Abstract
Influenza viruses have developed resistance to the current classes of drugs, which means they could eventually become more virulent and cause more mortality and hospitalization. Our study aims to investigate the antiviral activity of Rhazya stricta Decne leaves extract in vitro and search for new promising drugs from R. stricta identified compounds in silico. The study was performed in vitro by utilizing Madin-Darby Canine Kidney cell line (MDCK) as a substrate for the influenza virus and estimating the inhibition performance of the plant leaves extract. Additionally, in silico screening was conducted to explore the antiviral activity of R. stricta phytochemicals. We investigated the cytotoxicity of R. stricta leaves extract and its antiviral activity against influenza virus (A/Puerto Rico/8/34 (H1N1)) using the MTT assay. The mode of action of the plant leaves extract during the viral life cycle was tested using time-of-addition assay. In silico analyses were performed, including molecular docking, drug-likeness analysis, and toxicity risk assessment, to state the leading compounds to be developed into an anti-influenza virus drug. The 50% cytotoxicity concentration of the leaves extract was CC50: 184.6 µg/mL, and the 50% inhibition concentration was CI50: 19.71 µg\mL. The time of addition assay revealed that R. stricta leaves extract exerted its activity in the late step of the influenza virus replication cycle. In comparison to Oseltamivir, the leading compounds showed better binding affinity and can be developed into oral drugs with low toxicity risk. Isolation and purification of the leading compounds and testing their antiviral activity in vitro and in vivo are required.
Keywords: Plant extract, Cytopathic effect, Time of addition assay, Molecular docking.
1. Introduction
Influenza is a viral respiratory infection; in humans, it is caused by influenza A virus (genus Alphainfluenzavirus), influenza B virus (genus Betainfluenzavirus). Influenza viruses C and D (genus Gammainfluenzavirus and Deltainfluenzavirus are also known). The symptoms of influenza virus infection range from a mild upper respiratory tract infection with fever, sore throat, cough, and fatigue to severe and in some cases, lethal pneumonia caused by the influenza virus or secondary bacterial infection of the lower respiratory tract. Each year, influenza infects approximately 10%–20% of the world's population, resulting in 3–5 million hospitalizations, about 290 000–650 000 respiratory deaths, and an estimated annual economic burden of $87.1 billion in the United States alone (Krammer et al., 2018; WHO fact sheets, 2018, ICTV, 2022).
While vaccination is certainly the most preferred approach for influenza prevention, it is linked with various rates of protection due to inadequate absorption, mismatches with prevalent influenza virus strains, long manufacturing times in chicken eggs, and within-season loss of effectiveness (Holmes, et al., 2021).
Therefore, antivirals that specifically target the influenza virus functional or structural proteins are also critical for preventing or treating influenza infections. Antivirals against influenza are important in the treatment of hospitalized or critically ill influenza patients, as well as in the early phases of a pandemic when a compatible vaccine is unavailable (Hayden and Pavia, 2006).
Resistance of influenza virus to amantadine and the adamantane analogous drug rimantadine was quickly identified and shown to be associated with a single amino acid change at one of five sites in the M2 protein. Due to the development of resistance, adamantines are no longer effective against influenza type A (Whitley and Monto, 2020). The present gold standard for antiviral treatment of influenza includes various types of neuraminidase (NA) blockers, such as oseltamivir (Tamiflu), zanamivir, and peramivir. NA inhibitor-resistant influenza strains have emerged earlier as a result of drug treatment, emphasizing the need for the discovery of potential drugs targeting different viral gene products (Kormuth et al., 2020).
R. stricta is a member of the alkaloid-rich Apocynaceae family. Around 100 indole-type alkaloids have been isolated from its parts, and it is one of the most economically valuable medicinal plants found throughout arid South Asia and the Middle East. Leave extracts were traditionally used to treat a variety of ailments (Park et al., 2014, Gilani et al., 2007). It has been reported that the alkaloidal compounds possess multiple activities, including antitumor, antimicrobial, and antihypertensive (Roberts and Wink, 1998). The study objective is inspected the antiviral activity of R. stricta leaves extract against influenza virus and determine its inhibition mode.
2. Material and methods
In vitro:
2.1. Plant collection and extraction
Fresh aerial portions of the R. stricta plant were harvested from its natural environment in the Saudi Arabian desert along the Makkah-Jeddah Road. The plant leaves were assembled, rinsed under running water to eliminate dust, and left to dry at room temperature in the laboratory. A week later, the leaves were finely powdered and kept at room temperature for further extraction. We weighted 6 g of the powdered plant and added it to 1 L of sterile, distillated water, and the mixture was left in the shaker overnight. The next day, the extract was filtered using 0.22 µm sterile membrane filters and stored for one week in the refrigerator (Fig. 1).
Fig. 1.
A and B) Natural photos of R. stricta plant have been captured from the C) collection site at Jeddah-Makkah Road “ 21.544846089411394, 39.642608955967795 ” (color should be used).
2.2. Cells culturing
MDCK cell line was obtained from King Abdulaziz University Hospital. The cells were grown in Dulbecco's Modified Eagle Media (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin–streptomycin antibiotics at 37° in CO2 5% and humidified incubator. The cryopreservation procedure of the MDCK cells were carried out as follows: MDCK cells were grown to 90% confluency. Then, trypsin EDTA was added to detach the adherent cells. After 10 min, the detached cells were suspended at cryopreservation media consisting of (90% DMEM, 30% FBS, and 10% DMSO), then stored in liquid nitrogen for long-term preservation.
2.3. Virus propagation
MDCK cells were seeded into a T25 flask and supplemented with DMEM growth media. A day later, the cells had become 90% confluent monolayer. The culture media was removed, and the cells were washed with 2 mL of phosphate-buffered saline (PBS). The monolayer sheet was infected with 0.5 mL of influenza virus (A/Puerto Rico/8/1934) for 1 h. The unabsorbed virions were removed, and fresh DMEM media containing 2 µg\mL tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin was added. The infected cells are kept in the incubator for 2 days and monitored daily to observe the progress of the cytopathic effect (CPE). The cell culture supernatants were harvested and centrifuged to get rid of the cell’s debris. Finally, the purified supernatant, which contains the viral progeny, was split into aliquots and preserved in −70° freezer.
2.4. Virus titration (TCID50)
According to (Klimov et al., 2012), once the 25 T flask was 90% confluent, the cells were trypsinized and seeded into 96-well plate for 24 h. The next day, the growth media was removed, and the cells were washed with 200 µL of PBS to each well. In all wells except the first column of the 96-well plate, 100 µL of DMEM media containing 2 µg\mL (TPCK)-treated trypsin was added to all wells. The influenza virus stock vial was thawed and diluted 1:100 in influenza virus propagation media. 146 µL of the diluted virus was added to the first column of 96-well tissue culture plate, and ½ log10 dilutions was performed by transferring 46 µL between wells and disposing of the final 46 μL after the ninth column. The 96-well tissue culture plate was incubated for 3 days at 37 °C. The wells were observed daily for the presence or absence of CPE using an inverted microscope. The influenza virus titer was 10-4.25 TCID50/mL.
2.5. Cytotoxicity assay
The cytotoxicity assay was performed as described in the manufactory manual (Vybrant® MTT Cell Proliferation Assay Kit V-13154) with slight modifications. Cell suspension (100 μL) was seeded into 96 well microplate and immersed in DMEM growth media at 37° in CO2 5% humidified incubator. The growth media was removed after 24 h, and the 96 microplates were washed with 100 µL of PBS for each well. The adherent cells were treated with a two-fold serial dilution of the plant extract (3 mg/mL– 6 µg/mL) diluted with maintenance media (2% FBS) and incubated for 48 h to examine the cytotoxicity of the plant extract. After 48 h, the culture medium was replaced with fresh media, and 10 μL of MTT solution was added to each well and incubated for 4 h. To solubilize the formed formazan crystals, we removed all but 25 µL of medium from the wells, and 50 μL of DMSO was added to each well and incubated for 10 min. And the absorbance was read at 540 nm wavelength by (BioTak, Synergy HTX multi-mode microplate reader). Cell viability was calculated using this equation:
2.6. Antiviral activity
The antiviral activity of R. stricta leaves extract was determined as described in (Ding et al., 2017) with some modifications. MDCK cells were seeded into 96 microwell plates for 24 h with growth media. The consumed media was aspirated, and the plate was washed with 100 µL of PBS. Then, the cells were infected with 100 µL (100TCID50). After 1 h, the inoculum was removed, and the cells were washed with 100 µL of PBS to remove unabsorbed virions. Different concentrations of the leaves extract (100, 75, 50, and 25 µg\mL) dissolved in DMEM media containing 2 µg\mL (TPCK)-treated trypsin were added to the microplate. After incubation for 48 h, the MTT assay was performed as described previously. The anti-influenza virus drug (Oseltamivir) was used as positive control, and the antiviral activity was calculated according to this equation:
Odtv: The absorbance of the tested extract with virus-infected cells.
Odcv: The absorbance of the virus-infected cells only.
Odcd: the absorbance of the cells control.
2.7. Time of addition assay
To study the antiviral activity of R. stricta leaves extract at different periods of the viral life cycle. MDCK cells were cultured into 96 microwell plates for 1 day with complete growth media. The culture media was discarded, and the microwell plate was washed with 100 µL of PBS. Then, the cells were infected with 100 µL (100TCID50) for 2 h. The infected cells were treated with DMEM media containing 100 µg/mL of the plant extract and 2 µg\mL (TPCK)-treated at different periods of time: pre-infection: [-2 - (-1) h] and [(-1) – 0 h], during-infection [0 – 2 h], and post-infection [2–4 h] and [4 – 8 h]. In each step, the media was removed, and the wells were washed with PBS. After 48 h, the MTT assay was performed as described previously.
In silico study:
2.8. Molecular docking
We collected 204 compounds reported in the literature that were identified in R. stricta extract (Bukhari et al., 2017, Aziz et al., 2020, Al-Zharani et al., 2019, Mahmood et al., 2020, Marwat et al., 2012, Obaid et al., 2017, Nehdi et al., 2016). The compound data set was docked into the H1N1 Neuraminidase (NA) active site (PDB ID: 3TI6, Vavricka et al., 2011) using AutoDock Vina (Trott and Olson (2010)). The ligands were prepared by retrieving the chemical structures of the compounds as SDF files from the PubChem database and converting them to PDB format using Pymol (Schrodinger, 2010) or the canonical smiles converted by the National Cancer Institute structure file generator (https://cactus.nci.nih.gov/translate/) (Oellien and Nicklaus, 2004). The NA crystal structure was obtained from the protein data bank and was prepared by eliminating the co-crystallized ligand and water molecules. The graphical user interface software AutoDock 4.2 MGL tools1.5.6 (Morris et al., 2009) was utilized to add Gasteiger charges to the ligands and Kollman charges to the protein. The grid box was concentrated on the residues of the NA active site. The output docked complexes were visualized using Pymol (Schrodinger, 2010) and discovery studio visualizer (Biovia, 2017).
2.9. Drug-likeness and water solubility prediction
The SwissADME web server was utilized to predict the drug-likeness properties and water solubility for the best docked compounds. (Daina, et al., 2017). Lipinski's rule of five was applied to predict the drug-likeness characteristic of best docked compounds (Lipinski, et al., 2012), and water solubility prediction was obtained according to (Ali, et al., 2012) model.
2.10. Toxicity risk assessment
The ProToxII server was used to determine the relative toxicity of the leading compounds (Banerjee et al., 2018). The server received the Canonical Smiles of the compounds and produced predicted results indicating the 50% lethal dose (LD50) and toxicity class.
2.11. Statistical analysis
Virus titration was calculated according to the Reed-Muench method (Reed et al., 1938). The non-linear regression model was utilized to calculate IC50 and CC50 values by GraphPad prism 9.1.2 software (GraphPad Prism, version 9.1.2). The selectivity index (IS) value was calculated as the ratio of the CC50 to the IC50. The diagrams, including the standard error bars, were sketched by Microsoft Excel.
3. Results:
3.1. Cytotoxicity assay
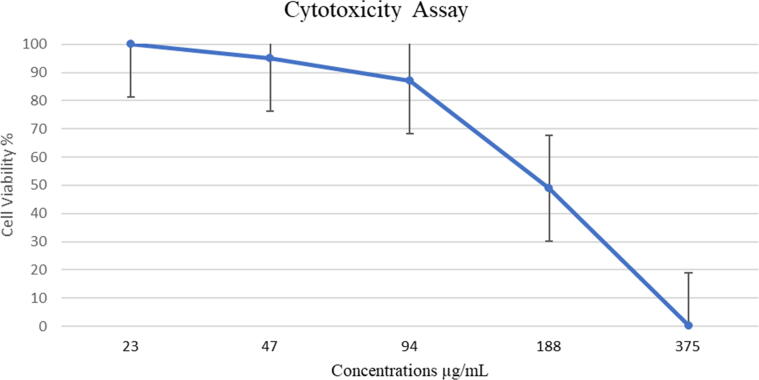
The four concentrations, ranging from 3 mg/mL to 375 μg/mL, were highly toxic (0% cell viability). A moderate toxic effect was observed on MDCK cells at the concentration of 187 µg/mL (49% cell viability), and the concentrations of 94 μg/mL and 47 μg/mL were slightly toxic (87% and 95% cell viability, respectively). There was no toxicity observed in any concentrations that were tested under 47 μg/mL, and the cytotoxicity concentration at which 50% of the cells are killed is CC50: 184.6 µg/mL (Fig. 2).
Fig. 2.
Cell viability of MDCK cells after treated with R. Stricta extract at two-fold dilution concentrations. The experiment was performed as one trial with three replicates.
3.2. Antiviral assay
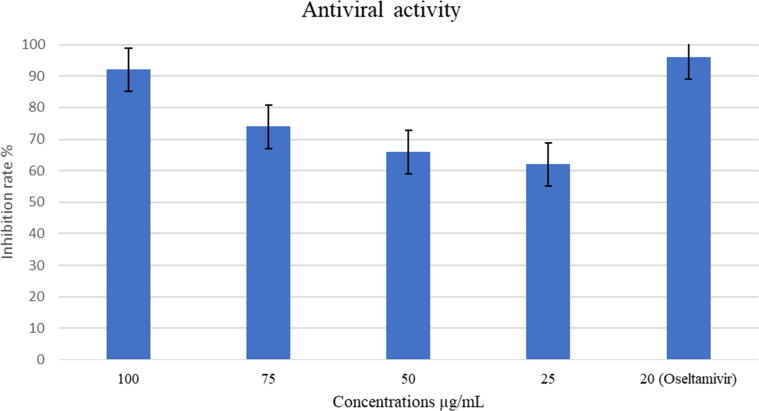
R. stricta leaves extract inhibited influenza virus (A/Puerto Rico/8/34 (H1N1) replication significantly, with IC50: 19.71 µg\mL. In comparison to the positive control (Oseltamivir), that suppressed influenza virus production with inhibition rate 96% at concentration of 20 µg\mL, the extract inhibition rate reached 92% at concentration of 100 µg/mL and 74% for 75 µg/mL. The concentration 50 µg/mL reached a 66% inhibition rate, and the 25 µg/mL concentration showed a 62% inhibition rate. The selectivity index (SI) value of R. Stricta leaves extract was 9.37. (Fig. 3 & Table 1).
Fig. 3.
Inhibition rate of R. stricta leaves extract against Influenza A virus (A/Puerto Rico/8/34 (H1N1)). The experiment was performed as one trial with four replicates.
Table 1.
Summarize of the statistical data, showed the confidence intervals (CI95), the coefficient of determination (R2) of the fitted regression line.
| CC50 | IC50 | SI | R2 | |
|---|---|---|---|---|
| R. Stricta extract | 184.6 µg/mL (CI95: 166.1–202.6) | 19.71 µg\mL (CI95: 6.826–37.25) | 9.37 | CC50: 0.9967 CI50: 0.9605 |
3.3. Time of addition experiments
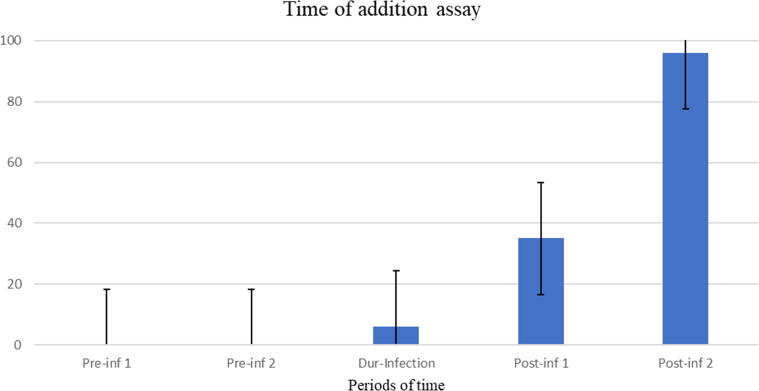
To understand the mode of action of the plant extract and its effect on the viral life cycle we performed an antiviral assay dependent on specific periods of time. R. stricta leaves extract did not inhibit the virus attachment to the receptor phase [-2 - (-1)] h and [(-1) – 0] h. The extract exerted insignificant viral entry activity (0–2 h) with 6% inhibition rate. The plant extract interferes with the virus replication phase (2–4 h) at a 35% inhibition rate. The high antiviral activity was observed in the late stage of the virus life cycle (4–8 h) with 96% inhibition rate. These findings imply that R. stricta leaves extract could contain compounds that inhibit the NA protein (Fig. 4).
Fig. 4.
Inhibition rate of R. Stricta leaves extract during different time intervals against Influenza A virus (A/Puerto Rico/8/34 (H1N1)). The horizontal axis represents the times intervals as: Pre-inf 1(-2 – −1h), Pre-inf 2 (-1 – 0 h), Dur-inf (0 – 1 h), Post-inf 1 (2 – 4 h), and Post-inf 2 (4 – 8 h). The experiment was performed as one trial with four replicates.
3.4. Molecular docking
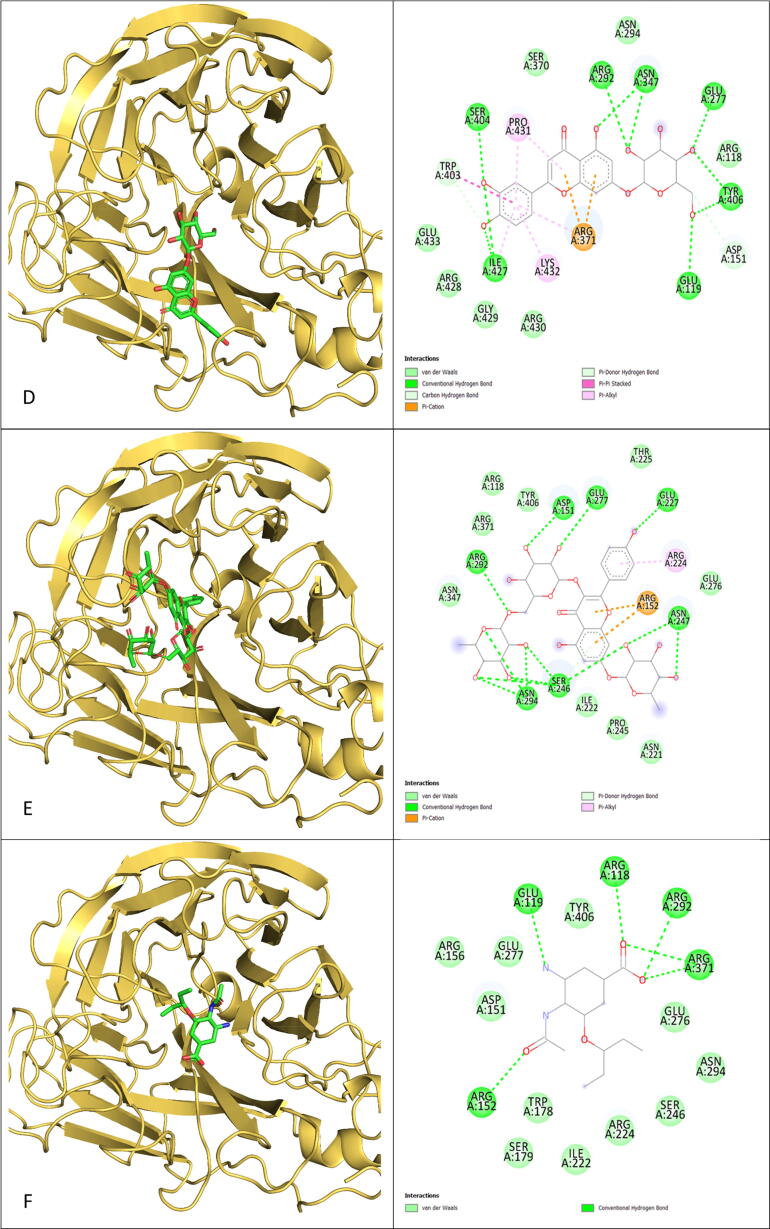
We established a cut-off value of 8.0 kcal/mol for the binding affinity of R. Stricta docked compounds and compared them to the oseltamivir-NA protein complex. Tight binding has been shown by 39 compounds to the sialic acid site of the NA protein. In comparison to oseltamivir, which had a binding energy of –6.6 kcal/mol, all 39 compounds had a lower affinity for the NA protein and established a different number of hydrogen bonds with the pocket residues (Table 2). In our docking investigation, Rutin, a flavonoid glycoside molecule, had the lowest binding energy (–10.6 kcal/mol), and its atoms were involved in 14 hydrogen bonds with the NA protein residues. Moreover, rutin induces electrostatic interactions such as van der Waals forces, as well as hydrophobic interactions such as the Pi-Alkyl interaction. The remaining 38 compounds docked remarkably well in the NA interaction pocket via a variety of bonds and interactions (Fig. 5).
Table 2.
Binding affinity and hydrogen bonds of the best docked compounds into the influenza virus NA protein active site.
| Compound | Binding affinity kcal/mol | Number of Hydrogen Bond |
|---|---|---|
| Rutin | −10.6 | 14 |
| Tetrahydrosecamine | −9.7 | 5 |
| 16-Hydrorhazisidine | −9.5 | 2 |
| Luteolin-7-glucoside | −9.1 | 10 |
| kaempferol rhamnoside rutinoside | −9.1 | 17 |
| Dihydrosecamine | −8.9 | 1 |
| 16 s,16′-Decarboxytetrahydrosecamine | −8.9 | 1 |
| Secamine | −8.8 | 2 |
| Presecamine | −8.7 | 4 |
| Strictosidine | −8.6 | 6 |
| Rhazizine | −8.6 | 5 |
| Quercetin-3-rhamnoside | −8.6 | 9 |
| Isoquercetin | −8.6 | 8 |
| Luteolin | −8.6 | 6 |
| Strictosamide | −8.5 | 5 |
| Rhazimol | −8.5 | 3 |
| CID: 9,603,606 | −8.5 | 2 |
| Antirhine | −8.4 | 3 |
| Leepacine | −8.4 | 3 |
| HR-1 | −8.4 | 1 |
| Apigenin | −8.4 | 4 |
| Quercetin | −8.3 | 5 |
| CID: 2,936,295 | −8.3 | 2 |
| leuconolam | −8.2 | 3 |
| Strictamine-N-oxide | −8.2 | 2 |
| Rhazicine | −8.2 | 6 |
| Strictisidine | −8.2 | 2 |
| Isorhazicine | −8.2 | 6 |
| kaempferol | −8.2 | 3 |
| 3-epi-Antirhine | −8.1 | 2 |
| Condylocarpine | −8.1 | 2 |
| Tetrahydroalstonine | −8.1 | 4 |
| Rhazinaline | −8.1 | 1 |
| Stricticine | −8.1 | 3 |
| Serpentine | −8.1 | 0 |
| Rhazinilam | −8.0 | 1 |
| Yohimbine | −8.0 | 2 |
| 2-Methoxy 1–2, dihydrorhazamine | −8.0 | 2 |
| Rhazine | −8.0 | 1 |
| Oseltamivir | −6.6 | 7 |
Fig. 5.
3D positions and 2D plotting of R. Stricta top 5 docked compounds into the NA protein active site were compared to the docked position of Oseltamivir. A) Rutin, B) Tetrahydrosecamine, C)16-Hydrorhazisidine, D) Luteolin-7-glucoside, E) kaempferol rhamnoside rutinoside, F) Oseltamivir. (Color should be used).
3.5. Drug-likeness and water solubility prediction
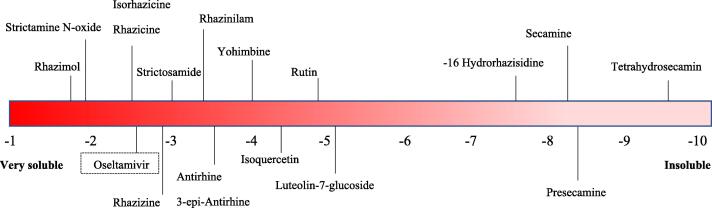
Drug-likeness analysis was performed to investigate the oral bioavailability of the compounds and was compared to the anti-influenza virus approved drug Oseltamivir. The parameters of the following compounds: Antirhine, Strictosamide, Leepacine, HR-1, Rhazimol, Rhazizine, Quercetin, Apigenin, Luteolin, CID: 9603606, CID: 2936295, Leuconolam, Strictamine N-oxide, Rhazicine, Strictisidine, kaempferol, 3-epi-Antirhine, Condylocarpine, Tetrahydroalstonine, Rhazinaline, Stricticine, Serpentine, Rhazinilam, Yohimbine, Rhazine, 2-Methoxy 1–2 dihydrorhazamine, Isorhazicine, and the positive control (Oseltamivir) were fitted into the Lipinski rule of five, which means these compounds could be administered orally. Tetrahydrosecamine, Presecamine, Strictosidine, Secamine, Dihydrosecamine, 16-Hydrorhazisidine, Decarboxytetrahydrosecamine, Quercetin-3-rhamnoside, Isoquercetin, Rutin, Luteolin-7-glucoside, and kaempferol rhamnoside Rutinoside violated the Lipinski rule of five, making it difficult for these compounds to be absorbed by the gastrointestinal tract. The unapplicable compounds may still be introduced through other routes, such as injection and inhalation (Table 3). We found from the water solubility prediction that only 6 molecules are predicted to be poorly soluble, and the rest are water-soluble or partially soluble, which makes these phytochemicals highly predicted to be constituents of the R. stricta leaves extract (Table 4, & Fig. 6).
Table 3.
Lipinski rules of five values of best docked compounds.
| Compound | Druglikeness |
||||
|---|---|---|---|---|---|
| 500 ≥ MW (g/mol) | 10 ≥ N or O | 5 ≥ NH or OH | 4.15 ≥ MLogP | Violation | |
| Antirhine | 296.41 | 3 | 2 | 2.63 | 0 |
| Tetrahydrosecamine | 680.92 | 8 | 1 | 4.42 | 2 |
| Presecamine | 676.89 | 8 | 1 | 5.03 | 2 |
| Strictosamide | 498.53 | 10 | 5 | −0.43 | 0 |
| Strictosidine | 530.57 | 11 | 6 | −0.59 | 3 |
| Leepacine | 350.41 | 5 | 1 | 2.21 | 0 |
| Secamine | 676.89 | 8 | 1 | 4.29 | 2 |
| Dihydrosecamine | 678.90 | 8 | 1 | 4.35 | 2 |
| 16-Hydrorhazisidine | 651.98 | 7 | 1 | 4.55 | 2 |
| HR-1 | 379.51 | 6 | 1 | 2.11 | 0 |
| Rhazimol | 363.51 | 5 | 1 | 2.11 | 0 |
| Rhazizine | 379.51 | 6 | 1 | 1.86 | 0 |
| Decarboxytetrahydrosecamine | 622.88 | 6 | 1 | 4.93 | 2 |
| Quercetin | 302.24 | 7 | 5 | −0.56 | 0 |
| Quercetin-3-rhamnoside | 448.38 | 11 | 7 | −1.84 | 2 |
| Isoquercetin | 464.38 | 12 | 8 | −2.59 | 2 |
| Rutin | 610.52 | 16 | 10 | −3.89 | 3 |
| Apigenin | 270.24 | 5 | 3 | 0.52 | 0 |
| Luteolin | 286.24 | 6 | 4 | −0.03 | 0 |
| Luteolin-7-glucoside | 448.38 | 11 | 7 | −2.10 | 2 |
| kaempferol rhamnoside rutinoside | 740.66 | 19 | 11 | −4.77 | 3 |
| CID: 9,603,606 | 285.36 | 4 | 1 | 1.68 | 0 |
| CID: 2,936,295 | 366.84 | 4 | 0 | 3.70 | 0 |
| Leuconolam | 326.39 | 5 | 2 | 1.76 | 0 |
| Strictamine N-oxide | 338.40 | 5 | 0 | 0.69 | 0 |
| Rhazicine | 368.43 | 6 | 2 | 1.80 | 0 |
| Strictisidine | 348.40 | 5 | 0 | 2.13 | 0 |
| kaempferol | 286.24 | 6 | 4 | −0.03 | 0 |
| 3-epi-Antirhine | 296.41 | 3 | 2 | 2.63 | 0 |
| Condylocarpine | 322.40 | 4 | 1 | 2.82 | 0 |
| Tetrahydroalstonine | 352.43 | 5 | 1 | 2.13 | 0 |
| Rhazinaline | 350.41 | 5 | 0 | 2.13 | 0 |
| Stricticine | 338.40 | 5 | 1 | 2.07 | 0 |
| Serpentine | 349.40 | 5 | 1 | 2.21 | 0 |
| Rhazinilam | 294.39 | 3 | 1 | 2.99 | 0 |
| Yohimbine | 354.44 | 5 | 2 | 2.21 | 0 |
| Rhazine | 354.44 | 5 | 0 | 2.70 | 0 |
| 2-Methoxy 1–2 dihydrorhazamine | 382.45 | 6 | 1 | 2.02 | 0 |
| Isorhazicine | 368.43 | 6 | 2 | 1.80 | 0 |
| Oseltamivir | 312.40 | 6 | 2 | 0.63 | 0 |
Table 4.
Solubility status and LogS values of the best docked compounds.
| Compound | Solubility | Log S |
|---|---|---|
| Antirhine | Soluble | −3.44 |
| Tetrahydrosecamine | Poorly soluble | −9.45 |
| Presecamine | Poorly soluble | −8.35 |
| Strictosamide | Soluble | −3.02 |
| Strictosidine | Soluble | −3.59 |
| Leepacine | Soluble | −2.58 |
| Secamine | Poorly soluble | −8.21 |
| Dihydrosecamine | Poorly soluble | −8.82 |
| 16-Hydrorhazisidine | Poorly soluble | −7.65 |
| HR-1 | Soluble | −2.25 |
| Rhazimol | Very soluble | −1.78 |
| Rhazizine | Soluble | −2.90 |
| Decarboxytetrahydrosecamine | Poorly soluble | −9.35 |
| Quercetin | Soluble | −3.91 |
| Quercetin-3-rhamnoside | Moderately soluble | −4.44 |
| Isoquercetin | Moderately soluble | −4.35 |
| Rutin | Moderately soluble | −4.87 |
| Apigenin | Moderately soluble | −4.59 |
| Luteolin | Moderately soluble | −4.51 |
| Luteolin-7-glucoside | Moderately soluble | −5.06 |
| kaempferol rhamnoside rutinoside | Moderately soluble | −4.69 |
| CID: 9,603,606 | Soluble | −3.39 |
| CID: 2,936,295 | Soluble | −3.90 |
| Leuconolam | Soluble | −2.06 |
| Strictamine N-oxide | Very soluble | −1.93 |
| Rhazicine | Soluble | −2.56 |
| Strictisidine | Soluble | −2.47 |
| kaempferol | Soluble | −3.86 |
| 3-epi-Antirhine | Soluble | −3.44 |
| Condylocarpine | Soluble | −2.85 |
| Tetrahydroalstonine | Soluble | −3.55 |
| Rhazinaline | Soluble | −2.11 |
| Stricticine | Soluble | −2.61 |
| Serpentine | Soluble | −3.91 |
| Rhazinilam | Soluble | −3.32 |
| Yohimbine | Soluble | −3.98 |
| Rhazine | Soluble | −2.49 |
| 2-Methoxy 1–2 dihydrorhazamine | Soluble | −2.89 |
| Isorhazicine | Soluble | −2.56 |
| Oseltamivir | Soluble | −2.60 |
Fig. 6.
Some of the best docked compounds distributed on Log S scale. (Color should be used).
3.6. Toxicity risk assessment
We utilized a computational approach to assess the toxicity of the lead compounds. The toxicity risk assessment values of the best docked compounds are shown in (Table 5). Based on the LD50 of the control (Oseltamivir), which was its 50% lethal dose 260 mg/kg in class 3 (50 < LD50 ≤ 300 mg/kg), we compared the R. stricta compounds. The class 5 (2000 < LD50 ≤ 5000 mg/kg) was include quercetin-3-rhamnoside, isoquercetin, rutin, luteolin-7-glucoside, kaempferol rhamnoside rutinoside, apigenin and kaempferol. The compounds: Antirhine, Tetrahydrosecamine, Presecamine, Strictosidine, Secamine, Dihydrosecamine, 16-Hydrorhazisidine, 16-Hydrorhazisidine, HR-1, Rhazimol, Rhazizine, Decarboxytetrahydrosecamine, CID: 9,603,606 and CID: 2936295, Leuconolam, 3-epi-Antirhine, and Rhazinilam were in the range of class 4 (300 < LD50 ≤ 2000 mg/kg). Strictosamide, Tetrahydroalstonine, Serpentine, Yohimbine, and Quercetin were placed in class 3 (50 < LD50 ≤ 300 mg/kg). Finally, in classes 1 and 2, leepacine, Strictamine-N-oxide, Rhazicine, Strictisidine, Isorhazicine, Condylocarpine, Rhazinaline, Stricticine, 2-Methoxy 1–2, dihydrorhazamine, and Rhazine were expected to be highly toxic.
Table 5.
LD50 of the R. Stricta compounds and their toxicity class.
| Compound | LD50 (mg/kg) | Toxicity Class |
|---|---|---|
| Antirhine | 760 | 4 |
| Tetrahydrosecamine | 1000 | 4 |
| Presecamine | 1500 | 4 |
| Strictosamide | 300 | 3 |
| Strictosidine | 620 | 4 |
| Leepacine | 1 | 1 |
| Secamine | 1000 | 4 |
| Dihydrosecamine | 1000 | 4 |
| 16-Hydrorhazisidine | 400 | 4 |
| HR-1 | 812 | 4 |
| Rhazimol | 1355 | 4 |
| Rhazizine | 933 | 4 |
| Decarboxytetrahydrosecamine | 1000 | 4 |
| Quercetin | 159 | 3 |
| Quercetin-3-rhamnoside | 5000 | 5 |
| Isoquercetin | 5000 | 5 |
| Rutin | 5000 | 5 |
| Apigenin | 2500 | 5 |
| Luteolin | 3919 | 5 |
| Luteolin-7-glucoside | 5000 | 5 |
| kaempferol rhamnoside rutinoside | 5000 | 5 |
| CID: 9,603,606 | 500 | 4 |
| CID: 2,936,295 | 500 | 4 |
| Leuconolam | 325 | 4 |
| Strictamine-N-oxide | 40 | 2 |
| Rhazicine | 1 | 1 |
| Strictisidine | 1 | 1 |
| Isorhazicine | 1 | 1 |
| kaempferol | 3919 | 5 |
| 3-epi-Antirhine | 760 | 4 |
| Condylocarpine | 28 | 2 |
| Tetrahydroalstonine | 300 | 3 |
| Rhazinaline | 1 | 1 |
| Stricticine | 1 | 1 |
| Serpentine | 198 | 3 |
| Rhazinilam | 484 | 4 |
| Yohimbine | 300 | 3 |
| 2-Methoxy 1–2, dihydrorhazamine | 1 | 1 |
| Rhazine | 1 | 1 |
| Oseltamivir | 260 | 3 |
In (Table 6) we eliminated the unexpected compounds to be developed into drugs based on the drug-likeness profile, and the toxicity risk prediction.
Table 6.
Summary of the computational study represents the best R. Stricta compounds that could be turned into modern drugs.
| Compound | Binding affinity (kcal/mol) | Violation To Lipinski rule of 5 | LD50 (mg/kg) |
|---|---|---|---|
| Rhazizine | −8.6 | 0 | 933 |
| Luteolin | −8.6 | 0 | 3919 |
| Strictosamide | −8.5 | 0 | 300 |
| Rhazimol | −8.5 | 0 | 1355 |
| CID: 2,936,295 | −8.5 | 0 | 500 |
| Antirhine | −8.4 | 0 | 760 |
| HR-1 | −8.4 | 0 | 812 |
| Apigenin | −8.4 | 0 | 2500 |
| CID: 9,603,606 | −8.3 | 0 | 500 |
| leuconolam | −8.2 | 0 | 325 |
| kaempferol | −8.2 | 0 | 3919 |
| 3-epi-Antirhine | −8.1 | 0 | 760 |
| Tetrahydroalstonine | −8.1 | 0 | 300 |
| Rhazinilam | −8.0 | 0 | 484 |
| Yohimbine | −8.0 | 0 | 300 |
| Oseltamivir | −6.6 | 0 | 260 |
4. Discussion
R. stricta is an economically valuable and medicinal plant that grows in arid South Asia and the Arabian Peninsula. Historically, leaves extract was used to treat a range of ailments, including syphilis, parasitic infections, hyperglycemia, and rheumatism, as well as the common cold. Lots of studies screened the phytochemical ingredients of R. stricta extract. Over a hundred alkaloids and several additional chemicals, including flavonoids and lipids, have been identified (Albeshri et al., 2021).
Current influenza virus research aims to develop new classes of drugs targeting distinct viral targets to expand the therapeutic options available and to minimize the risk of resistance emergence. Novel antiviral strategies are critical for providing the first line of defense against novel outbreaks and pandemics before the availability of specific vaccines (Goldhill et al., 2018).
To the best of our knowledge, no prior research investigating the cytotoxicity of R. stricta leaves extract on MDCK cell lines or the antiviral activity of the plant extract against influenza virus has been conducted to date. In the present study, the inhibitory activity of R. stricta leaves extract against influenza A/PuertoRico/8/1934 (H1N1) virus was investigated in vitro and followed by in silico study screening for potential active compounds that are present in the plant extract. The cytotoxicity of R. stricta leaves extract was observed to be dose dependent; the high concentrations reduced the cell viability. R. stricta leaves extract significantly inhibited the viral replication with IC50 value of 19.71 µg\mL and SI 9.37. This finding suggests that the plant extract contains phytochemicals that participate in suppressing viral replication.
The time of addition assay of one progeny of influenza virus replication revealed that, the extract exerted its most activity in the late phase of the influenza virus replication. Nevertheless, the extract demonstrated minor activity in the early stage of virus replication. Based on these results, we chose the NA protein as a potential target to dock R. stricta identified compounds into the protein cavity. NA protein are activated during the viral budding phase and helps release new virion progeny by cutting sialic acids from cell membrane receptors and carbohydrate side chains on budding virions (McAuley et al., 2019).
The molecular docking result revealed that 39 compounds, mostly alkaloids and flavonoids, exhibited a stronger bending affinity ranged (−10.6 – −8.0 kcal/mol). Some of these compounds are already examined against influenza virus such as; Apigenin, Luteolin, and Rutin (Liu, et al., 2008). Oppositely, Rhazizine, Rhazimol, Tetrahydroalstonine, etc., have never been tested in vitro.
Oral medication delivery is regarded as one of the most preferred routes of administration since it has been demonstrated to be convenient, less expensive, and highly adhered to by patients. Approximately 90% of active pharmaceuticals are administered orally (Indurkhya, et al., 2018).The exclusion of a pharmaceutical product from the market due to unanticipated adverse effects is one of the most dramatic incidents that may occur throughout the long procedures extending from development to marketing (Benigni, 2004). Therefore, we subjected the best docked compounds to drug-likeness and toxicity prediction. Inadequate absorbance or permeability is a concern event with compounds such as Tetrahydrosecamine and Strictosidine. High toxicity is predicted with compounds such as; Strictamine-N-oxide and Condylocarpine. Consequently, we excluded all the inapplicable computationally compounds and summarized them in (Table 6).
After we disputed all the tested compounds, we looked for their presence in the plant aqueous extract. We followed (Ali et al., 2012) model, which successfully predicted that 81% of data set contains 1265 different chemicals. Interestingly, we found 33 compounds are predicted to be water soluble, which indicates these compounds are more likely to be in the plant aqueous extract in moderate or high concentrations.
5. Conclusion
Accordingly, R. stricta extract could be a source of modern and effective antiviral agents and help in the viral resistance battle. We conducted a preliminary in vitro study followed by computational investigations reporting the antiviral potential of the plant extract. Further experimental research, including advanced in vitro methods and in vivo studies, is required to analyze the plant extract chemically and explicate the antiviral activity of the plant extract and its constituents.
6. Ethics approval statement
No ethics approval was required in this study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Abdulaziz Albeshri, Email: aalbishri0061@stu.kau.edu.sa.
Nabih A. Baeshen, Email: nabih_baeshen@hotmail.com.
Thamer A. Bouback, Email: tbouback@kau.edu.sa.
Abdullah A. Aljaddawi, Email: aaljaddawi@kau.edu.sa.
References
- Albeshri A., Baeshen N.A., Bouback T.A., Aljaddawi A.A. A review of rhazya stricta decne phytochemistry, bioactivities, pharmacological activities, toxicity, and folkloric medicinal uses. Plants. 2021;10:2508. doi: 10.3390/plants10112508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali J., Camilleri P., Brown M.B., Hutt A.J., Kirton S.B. Revisiting the general solubility equation: In silico prediction of aqueous solubility incorporating the effect of topographical polar surface area. J. Chem. Inf. Model. 2012;52:420–428. doi: 10.1021/ci200387c. [DOI] [PubMed] [Google Scholar]
- Al-Zharani M., Nasr F.A., Abutaha N., Alqahtani A.S., Noman O.M., Mubarak M., Wadaan M.A. Apoptotic induction and anti-migratory effects of Rhazya stricta fruit extracts on a human breast cancer cell line. Molecules. 2019;24:1–16. doi: 10.3390/molecules24213968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz, A.T., Alshehri, M.A., Alanazi, N.A., Panneerselvam, C., Trivedi, S., Maggi, F., Sut, S., Dall’Acqua, S., 2020. Phytochemical analysis of Rhazya stricta extract and its use in fabrication of silver nanoparticles effective against mosquito vectors and microbial pathogens. Sci. Total Environ. 700, 134443. https://doi.org/10.1016/j.scitotenv.2019.134443. [DOI] [PubMed]
- Banerjee, P., Eckert, A.O., Schrey, A.K., Preissner, R., 2018. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 46, W257–W263. https://doi.org/10.1093/nar/gky318. [DOI] [PMC free article] [PubMed]
- Benigni R. Computational prediction of drug toxicity: the case of mutagenicity and carcinogenicity. Drug Discov. Today Technol. 2004;1:457–463. doi: 10.1016/j.ddtec.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Biovia, D. S. 2017. Discovery studio visualizer. San Diego, CA, USA, 936.
- Bukhari N.A., Al-Otaibi R.A., Ibhrahim M.M. Phytochemical and taxonomic evaluation of Rhazya stricta in Saudi Arabia. Saudi J. Biol. Sci. 2017;24:1513–1521. doi: 10.1016/j.sjbs.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Cao Z., Cao L., Ding G., Wang Z., Xiao W. Antiviral activity of chlorogenic acid against influenza A (H1N1/H3N2) virus and its inhibition of neuraminidase. Sci. Rep. 2017;7(1) doi: 10.1038/srep45723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilani S.A., Kikuchi A., Shinwari Z.K., Khattak Z.I., Watanabe K.N. Phytochemical, pharmacological and ethnobotanical studies of Rhazya stricta Decne. Phyther. Res. 2007;21:301–307. doi: 10.1002/ptr.2064. [DOI] [PubMed] [Google Scholar]
- Goldhill D.H., Te Velthuis A.J.W., Fletcher R.A., Langat P., Zambon M., Lackenby A., Barclay W.S. The mechanism of resistance to favipiravir in influenza. Proc. Natl. Acad. Sci. U. S. A. 2018;115:11613–11618. doi: 10.1073/pnas.1811345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden F.G., Pavia A.T. Antiviral management of seasonal and pandemic influenza. J. Infect. Dis. 2006;194:119–126. doi: 10.1086/507552. [DOI] [PubMed] [Google Scholar]
- Holmes E.C., Hurt A.C., Dobbie Z., Clinch B., Oxford J.S., Piedra P.A. Understanding the impact of resistance to influenza antivirals. Clin. Microbiol. Rev. 2021;34:1–13. doi: 10.1128/CMR.00224-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indurkhya A., Patel M., Sharma P., Abed S.N., Shnoudeh A., Maheshwari R., Deb P.K., Tekade R.K. Influence of drug properties and routes of drug administration on the design of controlled release system. Dos. Form Des. Considerations. 2018;I:179–223. doi: 10.1016/B978-0-12-814423-7.00006-X. [DOI] [Google Scholar]
- International Committee on Taxonomy of Viruses. https://talk.ictvonline.org/ (accessed 21 May 2022).
- Klimov A., Balish A., Veguilla V., Sun H., Schiffer J., Lu X., Katz J.M., Hancock K. Influenza virus titration, antigenic characterization, and serological Mmethods for antibody detection. Methods Mol. Biol. 2012;865:25–51. doi: 10.1007/978-1-61779-621-0_3. [DOI] [PubMed] [Google Scholar]
- Kormuth K.A., Lakdawala S.S. Emerging antiviral resistance. Nat. Microbiol. 2020;5(1):4–5. doi: 10.1038/s41564-019-0639-7. [DOI] [PubMed] [Google Scholar]
- Krammer F., Smith G.J.D., Fouchier R.A.M., Peiris M., Kedzierska K., Doherty P.C., Palese P., Shaw M.L., Treanor J., Webster R.G., García-Sastre A. Influenza. Nat. Rev. Dis. Prim. 2018;4:1–21. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012;64:4–17. doi: 10.1016/j.addr.2012.09.019. [DOI] [PubMed] [Google Scholar]
- Liu A.L., Wang H.D., Lee S.M.Y., Wang Y.T., Du G.H. Structure-activity relationship of flavonoids as influenza virus neuraminidase inhibitors and their in vitro anti-viral activities. Bioorganic Med. Chem. 2008;16:7141–7147. doi: 10.1016/j.bmc.2008.06.049. [DOI] [PubMed] [Google Scholar]
- Mahmood R., Kayani W.K., Ahmed T., Malik F., Hussain S., Ashfaq M., Ali H., Rubnawaz S., Green B.D., Calderwood D., Kenny O., Rivera G.A., Mirza B., Rasheed F. Assessment of antidiabetic potential and phytochemical profiling of Rhazya stricta root extracts. BMC Complement. Med. Ther. 2020;20:1–17. doi: 10.1186/s12906-020-03035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwat S.K., Usman K., Shah S.S., Anwar N., Ullah I. A review of phytochemistry, bioactivities and ethno medicinal uses of Rhazya stricta Decsne (Apocynaceae) African J. Microbiol. Res. 2012;6:1629–1641. doi: 10.5897/ajmrx11.024. [DOI] [Google Scholar]
- McAuley J.L., Gilbertson B.P., Trifkovic S., Brown L.E., McKimm-Breschkin J.L. Influenza virus neuraminidase structure and functions. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehdi I.A., Sbihi H.M., Tan C.P., Al-Resayes S.I. Seed oil from Harmal (Rhazya stricta Decne) grown in Riyadh (Saudi Arabia): a potential source of δ-tocopherol. J. Saudi Chem. Soc. 2016;20:107–113. doi: 10.1016/j.jscs.2014.09.005. [DOI] [Google Scholar]
- Obaid A.Y., Voleti S., Bora R.S., Hajrah N.H., Omer A.M.S., Sabir J.S.M., Saini K.S. Cheminformatics studies to analyze the therapeutic potential of phytochemicals from Rhazya stricta. Chem. Cent. J. 2017;11:1–21. doi: 10.1186/s13065-017-0240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Ruhlman T.A., Sabir J.S.M., Mutwakil M.H.Z., Baeshen M.N., Sabir M.J., Baeshen N.A., Jansen R.K. Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids. BMC Genomics. 2014;15:1–18. doi: 10.1186/1471-2164-15-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- Roberts M.F., Wink M. Plenum Press; New York, London: 1998. Biochemistry, ecology, and medicinal applications: Alkaloids. [Google Scholar]
- Schrodinger, L. L. C. 2010. The PyMOL molecular graphics system. Version, 1(5), 0
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010 doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavricka C.J., Li Q., Wu Y., Qi J., Wang M., Liu Y., Gao F., Liu J., Feng E., He J., Wang J., Liu H., Jiang H., Gao G.F., Pekosz A. Structural and functional analysis of Laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS Pathog. 2011;7(10):e1002249. doi: 10.1371/journal.ppat.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley R.J., Monto A.S. Resistance of influenza virus to antiviral medications. Clin. Infect. Dis. 2020;71:1092–1094. doi: 10.1093/cid/ciz911. [DOI] [PubMed] [Google Scholar]
- World health orginaztion, 2018. Fect sheets. https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) / (accessed: 6 August 2021).
- GraphPad Prism version 9.1.2 for Windows, GraphPad Software, San Diego, California USA, www.graphpad.com (2021).
- Oellien, F., & Nicklaus, M. C. (2004). Online SMILES translator and structure file generator. National Cancer Institute, 29, 97-101. (https://cactus.nci.nih.gov/translate/). (accessed 20 June 2021)
References
Further Reading
- Prism, G. 2021. GraphPad Software version 9.1.2. La Jolla. California USA.