competition for growth between null mutants for these ORFs and wild-type bacteria in different growth media may also give hints as to specific functions.
However, if we use as a case study the family of thiol-disulfide oxidoreductases, we can see that this set of approaches would not be sufficient. One still needs to pose new biological-physiological questions, to carry out genetic selections to respond to these questions, and to probe deeper into the physiology of cells—in this case, the various ways in which electrons are passed around within and between subcellular compartment. Furthermore, the studies on arrays and on competition assume that we are already knowledgeable about the variety of environments and stresses that a particular bacteria encounters and that we should test. However, it is only in recent years that researchers have begun to go beyond the usual bacterial conditions for laboratory growth and to examine the behavior of bacteria in a broader array of environments. Studies on stationary phase (19) and response to different stresses (22, 27, 37), bacterial physiology during infection of hosts (21), and growth in replicas of natural habitats, such as the poor nutrient media of rivers (17), may still not have scratched very deeply beyond the surface into the hidden life of bacteria.
We suggest that the examination of this one system illustrates that the success of microbial genomics in allowing a description of an organism, including the function of its individual genes, requires a major and more innovative effort in traditional areas of bacteriology. Success would require, in addition to the new postgenomic technologies, enhanced efforts in ecology of bacteria, renewed studies in greater depth of bacterial physiology, and the use of genetic approaches to dissect fundamental physiological problems.
ACKNOWLEDGMENTS
We thank Eric Stewart, Roberto Kolter, Daniel Ritz, Hong-Ping Tian, and Federico Katzen for helpful comments and Eric Stewart for Fig. 1.
This work was supported by a grant from the National Institute of General Medical Sciences GM41883. J.B. is supported by an American Cancer Society Research Professorship, and F.Å. is supported by an EMBO Postdoctoral Fellowship.
Footnotes
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Andersen C L, Matthey-Dupraz A, Missiakas D, Raina S. A new Escherichia coli gene, dsbG, encodes a periplasmic protein involved in disulphide bond formation, required for recycling DsbA/DsbB and DsbC redox proteins. Mol Microbiol. 1997;26:121–132. doi: 10.1046/j.1365-2958.1997.5581925.x. [DOI] [PubMed] [Google Scholar]
- 2.Anfinsen C B, Haber E, Sela M, White F H., Jr The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci USA. 1961;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Åslund F, Berndt K D, Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 4.Åslund F, Ehn B, Miranda-Vizuete A, Pueyo C, Holmgren A. Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 is a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc Natl Acad Sci USA. 1994;91:9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell J C A, Lee J-O, Jander G, Martin N, Belin D, Beckwith J. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci USA. 1993;90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardwell J C A, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 7.Bedford E, Tabor S, Richardson C C. The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc Natl Acad Sci USA. 1997;94:479–484. doi: 10.1073/pnas.94.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belin P, Boquet P-L. Un second géne impliqué dans la formation des ponts disulfure de protéines localisées dans l’espace périplasmique de Escherichia coli. C R Acad Sci. 1993;361:469–473. [PubMed] [Google Scholar]
- 9.Bessette, P. H., J. J. Cotto, H. F. Gilbert, and G. Georgiou. In vivo and in vitro function of the E. coli periplasmic cysteine oxidoreductase DsbG. J. Biol. Chem., in press. [DOI] [PubMed]
- 10.Bron S, Bolhuis A, Tjalsma H, Holsappel S, Venema G, Van Dijl J M. Protein secretion and possible roles for multiple signal peptidases for precursor processing in bacilli. J Biotechnol. 1998;64:3–13. doi: 10.1016/s0168-1656(98)00099-6. [DOI] [PubMed] [Google Scholar]
- 11.Crooke H, Cole J. The biogenesis of c-type cytochromes in Escherichia coli requires a membrane-bound protein, DipZ, with a protein disulphide isomerase-like domain. Mol Microbiol. 1995;15:1139–1150. doi: 10.1111/j.1365-2958.1995.tb02287.x. [DOI] [PubMed] [Google Scholar]
- 12.Dailey F E, Berg H C. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1043–1047. doi: 10.1073/pnas.90.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debarbieux L, Beckwith J. The reductive enzyme thioredoxin 1 acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc Natl Acad Sci USA. 1998;95:10751–10756. doi: 10.1073/pnas.95.18.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derman A I, Beckwith J. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J Bacteriol. 1991;173:7719–7722. doi: 10.1128/jb.173.23.7719-7722.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derman A I, Prinz W A, Belin D, Beckwith J. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 16.Doublié S, Tabor S, Long A M, Richardson C C, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 17.Espinosa-Urgel M, Kolter R. Escherichia coli genes expressed preferentially in an aquatic environment. Mol Microbiol. 1998;28:325–332. doi: 10.1046/j.1365-2958.1998.00796.x. [DOI] [PubMed] [Google Scholar]
- 18.Fetrow J S, Godzik A, Skolnick J. Functional analysis of the Escherichia coli genome using the sequence-to-structure-to-function paradigm: identification of proteins exhibiting the glutaredoxin/thioredoxin disulfide oxidoreductase activity. J Mol Biol. 1998;282:703–711. doi: 10.1006/jmbi.1998.2061. [DOI] [PubMed] [Google Scholar]
- 19.Goodrich-Blair H, Uria-Nickelsen M, Kolter R. Regulation of gene expression in stationary phase. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman and Hall; 1996. pp. 571–583. [Google Scholar]
- 20.Gvakharia B O, Hanson E, Koonin E K, Mathews C K. Identification of a second functional glutaredoxin encoded by the bacteriophage T4 genome. J Biol Chem. 1996;271:15307–15310. doi: 10.1074/jbc.271.26.15307. [DOI] [PubMed] [Google Scholar]
- 21.Heithoff D M, Conner C P, Mahan M J. Dissecting the biology of a pathogen during infection. Trends Microbiol. 1997;5:509–513. doi: 10.1016/S0966-842X(97)01153-0. [DOI] [PubMed] [Google Scholar]
- 22.Hidalgo E, Demple B. Adaptive responses to oxidative stress: the soxRS and oxyR regulons. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman and Hall; 1996. pp. 435–452. [Google Scholar]
- 23.Holmgren A. Glutathione-dependent synthesis of deoxyribonucleotides: purification and characterization of glutaredoxin from Escherichia coli. J Biol Chem. 1979;254:3664–3671. [PubMed] [Google Scholar]
- 24.Jordan A, Åslund F, Pontis E, Reichard P, Holmgren A. Characterization of Escherichia coli NrdH—a glutaredoxin-like protein with a thioredoxin-like activity profile. J Biol Chem. 1997;272:18044–18050. doi: 10.1074/jbc.272.29.18044. [DOI] [PubMed] [Google Scholar]
- 25.Kamitani S, Akiyama Y, Ito K. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 1992;11:57–62. doi: 10.1002/j.1460-2075.1992.tb05027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurent T C, Moore E C, Reichard P. Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from E. coli. J Biol Chem. 1964;239:3436–3444. [PubMed] [Google Scholar]
- 27.Little J W. The SOS regulatory system. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman and Hall; 1996. pp. 453–480. [Google Scholar]
- 28.Lundström J, Krause G, Holmgren A. A Pro to His mutation in active site of thioredoxin increases its disulfide-isomerase activity 10-fold. New refolding systems for reduced or randomly oxidized ribonuclease. J Biol Chem. 1992;267:9047–9052. [PubMed] [Google Scholar]
- 29.Martin J L. Thioredoxin—a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 30.Martin, N. L., M. Kohli, G. A. Touchie, R. J. Kadner, and J. Beckwith. Characterization of PefS, a Salmonella typhimurium virulence plasmid-encoded analogue of the disulfide oxidoreductase DsbA. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 31.Metheringham R, Tyson K L, Crooke H, Missiakas D, Raina S, Cole J A. Effects of mutations in genes for proteins involved in disulphide bond formation in the periplasm on the activities of anaerobically induced electron transfer chains in Escherichia coli K12. Mol Gen Genet. 1996;253:95–102. doi: 10.1007/pl00013815. [DOI] [PubMed] [Google Scholar]
- 32.Michaelis S, Inouye H, Oliver D, Beckwith J. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J Bacteriol. 1983;154:366–374. doi: 10.1128/jb.154.1.366-374.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miranda-Vizuete A, Damdimopoulos A E, Gustafsson J-A, Spyrou G. Cloning, expression and characterization of a novel Escherichia coli thioredoxin. J Biol Chem. 1997;272:30841–30847. doi: 10.1074/jbc.272.49.30841. [DOI] [PubMed] [Google Scholar]
- 34.Miranda-Vizuete A, Rodríguez-Ariza A, Toribio F, Holmgren A, López-Barea J, Pueyo C. The levels of ribonucleotide reductase, thioredoxin, glutaredoxin 1, and GSH are balanced in Escherichia coli K12. J Biol Chem. 1996;271:19099–19103. doi: 10.1074/jbc.271.32.19099. [DOI] [PubMed] [Google Scholar]
- 35.Missiakas D, Georgopoulos C, Raina S. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci USA. 1993;90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Missiakas D, Georgopoulos C, Raina S. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 1994;13:2013–2020. doi: 10.1002/j.1460-2075.1994.tb06471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Missiakas D, Raina S, Georgopoulos C. Regulation of gene expression in Escherichia coli. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman and Hall; 1996. pp. 481–502. [Google Scholar]
- 38.Nikkola M, Gleason F K, Eklund H. Reduction of mutant phage T4 glutaredoxins by Escherichia coli thioredoxin reductase. J Biol Chem. 1993;268:3845–3849. [PubMed] [Google Scholar]
- 39.Peek J A, Taylor R K. Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc Natl Acad Sci USA. 1992;89:6210–6214. doi: 10.1073/pnas.89.13.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pigiet V, Schuster B J. Thioredoxin-catalyzed refolding of disulfide-containing proteins. Proc Natl Acad Sci USA. 1986;83:7643–7647. doi: 10.1073/pnas.83.20.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prinz W A, Åslund F, Holmgren A, Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 42.Rietsch A. Disulfide bond formation and isomerization in Escherichia coli. Ph.D. thesis. Cambridge, Mass: Harvard University; 1998. [Google Scholar]
- 43.Rietsch A, Belin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rietsch A, Bessette P, Georgiou G, Beckwith J. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol. 1997;179:6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodríguez-Peña J M, Alvarez I, Ibáñez M, Rotger R. Homologous regions of the Salmonella enteritidis virulence plasmid and the chromosome of Salmonella typhi encode thiol:disulphide oxidoreductases belonging to the DsbA thioredoxin family. Microbiology. 1997;143:1405–1413. doi: 10.1099/00221287-143-4-1405. [DOI] [PubMed] [Google Scholar]
- 46.Russel M, Holmgren A. Construction and characterization of glutaredoxin-negative mutants of Escherichia coli. Proc Natl Acad Sci USA. 1988;85:990–994. doi: 10.1073/pnas.85.4.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russel M, Model P. The role of thioredoxin in filamentous phage assembly. Construction, isolation and characterization of mutant thioredoxins. J Biol Chem. 1996;261:14997–15005. [PubMed] [Google Scholar]
- 48.Shevchik V E, Condemine G, Robert-Baudouy J. Characterization of DsbC, a periplasmic protein of Erwinia chrysanthemi and Escherichia coli with disulfide isomerase activity. EMBO J. 1994;13:2007–2012. doi: 10.1002/j.1460-2075.1994.tb06470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart E J, Åslund F, Beckwith J. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 1998;17:5543–5550. doi: 10.1093/emboj/17.19.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart, E. J., and J. Beckwith. 1998. Unpublished results.
- 51.Van Straaten M, Missiakas D, Raina S, Darby N J. The functional properties of DsbG, a thiol-disulfide oxidoreductase from the periplasm of Escherichia coli. FEBS Lett. 1998;428:255–258. doi: 10.1016/s0014-5793(98)00539-0. [DOI] [PubMed] [Google Scholar]
- 52.Zapun A, Missiakas D, Raina S, Creighton T E. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry. 1995;34:5075–5089. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]
- 53.Zheng M, Åslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]