Abstract
Inhibition of the NEDD8-activating enzyme (NAE), the key E1 enzyme in the neddylation cascade, has been considered an attractive anticancer strategy with the discovery of the first-in-class NAE inhibitor, MLN4924. In this study, we identified SOMCL-19-133 as a highly potent, selective, and orally available NAE inhibitor, which is an analog to AMP. It effectively inhibited NAE with an IC50 value of 0.36 nM and exhibited more than 2855-fold selectivity over the closely related Ubiquitin-activating enzyme (UAE). It is worth noting that treatment with SOMCL-19-133 prominently inhibited Cullin neddylation and delayed the turnover of a panel of Cullin-RING ligases (CRLs) substrates (e.g., Cdt1, p21, p27, and Wee1) at lower effective concentrations than that of MLN4924, subsequently caused DNA damage and Chk1/Chk2 activation, and thus triggered cell cycle arrest and apoptosis. Moreover, SOMCL-19-133 exhibited potent antiproliferative activity against a broad range of human tumor cell lines (mean IC50 201.11 nM), which was about 5.31-fold more potent than that of MLN4924. In vivo, oral delivery treatments with SOMCL-19-133, as well as the subcutaneous injection, led to significant tumor regression in mouse xenograft models. All of the treatments were well tolerated on a continuous daily dosing schedule. Compared with MLN4924, SOMCL-19-133 had a 5-fold higher peak plasma concentration, lower plasma clearance, and a 4-fold larger area under the curve (AUClast). In conclusion, SOMCL-19-133 is a promising preclinical candidate for treating cancers owing to its profound in vitro and in vivo efficacy and favorable pharmacokinetic properties.
Keywords: SOMCL-19-133, NAE inhibitor, MLN4924, NEDD8, Oral administration
Abbreviations: UAE, Ubiquitin-activating enzyme; NAE, NEDD8-activating enzyme; SAE, SUMO-activating enzyme; CRLs, Cullin-RING ligases; NEDD8, Neural precursor cell expressed developmentally down-regulated protein 8; UBL, Ubiquitin-like proteins
Introduction
Eukaryotic proteins can be modified by the attachment of various small molecules and proteins, such as ubiquitin and ubiquitin-like (UBL) proteins, which are essential for a wide range of cellular processes. Over a dozen structurally related UBL members, such as ubiquitin, NEDD8, ISG15, and SUMO, have been identified [1,2]. Among them, NEDD8 (Neural precursor cell expressed developmentally down-regulated protein 8) is closely related to ubiquitin and can similarly be conjugated to protein substrates in a process known as neddylation [3], [4], [5]. Similar to ubiquitination, neddylation covalently attaches NEDD8 to target proteins via NEDD8-specific E1-E2-E3 enzymes. Briefly, NEDD8 is first activated by an E1 NEDD8-activating enzyme (NAE, a heterodimer consisting of NAE1 and UBA3), transferred to one of two E2 enzymes (Ubc12/UBE2M; or UBE2F), finally conjugated by the E3 ligases to the targeted substrates [6,7].
The best-characterized neddylation substrates are the Cullin subunits of Cullin-RING ligases (CRLs), the largest subclass of ubiquitin E3 ligases that regulate the turnover of approximately 20% of proteins in cells [8,9]. Importantly, neddylation is essential for the CRLs ubiquitin ligase activity because it induces a conformational change in the Cullin-RING catalytic core and promotes the ubiquitylation of their protein substrates [10,11]. Therefore, the neddylation pathway plays a crucial role in protein degradation through the modulation of the ubiquitin-proteasome pathway.
Abnormal activation of the NEDD8 conjugation (neddylation) pathway is involved in the pathogenesis of cancer [12], [13], [14]. NAE, the only activation enzyme of the neddylation pathway, has recently been identified as an attractive target for cancer therapy [15], [16], [17]. NAE inhibitors have been shown to suppress tumor growth in a variety of preclinical models, including hematologic malignancies [18,19] and solid tumors [20], [21], [22]. Although scores of inhibitors targeting NAE have been developed, only the sulfonyl-adenosine-based compounds MLN4924 (also named Pevonedistat or TAK-924) and TAS4464 have been formally clinically developed as NAE inhibitors [15,23]. TAS4464 was identified as the most potent compound targeting NAE, however, the clinical trial was put on hold before phase Ⅱ due to serious liver function changes (NCT02978235). As the only clinical active compound, MLN4924 in combination with azacitidine for the treatment of patients with higher-risk myelodysplastic syndromes (MDS), chronic myelomonocytic leukemia (CMML), and low-blast acute myeloid leukemia (AML) have entered phase II/III clinical trials (NCT02610777, NCT03268954). However, the single-agent clinical research progress of MLN4924 is far behind. The treatment outcome of MLN4924 monotherapy by intermittent intravenous delivery is poor both in the animal models and in clinical patients [23,24]. Therefore, we sought to develop new compounds with improved anti-tumor potency than MLN4924.
Here, we report the identification of a novel AMP analog SOMCL-19-133, a highly potent, selective, and orally available inhibitor of NAE. Similar to our previous reported compound 26 [25], SOMCL-19-133 was obtained by scaffold hopping and subsequent optimization of the Head and Tail parts based on MLN4924. Compared with MLN4924, SOMCL-19-133 has a stronger anti-proliferative effect on a variety of tumor cells (IC50: 201.11 vs 646.72 nM, respectively), better pharmacokinetic (PK) properties under oral administration (AUClast: 810 vs 208 ng*h/mL), and showed profound in vivo anti-tumor efficacy.
Materials and methods
Synthesis of SOMCL-19-133
The compound 2-hydroxy-4-(7-(((1R,2S)-2-methoxy-2,3-dihydro-1H-inden-1-yl) amino)-3H- [1,2,3]triazolo [4,5-d]pyrimidin-3-yl)cyclopentyl)methyl sulfamate (SOMCL-19-133) was synthesized at the Shanghai Institute of Materia Medica at the Chinese Academy of Sciences. It was prepared as a white solid, according to the process described previously [26]. High Performance Liquid Chromatography (HPLC) analysis confirmed the purity to be ≥ 98%. 1H NMR (500 MHz, methanol-d4) δ 8.46 (s, 0.76H), 8.31 (s, 0.21H), 7.33 – 7.16 (m, 4H), 6.62 (d, J = 5.3 Hz, 0.23H), 6.05 (d, J = 5.4 Hz, 0.73H), 5.69– 5.60 (m, 1H), 4.62 – 4.57 (m, 1H), 4.43 (dd, J = 9.8, 7.4 Hz, 1H), 4.40 – 4.34 (m, 1H), 4.26 (dd, J = 9.8, 7.3 Hz, 1H), 3.43 – 3.36 (m, 1H), 3.27 – 3.11 (m, 2H), 2.98 – 2.89 (m, 1H), 2.68 – 2.60 (m, 1H), 2.51 – 2.44 (m, 1H), 2.38 – 2.31 (m, 2H). 13C NMR (126 MHz, MeOD) δ 157.70, 157.12, 156.17, 149.43, 142.15, 141.59, 129.29, 127.95, 126.19, 125.45, 83.52, 73.09, 70.53, 58.13, 57.88, 57.76, 44.80, 43.01, 36.77, 34.38. ESI ( [M - H]+) m/z 474.5. HRMS (ESI) calcd for C20H24N7O5S, 474.1565; found, 474.1578.
Drugs and antibodies
MLN4924 and MLN7243 were purchased from MedChemExpress (Monmouth Junction, NJ, USA). Drugs were dissolved in dimethyl sulfoxide (DMSO), aliquoted, and stored at -20°C.
Primary antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA) against the following: Ubc12 (#5641S); UBE2C (#14234S); Ubc9 (#4786S); Cul3 (#2759S); Cul4A (#2699S); NAE1/APPBP1 (#14321S); Cdt1 (#8064S); p27 (#3688S); p-Chk1-Ser317 (#2344S); Chk1 (#2360S); p-Chk2-T68 (#2661S); γH2AX (#2577L); Caspase-8 (#4790S); cleaved caspase-8 (#9748L); caspase-3 (#9662S); cleaved caspase-3 (#9661S); caspase-7 (#9492T); cleaved PARP (#5625S); BAK (#12105S); Noxa (#14766S); Puma (#4976S); BAD (#9292S); BID (#2002S); BCL-XL (#2764S). Antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, USA) against the following: UBA3 (sc-377272); Cul1 (sc-17775); Wee1 (sc-5285); p21 (sc-271610); Chk2 (sc-9604); PARP1 (sc-7150); BAX (sc-493); BCL-2 (sc-492); MCL-1 (sc-819). Antibody against Cul4B (C9995) was from MilliporeSigma (Burlington, Massachusetts, US). Antibody against GAPDH was purchased from Beyotime Biotechnology (Shanghai, China). The secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA, USA).
Cell culture
Human cell lines COLO 205, LNCaP, HGC-27, Jurkat, Clone E6-1, HCT 15, NCI-N87, NCI-H226, and SW620 were purchased from the Shanghai Institutes for Biological Sciences at the Chinese Academy of Sciences (Shanghai, China). NOMO-1 and SUIT-2 were purchased from the Japanese Collection of Research Bioresources Cell Bank (JCRB; Osaka, Japan). All other cell lines were from American Type Culture Collection (ATCC; Manassas, VA). The cells were authenticated by STR (short tandem repeats) DNA profiling and confirmed as negative for Mycoplasma contamination.
In vitro enzyme assays
The inhibition of the tested compounds on NAE or UAE enzymatic activity was determined by homogeneous time-resolved fluorescence (HTRF) assay as reported previously [15,27,28]. SOMCL-19-133 was serially diluted into a 384-well plate containing 50 mM HEPES, 5 mM MgCl2, 20 µM ATP, 250 µM L-glutathione, and 0.05% BSA, pH 7.5. A mixture containing 150 nM His-NEDD8, 80 nM GST-Ubc12, and 2 nM NAE recombinant enzymes (R&D Systems, USA) was then added. Reactions were incubated for 2 h at 27°C and stopped with stop/detection buffer (0.1 M HEPES, 20 mM EDTA, 410 mM KF, 0.05% Tween 20, pH 7.5, 0.25 µg/mL MAb Anti 6HIS-Eu, and 2 µg/mL MAb Anti GST-XL665 (CisBio International)). After incubation for 24 h, the plate was read on a PerkinElmer EnVision, using 330 nM excitation/620 nM emission and 330 nM excitation/665 nM emission. The inhibition rate (%) was calculated as follows:
The mean IC50 values were determined via the Logit method from three independent assays. A similar assay protocol was used to measure UAE enzymes.
Cellular thioester assay
Cellular E2-UBL thioester assays were performed as reported previously [15,23]. HCT-116 cells were treated with increasing concentrations of SOMCL-19-133, MLN4924, or 0.01% DMSO for 4 h. Whole-cell extracts were prepared and fractionated by SDS-PAGE under nonreducing conditions. The fractionated extracts were immunoblotted with primary antibodies to UBC12, UBE2C, and UBC9. All primary antibodies were used at 1:1000 dilution. The HRP-conjugated anti-mouse or anti-rabbit secondary antibodies were used at 1:5000 dilution, and blots were imaged using a Bio-Rad ChemiDoc Imaging system.
Western blotting
The standard western blotting protocol was used to measure the cellular level of the indicated proteins as described previously [29]. Briefly, cells treated with indicated drugs were lysed with 100 µL of SDS lysis buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 0.05% bromophenol blue, 10% glycerin, and 100 mM DTT) and boiled for 10 min. The lysates were separated by SDS-PAGE on 10% acrylamide gels at 150 V for 1 h, transferred by Trans-Blot Turbo (Bio-Rad), and blocked with TBST 5% milk. The membranes were incubated with primary antibodies at 1:1000, followed by secondary anti-rabbit-HRP or anti-mouse-HRP. The signal was analyzed using a Bio-Rad ChemiDoc Imaging system. All blots were derived from the same experiment and processed in parallel.
Flow cytometry
Flow cytometry was conducted as described previously [30]. Following treatment with SOMCL-19-133, MLN4924 or 0.01% of DMSO for the indicated time, HCT-116 and RKO cells were prepared for the analysis of cell cycle distribution or apoptosis. For cell cycle analysis, cells were incubated with indicated drugs for 24 h, fixed with 70% ethanol overnight at 4°C, followed by incubating with RNase A for 15 min, then stained with propidium iodide (Beyotime; Shanghai, China) for 30 min. For apoptosis analysis, cells were incubated with indicated drugs for 48 h, collected, and stained with FITC Annexin V and propidium iodide according to the manufacturer's protocols (Apoptosis Detection Kit I, #556547; BD Pharmingen). All data were collected using a FACS Calibur instrument (BD Biosciences; USA) and analyzed with FlowJo software.
Cell viability assay
Cells were plated at a density of 1,000-8,000 cells per well in 96-well plates before treatment with SOMCL-19-133 or MLN4924 at varying concentrations for 3 days. To measure cell viability, sulforhodamine B (SRB) and CCK-8 assays were performed as described earlier [30]. The inhibition rate (%) was calculated as follows:
The IC50 value was calculated using the Logit method.
Clonogenic assay
Cells were seeded at a density of 1×103 in a 6-well plate and treated with indicated doses of SOMCL-19-133, MLN4924 (0.01 and 0.03 µM), or 0.01% of DMSO. After 11 days of culture, the colonies were fixed, stained with SRB, and the optical density value was measured at 560 nM using a Spectra-MAX190 microplate reader (Molecular Devices, CA, USA).
In vivo studies
In vivo studies were carried out following the Animal Care and Use Committee of Shanghai Institute of Materia Medica and performed according to the institutional ethical guidelines.
HCT-116 and MV-4-11 xenografts (s.c.). 5-week female BALB/c nude mice were subcutaneously implanted with tumor fragments at the right armpit. As the tumor size reached 90-110 mm3, mice were randomized into the vehicle group (n=12) and treatment groups (n=6). Mice were dorsal subcutaneously injected with SOMCL-19-133, MLN4924, or vehicle (10% 2-Hydroxypropyl-β-cyclodextrin) once a day (QD) consecutively or twice daily (BID; 5on/5off) for two cycles. Recorded the tumor volume twice a week and evaluated the anti-tumor efficacy by calculating the relative growth rate (T/C; T is the treatment group and C is the control group) on the last day. Statistical significance was assessed by using one-way ANOVA and Dunnett's multiple comparison tests to compare the significant difference (P < 0.05) between vehicle and treatment groups.
NCI-H1975 xenograft (s.c.). Freshly dissected NCI-H1975 fragments were subcutaneously implanted onto the mice's right flank. Randomized into groups as tumor size reached approximately 120 mm3. Mice were treated with 3 mg/kg SOMCL-19-133 daily for 14 days and were sacrificed 31 days after withdrawal of the drug. Determined the statistical significance by using Student's t-test.
RKO and MV-4-11 xenografts (p.o.). Mice were treated with vehicle, various doses (2.5, 5, 10 mg/kg) of SOMCL-19-133 (oral administration; QD) or 120 mg/kg MLN4924 (intravenous injection; twice a week) for 18-21 days. Percent inhibition was calculated from the final average tumor volume at the end of the study. One-way ANOVA was used to evaluate the therapeutic difference.
Pharmacokinetic studies
Male Sprague-Dawley (SD) rats (200-250 g) fasted for 12 h were administrated with SOMCL-19-133 or MLN4924 at 1 mg/kg i.v. in DMSO/EtOH/PEG300/NaCl (5/5/40/50, v/v/v/v) or 3 mg/kg p.o. in DMSO/0.5% HPMC (5/95, v/v). Blood was collected at the indicated times, processed, and analyzed by LC/MS/MS method. Each time point is the average value of three animals and the data were analyzed by using the non-compartmental method (Phoenix, version 1.3; Pharsight, Mountain View, CA) to derive pharmacokinetic parameters.
Statistical analyses
All statistical data were assessed via Graphpad Prism 7.0 and presented as mean ± SEM without special statements. *, P < 0.05; **, P < 0.01; ***, P < 0.001, P < 0.05 was considered to be statistically significant.
Results
Selectivity and potency of SOMCL-19-133 as a NEDD8-activating enzyme inhibitor
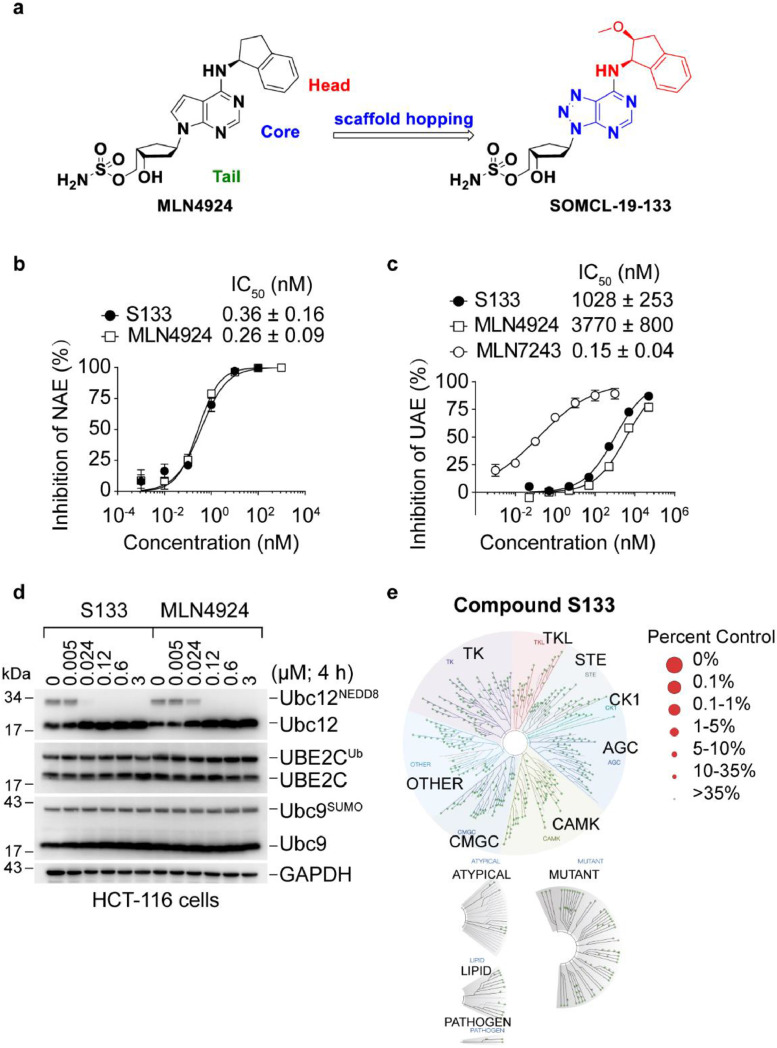
To improve the cellular activity of MLN4924, the competitive structural optimization was conducted, first by scaffold hopping to screen a small set of compounds bearing different central cores, followed by optimization of the Head and Tail structural motifs, leading to the identification of the new candidate compound SOMCL-19-133 bearing a distinct triazolopyrimidine framework. (Figure 1a). The IC50 values of SOMCL-19-133 and MLN4924 against NAE were 0.36 ± 0.16 nM and 0.26 ± 0.09 nM, respectively (Figure 1b), whereas the former displayed more than 2855-fold selectivity towards the closely related UAE (IC50: 1028 ± 253 nM) (Figure 1c). In comparison, the IC50 value of TAS4464 against NAE was 0.004 ± 0.0008 nM (Supplemental Figure 1a). Next, we characterized the selectivity of SOMCL-19-133 in colon cancer HCT-116 cells using a panel of UBL-thioester charging assays that evaluate the levels of E2-UBL thioester products by western blot. Consistent with the results using purified NAE, SOMCL-19-133 showed highly potent inhibition of NAE capable of activating neddylation, as indicated by the decreases in the levels of NEDD8 charged Ubc12. As shown in Figure 1d, SOMCL-19-133 selectively reduced cellular Ubc12NEDD8, but the levels of UBE2CUb and Ubc9SUMO in HCT-116 cells were unaffected (Figure 1d). Therefore, SOMCL-19-133 showed strong selectivity over the ubiquitin and SUMO pathways. Finally, to exclude the off-target activities, we screened SOMCL-19-133 against a panel of 468 kinases and their mutants using KINOMEScan screening at a concentration of 1 µM. SOMCL-19-133 did not demonstrate detectable inhibition or stimulation of all of the using enzymes (Figure 1e). These results indicate that SOMCL-19-133 selectively and functionally inhibits NAE.
Figure 1.
SOMCL-19-133 demonstrates potent and selective inhibition of NAE. (a) Chemical structures of MLN4924 and SOMCL-19-133. (b and c) Dose-response curves of enzymatic inhibition by SOMCL-19-133. (b) NAE; (c) UAE. NAE inhibitor MLN4924 and UAE inhibitor MLN7243 were used as the positive control, respectively. Data are represented as mean ± SD from three independent experiments. S133 is short for SOMCL-19-133. (d) Representative western blot analysis showing the effect of SOMCL-19-133 on Ubc12NEDD8, UBC10Ub, and Ubc9SUMO thioester levels. HCT-116 cells were treated with different concentrations of SOMCL-19-133 and MLN4924 for 4 h. Cell lysates were separated by non-reducing SDS-PAGE and immunoblotted using the indicated antibodies. The experiment was repeated three times. (e) Kinase affinity assay of SOMCL-19-133 against 468 kinases and some of their mutants at 1 µM with DiscoveRx KINOMEScan screening platform and presented as a TREEspot interaction map.
SOMCL-19-133 induced accumulation of CRLs substrates and triggered cell cycle arrest and apoptosis
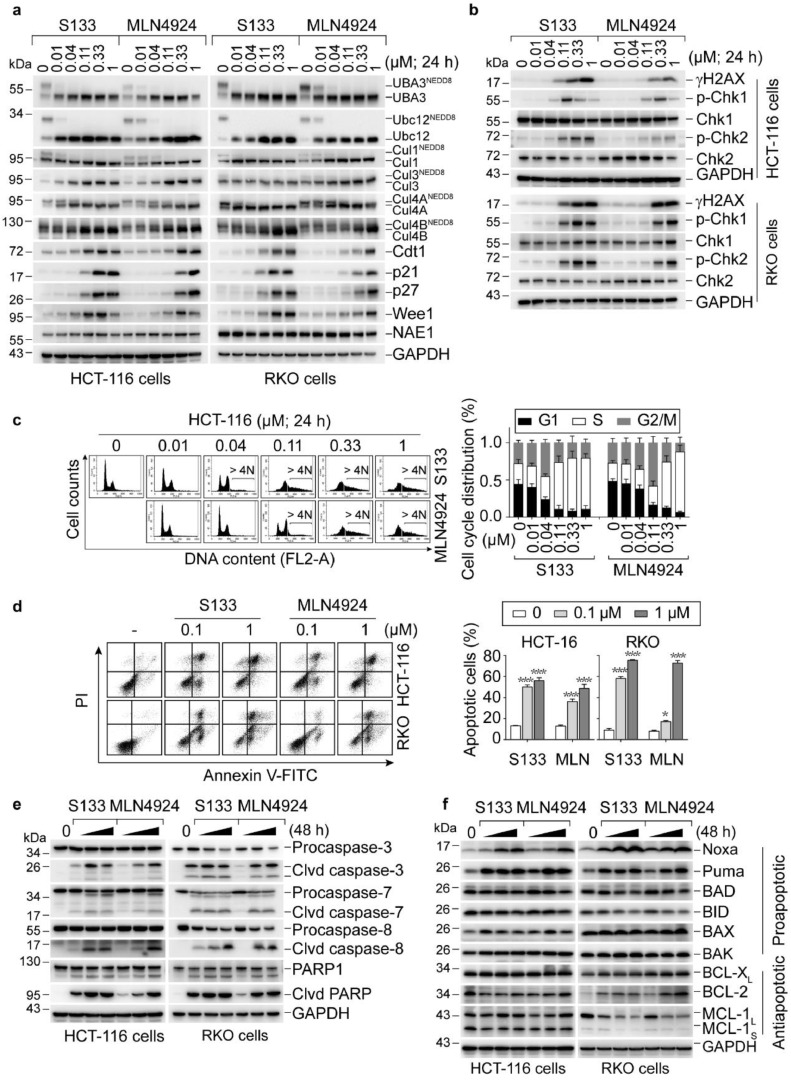
NAE inhibitors have been shown to selectively inhibit cullin neddylation, inactivate CRLs that cause the accumulation of CRLs substrates, and trigger multiple biological responses [15,31]. We then evaluated the effects of SOMCL-19-133 on the protein levels of various CRLs substrates in HCT-116 and RKO cells using western blotting. A 24-h treatment with SOMCL-19-133 (dose range, 0.01–1 µM) induced a concentration-dependent reduction of the levels of UBA3NEDD8, Ubc12NEDD8, Cul1NEDD8, Cul3NEDD8, Cul4ANEDD8, and Cul4BNEDD8 and intensively increased the CRLs substrates Cdt1, p21, p27, and Wee1 (Figure 2a). Notably, SOMCL-19-133 is more effective than MLN4924 in blocking intracellular neddylation modification in HCT-116 and RKO cells.
Figure 2.
SOMCL-19-133 inhibits the NEDD8 pathway, which leads to DNA damage response, cell cycle arrest, and apoptosis. (a) Representative western blot showing the levels of UBA3NEDD8, Ubc12NEDD8, CulNEDD8 conjugates, and CRLs substrates in SOMCL-19-133 or MLN4924 treated cells. (b) SOMCL-19-133 induced a dose-dependent increase in DNA damage. HCT-116 and RKO cells were treated with various concentrations of SOMCL-19-133 or MLN4924 for 24 h. Induction of γH2AX, p-Chk1 (Ser317), and p-Chk2 (T68) was determined by western blot. (c and d) Effects of SOMCL-19-133 on cell cycle distribution and apoptosis. Cells were treated with SOMCL-19-133, MLN4924, or Control for (c) 24 h or (d) 48 h, and (c) DNA profiles were analyzed by PI staining-based flow cytometry; > 4N represents cells with greater than tetraploid DNA content. >4N cells were not included in the cell cycle distribution analysis. (d) Apoptotic cells were detected by Annexin V-FITC/PI staining-based flow cytometry. MLN represents MLN4924. Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. (e) The levels of cleaved (Clvd) caspase-3, cleaved caspase-7, cleaved caspase-8, and cleaved PARP were assessed by western blot in cells treated with various doses (0, 0.1, 0.3, 1 µM) of SOMCL-19-133, MLN4924 for 48 h. (f) Western blot analysis of Noxa, Puma, BAD, BID, BAX, BAK, BCL-XL, BCL-2, and MCL-1 in cells treated with SOMCL-19-133 or MLN4924 at 0, 0.1, 0.3, 1 µM for 48 h. All of the experiments were repeated three times.
As overexpression of Cdt1 is known to cause DNA re-replication and trigger DNA damage response (DDR) [15,32]. Thus, we tested whether checkpoint activation occurred under these conditions. Indeed, the treatments with SOMCL-19-133, just as with MLN4924, caused the accumulation of DNA double-strand breaks marked by the increased levels of γH2AX (Figure 2b) in both HCT-116 and RKO cells. These results were further supported by the enhanced protein levels of p-S317-Chk1 and p-T68-Chk2 (Figure 2b).
We further analyzed whether CRLs substrates accumulation and checkpoint activation in response to SOMCL-19-133 treatment was accompanied by cell cycle dysregulation. As expected, an increase in cells in the G2/M phase was observed at 24 h with 40 nM SOMCL-19-133 treatment (Figure 2c). Moreover, the S phase cells increased at the higher doses, accompanied by an increase in the polyploidy population (DNA content > 4N) (Figure 2c and Supplemental Figure 2). Apoptosis is always accompanied by aberrant cell cycle progression. We then assessed the apoptotic effects of SOMCL-19-133 on HCT-116 and RKO cells using Annexin V-FITC/PI labeling flow cytometry. As shown in Figure 2d, 0.1 (P < 0.001) and 1 (P < 0.001) µM SOMCL-19-133 induced profound apoptosis in both cell lines at 48 h post-treatment. This apoptotic induction capacity was further confirmed by western blotting analysis of apoptosis-related proteins. As shown in Figure 2e, treatment with SOMCL-19-133 intensively increased the expression of cleaved caspase-3, -7, -8, and PARP1. The BH3-only protein Noxa has been reported to contribute to MLN4924-induced apoptosis [33]. Consistently, immunoblotting for pro- and anti-apoptotic Bcl-2 family members in cells treated with SOMCL-19-133 revealed a concentration-dependent increase in the BH3-only proteins Noxa and Puma, while BAD, BID, BAX, BAK, and anti-apoptotic proteins, including BCL-2, BCL-XL, and MCL-1, were relatively unchanged (Figure 2f). The results indicate that SOMCL-19-133 profoundly elicits CRLs substrates accumulation, DNA damage response, cell cycle arrest, and apoptosis in colon cancer cells.
Anti-proliferative activity of SOMCL-19-133 in human cancer cells
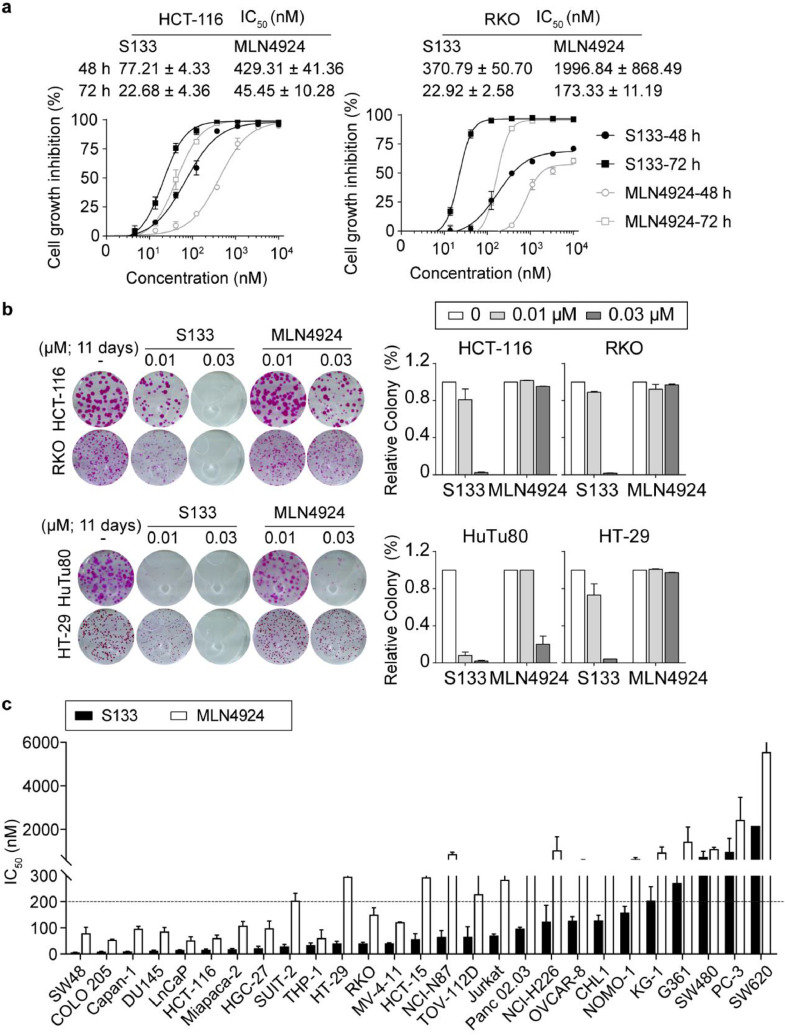
NAE inhibition leads to DNA damage accumulation, impaired cell cycle progression, and induction of cell apoptosis; therefore, we hypothesized that SOMCL-19-133 would exert a potent antiproliferative activity against human cancer cells. To this end, we evaluated the growth inhibitory effect of SOMCL-19-133 in the colon cancer HCT-116 and RKO cells by treatment with serial dilutions of SOMCL-19-133 (0.001–10 µM) using SRB assays. The growth inhibition of the cells was determined at two distinct time points (48 and 72 h) after exposure to the indicative drugs. Indeed, SOMCL-19-133 treatment showed marked time- and dose-dependent inhibition of cell proliferation in both cell lines (Figure 3a). These cell lines were also profoundly sensitive to TAS4464 (Supplemental Figure 1b). Furthermore, the clonogenic assay showed that SOMCL-19-133 was effective in targeting a variety of colon cancer cell lines regardless of different genetic backgrounds (Figure 3b).
Figure 3.
SOMCL-19-133 displays broad-spectrum inhibitory effects on the viability of a panel of human cancer cell lines. (a) Dose-response curves of SOMCL-19-133 and MLN4924-treated HCT-116 and RKO cells. Data are presented as mean ± SEM, n = 3. (b) Colony-forming efficiency of HCT-116, RKO, HuTu80, and HT-29 cells treated with SOMCL-19-133, MLN4924, or Control for 11 days. Data are mean ± SEM, n = 3. (c) IC50 values of SOMCL-19-133 and MLN4924 against a panel of human tumor cell lines were assessed by using SRB or CCK8 assay. A panel of 28 human cancer cell lines was treated with SOMCL-19-133 or MLN4924 at a series of different concentrations (0.001-20 µM), and the viable cells are measured after 72 h of treatment. Data were presented as mean ± SEM, n = 3.
In recent years, NAE inhibitors have been investigated in the treatment of hematologic malignancies [18,34,35]. In addition, NAE inhibitors have demonstrated potential efficacy in solid tumors, including colon [36], breast [21,37], pancreas [38], clear cell renal cell carcinoma [39], [40], [41], and ovarian cancer [42]. Thus, we assessed the antiproliferative effect of SOMCL-19-133 using a panel of 28 cancer cell lines derived from different cancer types across solid and hematological tumors. As a result, the data showed that SOMCL-19-133 treatment induced substantial cell-killing effects on almost all the tested cell lines, with variable IC50 values that ranged from 0.006 µM to 2.16 µM (Table 1). Many tumor cell lines demonstrated IC50 values in the double-digit nanomolar range, and 82% (23/28) of cell models were especially sensitive to the treatment of SOMCL-19-133 (IC50 < 200 nM) (Figure 3c and Table 1). Furthermore, its cell-killing effects were on average 5.31-fold greater than that of MLN4924 in all the examined cancer cell lines (Table 1). These results indicate that SOMCL-19-133 demonstrated substantial antiproliferative effects in solid tumor cells, including colorectal-, pancreatic-, gastric-, and prostate-derived cell lines (Table 1).
Table 1.
Inhibitory concentration (IC50) values of cancer cell lines in response to NAE inhibitors SOMCL-19-133 and MLN4924 treatments.
| No. | Cell lines | Types | IC50 (mean ± SD) (nM) |
IC50 (4924)/ IC50 (S133) | |
|---|---|---|---|---|---|
| SOMCL-19-133 | MLN4924 | ||||
| 1 | THP-1 | Leukemia | 33.59 ± 8.48 | 60.15 ± 32.24 | 1.79 |
| 2 | MV-4-11 | Leukemia | 40.83 ± 2.21 | 121.17 ± 1.79 | 2.97 |
| 3 | Jurkat, Clone E6-1 | Leukemia | 70.43 ± 6.19 | 283.83 ± 95.63 | 4.03 |
| 4 | NOMO-1 | Leukemia | 159.29 ± 23.33 | 626.87 ± 77.82 | 3.94 |
| 5 | KG-1 | Leukemia | 204.37 ± 53.17 | 935.65 ± 257.77 | 4.58 |
| 6 | SW48 | Colon cancer | 5.59 ± 1.16 | 79.54 ± 23.09 | 14.23 |
| 7 | COLO 205 | Colon cancer | 8.75 ± 1.69 | 54.23 ± 2.47 | 6.20 |
| 8 | HCT-116 | Colon cancer | 15.71 ± 3.78 | 60.78 ± 11.74 | 3.87 |
| 9 | HT-29 | Colon cancer | 40.22 ± 8.54 | 296.27 ± 32.30 | 7.37 |
| 10 | RKO | Colon cancer | 40.23 ± 5.12 | 150.50 ± 26.38 | 3.74 |
| 11 | HCT 15 | Colon cancer | 56.88 ± 20.93 | 293.65 ± 80.29 | 5.16 |
| 12 | SW480 | Colon cancer | 732.48 ± 258.11 | 1091.82 ± 91.73 | 1.49 |
| 13 | SW620 | Colon cancer | 2164.76 ± 221.23 | 5552.47±2367.56 | 2.56 |
| 14 | Capan-1 | Pancreatic cancer | 9.44 ± 0.95 | 96.48 ± 10.06 | 10.22 |
| 15 | MIA PaCa-2 | Pancreatic cancer | 17.98 ± 3.18 | 108.77 ± 16.05 | 6.05 |
| 16 | SUIT-2 | Pancreatic cancer | 29.22 ± 7.63 | 203.71 ± 29.05 | 6.97 |
| 17 | Panc 02.03 | Pancreatic cancer | 97.25 ± 5.67 | 355.91 ± 39.08 | 3.66 |
| 18 | HGC-27 | Gastric cancer | 22.03 ± 7.44 | 98.53 ± 28.33 | 4.47 |
| 19 | NCI-N87 | Gastric cancer | 65.08 ± 24.18 | 864.58 ± 77.52 | 13.28 |
| 20 | DU 145 | Prostate cancer | 12.55 ± 2.79 | 86.45 ± 15.13 | 6.89 |
| 21 | LNCaP | Prostate cancer | 15.24 ± 0.99 | 52.48 ± 13.36 | 3.44 |
| 22 | PC-3 | Prostate cancer | 962.55 ± 624.98 | 2434.10 ± 1046.08 | 2.53 |
| 23 | TOV-112D | Ovarian cancer | 66.00 ± 39.15 | 228.76 ± 103.56 | 3.47 |
| 24 | OVCAR-8 | Ovarian cancer | 127.15 ± 15.72 | 540.59 ± 64.15 | 4.25 |
| 25 | CHL-1 | Melanoma | 128.53 ± 19.75 | 320.46 ± 121.92 | 2.49 |
| 26 | G-361 | Melanoma | 272.36 ± 182.09 | 1427.71 ± 683.93 | 5.24 |
| 27 | NCI-H1975 | NSCLC | 108.47±22.77 | ND | NA |
| 28 | NCI-H226 | NSCLC | 124.18 ± 62.36 | 1035.87 ± 624.15 | 8.34 |
| Average | 201.11 | 646.72 | 5.31 | ||
The IC50 (the half-maximal inhibitory concentration) values were determined from cell viability assays (SRB or CCK-8). Repeated the experiment three times and expressed the data as mean ± SD. ND. not determined; NA, not applicable.
The antiproliferative effects of SOMCL-19-133 are likely to be driven by apoptosis, as apoptotic cell death markers (cleaved caspase-3, -7, -8, and cleaved PARP) were detectable following treatment (see Figure 2e). These results showed that SOMCL-19-133 has potent in vitro antitumor activity in a broad range of malignancies, including leukemia, colon, pancreas, gastric, and prostate.
SOMCL-19-133 inhibited tumor growth in multiple xenograft models
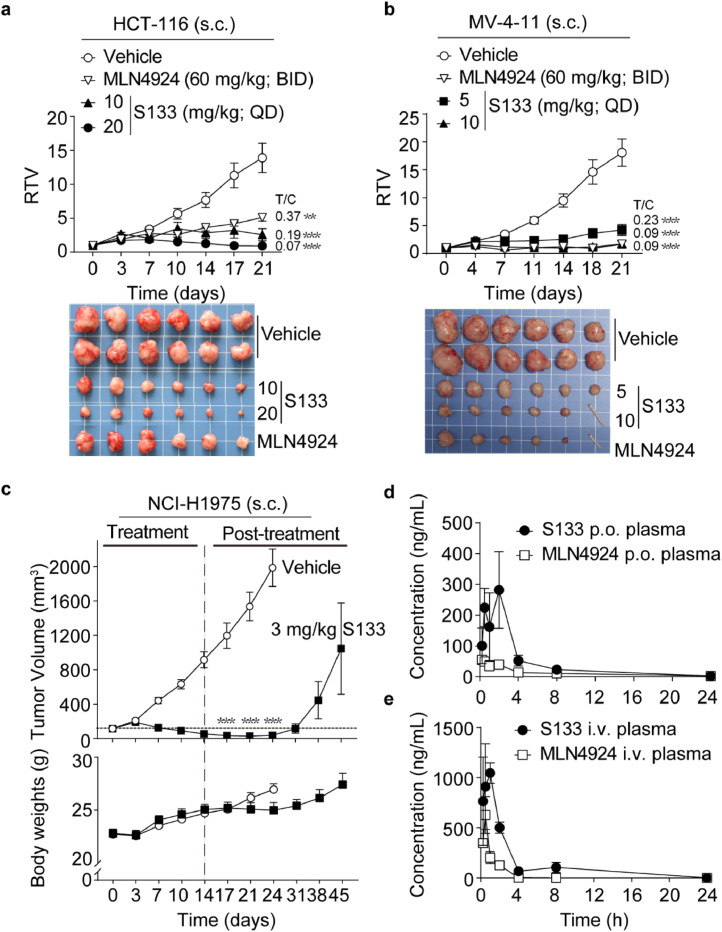
The antitumor activity of SOMCL-19-133 was determined with a variety of sensitive (IC50<200 nM) cell line-derived xenograft (CDX) tumor models that represent both solid and hematological cancers. SOMCL-19-133 treatment of three tumor subcutaneous xenograft models, human colorectal cancer HCT-116 cell xenografts, human leukemia MV-4-11 cell xenografts, and human non-small cell lung cancer NCI-H1975 cell xenografts, is shown in Figure 4. Previous studies have shown that MLN4924 was administered on a BID schedule 5on/5off by s.c. at 60 mg/kg significantly inhibited HCT-116 tumor xenograft growth [15]. To evaluate the anti-tumor activity of SOMCL-133, we administered the compound by s.c. once daily (QD) to mice bearing xenografts, and MLN4924, on a BID schedule, served as a positive control. The inhibition of tumor growth (T/C) was calculated on the last day of treatment.
Figure 4.
In vivo efficacy of SOMCL-19-133 by subcutaneous injection and the plasma drug concentrations-time profiles in SD rats. (a and b) The inhibitory effects of SOMCL-19-133 on (a) human colon cancer HCT-116 and (b) human leukemia MV-4-11 xenograft models. Mice bearing subcutaneous xenografts were given with the indicated dose of SOMCL-19-133 by s.c. injection once daily, or MLN4924 (60 mg/kg, BID, 5on/5off) for 21 days (n = 6). The tail indicated the tumor disappeared in that mouse. (c) The antitumor effects of SOMCL-19-133 on human non-small cell lung cancer NCI-H1975 in vivo model. Mice bearing with tumors were dosed with SOMCL-19-133 (3 mg/kg; s.c.) daily for 14 days (n = 6) and were sacrificed on day 24 (vehicle group) or day 45 (SOMCL-19-133-treated group) after withdrawal of the drug. (d) Plasma concentration-time profile of SOMCL-19-133 and MLN4924 at 3 mg/kg p.o. in SD rats, n=3. (e) Plasma concentration-time profile of SOMCL-19-133 and MLN4924 at 1 mg/kg i.v. in SD rats, n=3. Data were represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
In the HCT-116 xenograft model, significant inhibition of tumor growth was observed when SOMCL-19-133 was administered at 10 mg/kg and 20 mg/kg (T/C = 0.19, P < 0.001 and 0.07, P < 0.001, respectively) on a QD dosing schedule, when compared to the vehicle-treated group. These effects were also preferable to MLN4924 treatment (T/C = 0.37, P = 0.005) (Figure 4a). In the leukemia MV-4-11 xenograft model, rapid tumor shrinkage with partial or complete remissions were observed in tumor-bearing mice after repeated daily treatment with 5 mg/kg and 10 mg/kg SOMCL-19-133 (T/C = 0.23, P < 0.001; T/C = 0.09, P < 0.001; respectively) (Figure 4b). Among the preclinical models tested, NCI-H1975 is the CDX xenograft most sensitive to in vivo SOMCL-19-133 treatment. In this model, daily administration of 3 mg/kg SOMCL-19-133 for 14 days led to nearly complete tumor regression, and all the treated animals remained tumor-free for over 10 days after withdrawal of the drug (Figure 4c). Therefore, SOMCL-133, at a low dose, significantly inhibited tumor growth in NCI-H1975-bearing mice. All of the tested dosages of drugs were well tolerated during the treatment period, and no significant weight loss was observed in the treatment groups (Figure 4c and Supplementary Figure 3).
Pharmacokinetic studies of SOMCL-19-133
To further evaluate the developability of SOMCL-19-133, we tested the plasma pharmacokinetic (PK) properties of SOMCL-19-133 in comparison with MLN4924 in SD rats after oral and intravenous administration. The drug concentration-versus-time profiles were shown in Figure 4d, e, and pharmacokinetic parameters were listed in Table 2. Following oral (3 mg/kg) administration in mice, SOMCL-19-133 demonstrated an almost 4-fold increase in drug exposure (AUClast: 810 vs. 208 ng*h/mL) and more than a 5-fold increase in Cmax (350 vs. 64 ng/mL) compared to that of oral MLN4924. Following intravenous (1 mg/kg) administration, SOMCL-19-133 similarly exhibited more than a 4-fold increase in drug exposure (AUClast: 2500 vs. 560 ng*h/mL) compared with MLN4924. In contrast, SOMCL-19-133 displayed a lower systemic plasma clearance (CL: 5.33 vs. 29.7 mL/min/kg) than that of MLN4924. Moreover, SOMCL-19-133 showed moderate terminal half-lives (T1/2) with a bioavailability of 10.8%.
Table 2.
Pharmacokinetic parameters of SOMCL-19-133 and MLN4924 in SD rats plasma after oral and intravenous administration.
| Drugs | Dose(mg/kg) | Route | T1/2(h) | Tmax(h) | Cmax(ng/mL) | AUClast(ng*h/mL) | Vss_obs(mL/kg) | CL(mL/min/kg) | F(%) |
|---|---|---|---|---|---|---|---|---|---|
| S133 | 3 | p.o. | 3.14 | 1.17 | 350 | 810 | NA | NA | 10.8 |
| 1 | i.v. | 5.68 | NA | NA | 2500 | 1582 | 5.33 | NA | |
| MLN4924 | 3 | p.o. | 5.50 | 0.92 | 64 | 208 | NA | NA | 12.4 |
| 1 | i.v. | 0.71a | NA | NA | 560a | 1783a | 29.70a | NA |
The data were collected and measured from three SD rats, and analyzed by using the non-compartmental method. p.o., oral administration; i.v., intravenous injection; NA, not applicable. aData are from reference [25].
Potent antitumor activity of oral SOMCL-19-133 in vivo
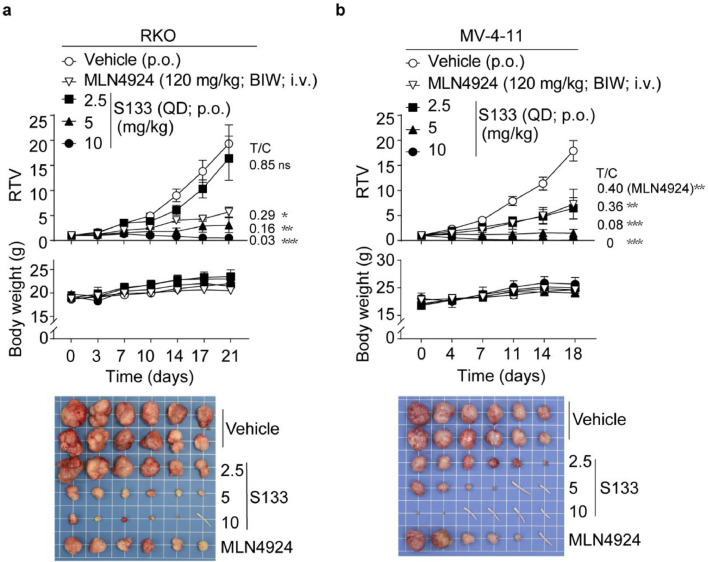
Compared with MLN4924, either oral delivery or intravenous injection of SOMCL-19-133 had a higher plasma drug exposure, and SOMCL-19-133 is 5.31 folds cytotoxic than MLN4924, we hypothesized that SOMCL-19-133 is active by oral gavage, a preferable route of administration than intravenous injection. Thus the therapeutic efficacy of oral SOMCL-19-133 was tested in nude mice bearing NAE inhibitors-sensitive RKO and MV-4-11 xenografts derived from solid tumor and hematologic malignancy, respectively.
Since MLN4924 was administrated intermittently to patients by intravenous infusion in the clinical trial, the regimen for MLN4924 was determined at 120 mg/kg twice weekly (BIW) by i.v. injection as previously described [23]. SOMCL-19-133 was orally delivered to mice at 2.5, 5, and 10 mg/kg daily (QD, p.o.). As shown in Figure 5a, SOMCL-19-133 significantly inhibited the growth of RKO xenografts in a dose-dependent manner, yielding T/C values of 0.16 (P = 0.004) and 0.03 (P < 0.001) at doses of 5 and 10 mg/kg, respectively. Similarly, oral administration of SOMCL-19-133 at 10 mg/kg for 18 days significantly inhibited the growth of MV-4-11 xenografts in mice, with four out of six mice achieving a complete remission (Figure 5b). Tumor reduction after treatment with SOMCL-19-133 at 5 (T/C = 0.08, P < 0.001) and 10 mg/kg (T/C = 0, P < 0.001) daily were much greater than that of MLN4924. These treatments were well tolerated, with no significant weight loss or toxic symptoms in the treatment groups (Figure 5a and b). These data indicate that at tolerable doses, SOMCL-19-133 is an orally effective NAE inhibitor with antitumor activity in mice.
Figure 5.
Oral administration of SOMCL-19-133 suppresses the growth of RKO and MV-4-11 xenografts in nude mice. (a and b) The effects of oral administration of SOMCL-19-133 on (a) human colon cancer RKO and (b) human leukemia MV-4-11 xenografts. Mice bearing subcutaneous xenografts were dosed with SOMCL-19-133 orally daily, or MLN4924 (120 mg/kg; i.v.) twice a week for (a) 21 days or (b) 18 days (n = 6). The tail indicated the tumor disappeared in that mouse.
Discussion
Here, we have described a novel, potent, and orally available NAE inhibitor SOMCL-19-133. Similar to other NAE inhibitors, SOMCL-19-133 blocked NAE catalytic activity at the first step of the neddylation pathway. At the cellular level, SOMCL-19-133 inhibited neddylation and decreased the turnover of CRLs substrates Cdt1, p21, p27, and Wee1 with lower effective concentrations than that of MLN4924. It showed strong antitumor activity against many kinds of cancer cell lines across solid and hematological tumors in vitro. Compared with MLN4924, this compound had a broader spectrum of cell-killing activity, lower systemic plasma clearance, and significantly higher plasma drug exposure, this allowed for oral administration once a day. The xenograft mouse models showed that all the schedules of SOMCL-19-133 (subcutaneous injection or oral delivery) significantly reduced tumor burden, compared with the control. These therapeutic effects were achieved with tolerable toxicity.
With IC50 values of 0.36 nM and 1.03 µM against NAE and UAE, respectively, SOMCL-19-133 is a potent and selective small-molecule NAE inhibitor. We were surprised by the significantly greater cytotoxicity in a wide range of cell lines, compared with MLN4924, which has comparable potency against NAE catalytic activity. We initially suspected that SOMCL-19-133 might have activity other than NAE inhibition, and this activity may be responsible for or contributing to the cytotoxicity. However, the KINOMEScan screening assay revealed SOMCL-19-133 had no activity against any of the kinases tested. In the cellular systems, the result showed that SOMCL-19-133 was able to suppress the activity of NAE but not the closely related UAE and SAE. We also observed that the antiproliferation potency of SOMCL-19-133 in both HCT-116 and RKO cells was stronger than that of MLN4924 under the same conditions, and it was time- and dose-dependent. These results suggest that its remarkable cytotoxic properties in cancer cells are likely a direct result of its ability to inhibit NAE. The improvement of growth inhibition may be due to the increase in solubility or membrane permeability which is needed to be verified.
NAE inhibitors significantly suppress the proliferation of cancer cells via the induction of DNA re-replication, cell cycle arrest, and apoptosis [15]. Indeed, SOMCL-19-133 induced the accumulation of a panel of tumor-suppressive Cullin-RING ligase substrates (e.g., Cdt1, p21, p27, and Wee1). Notably, SOMCL-19-133 induced more prominent DNA damage (marked by γH2AX and p-Chk1), greater cell cycle arrest, and apoptosis in HCT-116 and RKO cells than MLN4924. Consistent with this, SOMCL-19-133 resulted in 5-fold more potent cell-killing effects than MLN4924 in the tested cancer cell lines. These data further supported that SOMCL-19-133 exerts its effects through NAE inhibition.
NAE inhibitors are emerging as an exciting new class of potential anticancer agents for the treatment of solid and hematological malignancies [43]. However, stand-alone clinical trials of these inhibitors have been unsuccessful, due to insufficient clinical benefits and difficulty in defining suitable patient populations [44]. Compared with MLN4924, SOMCL-19-133 has enhanced antitumor activity resulting from increased blockage of neddylation modification. Consistently, SOMCL-19-133 demonstrated substantial antitumor activity in the preclinical colon and non-small cell lung cancer models (e.g., HCT-116 and NCI-H1975), implying SOMCL-19-133 may demonstrate clinical benefit in patients with advanced solid malignancies. Our preclinical data also suggested that a potential therapeutic window may exist for SOMCL-19-133 treatment. Mice tolerated SOMCL-19-133 with repeated dosing at 20 mg/kg s.c. or 10 mg/kg p.o., which yielded robust antitumor activity with minimal bodyweight loss. Of the 28 cell lines tested, SW620, PC-3, and SW480 cells were much more resistant to the compounds. However, little is known about the molecular basis defining the efficacy of NAE inhibitors. Nevertheless, neddylation biomarkers should be further developed to increase the therapeutic efficacy of SOMCL-19-133.
First-generation NAE inhibitors, such as MLN4924, are administered by intermittent intravenous injections (Int-IV). Notably, oral administration of SOMCL-19-133 with a continuous daily dosing schedule displayed limited toxicity and favorable efficacy in vivo. In addition, the overall PK profile of SOMCL-19-133 was better than that of MLN4924. At the same dose, it attained a 5-fold higher Cmax and 4-fold higher AUClast than that of MLN4924 by oral delivery. As a result, daily oral treatment of SOMCL-19-133 led to tumor stasis in established RKO xenografts and MV-4-11 xenografts, which was more effective than MLN4924 administered via tail vein injection (i.v.). Therefore, the oral administration of SOMCL-19-133 might lead to an improved therapeutic window and provide more convenient clinical use, compared to other NAE inhibitors that require i.v. injections.
In conclusion, the discovery and characterization of SOMCL-19-133 as a potent, selective, and orally bioavailable NAE inhibitor provided a new strategy for cancer treatment.
Author contributions
L.N. Zhou and J.X. He designed the research; L.N. Zhou performed the experiments and analyzed the data; C.D. Xiong designed and synthesized the compound; X.B. Bao, L.N. Zhou, X.J. Huan, and T.Y. Wang conducted the in vivo experiments; S.S. Song and J.X. He performed the early screening and identification of the compound; L.N. Zhou, Y.J. Cheng, S.S. Song, and X.J. Huan carried out the antitumor spectrum experiments of the compound; L.N. Zhou and J.X. He drafted the manuscript; A. Zhang, Z.H. Miao, and J.X. He supervised the study.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (82073875 to J.X. He and 82073865 to Z.H. Miao), the Chinese Academy of Sciences (29201731121100101 to J.X. He and XDA12020104, XDA12020109 and CASIMM0120185003 to Z.H. Miao), the Shanghai Rising-Star Program (19QA1410900 to J.X. He), and the State Key Laboratory of Drug Research.
Footnotes
No potential conflicts of interest were disclosed.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100823.
Contributor Information
Ao Zhang, Email: ao6919zhang@sjtu.edu.cn.
Ze-Hong Miao, Email: zhmiao@simm.ac.cn.
Jin-Xue He, Email: jinxue_he@simm.ac.cn.
Appendix. Supplementary materials
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson KD. The discovery of ubiquitin-dependent proteolysis. Proc Nat Acad Sci USA. 2005;102(43):15280–15282. doi: 10.1073/pnas.0504842102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23(11):1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 4.Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422(6929):330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- 5.Bohnsack RN, Haas AL. Conservation in the mechanism of nedd8 activation by the human AppBp1-Uba3 heterodimer. J Biol Chem. 2003;278(29):26823–26830. doi: 10.1074/jbc.M303177200. [DOI] [PubMed] [Google Scholar]
- 6.Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, Suzuki H, Shimbara N, Hidaka Y, Osaka F, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20(15):4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong LM, Yeh ETH. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274(17):12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 8.Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov. 2014;13(12):889–903. doi: 10.1038/nrd4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125(Pt 3):531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of Cullin-RING ligases: conformational control of conjugation. Cell. 2008;134(6):995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32(1):21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang YL, Ni J, Hsu PC, Mao JH, Hsieh D, Xu A, Chan G, Au A, Xu Z, Jablons DM, et al. Cul4A overexpression associated with Gli1 expression in malignant pleural mesothelioma. J Cell Mol Med. 2015;19(10):2385–2396. doi: 10.1111/jcmm.12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia L, Sun Y. SCF E3 ubiquitin ligases as anticancer targets. Curr Cancer Drug Targets. 2011;11(3):347–356. doi: 10.2174/156800911794519734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated Cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15(12):3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 15.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 16.Lukkarila JL, da Silva SR, Ali M, Shahani VM, Xu GW, Berman J, Roughton A, Dhe-Paganon S, Schimmer AD, Gunning PT. Identification of NAE inhibitors exhibiting potent activity in leukemia cells: exploring the structural determinants of NAE specificity. ACS Med Chem Lett. 2011;2(8):577–582. doi: 10.1021/ml2000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee HW, Nam SK, Choi WJ, Kim HO, Jeong LS. Stereoselective synthesis of MLN4924, an inhibitor of NEDD8-activating enzyme. J Org Chem. 2011;76(9):3557–3561. doi: 10.1021/jo2001897. [DOI] [PubMed] [Google Scholar]
- 18.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116(9):1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 19.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O'Dwyer M, Nawrocki ST, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115(18):3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Han S, Wilder-Romans K, Sun GY, Zhu H, Liu X, Tan M, Wang G, Feng FY, Sun Y. Neddylation inactivation represses androgen receptor transcription and inhibits growth, survival and invasion of prostate cancer cells. Neoplasia. 2020;22(4):192–202. doi: 10.1016/j.neo.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naik SK, Lam EWF, Parija M, Prakash S, Jiramongkol Y, Adhya AK, Parida DK, Mishra SK. NEDDylation negatively regulates ERRβ expression to promote breast cancer tumorigenesis and progression. Cell Death Dis. 2020;11(8):703. doi: 10.1038/s41419-020-02838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McHugh A, Fernandes K, Chinner N, Ibrahim AFM, Garg AK, Boag G, Hepburn LA, Proby CM, Leigh IM, Saville MK. The identification of potential therapeutic targets for cutaneous squamous cell carcinoma. J Invest Dermatol. 2020;140(6) doi: 10.1016/j.jid.2019.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshimura C, Muraoka H, Ochiiwa H, Tsuji S, Hashimoto A, Kazuno H, Nakagawa F, Komiya Y, Suzuki S, Takenaka T, et al. TAS4464, a highly potent and selective inhibitor of NEDD8-activating enzyme, suppresses neddylation and shows antitumor activity in diverse cancer models. Mol Cancer Ther. 2019;18(7):1205–1216. doi: 10.1158/1535-7163.MCT-18-0644. [DOI] [PubMed] [Google Scholar]
- 24.Sarantopoulos J, Shapiro GI, Cohen RB, Clark JW, Kauh JS, Weiss GJ, Cleary JM, Mahalingam D, Pickard MD, Faessel HM, et al. Phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with advanced solid tumors. Clin Cancer Res. 2016;22(4):847–857. doi: 10.1158/1078-0432.CCR-15-1338. [DOI] [PubMed] [Google Scholar]
- 25.Xiong C, Zhou L, Tan J, Song S, Bao X, Zhang N, Ding H, Zhao J, He JX, Miao ZH, et al. Development of potent NEDD8-activating enzyme inhibitors bearing a pyrimidotriazole scaffold. J Med Chem. 2021;64(9):6161–6178. doi: 10.1021/acs.jmedchem.1c00242. [DOI] [PubMed] [Google Scholar]
- 26.Zhang A, Miao Z-H, Xiong C-D, Song S-S, He J-X. Triazolopyrimidine compounds as NAE inhibitors and their preparation, pharmaceutical compositions and use in the treatment of cancer. WO 2020249109 A1. 2020.
- 27.Xiong C, Zhou L, Tan J, Song S, Bao X, Zhang N, Ding H, Zhao J, He JX, Miao ZH, et al. Development of potent NEDD8-activating enzyme inhibitors bearing a pyrimidotriazole scaffold. J Med Chem. 2021 doi: 10.1021/acs.jmedchem.1c00242. [DOI] [PubMed] [Google Scholar]
- 28.Chen JJ, Tsu CA, Gavin JM, Milhollen MA, Bruzzese FJ, Mallender WD, Sintchak MD, Bump NJ, Yang X, Ma J, et al. Mechanistic studies of substrate-assisted inhibition of ubiquitin-activating enzyme by adenosine sulfamate analogues. J Biol Chem. 2011;286(47):40867–40877. doi: 10.1074/jbc.M111.279984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen HD, Chen CH, Wang YT, Guo N, Tian YN, Huan XJ, Song SS, He JX, Miao ZH. Increased PARP1-DNA binding due to autoPARylation inhibition of PARP1 on DNA rather than PARP1-DNA trapping is correlated with PARP1 inhibitor's cytotoxicity. Int J Cancer. 2019;145(3):714–727. doi: 10.1002/ijc.32131. [DOI] [PubMed] [Google Scholar]
- 30.Yuan B, Ye N, Song SS, Wang YT, Song Z, Chen HD, Chen CH, Huan XJ, Wang YQ, Su Y, et al. Poly(ADP-ribose)polymerase (PARP) inhibition and anticancer activity of simmiparib, a new inhibitor undergoing clinical trials. Cancer Lett. 2017;386:47–56. doi: 10.1016/j.canlet.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010;70(24):10310–10320. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovejoy CA, Lock K, Yenamandra A, Cortez D. DDB1 maintains genome integrity through regulation of Cdt1. Mol Cell Biol. 2006;26(21):7977–7990. doi: 10.1128/MCB.00819-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knorr KLB, Schneider PA, Meng XW, Dai H, Smith BD, Hess AD, Karp JE, Kaufmann SH. MLN4924 induces Noxa upregulation in acute myelogenous leukemia and synergizes with Bcl-2 inhibitors. Cell Death Differ. 2015;22(12):2133–2142. doi: 10.1038/cdd.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekeres MA, Watts J, Radinoff A, Sangerman MA, Cerrano M, Lopez PF, Zeidner JF, Campelo MD, Graux C, Liesveld J, et al. Randomized phase 2 trial of pevonedistat plus azacitidine versus azacitidine for higher-risk MDS/CMML or low-blast AML. Leukemia. 2021;35(7):2119–2124. doi: 10.1038/s41375-021-01125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swords RT, Coutre S, Maris MB, Zeidner JF, Foran JM, Cruz J, Erba HP, Berdeja JG, Tam W, Vardhanabhuti S, et al. Pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, combined with azacitidine in patients with AML. Blood. 2018;131(13):1415–1424. doi: 10.1182/blood-2017-09-805895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Picco G, Petti C, Sassi F, Grillone K, Migliardi G, Rossi T, Isella C, Di Nicolantonio F, Sarotto I, Sapino A, et al. Efficacy of NEDD8 pathway inhibition in preclinical models of poorly differentiated, clinically aggressive colorectal cancer. J Natl Cancer Inst. 2017;109(2) doi: 10.1093/jnci/djw209. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, Cui D, Bi Y, Shu J, Xiong X, Zhao Y. AKT inhibitor MK-2206 sensitizes breast cancer cells to MLN4924, a first-in-class NEDD8-activating enzyme (NAE) inhibitor. Cell Cycle. 2018;17(16):2069–2079. doi: 10.1080/15384101.2018.1515550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Zhou W, Li L, Wu J, Liu X, Zhao L, Jia L, Sun Y. Inhibition of neddylation modification sensitizes pancreatic cancer cells to gemcitabine. Neoplasia. 2017;19(6):509–518. doi: 10.1016/j.neo.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong S, Si Y, Yu H, Zhang L, Xie P, Jiang W. MLN4924 (Pevonedistat), a protein neddylation inhibitor, suppresses proliferation and migration of human clear cell renal cell carcinoma. Sci Rep. 2017;7(1):5599. doi: 10.1038/s41598-017-06098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Wang S, Zhang W, Wang X, Liu X, Liu L, Li L, Liang Y, Yu J, Jeong LS, et al. Targeting neddylation pathway with MLN4924 (Pevonedistat) induces NOXA-dependent apoptosis in renal cell carcinoma. Biochem Biophys Res Commun. 2017;490(4):1183–1188. doi: 10.1016/j.bbrc.2017.06.179. [DOI] [PubMed] [Google Scholar]
- 41.Xu B, Deng Y, Bi R, Guo H, Shu C, Shah NK, Chang J, Liu G, Du Y, Wei W, et al. A first-in-class inhibitor, MLN4924 (pevonedistat), induces cell-cycle arrest, senescence, and apoptosis in human renal cell carcinoma by suppressing UBE2M-dependent neddylation modification. Cancer Chemother Pharmacol. 2018;81(6):1083–1093. doi: 10.1007/s00280-018-3582-z. [DOI] [PubMed] [Google Scholar]
- 42.Jazaeri AA, Shibata E, Park J, Bryant JL, Conaway MR, Modesitt SC, Smith PG, Milhollen MA, Berger AJ, Dutta A. Overcoming platinum resistance in preclinical models of ovarian cancer using the neddylation inhibitor MLN4924. Mol Cancer Ther. 2013;12(10):1958–1967. doi: 10.1158/1535-7163.MCT-12-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. The NEDD8 conjugation pathway and its relevance in cancer biology and therapy. Genes Cancer. 2010;1(7):708–716. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto N, Shimizu T, Yonemori K, Kitano S, Kondo S, Iwasa S, Koyama T, Sudo K, Sato J, Tamura K, et al. A first-in-human, phase 1 study of the NEDD8 activating enzyme E1 inhibitor TAS4464 in patients with advanced solid tumors. Invest New Drugs. 2021 doi: 10.1007/s10637-020-01055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.