Abstract
Until now, identification of components of the flagellar protein export apparatus has been indirect. We have now identified these components directly by establishing whether mutants defective in putative export components could translocate export substrates across the cytoplasmic membrane into the periplasmic space. Hook-type proteins could be exported to the periplasm of rod mutants, indicating that rod protein export does not have to precede hook-type protein export and therefore that both types of proteins belong to a single export class, the rod/hook-type class, which is distinct from the filament-type class. Hook-capping protein (FlgD) and hook protein (FlgE) required FlhA, FlhB, FliH, FliI, FliO, FliP, FliQ, and FliR for their export to the periplasm. In the case of flagellin as an export substrate, because of the phenomenon of hook-to-filament switching of export specificity, it was necessary to use temperature-sensitive mutants and establish whether flagellin could be exported to the cell exterior following a shift from the permissive to the restrictive temperature. Again, FlhA, FlhB, FliH, FliI, and FliO were required for its export. No suitable temperature-sensitive fliQ or fliR mutants were available. FliP appeared not to be required for flagellin export, but we suspect that the temperature-sensitive FliP protein continued to function at the restrictive temperature if incorporated at the permissive temperature. Thus, we conclude that these eight proteins are general components of the flagellar export pathway. FliJ was necessary for export of hook-type proteins (FlgD and FlgE); we were unable to test whether FliJ is needed for export of filament-type proteins. We suspect that FliJ may be a cytoplasmic chaperone for the hook-type proteins and possibly also for FliE and the rod proteins. FlgJ was not required for the export of the hook-type proteins; again, because of lack of a suitable temperature-sensitive mutant, we were unable to test whether it was required for export of filament-type proteins. Finally, it was established that there is an interaction between the processes of outer ring assembly and of penetration of the outer membrane by the rod and nascent hook, the latter process being of course necessary for passage of export substrates into the external medium. During the brief transition stage from completion of rod assembly and initiation of hook assembly, the L ring and perhaps the capping protein FlgD can be regarded as bona fide export components, with the L ring being in a formal sense the equivalent of the outer membrane secretin structure of type III virulence factor export systems.
Many species of bacteria, including Salmonella, swim by means of flagella. The flagellum consists of the motor structure and basal body, which are embedded in the cell surface, and the hook and the helical filament, both of which are external to the cell. If we define the term “external structure” in an even more restrictive sense to mean lying beyond the cytoplasmic membrane, there are at least nine such structures: the basal body rod, the periplasmic P ring, the outer membrane L ring, the hook, the hook cap, two hook-filament junction zones, the filament, and the filament cap. The proteins corresponding to these substructures are listed in Table 1. All of these proteins have to cross the cytoplasmic membrane, and several of them must cross the outer membrane as well. Also included in Table 1 is FliE, a basal body protein (27) whose precise location and function are not known but which is postulated to interface with, or even be part of, the rod (16).
TABLE 1.
External flagellar substructures and the proteins from which they are composed
| Substructure | Protein(s) |
|---|---|
| MS ring/rod junction? | FliE |
| Proximal rod | FlgB, FlgC, FlgF |
| Distal rod | FlgG |
| Periplasmic P ring | FlgI |
| Outer membrane L ring | FlgH |
| Hook | FlgE |
| Hook cap (scaffolding protein) | FlgD |
| First hook-filament junction zone | FlgK |
| Second hook-filament junction zone | FlgL |
| Filament (helical propeller) | FliC (flagellin) |
| Filament cap | FliD |
Over the years, several observations have made it clear that export of these flagellar proteins occurs by a rather unusual process. The first was the demonstration that the addition of new monomers of flagellin to a growing flagellar filament occurs at the distal end, i.e., the end furthest from the cell surface (5, 12). The second was that in the process of flagellar morphogenesis, substructures are successively added from most cell proximal to most cell distal (13, 16, 31, 32). Taken together, these observations indicate a successive addition of protein subunits from most cell proximal to most cell distal. A third observation was that the flagellar filament is not a solid cylinder but has a central channel (22, 26, 34); the same has subsequently been found to be true of the hook (25) and the rod (33). A fourth observation was that all exported proteins except those of the P and L rings (8, 10) do not undergo signal peptide cleavage during the export process (7, 9) and therefore cannot be proceeding by the general secretory pathway.
These observations led to a general acceptance of the idea that subunits reach their assembly point by traveling down the channel. This, however, did not explain how the subunits enter the channel in the first place, i.e., how they cross the plane of the cytoplasmic membrane. A specialized export apparatus seemed to be called for. In this study, we have not investigated the location or structure of the apparatus. Suffice to say that a reasonable hypothesis, which is beginning to gain experimental support (6), is that the apparatus exists within a central pore in the MS ring of the basal body; this ring is located in the cytoplasmic membrane and therefore is in a position to deliver exported substrates to the lumen of the growing flagellar structure.
The aim of this study was to positively identify the components of the export apparatus. There is currently suggestive evidence for several such components but little if any data in the way of direct proof. One line of evidence is a default one. The flagellar gene system has been studied exhaustively, and the functions of many of the more than 40 genes are known (see, e.g., reference 21). These functions include structural ones (hook protein, flagellin, etc.) and regulatory ones (sigma factor, anti-sigma factor, etc.). After assignment of these functions there was a residue of around 10 genes of unknown function, and as the probable existence of a specialized export apparatus became increasingly apparent, it was natural to consider these unassigned genes as candidates for encoding the apparatus.
An experiment involving the ability of various temperature-sensitive mutants to regrow sheared filaments at the restrictive temperature (35) suggested that three of these genes, flhA, fliH, and fliI, might indeed be involved in the export process. Studies of flagellar morphogenesis (13, 16, 17) are also consistent with several of these genes being involved in export, but they do not clearly distinguish between the issues of export and assembly, or of export across the cytoplasmic membrane and export across the outer membrane.
Finally, it has become evident in recent years that export of a variety of virulence factors by pathogenic bacteria has characteristics in common with export of flagellar proteins, and they now carry the common designation of type III export pathways (reviewed in reference 11). Most significantly, the proteins thought to be involved in flagellar protein export and those thought to be involved in virulence factor export show a high degree of similarity. Thus, the tentative assignment for one system lends credence to the tentative assignment for the other. This, however, does not constitute proof.
We therefore felt that direct evidence was needed and decided to make our criterion for export as simple as possible: is the presence of a given protein required for a given flagellar protein substrate to cross the cytoplasmic membrane? As far as flagellar structure was concerned, the only other condition besides integrity of the export apparatus itself was the integrity of the MS ring and the C ring, since they are presumed to provide a physical support structure for the export apparatus.
In the case of flagellin (and presumably the other filament-type proteins; see below), the simple approach of measuring periplasmic content was not possible because the export apparatus will not export these proteins until it has received a signal to switch its specificity, and this signal is generated only upon completion of an external structure, the hook. We were therefore forced to adopt a modified temperature shift protocol and measure the content of the cell exterior.
A question related to the issue of identification of export components is that of classes of export substrates. It is well established that hook-type proteins, i.e., the hook protein itself (FlgE) and the hook-capping protein (FlgD), belong to a different export class than the filament-type proteins, i.e., flagellin (FliC), the hook-filament junction proteins (FlgK and FlgL), the filament-capping protein (FliD), and the anti-sigma factor (FlgM). By defining these classes, we mean that there is an ordered export of hook-type proteins followed by export of filament-type proteins and that the switch in export specificity, mediated by FlhB, occurs at a well-defined point in the export/assembly process (20, 36). However, it has not been clear whether the rod proteins (FlgB, FlgC, FlgF, FlgG, and possibly FliE) belong to a distinct class from the hook-type proteins. This is a question of intrinsic interest, and it was also important in the present study in terms of finding a positive control for export of substrates to the periplasm.
Since export across the outer membrane, at least once it has been fully penetrated by the nascent structure, probably consists simply of diffusion of subunits through a passive channel, it should probably not be thought of as part of the primary export process, nor should structures such as the rod, hook, or filament be thought of as components of the export apparatus. However, we have found that the events immediately surrounding the stage of rod completion and hook initiation have some very interesting features that suggest a genuine export process and a genuine export structure, which in some regards resembles the secretin structure associated with export of virulence factors by the type III pathways.
MATERIALS AND METHODS
Bacterial strains, plasmids and media.
The strains and plasmids used in this study are listed in Table 2. Luria broth (LB) contained 10 g of Bacto Tryptone (Difco, Detroit, Mich.), 5 g of yeast extract, and 5 g of NaCl per liter. M9 medium contained (per liter) 100 ml of 10× M9 salts (10 g of NH4Cl, 59 g of Na2HPO4, 30 g of KH2PO4, 5 g of NaCl), 1 ml of 0.1 M CaCl2, 1 ml of 1 M MgSO4, 20 ml of 50% glycerol, and 1 mg of vitamin B1 per ml. Ampicillin was added to the medium at a final concentration of 100 μg/ml.
TABLE 2.
Strains and plasmids used
| Strain or plasmid | Relevant characteristic | Source or reference |
|---|---|---|
| Salmonella strains | ||
| SJW1446 | flgA | 29 |
| SJW1525 | flgB | 29 |
| SJW156 | flgD | 29 |
| SJW157 | flgD | 29 |
| SJW1353 | flgE | 29 |
| SJW1444 | flgF | 29 |
| SJW1378 | flgG | 29 |
| SJW1469 | flgH | 29 |
| SJW1351 | flgI | 29 |
| SJW1437 | flgJ | 29 |
| KK1334 | flgK | 18 |
| SJW1103GK | flgK::Tn10 | This study |
| SJW1364 | flhA | 19 |
| SJW2201GK | flhA(Ts) flgK::Tn10 | This study |
| SJW1383 | flhB | 19 |
| MY666GK | flhB(Ts) flgK::Tn10 | This study |
| SJW1371 | fliE | 29 |
| SJW1429 | fliH | 37 |
| MY671GK | fliH(Ts) flgK::Tn10 | This study |
| SJW2702 | fliI | 37 |
| MY640GK | fliI(Ts) flgK::Tn10 | This study |
| SJW104 | fliJ | 37 |
| SJW1357 | fliO | 37 |
| MY621GK | fliO(Ts) flgK::Tn10 | This study |
| SJW1401 | fliP | 37 |
| SJW2215GK | fliP(Ts) flgK::Tn10 | This study |
| SJW180 | fliQ | 37 |
| SJW161 | fliR | 37 |
| Plasmids | ||
| pUC18 | Plasmid for β-lactamase | |
| pDIC7 | pTrc99B/FliC | 1, 4 |
| pMKGD1 | pTrc99A/FlgD | 24 |
Preparation of the periplasmic contents and culture supernatants.
To examine FlgD and FlgE export, pUC18 was introduced into various nonflagellate (Fla−) mutants (in order to generate β-lactamase as a control for the periplasmic fraction). These transformants were incubated at 37°C in M9 medium containing ampicillin. When the cells reached an optical density at 600 nm of 1.0 to 1.2, 1.5 ml of culture was centrifuged and the cells and culture supernatants were collected separately. The cells were washed twice with TN buffer (10 mM Tris-HCl [pH 8.0], 0.3 M NaCl) and then suspended in 400 μl of sucrose buffer (20% sucrose, 100 mM Tris-HCl [pH 8.0], 0.5 mM EDTA) and incubated at room temperature for 20 min. After centrifugation, the cells were resuspended in 750 μl of 0.5 mM MgCl2 and placed on ice for 10 min to release the periplasmic fraction. The proteins in the supernatant and periplasmic fractions were precipitated by 10% trichloroacetic acid and suspended in Tris-saturated sodium dodecyl sulfate (SDS) loading buffer.
Antibodies.
Polyclonal antibodies were used for detection of FlgD, FlgE, FliC, and anti-β-lactamase (the latter from 5Prime→3Prime, Berkeley, Calif.).
Immunoblotting.
After the proteins in each fraction had been separated by SDS-polyacrylamide gel (normally 12% acrylamide) electrophoresis (PAGE), they were transferred onto a nitrocellulose membrane (Schleicher & Schull, Keene, N.H.) with a transblotting apparatus (Hoefer, San Francisco, Calif.). The membrane was then blocked for 1 h with Tris-buffered saline plus 0.1% Tween 20 and 5% nonfat dry milk and probed with polyclonal anti-β-lactamase, anti-FlgD, anti-FlgE, or anti-FliC. Immunodetection was performed with an enhanced chemiluminescence immunoblotting detection kit (Amersham International, Little Chalfont, United Kingdom).
Temperature shift-up experiments.
Temperature-sensitive Fla− mutants, which also contained a flgK::Tn10 mutation, were transformed with pDIC7 (a pTrc99B-based plasmid encoding fliC [4]) and were grown overnight at 30°C in 1 ml of LB containing ampicillin; 30 μl of the overnight culture was inoculated into 1.5 ml of fresh LB containing ampicillin and grown at 30°C to an optical density at 600 nm of 0.8. Then 1.5 ml of the culture was transferred to the sampling tube, washed twice with 500 μl of M9 medium containing ampicillin and 20 μg of each of the common amino acids except methionine per ml, and resuspended in 300 μl of the same medium; 150 μl of this culture was incubated at 30°C for 20 min, and the remainder was incubated at 42°C for the same length of time. The cells were then labeled with [35S]methionine for 5 min each at 30 and 42°C and incubated in the presence of excess unlabeled methionine for a further 20 min. The samples were fractionated into whole cells and culture supernatant as before. After immunoprecipitation of the cell lysates and culture supernatants with polyclonal anti-FliC antibody, the labeled proteins in each sample were separated by SDS-PAGE and analyzed by autoradiography.
RESULTS
Choice of export substrates.
From the known exported flagellar proteins (Table 1), we chose as substrates for our analysis of the export apparatus the following proteins: FlgD (hook-capping protein) and FlgE (hook protein), both representative of the hook protein class, and flagellin (representative of the filament protein class).
Choice of putative export components.
Initially, we confined ourselves to those proteins (FlhA, FlhB, FliH, FliI, FliO, FliP, FliQ, and FliR) which there was already reason to suspect were members of the export apparatus. Later, we extended the list to include proteins (FliE, FliJ, and FlgJ) whose function we felt was not well enough understood to warrant excluding them from a possible role in export. As controls, we examined the presumed need (for structural reasons) for the MS ring protein, FliF, and for the switch proteins of the cytoplasmic C ring, FliG, FliM, and FliN. Finally, we examined the role of the periplasmic P-ring protein and the outer membrane L-ring protein in the process of export across the outer membrane.
Protocol for determining whether a putative export component was required for export of a given export substrate.
The basic idea for the hook-type proteins was to test whether they would cross from the cytoplasm to the periplasm in a strain with a defect in a suspected export component. After the cells were washed, the periplasmic fraction was released by sucrose osmotic shock. The protein content was concentrated by trichloroacetic acid precipitation, and the samples were subjected to SDS-PAGE, electrotransferred to nitrocellulose membrane, and probed with the relevant antibody. In some cases, where the signal was weak or simply to confirm the immunoblotting data, we also carried out radiolabeling experiments.
Hook-type proteins can be exported in the absence of rod protein export.
It is well established that a major switch in the specificity of substrates being exported occurs during flagellar morphogenesis when the hook has reached its mature length. At that point, an integral membrane protein, FlhB, switches the specificity of substrates that it will accept from the hook-type proteins to the filament-type proteins (20, 36). One can readily rationalize this event, since flagellin is by far the most abundant exported substrate, and premature export would probably have severe logistical consequences. Less clear is whether a similar switch should occur at an earlier stage of morphogenesis, upon completion of the rod.
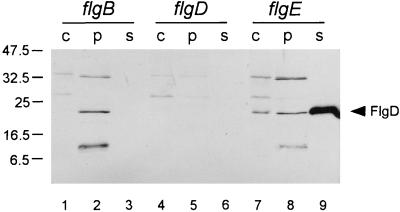
To test this, we examined whether hook-type proteins could be exported to the periplasm in a rod mutant. As illustrated by the immunoblot in Fig. 1, a representative hook-type protein, FlgD, was detected by anti-FlgD antibody in the periplasmic fraction of a rod mutant (flgB; lane 2) and a hook protein mutant (flgE; lane 8). Control experiments using anti β-lactamase antibody yielded a strong signal that was evenly balanced in the periplasmic fractions of all samples (data not shown). In the case of the flgE mutant, significant amounts of FlgD were detected in the supernatant. Similar results were obtained with other rod mutants (flgF and flgG) and with anti-FlgE instead of anti-FlgD (data not shown).
FIG. 1.
Immunoblot, using polyclonal anti-FlgD antibody, of fractionated whole-cell material from various mutants as indicated above the lanes. c, whole cells; p, periplasmic fraction; s, culture supernatant. Positions of molecular mass markers are shown in kilodaltons to the left; the position of FlgD (whose deduced molecular mass is 24 kDa) is indicated at the right. Bands at higher apparent molecular mass are nonspecific cross-reacting material; the band at lower apparent molecular mass is probably a degradation product of FlgD.
We conclude that the rod proteins and the hook-type proteins belong to a single export class; i.e., there is no specificity switch such as occurs between the hook-type proteins and the filament-type proteins. We propose that this single class be named the rod/hook-type export class.
Components necessary for export of FlgD, the hook-capping protein.
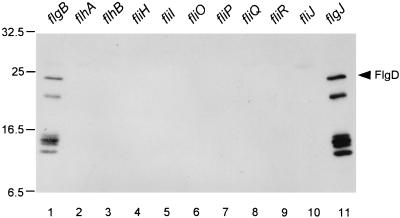
Using the same protocol, we tested for the presence of FlgD in the periplasmic fraction of a variety of other mutants (Table 3 and Fig. 2). It was detected in the periplasmic fraction of the flgB rod mutant (lane 1; see above) and of a strain with a mutation in flgJ (lane 11) but not in the periplasmic fractions of strains with mutations in flhA (lane 2), flhB (lane 3), fliH (lane 4), fliI (lane 5), fliO (lane 6), fliP (lane 7), fliQ (lane 8), fliR (lane 9), and fliJ (lane 10). The same results were obtained when the hook protein FlgE was used as the export substrate (data not shown).
TABLE 3.
Export of protein substrates of various mutant strains
| Export substrate | Phenotype of strain with indicated genotypea
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| flgB/flgKb | flhA | flhB | fliH | fliI | fliO | fliP | fliQ | fliR | fliJ | flgJ | |
| FlgDc | +d | −d | −d | − | − | − | − | − | − | − | + |
| FliCe | +d | −d | − | − | − | − | [+] | NT | NT | NT | NT |
[+], probably a false positive resulting from the nature of the temperature-sensitive mutation; NT, not tested because of lack of a suitable temperature-sensitive mutant.
A flgB mutant was used as a positive control for the hook-type proteins, FlgD and FlgE; a flgK mutant was used for FliC (flagellin). See the text.
Export to the periplasmic compartment.
Confirmation of results originally obtained by Minamino and Kutsukake (24).
Export to the external compartment in a temperature-shift protocol with temperature-sensitive strains. See the text.
FIG. 2.
Immunoblot, using polyclonal anti-FlgD antibody, of the periplasmic fraction from various mutants as indicated above the lanes. Positions of molecular mass markers are shown in kilodaltons to the left; the position of FlgD is indicated at the right. The bands at lower apparent molecular mass are probably degradation products of FlgD.
Components necessary for export of FliC, flagellin.
Flagellin presents a special case, since the switch in export specificity from hook-type proteins to filament-type proteins must occur before flagellin can be exported, and this switch is dependent on completion of the hook. Since we were now dealing with the external compartment rather than the periplasmic compartment, we used a flgK (first hook-filament junction protein; hook-associated protein 1) mutant as a control. This permitted assay of export of monomeric flagellin without the complication of filament assembly. The cells were grown at the permissive temperature of 30°C and shifted to the restrictive temperature of 42°C, and then export of flagellin to the external medium was monitored. This protocol assumes that preexisting copies of a temperature-sensitive component will become nonfunctional upon the temperature shift. This assumption may not always be justified, since a component tightly constrained by surrounding structure might continue to function normally, as we in fact found with FliP (see below).
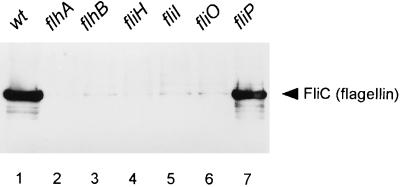
We had in our collection temperature-sensitive strains with mutations in flhA, flhB, fliH, fliI, fliO, and fliP but not in fliQ, fliR, fliJ, and flgJ. We found flagellin in the culture supernatant of the control strain (wild type except for the flgK mutation) (Table 3 and Fig. 3, lane 1) but not in the supernatants of flhA, flhB, fliH, fliI, and fliO mutants (Table 3 and Fig. 3, lanes 2 to 6). Unexpectedly, we found flagellin in the supernatant of the fliP mutant (lane 7). However, in this strain (which is not the same one used for Fig. 2), the mutation (V42F) was at a position which would place it in the first membrane span of the mature protein, following signal peptide cleavage. (As a prokaryotic cytoplasmic membrane protein, FliP is unusual in undergoing such cleavage [28].) We suspect that this residue may be so highly constrained in the membrane that it retains function following the temperature upshift and so yields a false-positive result. We think it likely that FliP is needed for export of flagellin, as it is for other export substrates.
FIG. 3.
Autoradiogram of sample precipitated with polyclonal anti-FliC antibody of the supernatant fraction from the wild type (except for the flgK::Tn10 mutation; wt) and various mutants in temperature shift experiments with temperature-sensitive mutants (see the text). In addition to the mutation of interest, all strains contain a flgK::Tn10 mutation to prevent filament assembly. The position of FliC is indicated.
The MS ring and the cytoplasmic C rings are required for export of all substrates.
Morphological assembly studies (13, 16, 17) have indicated that the MS ring (FliF) and the cytoplasmic C ring (FliG, FliM, and FliN) are necessary for flagellar assembly to proceed to the external structures such as the rod. Consistent with this finding, we found that export to the periplasm of the substrates tested did not occur in fliF, fliG, fliM, and fliN mutants (data not shown). However, we interpret these results to mean that the MS and C rings provide the necessary structural environment for the export apparatus, not that they are themselves directly involved in export.
FliE is a prerequisite for hook-type protein export.
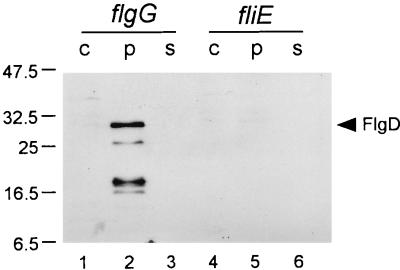
FliE is a structural component of the basal body, but its location and function are not known. When the periplasmic fraction of a fliE mutant was examined with anti-FlgD antibody, no FlgD was detected (Fig. 4, lane 5).
FIG. 4.
Immunoblot, using polyclonal anti-FlgD antibody, of fractionated whole-cell material from a flgG (rod) mutant and a fliE mutant. c, whole cells; p, periplasmic fraction; s, culture supernatant. Molecular mass markers are shown in kilodaltons to the left; the position of FlgD is indicated at the right. The gel used contained 15% acrylamide.
Export of hook-type proteins in outer ring (flgA, flgH, and flgI) mutants.
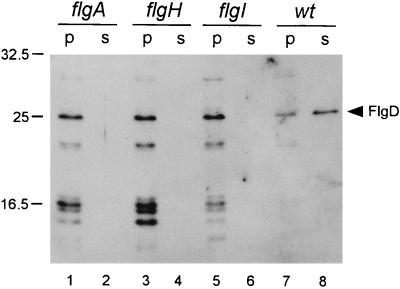
Kubori et al. (16) found that flgD mutants were capable of assembling L rings onto the rod but far less so than wild-type cells, suggesting that the processes of L-ring assembly and addition of the hook-capping protein, FlgD, to a mature rod were somehow interconnected. This led us to question whether the interconnection might be reciprocal; in other words, might the existence of an L ring be important or necessary for export of hook-type proteins across the outer membrane? We therefore examined mutants defective in flgA (necessary for P-ring assembly), flgI (structural gene for the P ring, which is required for L-ring assembly), and flgH (structural gene for the L ring). The periplasmic and culture supernatant fractions from these mutants were subjected to immunoblotting with anti-FlgD antibody. FlgD was detected in the periplasmic fraction but not in the culture supernatant fraction (Fig. 5). The same result was obtained with FlgE (data not shown).
FIG. 5.
Immunoblot, using polyclonal anti-FlgD antibody, of the periplasmic (p) and supernatant (s) fractions from various mutants with defects in outer ring production and from the wild-type control (wt). Positions of molecular mass markers are shown in kilodaltons to the left; the position of FlgD is indicated at the right.
Export of hook protein is impaired in flgD mutants.
If the FlgD protein is important for L-ring formation, and if L-ring formation is important for export, the presence of a mutation in flgD might impair export of the other hook-type protein, FlgE. Consistent with this idea, we found that most flgD mutants resulted in FlgE accumulating exclusively in the periplasm. However, several strains with point mutations, such as SJW157, resulted in approximately equal amounts of FlgE in the periplasm and the external medium (data not shown), confirming results reported previously by Ohnishi et al. (29).
DISCUSSION
The rod and hook proteins belong to a single export class.
It was known from previous work that the hook protein FlgE and its capping protein FlgD belonged to a different export class from the filament-type proteins (the hook-filament junction proteins FlgK and FlgL, flagellin [FliC], the filament-capping protein FliD, and the anti-sigma factor FlgM). In other words, export of the hook-type proteins proceeds at an earlier stage, during which export of the filament-type proteins is blocked because of the specificity of acceptance of substrates by the export component FlhB. Subsequently, the specificity is reversed such that hook-type proteins are rejected as export substrates and filament-type proteins are accepted. Since rod assembly precedes hook assembly, it seemed reasonable to ask whether the rod proteins might also represent a distinct export class. We found that hook-type proteins can be exported regardless of whether rod protein export is occurring. Thus, the rod proteins and the hook-type proteins appear to belong to the same export class, which we call the rod/hook-type class of export substrates.
This leaves another question, which is how rod proteins get assembled ahead of the hook-type proteins, if they can be exported simultaneously. This is a general question within a given export class and one to which we do not know the answer. Possibly some export substrates are kinetically accepted ahead of others. We know, for example, that interference can occur between exported substrates, since overproduction of FlgG has a dominant negative effect on flagellar assembly (data not shown). Alternatively, the sorting may occur at the assembly level so that exported hook protein is left unassembled while the rod structure is under construction.
The flagellum-specific export apparatus contains at least eight general components.
In this study, we have established that all eight of the suspected major components of the flagellum-specific protein export apparatus—FlhA, FlhB, FliH, FliI, FliO, FliP, FliQ, and FliR—are indeed components of this apparatus.
Charkowski et al. (2) have identified groups of Hrc proteins as being components of the type III export pathway within the hrp system of Pseudomonas syringae. They identified either or both of HrcV (equivalent to FlhA) and HrcN (FliI) and one or more of HrcR (FliP), HrcS (FliQ), HrcT (FliR), and HrcU (FlhB) as components of the pathway and (in a separate experiment) uniquely identified HrcU (FlhB) as such. Their results and ours are in agreement.
Without the knowledge gained in the present study, it would have been difficult to proceed with certainty to a more detailed understanding of the flagellar export apparatus and the export process. They appear all to be general components, i.e., to be required for export of all flagellar proteins that use the flagellum-specific pathway.
In this regard, we tested as substrates both examples of hook-type proteins (FlgD and FlgE) and a representative filament-type protein (flagellin [FliC]). Regrettably, we were unable to test FliE and any of the four rod proteins because of difficulties in detection, even when we had tagged them with the FLAG epitope. In terms of the apparatus components, there were in the case of flagellin export four proteins (FliQ, FliR, FliJ, and FlgJ) that for technical reasons we could not test; there was also one (FliP) for which the result obtained (that it was not needed for export) is almost certainly artifactual. Nonetheless, we feel confident in presuming that FlhA, FlhB, FliH, FliI, FliO, FliP, FliQ, and FliR are general components, without which no export of any kind can occur. This presumption is reinforced by the fact that most of these proteins have homologs in type III pathways for export of virulence factors.
FliJ is an export component, necessary for export of at least some substrates.
FliJ, a cytoplasmic protein, was found to be necessary for export of at least FlgD and FlgE. A plausible role would be as a cytoplasmic chaperone, as has been suggested by Stephens et al. (30) in the case of fliJ of Caulobacter crescentus, since it shares some of the general properties of other chaperones: small size, high charge density, and high predicted α-helical content. We were unable to test whether it is necessary for export of flagellin, but we suspect not, since the role of a flagellin-specific chaperone has been postulated for another protein, FliS (39). Interestingly, fliJ has a leaky mutant phenotype (38), so that export of its putative substrates can occur with low probability in its absence; this is a feature that has also been noted with fliS. We are currently trying to obtain evidence for physical interactions between FliJ and the proteins whose export it facilitates.
FliE, a basal-body component, is required for export of some substrates.
FliE is a basal body protein whose location is not known with certainty but which is postulated to be a component of the rod (17). We found that it was required for the export of at least two other substrates, the hook protein FlgE and the hook-capping protein FlgD. The reason for this requirement is not known. One possibility could be that it interacts with the periplasmic domains of integral membrane components such as FliO or FliP and facilitates passage of the other substrates into the periplasmic space.
FlgJ is essential for rod assembly but not for export of hook-capping protein or hook protein.
Until this study, essentially nothing was known about the FlgJ protein. Although not needed for export of the substrates tested, it is necessary for the flagellar assembly process, since flgJ mutants lack a rod structure (13, 16). One possibility would be that it functions as a scaffolding protein for rod assembly, in a similar fashion as FlgD does for hook assembly. Another would be that it interacts with the peptidoglycan layer. Consistent with the latter suggestion, Dijkstra and Keck (3) have noted that the FlgJ sequence contains a muramidase consensus motif.
The flagellar export apparatus.
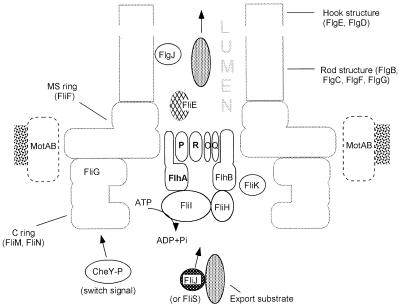
Although the primary purpose of this study was to identify export apparatus components and the range of export substrates that they each handle, it is perhaps worthwhile to make a few comments concerning the possible locations and functions of the various components (Fig. 6). The proposed location of the membrane-associated components (FlhA, FlhB, FliO, FliP, FliQ, and FliR) is in a central pore within the MS ring of the flagellar basal body. There is physical evidence that such a pore exists (14), and there is evidence that the FliP and FliR proteins are associated with the basal body (6). We have recently established that FlhA is also associated with the basal body (23) and are currently seeking equivalent evidence for FlhB, FliO, and FliQ. FlhA and FlhB have large soluble domains that presumably protrude into the cytoplasm within the cavity that lies at the center of the cytoplasmic C ring of the motor. We suggest that export substrates, with the aid of possible cytosolic chaperones such as FliJ or FliS, are brought into contact with FliI (an ATPase) and FliH (function unknown), which then interface with the soluble domains of FlhA and FlhB and release their substrates. These substrates are then translocated across the plane of the membrane through a complex consisting of FliO, FliP, FliQ, FliR, and the transmembrane domains of FlhA and FlhB; the point at which ATP hydrolysis occurs is not known. FliK in some fashion enables FlhB to recognize when hook assembly is complete and to switch its export specificity to filament-class substrates. FliE may be located at the periplasmic side of the lumen and facilitate transfer of the export substrates into the lumen. The role of FlgJ is unclear; it does not seem to play a role in the export process but may participate in some aspect of assembly of subunits once they have been exported.
FIG. 6.
Hypothetical model for the flagellar protein export apparatus (see the text for further details). This study has shown that FlhA, FlhB, FliO, FliP, FliQ, FliR, FliH, and FliI are components of this apparatus. FlhA, FlhB, FliO, FliP, FliQ, and FliR are all integral membrane proteins. Of these, FlhA, FliP, and FliR (shown in boldface) have been shown to be physically associated with the flagellar basal body (6, 23); FlhB, FliO, and FliQ may be also. It is suggested that these proteins may be located in a pore within the basal body MS ring. Two soluble components, FliH and FliI (an ATPase) may receive the export substrates from cytoplasmic chaperones (such as FliJ?) and deliver them in an energy-dependent process to the membrane-associated structure, which then translocates them to the channel or lumen in the nascent structure. FliK is involved in control of the length of the flagellar hook. FliE appears to be needed for the export of other substrates, but it is not known whether it is itself an export substrate. FlgJ is needed for assembly of the rod but does not appear to be involved in the export process. The representation of the rod and hook structures is not to scale and is highly schematic.
The basal body L ring can be regarded as a component of the export apparatus at the stage of rod completion and hook initiation.
We thought initially that our definition of the flagellar export apparatus should be confined to components that were responsible for translocating export substrates across the plane of the cytoplasmic membrane. Our rationale for using this restrictive definition was that subsequent events were merely diffusion down a tunnel. However, at one critical point in assembly, when the periplasmic rod has just been completed and the external hook is just being initiated, the structure has to penetrate the outer membrane; only after that has happened does the simple diffusion model once again apply.
The results from this study make it clear that the L ring is essential, or at least very important, for this penetration process and, further, that assembly of the L ring is to a considerable extent dependent on one of the export substrates themselves, namely, the hook-capping protein FlgD. We therefore suggest that for a brief period of time there is an additional element in the export apparatus, consisting of either the L ring alone or an L ring-FlgD complex. (The need for the P ring is probably a result of the fact that L-ring assembly appears to be dependent on preassembly of the P ring [16], not because the P ring participates directly in penetration of the outer membrane.)
In type III secretion systems for virulence factors there exists in the outer membrane a large homomultimeric annular complex made of molecules called secretins. The function of this complex is believed to be to permit the passage of export substrates across the outer membrane. Also, a recent ultrastructural study of Salmonella has revealed a needle-like structure that contains known components of the type III virulence factor export system (15). Among these components was InvG, which is believed to be a secretin and whose homolog, HrcC of P. syringae, is known to be necessary for substrate to pass the outer membrane (2). It seems reasonable to postulate that InvG comprises the outer ring that is seen in the needle-like structure. We suggest that the L ring of the flagellar basal body may transiently correspond to a secretin-like annulus, in addition to its long-term function as part of a bushing to stabilize flagellar rotation.
ACKNOWLEDGMENTS
We thank Kazuhiro Kutsukake for the gift of unpublished plasmids, Shigeru Yamaguchi for the gift of unpublished strains and for unpublished information about the leakiness of fliJ mutants, and Gabriele Miller and May Kihara for technical assistance.
This work was supported by USPHS grant AI12202.
ADDENDUM IN PROOF
Our understanding of the role of FlgJ in flagellar assembly has significantly advanced with the concurrent publication of a paper by Nambu et al. (T. Nambu, T. Minamino, R. M. Macnab, and K. Kutsukake, J. Bacteriol. 181:1555–1561, 1999).
REFERENCES
- 1.Amann E, Ochs B, Abel K-J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–314. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 2.Charkowski A O, Huang H-C, Collmer A. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dijkstra A J, Keck W. Peptidoglycan as a barrier to transenvelope transport. J Bacteriol. 1996;178:5555–5562. doi: 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doi, H., and K. Kutsukake. Unpublished data.
- 5.Emerson S U, Tokuyasu K, Simon M I. Bacterial flagella: polarity of elongation. Science. 1970;169:190–192. doi: 10.1126/science.169.3941.190. [DOI] [PubMed] [Google Scholar]
- 6.Fan F, Ohnishi K, Francis N R, Macnab R M. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- 7.Homma M, DeRosier D J, Macnab R M. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J Mol Biol. 1990;213:819–832. doi: 10.1016/S0022-2836(05)80266-9. [DOI] [PubMed] [Google Scholar]
- 8.Homma M, Komeda Y, Iino T, Macnab R M. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol. 1987;169:1493–1498. doi: 10.1128/jb.169.4.1493-1498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homma M, Kutsukake K, Hasebe M, Iino T, Macnab R M. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1990;211:465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- 10.Homma M, Ohnishi K, Iino T, Macnab R M. Identification of flagellar hook and basal body gene products (FlaFV, FlaFVI, FlaFVII, and FlaFVIII) in Salmonella typhimurium. J Bacteriol. 1987;169:3617–3624. doi: 10.1128/jb.169.8.3617-3624.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iino T. Polarity of flagellar growth in Salmonella. J Gen Microbiol. 1969;56:227–239. doi: 10.1099/00221287-56-2-227. [DOI] [PubMed] [Google Scholar]
- 13.Jones C J, Macnab R M. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J Bacteriol. 1990;172:1327–1339. doi: 10.1128/jb.172.3.1327-1339.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katayama E, Shiraishi T, Oosawa K, Baba N, Aizawa S-I. Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella typhimurium as determined by stereo-photogrammetry of quick-freeze deep-etch replica images. J Mol Biol. 1996;255:458–475. doi: 10.1006/jmbi.1996.0038. [DOI] [PubMed] [Google Scholar]
- 15.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J E, Aizawa S-I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 16.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S-I. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol. 1992;226:433–446. doi: 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- 17.Kubori T, Yamaguchi S, Aizawa S-I. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J Bacteriol. 1997;179:813–817. doi: 10.1128/jb.179.3.813-817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutsukake, K. Unpublished data.
- 19.Kutsukake K, Iino T. Refined genetic analysis of the region II che mutants in Salmonella typhimurium. Mol Gen Genet. 1985;199:406–409. doi: 10.1007/BF00330750. [DOI] [PubMed] [Google Scholar]
- 20.Kutsukake K, Minamino T, Yokoseki T. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J Bacteriol. 1994;176:7625–7629. doi: 10.1128/jb.176.24.7625-7629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 22.Mimori Y, Yamashita I, Murata K, Fujiyoshi Y, Yonekura K, Toyoshima C, Namba K. The structure of the R-type straight flagellar filament of Salmonella at 9 Å resolution by electron cryomicroscopy. J Mol Biol. 1995;249:69–87. doi: 10.1006/jmbi.1995.0281. [DOI] [PubMed] [Google Scholar]
- 23.Minamino, T. Unpublished data.
- 24.Minamino, T., and K. Kutsukake. Unpublished data.
- 25.Morgan D G, Macnab R M, Francis N R, DeRosier D J. Domain organization of the subunit of the Salmonella typhimurium flagellar hook. J Mol Biol. 1993;229:79–84. doi: 10.1006/jmbi.1993.1009. [DOI] [PubMed] [Google Scholar]
- 26.Morgan D G, Owen C, Melanson L A, DeRosier D J. Structure of bacterial flagellar filaments at 11 Å resolution: packing of the α-helices. J Mol Biol. 1995;249:88–110. doi: 10.1006/jmbi.1995.0282. [DOI] [PubMed] [Google Scholar]
- 27.Müller V, Jones C J, Kawagishi I, Aizawa S-I, Macnab R M. Characterization of the fliE genes of Escherichia coli and Salmonella typhimurium and identification of the FliE protein as a component of the flagellar hook-basal body complex. J Bacteriol. 1992;174:2298–2304. doi: 10.1128/jb.174.7.2298-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohnishi K, Fan F, Schoenhals G J, Kihara M, Macnab R M. The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J Bacteriol. 1997;179:6092–6099. doi: 10.1128/jb.179.19.6092-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohnishi K, Ohto Y, Aizawa S-I, Macnab R M, Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol. 1994;176:2272–2281. doi: 10.1128/jb.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens C, Mohr C, Boyd C, Maddock J, Gober J, Shapiro L. Identification of the fliI and fliJ components of the Caulobacter flagellar type III protein secretion system. J Bacteriol. 1997;179:5355–5365. doi: 10.1128/jb.179.17.5355-5365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T, Iino T, Horiguchi T, Yamaguchi S. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J Bacteriol. 1978;133:904–915. doi: 10.1128/jb.133.2.904-915.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki T, Komeda Y. Incomplete flagellar structures in Escherichia coli mutants. J Bacteriol. 1981;145:1036–1041. doi: 10.1128/jb.145.2.1036-1041.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas, D., D. Morgan, and D. DeRosier. Personal communications.
- 34.Trachtenberg S, DeRosier D J. Three-dimensional structure of the frozen-hydrated flagellar filament. The left-handed filament of Salmonella typhimurium. J Mol Biol. 1987;195:581–601. doi: 10.1016/0022-2836(87)90184-7. [DOI] [PubMed] [Google Scholar]
- 35.Vogler A P, Homma M, Irikura V M, Macnab R M. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol. 1991;173:3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams A W, Yamaguchi S, Togashi F, Aizawa S-I, Kawagishi I, Macnab R M. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi, S. Unpublished data.
- 38.Yamaguchi, S. Personal communication.
- 39.Yokoseki T, Kutsukake K, Ohnishi K, Iino T. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology. 1995;141:1715–1722. doi: 10.1099/13500872-141-7-1715. [DOI] [PubMed] [Google Scholar]