Abstract
Background
The current high mortality rate of female breast cancer (BC) patients emphasizes the necessity of identifying powerful and reliable prognostic signatures in BC patients. Epithelial-mesenchymal transition (EMT) was reported to be associated with the development of BC. The purpose of this study was to identify prognostic biomarkers that predict overall survival (OS) in female BC patients by integrating data from TCGA database.
Method
We first downloaded the dataset in TCGA and identified gene signatures by overlapping candidate genes. Differential analysis was performed to find differential EMT-related genes. Univariate regression analysis was then performed to identify candidate prognostic variables. We then developed a prognostic model by multivariate analysis to predict OS. Calibration curves, receiver operating characteristics (ROC) curves, C-index, and decision curve analysis (DCA) were used to test the veracity of the prognostic model.
Result
In this study, we identified and validated a prognostic model integrating age and six genes (CD44, P3H1, SDC1, COL4A1, TGFβ1, and SERPINE1). C-index values for BC patients were 0.672 (95% CI 0.611–0.732) and 0.692 (95% CI 0.586–0.798) in the training cohort and test set, respectively. The calibration curve and the DCA curve show the good predictive performance of the model.
Conclusion
This study offered a robust predictive model for OS prediction in female BC patients and may provide a more accurate treatment strategy and personalized therapy in the future.
1. Introduction
Breast cancer is one of the most prevalent malignancies in women worldwide and the leading cause of most cancer-related deaths, although early-stage BC is considered curable [1, 2]. In 2018, BC was the most commonly diagnosed cancer (24.2% of all cancer cases) and the leading cause of cancer-related deaths (15% of all cancer deaths) in women worldwide. Among these, metastatic BC accounted for more than 90% of BC-related deaths [3]. At present, the main treatment strategies for BC include surgery, chemotherapy, radiotherapy, immunotherapy, and hormonal therapy [4]. Although nanomedicine has been developed this year to target progesterone and estrogen receptors (PR and ER), human epidermal growth factor receptor 2 (HER2), and microRNA (miRNAs) and long chain non-coding RNAs, the incidence of BC remains high, with previous studies suggesting that the number of new cases worldwide will be 2,261,419 women in 2020, and this number is expected to increase to 30.2 million by 2040 [5, 6].
Epithelial-mesenchymal transition (EMT) is widely known to occur during mammalian development, wound healing, and cancer metastasis [7]. In recent years, EMT has received increasing attention for its role in cancer drug resistance [8]. Many studies have shown that EMT is associated with tumorigenesis, invasion, metastasis, and resistance to treatment, especially in BC [9, 10]. Saotome et al. demonstrated that GATA3 truncation mutants affected ductal BC development by altering EMT-related gene expression through partial motif recognition in luminal BC cells [11]. Parthasarathi and his colleagues found that EMT-related genes were associated with dysregulated ion channels in BC-associated tumorigenesis and could potentially be used to determine the prognosis of BC patients. Therefore, in this study, we evaluated the relevance of the EMT genes in female BC patients to explore the mechanisms of EMT in BC [12].
The new 8th edition of a related Union for International Cancer Control (UICC) and American Joint Committee on Cancer (AJCC) publication updates the description of BC staging for tumor lymph node metastasis (TNM) [13]. Yet, it is not sufficient to simply predict the prognosis of BC based on the TNM staging system. Some of the factors that influence BC include age, genes, reproductive factors, estrogen, and lifestyle [14]. Hence, a multifactorial predictive model is essential. Predictive modeling is a more advanced approach as it can be visualized using a nomogram and it can estimate individualized risk based on a more comprehensive set of gene signatures and clinical characteristics. In previous studies, we constructed a clinical prediction model based on the clinical data of metastatic colon cancer patients extracted from the SEER database. The nomogram developed with high prognosis prediction accuracy to evaluate the 1-, 3-, and 5-year survival of metastatic colon cancer patients, which will help clinical decision-making of metastatic colon cancer patients after surgery and individualized treatment [15].
In this study, we identified EMT-related genes with independent prognostic value to establish a prognostic model for predicting the overall survival (OS) at 1-, 3-, and 5-year of female BC patients and generating new insights about BC progression.
2. Materials and Methods
2.1. Data Collection
We downloaded gene expression data from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov) of 1109 BC patients and 113 nontumor breast tissues. Clinical data were also acquired, but the clinical data of 12 male BC patients were removed because the study population in this paper was female. EMT-related genes were collected from the Molecular Signature database v7.1 (MSigDB) (http://www.broad.mit.edu/gsea/msigdb/).
2.2. Identification of Differentially Expressed EMT-Related Genes
Combining the gene expression data obtained from TCGA database with EMT-related genes by using the “edgeR” package of R software, the expression data of the target genes could be obtained. After that, the “limma” package was utilized to derive differentially expressed EMT-related genes according to False Discovery Rate (FDR) values less than 0.05 and the absolute value of fold change above 1.
2.3. Statistical Analysis
2.3.1. Univariate Cox Regression Analysis for Independent Prognostic Factors
The expression matrix of the obtained EMT-related genes was further analyzed by incorporating the matrix with the survival time and survival status. Based on previous studies, age had an impact on the prognosis of female BC patients, so we included age as a study variable [16]. Using the “caret” package in R software (version 4.1.0) to randomly divide the overall cohort into two groups in the ratio of 7 : 3. The subgroup containing 70% of female BC patients was used to construct the prediction model, while the remaining 30% of patients were examined for the accuracy and reliability of the model. Also, the whole cohort was used as the overall internal validation set. The basic values of patients were listed (Table S1).
Univariate Cox regression analysis was used to screen for independent prognostic factors. Factors with a cutoff value of P < 0.1 were defined as candidates associated with OS.
2.3.2. Prognostic Nomogram Construction
The genes filtered by univariate Cox regression were then analyzed in the multivariate Cox regression for the risk scoring model. The risk score for each patient can be calculated by the following formula: risk score = Exp(x1)∗β1 + Exp(x2)∗β2 + ⋯+Exp(xn)∗βn, where n is the number of selected variables, Exp is the expression level of the variable, and β is the regression coefficient of the variable. Then, the nomogram was developed using R software. According to the scores calculated from the nomogram, the patient's OS at 1, 3, and 5 years can be predicted. Subsequently, according to the median risk scores, patients with risk scores greater than the median value were divided into the high-risk group and otherwise into the low-risk group.
2.3.3. Prognostic Nomogram Evaluation and Validation
In order to improve the reliability of the prediction model and thus its clinical application, 30% of the patients and the overall cohort were used as an internal validation cohort to test the validity of the prediction model.
The discriminative power of the nomogram was calculated using the concordance index (C-index). We also measured the area under the curve (AUC) at 1, 3, and 5 years, which was derived from a ROC analysis. The C-indexes and AUCs take values ranging from 0.5 to 1.0, where 1.0 represents the perfect ability to correctly distinguish the results from the model and 0.5 represents random chance. The calibration curve of the nomogram was evaluated graphically by plotting the ratio of the predicted probability to the observed ratio of the nomogram. Overlapping with the reference line indicated that the model was perfectly consistent. Finally, decision curve analysis was performed to evaluate the clinical benefits. A flow chart of the study process of this article was presented (Figure 1).
Figure 1.
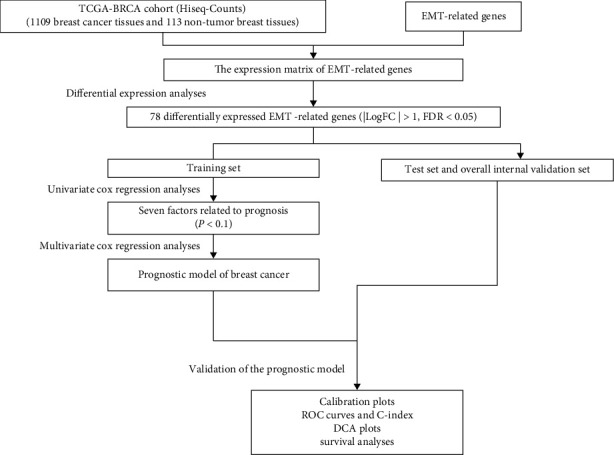
Flow chart of this study.
3. Results
3.1. Identification of Differentially Expressed EMT-Related Genes
To describe our study more clearly, we developed a flowchart of the analysis procedure. First, we obtained data from TCGA database for 1109 tumor tissues and 113 nontumor tissues. After taking intersection with EMT-related genes, a matrix of 200 EMT-related genes (Table S2) expression values was acquired. Then, after differential analyses, a total of 78 differentially expressed EMT-related genes were identified. (logFC > 1 or logFC < −1, FDR < 0.05). The results were expressed in heat maps and volcano plots (Figures 2(a) and 2(b)).
Figure 2.

Heat map (a) and volcano map (b) of differentially expressed gene related to EMT.
3.2. Prognostic Nomogram Construction
Since age is associated with prognosis in female BC patients, we included age and 78 differentially expressed genes in univariate Cox regression to investigate the correlation between the included variables and prognostic value in BC patients and finally identified seven variables significantly associated with OS in BC patients at P value < 0.1 (Figure 3). The model was then constructed with age, CD44, P3H1, SDC1, COL4A1, TGFβ1, and SERPINE1 by multivariate Cox regression: risk score = (0.030655∗age level) + (3.35E − 05∗expression level of CD44) + (0.000142∗expression level of P3H1) + (3.76E − 05∗expression level of SDC1) + (1.60E − 05∗COL4A1 expression level) + (2.04E − 05∗TGFβ1 expression level) + (0.000247∗SERPINE1 expression level) (Table 1).
Figure 3.
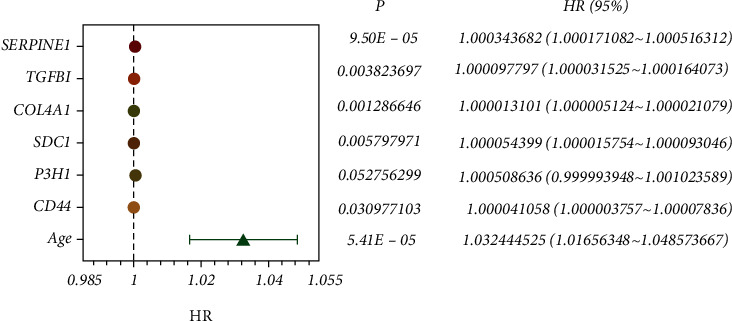
Forest plot analyzed by univariate Cox regression.
Table 1.
Genes contained in the prognostic model of breast cancer.
| Factors | coef | HR | HR_95L | HR_95U | P |
|---|---|---|---|---|---|
| Age | 0.030655 | 1.031129 | 1.015051 | 1.047463 | 0.000132 |
| CD44 | 3.35E − 05 | 1.000033 | 0.999994 | 1.000073 | 0.098155 |
| P3H1 | 0.000142 | 1.000142 | 0.999283 | 1.001002 | 0.74605 |
| SDC1 | 3.76E − 05 | 1.000038 | 0.999966 | 1.000109 | 0.304272 |
| COL4A1 | 1.60E − 05 | 1.000016 | 1.000008 | 1.000024 | 0.000149 |
| TGFBI | 2.04E − 05 | 1.00002 | 0.999884 | 1.000156 | 0.768605 |
| SERPINE1 | 0.000247 | 1.000247 | 0.999911 | 1.000583 | 0.149765 |
The nomogram was then constructed and consisted of a total of seven variables (Figure 4), and the total score could be obtained by summing the scores of each variable. The total score can be used to predict the survival rate of individual patients at 1, 3, and 5 years. For example, a BC patient aged 65 years (20 points) with CD44 expression of 0 (20 points), P3H1 expression of 0 (21 points), SDC1 expression of 0 (20 points), COL4A1 expression of 80000 (43 points), TGFβ1 expression of 0 (21 points), and SERPINE1 expression of 0 (21 points) gets a sum-point of 166, corresponding to predicted 1-, 3-, and 5-year OS of 94.8%, 76.0%, and 57.4%, respectively.
Figure 4.
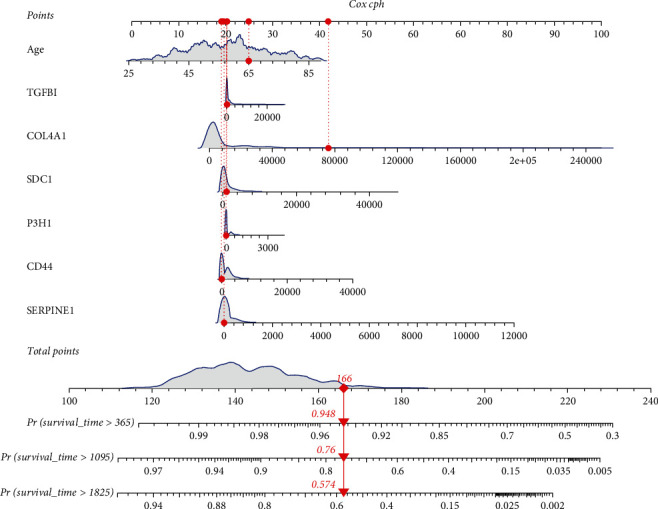
Nomogram for predicting 1-, 3-, and 5-year overall survival (OS) for BC patients in the training cohort.
Patients in TCGA group were divided into a low-risk group and a high-risk group using the median risk score as the threshold value. Figures 5(a), 5(c), and 5(e) show the distribution of the risk scores of BC patients from high to low in the training set, the internal validation set, and the overall internal validation set. The relationship between risk score and patient survival time in the training set, test set, and overall internal validation set is also shown (Figures 5(b), 5(d), and 5(f)). Patients with high-risk scores tended to have poorer clinical outcomes compared with those with low-risk scores. The survival analyses indicated the high-risk group had worse OS than that of the high-risk group (P < 0.05) (Figures 6(a)–6(c)).
Figure 5.
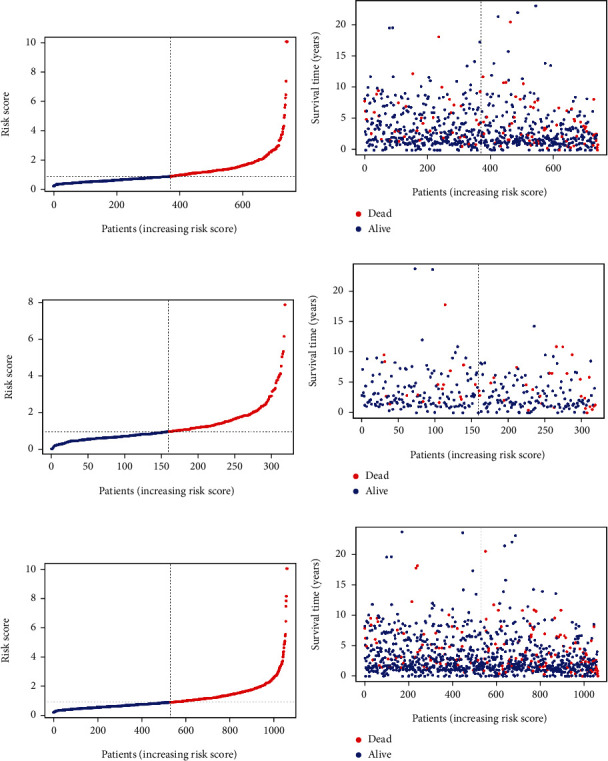
(a, c, e) Distribution of risk score in patients with BC. The black dotted line serves as the dividing line between the high-risk group and the low-risk group. (b, d, f) Diagram of the relationship between risk score and patient survival time. The result of (a, b) is based on training set, the result of (c, d) is based on test set, and the result of (e, f) is based on the overall internal validation set.
Figure 6.

Overall survival (OS) Kaplan-Meier curves for patients in the low- and high-risk groups: (a) training set; (b) test set; (c) overall internal validation set.
3.3. Nomogram Calibration and Validation
The small angle between the survival probability and the actual survival outcome in the calibration curve indicates a strong agreement between them (Figure 7).
Figure 7.
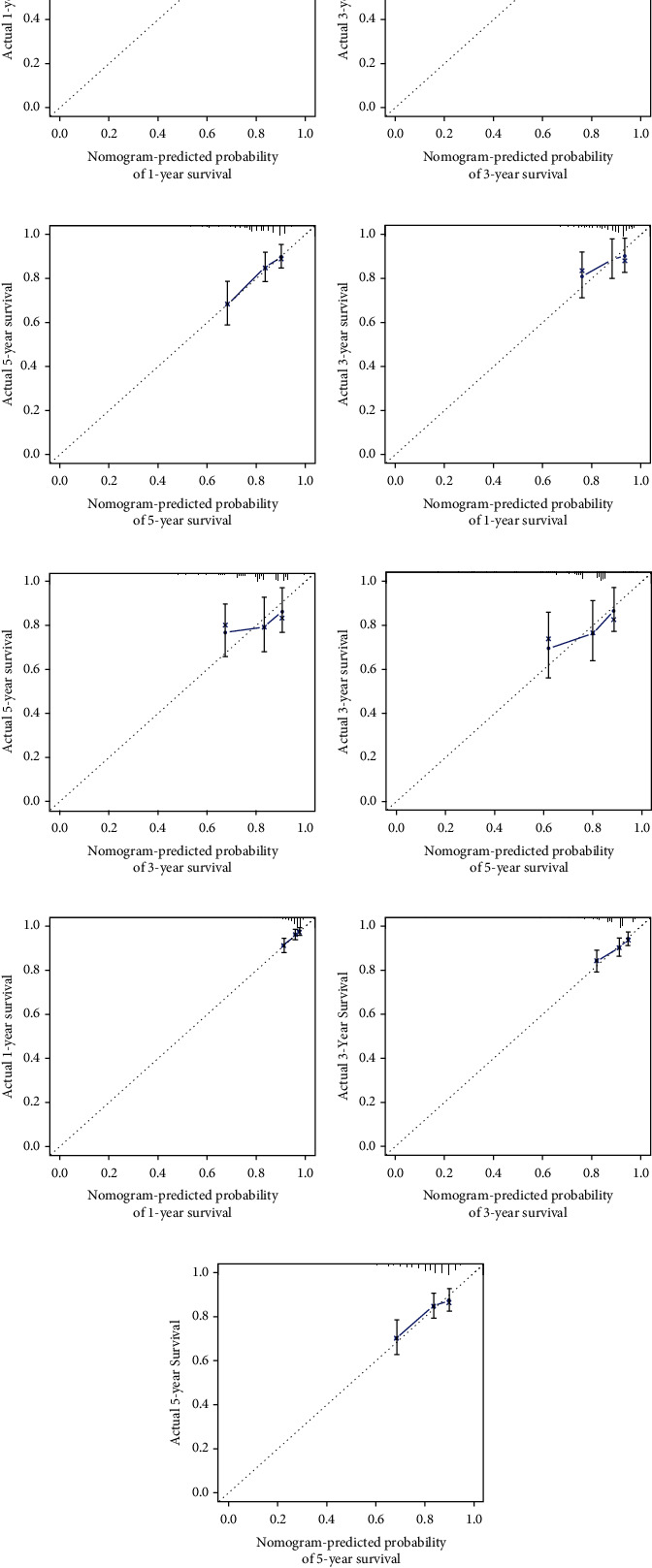
(a–c) Calibration plots to predict 1-, 3-, and 5-year overall survival (OS) in the training set; (d–f) calibration plots to predict 1-, 3-, and 5-year; overall survival (OS) in the test set; (g–i) calibration plots to predict 1-, 3-, and 5-year overall survival (OS) in the overall internal validation set.
The C-index values for BC patients were 0.672 (95% CI 0.611–0.732), 0.692 (95% CI 0.586–0.798), and 0.679 (95% CI 0.626–0.732) in the training cohort, test set, and overall internal validation set, respectively. The time-dependent ROC curves were used to measure the sensitivity and specificity of the prediction model for predicting OS. Significantly, the AUC values were all greater than 0.63, except for the overall internal validation set with a 5-year predicted survival rate of 0.56, indicating that the model has high survival outcome prediction performance (Figure 8). The DCA curves also revealed better clinical applications for the risk scoring model (Figure 9).
Figure 8.
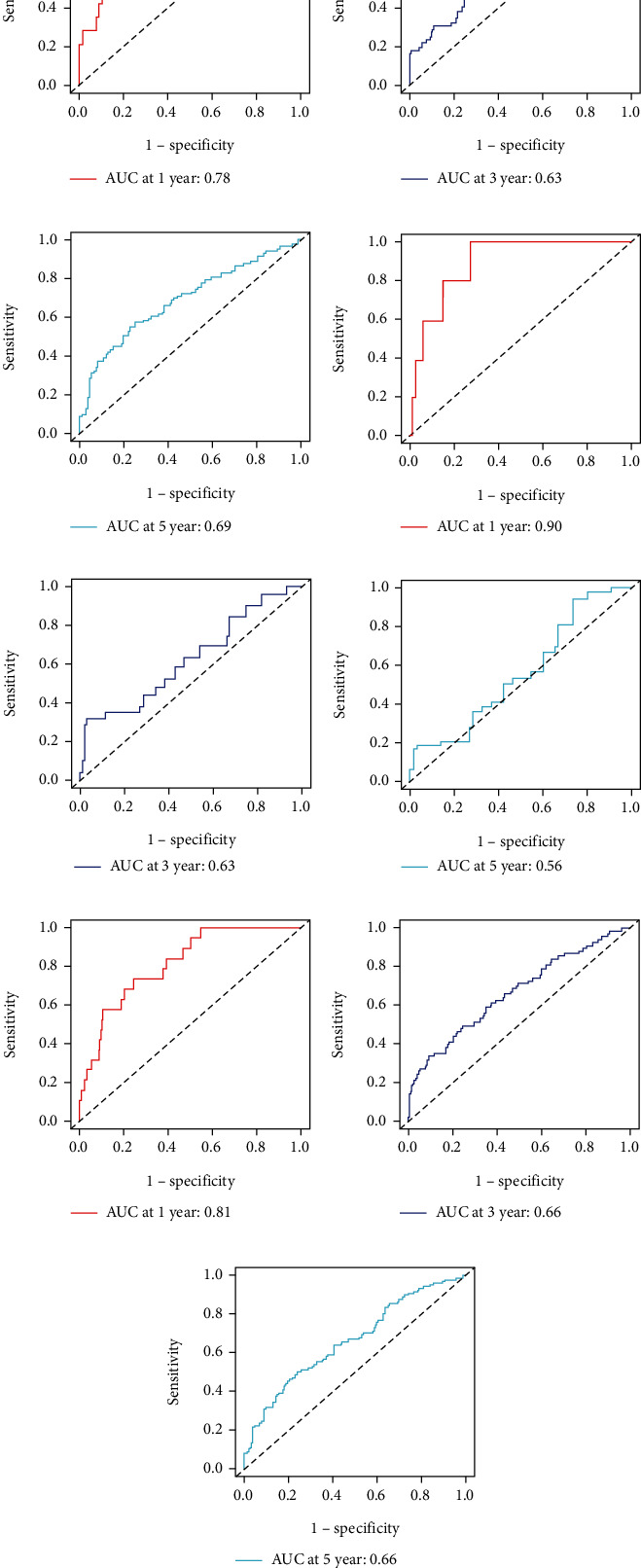
(a–c) ROC curves to predict 1-, 3-, and 5-year overall survival (OS) in the training set; (d–f) ROC curves to predict 1-, 3-, and 5-year; overall survival (OS) in the test set; (g–i) ROC curves to predict 1-, 3-, and 5-year overall survival (OS) in the overall internal validation set.
Figure 9.

(a–c) DCA analysis predicting 1-, 3-, and 5-year overall survival (OS) in the training set; (d–f) DCA analysis predicting 1-, 3-, and 5-year; overall survival (OS) in the test set; (g–i) DCA analysis predicting 1-, 3-, and 5-year overall survival (OS) in the overall internal validation set.
The results based on C-index, ROC curves, calibration curves, and DCA curves indicated that the nomogram in our study demonstrated favorable predictive accuracy for the survival prognosis of female BC patients.
4. Discussion
BC is one of the most common cancers in females, with over 1,300,000 new cases and 450,000 deaths worldwide each year [16, 17]. Treatment of BC has advanced considerably, mainly through surgery, neoadjuvant chemotherapy, adjuvant chemotherapy, radiotherapy, systemic therapy, targeted therapy, and so on, with initial conventional surgery no longer being the best option for all patients [1]. However, BC remains one of the leading causes of cancer deaths in women worldwide, largely due to delayed diagnosis and unsuccessful treatment strategies [18]. Therefore, it is crucial to diagnose BC at an early stage and propose a personalized treatment plan based on the characteristics of the women patient's condition to predict their prognosis.
The TNM staging system is still the most widely used prognosis method to predict the survival of patients with BC. Although the American Joint Committee on Cancer (AJCC) updated BC staging in 2016 to include T, N, M, tumor grade, and expression of estrogen and progesterone receptors and HER2 [19], the current TNM staging system still has its undeniable deficiencies. For instance, it does not take into account other pathophysiological characteristics of the patient that have an impact on the prognosis of the tumor: age, gender, exercise, and overweight [20–22]. In addition, gene signature is an important factor in determining the prognosis of BC patients, as BC is a highly heterogeneous disease with different subtypes with different biological, molecular, and clinical processes. Gene expression profiling can identify genetic features to predict prognosis and guide the use of adjuvant therapy [23]. Among others, EMT genes regulate tumor proliferation, invasion, and metastasis [24, 25]. There are many prognostic models on BC; however, this is the first gene signature constructed by EMT-related genes. Moreover, considering the role of age and gender in the onset and progression of BC, we chose to study the prognosis of patients in women and used age as one of the predictors. Compared to previous studies, this nomogram was more accurate.
EMT is a cellular process in which cells lose their epithelial characteristics and acquire mesenchymal characteristics, such as quiescent adnexal cells gaining the ability to migrate [26]. EMT has been associated with a variety of tumor functions, including tumor initiation, malignant progression, tumor stemness, tumor cell migration, intravascular infiltration, metastasis, and resistance to therapy [9]. Most notably in this context, previous studies have shown that both cancer stem cell-like properties and drug resistance are associated with EMT [27]. Given the close link between oncogenic signaling and EMT blockers, EMT has emerged as a therapeutic target or goal in cancer therapy [28]. The relationship between EMT-related genes and breast cancer is also increasingly being investigated by researchers. The major focus of current studies is the regulatory mechanisms and therapeutic approaches of EMT for breast cancer in metastasis and invasion, mainly including miRNA and signaling pathways such as Wnt, Notch, TNF-α, NF-κB, and RTK. Investigators suppress breast cancer by attempting to therapeutically target or inhibit key/auxiliary players in these pathways [8, 29–31]. Most notably, upregulation of programmed death ligand 1 (PD-L1) expression is associated with EMT cell phenotype activation, and the control of the interaction between p53 and EMT master regulators is of importance in breast cancer. These two mechanisms have also been studied in other types of cancer and play a key role in the development and metastasis of cancer [30, 32].
This study was based on TCGA database. Differential analysis was firstly performed to find differential EMT-related genes. Univariate regression analysis was then conducted to identify candidate prognostic variables. We then developed a prognostic model by multivariate analysis to predict OS. Calibration curves, receiver operating characteristics (ROC) curves, C-index, and decision curve analysis (DCA) were used to test the veracity of the prognostic model. In addition to the training cohort of 70% BC patients, the remained cohort was treated as the test set. In the end, we derived that patient's age, CD44, P3H1, SDC1, COL4A1, TGFβ1, and SERPINE1 were independent prognostic factors for overall survival in female BC patients and constructed predictive models. The accuracy of the model has also been verified using various methods.
In accordance with our findings, in stage I and IV BC tumors, excess mortality increased linearly with age [33]. Recent studies have shown that a novel positive feedback loop between IL1β and CD44 promoted malignant progression in triple-negative BC (TNBC) and that CD44 was a potential target for inhibiting PD-L1 function in TNBC [34, 35]. Sayyad et al. demonstrated the role of Sdc1 in promoting brain metastasis in BC [36]. Several studies have demonstrated that COL4A1 expression could be used as a biomarker for superior prognosis in BC patients receiving neoadjuvant chemotherapy [37], while epigallocatechin-3-gallate (EGCG) exerted antitumor effects by restoring nine key genes, including COL4A1, in myeloid-derived suppressor cells (MDSCs) [37]. TGFβ1-activated cancer-associated fibroblasts (CAFs) promote BC growth and metastasis in part through autophagy [38]. The evolutionary branch E member 1 (SERPINE1) is a molecule involved in a variety of human malignancies. Zhang et al. showed that SERPINE1 served as an oncogene for PTX resistance in BC, and Xu et al. identified a functional pathway linking miR-1185-2-3p, GOLPH3L, and SERPINE1, which played an essential role in glucose metabolism in BC. Both of their studies revealed that it may serve as a possible target for the treatment of BC [39, 40]. No studies have yet explored the mechanisms by which P3H1 affects BC development, progression, and metastasis, but an algorithm-based meta-analysis of genome-wide and proteomic data identified P3H1 as a potential biomarker for CRC. Our study indicates a direction of research for subsequent basic studies [41].
In this endeavor, some limitations need to be acknowledged. To begin with, the population races in TCGA database are primarily limited to whites and blacks, and extrapolation of findings to other racial groups needs to be validated. Second, a robust nomogram should be externally validated across cohorts; therefore, our nomogram needs to be further validated in multicenter clinical trials and prospective studies. Finally, some of the genes identified in this paper are relatively rarely reported in the academic literature. Therefore, more evidence including sample collection with complete experimental and clinical information should be performed for future validation is needed to elucidate the intrinsic association between age and six-gene signature and prognosis of BC patients.
However, our study also has some advantages. To our knowledge, this is the first study to additionally combine age as a prognostic variable with EMT-related genes to predict the prognosis of BC patients. Prognostic models may predict patient prognosis more accurately than conventional indicators.
In conclusion, we have developed and validated a relatively effective predictive model to predict the survival outcome of female BC patients at 1, 3, and 5 years. The accuracy and reliability of the prognostic model have also been verified. The results of our research need to be further validated in subsequent clinical practice.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 82074347) and the Innovation Project of Guangxi Graduate Education (No. YCBZ2021048).
Data Availability
The research article data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Wei Xue and Chenyu Sun performed the study, collected data, and wrote the manuscript. Hui Yuan, Xin Yang, Qiuping Zhang, and Yunnuo Liao participated in analyzing data and prepared the tables and figures. Hongwei Guo designed the research study and revised the manuscript. All authors read and approved the final manuscript. Wei Xue and Chenyu Sun contributed equally to this work.
Supplementary Materials
Supplementary Table S1: basic values of patients. Supplementary Table S2: EMT-related gene list.
References
- 1.Harbeck N., Gnant M. Breast cancer. Lancet . 2017;389(10074, article S0140673616318918):1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 2.Liang Y., Zhang H., Song X., Yang Q. Metastatic heterogeneity of breast cancer: molecular mechanism and potential therapeutic targets. Seminars in Cancer Biology . 2020;60, article S1044579X1930063X:14–27. doi: 10.1016/j.semcancer.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Xu X., Zhang M., Xu F., Jiang S. Wnt signaling in breast cancer: biological mechanisms, challenges and opportunities. Molecular Cancer . 2020;19(1, article 1276):p. 165. doi: 10.1186/s12943-020-01276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui G., Wu J., Lin J., et al. Graphene-based nanomaterials for breast cancer treatment: promising therapeutic strategies. Journal of Nanobiotechnology . 2021;19(1):p. 211. doi: 10.1186/s12951-021-00902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boix-Montesinos P., Soriano-Teruel P. M., Armiñán A., Orzáez M., Vicent M. J. The past, present, and future of breast cancer models for nanomedicine development. Advanced Drug Delivery Reviews . 2021;173, article S0169409X2100096X:306–330. doi: 10.1016/j.addr.2021.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yardım-Akaydin S., Karahalil B., Baytas S. N. New therapy strategies in the management of breast cancer. Drug Discovery Today . 2022;27(6):S1359–S6446. doi: 10.1016/j.drudis.2022.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Bakir B., Chiarella A. M., Pitarresi J. R., Rustgi A. K. EMT, MET, plasticity, and tumor metastasis. Trends in Cell Biology . 2020;30(10, article S0962892420301446):764–776. doi: 10.1016/j.tcb.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo J., Yao J. F., Deng X. F., et al. 14, 15-EET induces breast cancer cell EMT and cisplatin resistance by up-regulating integrin αvβ3 and activating FAK/PI3K/AKT signaling. Journal of Experimental & Clinical Cancer Research: CR . 2018;37(1, article 694):p. 23. doi: 10.1186/s13046-018-0694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pastushenko I., Blanpain C. EMT transition states during tumor progression and metastasis. Trends in Cell Biology . 2019;29(3, article S0962892418302010):212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Saotome M., Poduval D. B., Nair R., Cooper M., Takaku M. GATA3 truncation mutants Alter EMT related gene expression via partial motif recognition in luminal breast cancer cells. Frontiers in Genetics . 2022;13:p. 820532. doi: 10.3389/fgene.2022.820532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parthasarathi K., Mandal S., Singh S., et al. In silico analysis of ion channels and their correlation with epithelial to mesenchymal transition in breast cancer. Cancers . 2022;14(6):p. 1444. doi: 10.3390/cancers14061444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cserni G., Chmielik E., Cserni B., Tot T. The new TNM-based staging of breast cancer. Virchows Archiv: an International Journal of Pathology . 2018;472(5, article 2301):697–703. doi: 10.1007/s00428-018-2301-9. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y. S., Zhao Z., Yang Z. N., et al. Risk factors and preventions of breast cancer. International Journal of Biological Sciences . 2017;13(11):1387–1397. doi: 10.7150/ijbs.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao L., Towe C. W., Shenk R., Stabellini N., Amin A. L., Montero A. J. A comparison of local therapy alone with local plus systemic therapy for stage I pT1aN0M0 HER2+ breast cancer: a National Cancer Database analysis. Cancer . 2022;128(13):2433–2440. doi: 10.1002/cncr.34200. [DOI] [PubMed] [Google Scholar]
- 15.Tai Q., Xue W., Li M., et al. Survival nomogram for metastasis colon cancer patients based on SEER database. Frontiers in Genetics . 2022;13:p. 832060. doi: 10.3389/fgene.2022.832060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagegni N. A., Peterson L. L. Age-related disparities in older women with breast cancer. Advances in Cancer Research . 2020;146:23–56. doi: 10.1016/bs.acr.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y., Du S., Liu J., et al. Diagnosis and prognosis of breast cancer by high-performance serum metabolic fingerprints. Proceedings of the National Academy of Sciences of the United States of America . 2022;119(12, article e2122245119) doi: 10.1073/pnas.2122245119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jokar N., Velikyan I., Ahmadzadehfar H., et al. Theranostic approach in breast cancer. Clinical Nuclear Medicine . 2021;46(8):e410–e420. doi: 10.1097/RLU.0000000000003678. [DOI] [PubMed] [Google Scholar]
- 19.Edge S. B., Hortobagyi G. N., Giuliano A. E. New and important changes in breast cancer TNM: incorporation of biologic factors into staging. Expert Review of Anticancer Therapy . 2019;19(4):309–318. doi: 10.1080/14737140.2019.1582335. [DOI] [PubMed] [Google Scholar]
- 20.Latha N. R., Rajan A., Nadhan R., et al. Gene expression signatures: a tool for analysis of breast cancer prognosis and therapy. Critical Reviews in Oncology/Hematology . 2020;151, article S1040842820301025:p. 102964. doi: 10.1016/j.critrevonc.2020.102964. [DOI] [PubMed] [Google Scholar]
- 21.de Roon M., May A. M., McTiernan A., et al. Effect of exercise and/or reduced calorie dietary interventions on breast cancer-related endogenous sex hormones in healthy postmenopausal women. Breast Cancer Research: BCR . 2018;20(1, article 1009):p. 81. doi: 10.1186/s13058-018-1009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiralerspong S., Goodwin P. J. Obesity and breast cancer prognosis: evidence, challenges, and opportunities. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology . 2016;34(35):4203–4216. doi: 10.1200/JCO.2016.68.4480. [DOI] [PubMed] [Google Scholar]
- 23.Kwa M., Makris A., Esteva F. J. Clinical utility of gene-expression signatures in early stage breast cancer. Clinical Oncology . 2017;14(10):595–610. doi: 10.1038/nrclinonc.2017.74. [DOI] [PubMed] [Google Scholar]
- 24.Sun L., Shi C., Liu S., et al. Overexpression of NuSAP1 is predictive of an unfavourable prognosis and promotes proliferation and invasion of triple-negative breast cancer cells via the Wnt/β-catenin/EMT signalling axis. Gene . 2020;747, article S0378111920303267:p. 144657. doi: 10.1016/j.gene.2020.144657. [DOI] [PubMed] [Google Scholar]
- 25.Wang H., Guo S., Kim S. J., et al. Cisplatin prevents breast cancer metastasis through blocking early EMT and retards cancer growth together with paclitaxel. Theranostics . 2021;11(5):2442–2459. doi: 10.7150/thno.46460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aiello N. M., Kang Y. Context-dependent EMT programs in cancer metastasis. The Journal of Experimental Medicine . 2019;216(5):1016–1026. doi: 10.1084/jem.20181827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan G., Liu Y., Shang L., Zhou F., Yang S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer communications (London, England) . 2021;41(3):199–217. doi: 10.1002/cac2.12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho E. S., Kang H. E., Kim N. H., Yook J. I. Therapeutic implications of cancer epithelial-mesenchymal transition (EMT) Archives of Pharmacal Research . 2019;42(1, article 1108):14–24. doi: 10.1007/s12272-018-01108-7. [DOI] [PubMed] [Google Scholar]
- 29.Kotiyal S., Bhattacharya S. Breast cancer stem cells, EMT and therapeutic targets. Biochemical and Biophysical Research Communications . 2014;453(1, article S0006291X14016957):112–116. doi: 10.1016/j.bbrc.2014.09.069. [DOI] [PubMed] [Google Scholar]
- 30.Messeha S. S., Zarmouh N. O., Soliman K. Polyphenols modulating effects of PD-L1/PD-1 checkpoint and EMT-mediated PD-L1 overexpression in breast cancer. Nutrients . 2021;13(5, article nu13051718):p. 1718. doi: 10.3390/nu13051718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Wang Y. W., Chen X., et al. MicroRNA-4472 promotes tumor proliferation and aggressiveness in breast cancer by targeting RGMA and inducing EMT. Clinical Breast Cancer . 2020;20(2, article S152682091930669X):e113–e126. doi: 10.1016/j.clbc.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Parfenyev S., Singh A., Fedorova O., Daks A., Kulshreshtha R., Barlev N. A. Interplay between p53 and non-coding RNAs in the regulation of EMT in breast cancer. Cell Death & Disease . 2021;12(1, article 3327):p. 17. doi: 10.1038/s41419-020-03327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cluze C., Colonna M., Remontet L., et al. Analysis of the effect of age on the prognosis of breast cancer. Breast Cancer Research and Treatment . 2009;117(1, article 222):121–129. doi: 10.1007/s10549-008-0222-z. [DOI] [PubMed] [Google Scholar]
- 34.Jang J. H., Kim D. H., Lim J. M., et al. Breast cancer cell-derived soluble CD44 promotes tumor progression by triggering macrophage IL1β production. Cancer Research . 2020;80(6):1342–1356. doi: 10.1158/0008-5472.CAN-19-2288. [DOI] [PubMed] [Google Scholar]
- 35.Kong T., Ahn R., Yang K., et al. CD44 promotes PD-L1 expression and its tumor-intrinsic function in breast and lung cancers. Cancer Research . 2020;80(3):444–457. doi: 10.1158/0008-5472.CAN-19-1108. [DOI] [PubMed] [Google Scholar]
- 36.Sayyad M. R., Puchalapalli M., Vergara N. G., et al. Syndecan-1 facilitates breast cancer metastasis to the brain. Breast Cancer Research and Treatment . 2019;178(1, article 5347):35–49. doi: 10.1007/s10549-019-05347-0. [DOI] [PubMed] [Google Scholar]
- 37.Wang S. M., Chen P. M., Sung Y. W., Huang W. C., Huang H. S., Chu P. Y. Effect of COL4A1 expression on the survival of neoadjuvant chemotherapy breast cancer patients. Journal of Oncology . 2020;2020:8. doi: 10.1155/2020/5209695.5209695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang M., Fu M., Wang J., et al. TGF-β1-activated cancer-associated fibroblasts promote breast cancer invasion, metastasis and epithelial-mesenchymal transition by autophagy or overexpression of FAP-α. Biochemical Pharmacology . 2021;188, article S0006295221001234:p. 114527. doi: 10.1016/j.bcp.2021.114527. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Q., Lei L., Jing D. Knockdown of SERPINE1 reverses resistance of triple-negative breast cancer to paclitaxel via suppression of VEGFA. Oncology Reports . 2020;44(5):1875–1884. doi: 10.3892/or.2020.7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y., Chen W., Liang J., et al. The miR-1185-2-3p-GOLPH3L pathway promotes glucose metabolism in breast cancer by stabilizing p53-induced SERPINE1. Journal of Experimental & Clinical Cancer Research: CR . 2021;40(1, article 1767):p. 47. doi: 10.1186/s13046-020-01767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gawel D. R., Lee E. J., Li X., et al. An algorithm-based meta-analysis of genome- and proteome-wide data identifies a combination of potential plasma biomarkers for colorectal cancer. Scientific Reports . 2019;9(1, article 51999):p. 15575. doi: 10.1038/s41598-019-51999-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: basic values of patients. Supplementary Table S2: EMT-related gene list.
Data Availability Statement
The research article data used to support the findings of this study are included within the article.


