Abstract
We have recently described the expression of two pili of different lengths on the surface of Legionella pneumophila (B. J. Stone and Y. Abu Kwaik, Infect. Immun. 66:1768–1775, 1998). Production of long pili requires a functional pilEL locus, encoding a type IV pilin protein. Since type IV pili in Neisseria gonorrhoeae are associated with competence for DNA transformation, we examined the competence of L. pneumophila for DNA transformation under conditions that allowed the expression of type IV pili. We show that L. pneumophila is naturally competent for DNA transformation by isogenic chromosomal DNA and by plasmid DNA containing L. pneumophila DNA. Many different L. pneumophila loci are able to transform L. pneumophila after addition of plasmid DNA, including gspA, ppa, asd, and pilEL. The transformation frequency is reduced when competing DNA containing either L. pneumophila DNA or vector sequences is added to the bacteria, suggesting that uptake-specific sequences may not be involved in DNA uptake. Competence for DNA transformation correlates with expression of the type IV pili, and a pilEL mutant defective in expression of type IV pili is not competent for DNA transformation. Complementation of the mutant for competence is restored by the reintroduction of a cosmid that restores production of type IV pili. Minimal competence is restored to the mutant by introduction of pilEL alone. We conclude that competence for DNA transformation in L. pneumophila is associated with expression of the type IV pilus and results in recombination of L. pneumophila DNA into the chromosome. Since expression of type IV pili also facilitates attachment of L. pneumophila to mammalian cells and protozoa, we designated the type IV pili CAP (for competence- and adherence-associated pili).
Legionella pneumophila is the causative agent of Legionnaires’ disease. Infection by L. pneumophila occurs after introduction of bacteria from an environmental source into a human host after inhalation of contaminated aerosols (see references 1 and 5 for recent reviews). The bacteria are then able to replicate within phagosomes of mononuclear phagocytes and possibly epithelial cells (36, 37). Multiple bacterial factors appear to be required for attachment of L. pneumophila to protozoan and mammalian cells. Complement-dependent and complement-independent mechanisms have been described for attachment of L. pneumophila to macrophages and lung fibroblasts (10, 22, 40). Mutants defective in attachment to both protozoan and mammalian cells have recently been described (19–21, 26).
One of the L. pneumophila attachment factors that has been recently described is the pili (50). Expression of pili by L. pneumophila was originally identified by electron microscopy (41, 42). The pili can be categorized as short (0.1 to 0.6 μm) or long (0.8 to 1.5 μm) (50). Expression of long pili on the bacterial surface is abolished by an insertion mutation in the pilEL locus, encoding a type IV pilin protein (50). The pilEL mutant has a 50% reduction in attachment to the protozoan Acanthamoebae polyphaga and a 50 to 65% reduction in attachment to human alveolar epithelial cells and U937 macrophage-like cells.
In addition to L. pneumophila, type IV pilin genes have been identified in a number of pathogenic bacteria, such as Neisseria gonorrhoeae (35), Neisseria meningitidis (39), Pseudomonas aeruginosa (28), enteropathogenic Escherichia coli (15), and Vibrio cholerae (17, 46). Type IV pili are characterized by a region of homology at the amino-terminal end of the amino acid sequence (27, 52). Protein components involved in biogenesis of type IV pili have been implicated in protein secretion, twitching motility, and filamentous phage assembly (27, 52). In N. gonorrhoeae, factors required for type IV pilus expression on the bacterial surface are also required for natural competence for DNA transformation (for a recent review, see reference 18).
Genetic exchange between naturally competent bacteria has been described for a number of bacteria, including N. gonorrhoeae (49), Bacillus subtilis (54), Haemophilus influenzae (9), Streptococcus pneumoniae (8), Acinetobacter calcoaceticus (29), and others (see reference 33 for a review). Competence for transformation can be constitutive, as is the case for N. gonorrhoeae (49). Competence can also be induced as a function of growth conditions, such as in H. influenzae (48). Natural transformation requires uptake of extracellular DNA prior to incorporation of the DNA in the bacterial genetic repertoire. DNA uptake in the gram-negative bacteria N. gonorrhoeae (16, 23) and H. influenzae (47) is recognition sequence dependent while DNA uptake in the gram-positive bacteria B. subtilis and S. pneumoniae (reviewed in reference 33) and in the gram-negative bacterium A. calcoaceticus (32, 38) is recognition sequence independent. Processing of DNA prior to transformation (reviewed in reference 33) allows entry of linear DNA molecules into the bacterium in a single-stranded form prior to recombination into the chromosome in a recA-dependent manner. Circular DNA molecules are rendered linear prior to similar processing and recombination, although reconstitution of replicating plasmids can also occur.
In this report, we describe the phenomenon of natural competence for DNA transformation in L. pneumophila. Transformation of L. pneumophila occurs after addition of exogenous DNA, either L. pneumophila chromosomal DNA or plasmid DNA containing L. pneumophila DNA. Transformation is dependent upon recombination of donor DNA into the chromosome. Similar to the case for N. gonorrhoeae (11, 14, 49), transformation in L. pneumophila is induced under conditions similar to those for expression of type IV pili. Transformation in L. pneumophila requires the type IV pilin locus (pilEL), which is also essential for expression of type IV pili.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The L. pneumophila serogroups and strains, Legionella species, and E. coli strain used in this study are listed in Table 1. Strain BS200 is a spontaneous nalidixic acid-resistant (Nalr) mutant generated from passage of strain AA100 on buffered charcoal-yeast extract (BCYE) agar supplemented with 30 μg of nalidixic acid per ml. The plasmids used in this study are also described in Table 1. Plasmid pBC-K was derived from pBC SK+ by insertion of the Kanr gene from pUC4K into the EcoRI site. Cosmid pCC3 was isolated from a chromosomal library of L. pneumophila AA100 that contains approximately 45 kb of L. pneumophila DNA.
TABLE 1.
Bacterial strains and plasmids
| Species, strain, serogroup, or plasmid | Relevant properties | Reference(s) or source |
|---|---|---|
| Bacteria | ||
| L. pneumophila | ||
| AA100 | 3 | |
| AA400 | AA100; asd::Kanr | 25 |
| BS100 | AA100; pilEL::Kanr | 50 |
| BS200 | AA100; Nalr | This study |
| Serogroup 1 | pilEL+ | 50 |
| Serogroup 2 | pilEL+ | 50 |
| Serogroup 3 | pilEL+ | 50 |
| Serogroup 4 | pilEL+ | 50 |
| Serogroup 5 | pilEL+ | 50 |
| Serogroup 6 | pilEL+ | 50 |
| Serogroup 7 | pilEL+ | 50 |
| Serogroup 8 | pilEL+ | 50 |
| Serogroup 9 | pilEL+ | 50 |
| Serogroup 10 | pilEL+ | 50 |
| Serogroup 11 | pilEL+ | 50 |
| Serogroup 12 | pilEL+ | 50 |
| Serogroup 13 | pilEL+ | 50 |
| L. wadsworthii | pilEL+ | 50 |
| L. santicrucis | pilEL+ | 50 |
| L. cherii | pilEL+ | 50 |
| L. longbeachae | pilEL | 50 |
| L. gormanii | pilEL | 50 |
| L. micdadei | pilEL | 50 |
| E. coli DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 24 |
| Plasmids | ||
| pBC SK+ | ColE1 origin, lacZα, Cmr | Stratagene (La Jolla, Calif.) |
| pBC-K | pBC SK+; Cmr Kanr | This study |
| pUC4K | pBR322 origin; Ampr Kanr; source for the Kanr marker | Pharmacia Biotech (Piscataway, N.J.) |
| pBJ120 | pBC SK+; CmrpilEL+ | 50 |
| pBJ114 | pBC SK+; CmrpilEL::Kanr | 50 |
| pBOC20 | oriV oriT sacB Cmr | 6 |
| pBJ115 | pBOC20; CmrpilEL::Kanr | 50 |
| pGS-K | pBOC20; CmrgspA::Kanr | 4 |
| pJA2A-K2 | pBOC20; Cmrppa::Kanr | 2 |
| pOHBOC1 | pBOC20; Cmrasd+ | 25 |
| pOHBOC1-K | pBOC20; Cmrasd::Kanr | 25 |
| pOHBOC2 | pBOC20; Cmrasd+ | 25 |
| pTLP6 | oriV oriT rpsL cos′ Cmr | 7, 34 |
| pCC3 | pTLP6; CmrpilEL+ | 50 |
DNA preparation.
Chromosomal DNAs from L. pneumophila and E. coli strains were isolated as previously described (45) or by using a Puregene DNA isolation kit (Gentra Systems, Inc., Minneapolis, Minn.) as specified by the manufacturer. Plasmid DNA was isolated by using a QIAGEN (Chatsworth, Calif.) Midiprep kit. DNA preparations were resuspended in sterile distilled water and vortexed prior to use.
Bacterial culture and induction of competence.
L. pneumophila strains obtained from freezer stocks were used directly or grown on BCYE agar for 48 h at 37°C prior to culture for induction of competence or examination of pilus expression by electron microscopy. Bacteria were grown in 5 ml of buffered yeast extract (BYE) broth without shaking at 37°C for 4 days in 15-ml capped plastic tubes. Prior to addition of chromosomal DNA or plasmid DNA, 4.6 ml of BYE broth was removed from the culture without disturbing the bacteria settled at the bottom of the culture tube. DNA was added to the bacterial culture (unless otherwise specified) to a final concentration of 400 μg of chromosomal DNA per ml or 40 μg of plasmid DNA per ml in a final volume of 0.5 ml, gently mixed, and returned to 37°C for an additional 2 days. Viable counts were determined for total bacteria on BCYE agar and for transformed bacteria on BCYE agar containing the appropriate antibiotics (20 μg of kanamycin per ml, 2.5 μg of chloramphenicol per ml, or 30 μg of nalidixic acid per ml).
L. pneumophila AA400 was grown in the presence of 40 μg of diaminopimelic acid (DAP) per ml. DAP was also added during transformation of AA100 with pOHBOC1-K and during growth for total viable counts. L. pneumophila cultures grown under alternate growth conditions were all started from L. pneumophila cultures grown on plates. Bacteria were resuspended in 1 ml of sterile water at approximately 109 CFU/ml, and 20 μl (in BYE broth) or 200 μl (in water) was added to each 5-ml culture. Cultures were incubated at 24, 30, or 37°C. Aerated cultures and cultures grown on agar prior to electron microscopic examination or addition of DNA were grown for 2 days at 37°C. Transformation experiments performed in the presence of competing plasmid DNA were performed as described above except that 4 or 80 μg of competing plasmid DNA per ml was added to the bacteria and gently mixed just prior to addition of 4 μg of pBJ114 DNA per ml.
Data analysis.
The transformation frequency was calculated as the number of antibiotic-resistant CFU per milliliter/total CFU per milliliter. For transfer of the asd gene into AA400, transformation frequency was measured as CFU per milliliter for cells grown on BCYE without DAP/CFU per milliliter for cells grown on BCYE with DAP. All experiments were performed with duplicate samples and performed two or more times, depending on the degree of variability. Data presented are from representative experiments, and error ranges are calculated from duplicate samples.
RESULTS
L. pneumophila is naturally competent for DNA transformation.
We have recently described the expression of type IV pili by L. pneumophila. Since type IV pili have been associated with natural DNA transformation in N. gonorrhoeae (11), L. pneumophila was examined for competence after growth of the bacteria under conditions which promoted expression of pili (50). Bacteria were grown without aeration for 4 days in BYE medium (as described in Materials and Methods). DNA transformation was first attempted with chromosomal DNA isolated from strain BS200, a spontaneous Nalr isogenic mutant of L. pneumophila AA100. The ability of L. pneumophila wild-type strain AA100 to become Nalr was examined after addition of BS200 chromosomal DNA. The bacteria and DNA were allowed to incubate for 2 days prior to enumeration of transformants. The Nalr phenotype from BS200 was transferred to AA100 at a frequency of approximately 1.4 × 10−6 ± 7.1 × 10−8 with the addition of 400 μg of chromosomal DNA per ml. Incubation of bacteria with DNA for less than 2 days resulted in a decreased transformation frequency (data not shown). In contrast, addition of the same amount of chromosomal DNA from AA100 to strain AA100 did not result in detection of any transformants (>10−9).
DNA transformation of an alternative resistance marker was attempted. The transformation frequency of isogenic chromosomal DNA containing a Kanr marker, which is 1.7 kb in size, was determined. The Kanr marker was previously introduced into L. pneumophila AA100 by homologous recombination to generate strain BS100 and is present within pilEL, the type IV pilin locus (50). The transformation frequency after addition of BS100 chromosomal DNA to AA100 was 5.8 × 10−7 ± 5.0 × 10−7, which is comparable to the frequency of transformation after addition of BS200 DNA (Nalr). Addition of AA100 chromosomal DNA to strain AA100 did not result in any detectable transformation (<10−9). In all transformation experiments, variability (up to 10-fold) between experiments with regard to transformation frequency was seen. However, the competence phenotype was consistent in all experiments. Therefore, results shown are representatives of a typical experiment.
Transformation requires L. pneumophila sequences.
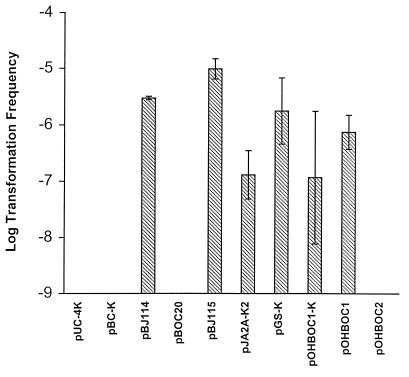
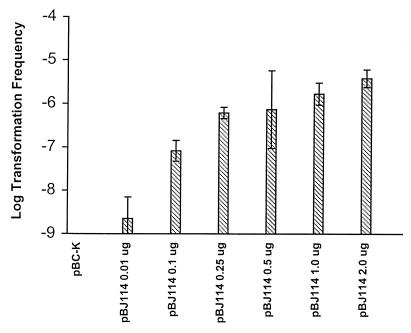
The ability of plasmids obtained from an E. coli background to generate transformants of L. pneumophila was determined to examine potential barriers to uptake of exogenous DNA. All plasmids were obtained from the same E. coli strain prior to transformation experiments to eliminate differences in the methylation status of the DNA, which may affect transformation frequency. Plasmids obtained from E. coli are able to replicate autonomously in L. pneumophila after introduction by electroporation (data not shown). Plasmid pBJ114 contains approximately 2 kb of L. pneumophila chromosomal DNA (including the pilEL open reading frame) with a Kanr marker in the same location within pilEL as in strain BS100. The transformation frequency was determined for transfer of the Kanr marker. pBJ114 added to L. pneumophila AA100 at 4 μg/ml resulted in a transformation frequency of 3 × 10−5 (Fig. 1). The transformation frequency was dose dependent (Fig. 2) and saturable, as addition of more than 4 μg of plasmid DNA per ml did not increase the transformation frequency (data not shown). However, plasmids pUC4K (the original source of the Kanr marker) and pBC-K (containing the same vector as pBJ114 and the Kanr marker), which have no L. pneumophila DNA, were unable to transfer to AA100 a Kanr phenotype (Fig. 1). Therefore, the presence of L. pneumophila DNA is required for transformation after addition of exogenous plasmid DNA.
FIG. 1.
Transformation of L. pneumophila with plasmid DNA. Transformation frequency was measured after addition, at 4 μg/ml, of plasmid DNA from pUC4K, pBC-K, pBJ114 (pilEL::Kanr), pBJ115 (pilEL::Kanr), pJA2A-K2 (ppa::Kanr), pGS-K (gspA::KanR), or pOHBOC1-K (asd::Kanr) to L. pneumophila AA100. The number of chloramphenicol-resistant transformants was measured after addition of pBOC20. Loss of auxotrophy for DAP was measured after addition of 4 μg of pOHBOC1 or pOHBOC2 plasmid DNA per ml to L. pneumophila AA400. The limit of detection for transformation frequency was 10−9. Error bars indicate standard deviations.
FIG. 2.
DNA concentration dependence of transformation with plasmid DNA. Transformation frequency was measured after addition of the indicated amount of plasmid DNA. The limit of detection for transformation frequency was 10−9. Error bars indicate standard deviations.
Transformation of L. pneumophila DNA leads to recombination with the chromosome.
The inability of plasmids pUC4K and pBC-K to transfer a Kanr phenotype to L. pneumophila is not due to an inherent inability of L. pneumophila to maintain these plasmids, since they can be electroporated into L. pneumophila and subsequently maintained. Since the replicative status of plasmid DNA after uptake by competent L. pneumophila was unknown, the ability of competent L. pneumophila to maintain plasmid DNA was determined. Our results showed that no Cmr or Kanr transformants were obtained after addition of exogenous pBC-K DNA. Interestingly, transformants obtained after addition of pBJ114 DNA to AA100 were able to grow in the presence of kanamycin but not chloramphenicol, suggesting a recombination event between the L. pneumophila DNA from pBJ114 and the chromosome. Southern analysis of chromosomal DNA obtained from 17 Kanr transformants and probed with pilEL indicated replacement of the pilEL locus with the pilEL::Kanr locus from pBJ114 (data not shown). Therefore, transformation of L. pneumophila with pBJ114 is related to the presence of L. pneumophila chromosomal DNA sequences and resulted in homologous recombination of the DNA into the chromosome.
Uptake of DNA by N. gonorrhoeae and H. influenzae requires the presence of specific sequences within the DNA (16, 23, 47), while S. pneumoniae and B. subtilis do not require the presence of recognition sequences for DNA uptake (33). To help determine if specific L. pneumophila DNA sequences were required for uptake of DNA, transformation of L. pneumophila with plasmid DNA was measured in the presence of competing DNA. Plasmid pBJ114 (4 μg/ml) was added to L. pneumophila after addition of competing DNA. Plasmids pBJ120 (containing the pilEL locus) and pBC-K (containing no L. pneumophila DNA) were able to reduce the frequency of transformation in a dose-dependent manner. Addition of pBC-K or pBJ120 reduced transformation equally well. Addition of 4 μg of DNA per ml reduced transformation frequency 5- to 10-fold, while 80 μg of plasmid DNA per ml reduced transformation 10- to 100-fold. Addition of 400 μg of chromosomal DNA from L. pneumophila AA100 or E. coli DH5α per ml completely abolished transformation. Because both plasmids and both sources of chromosomal DNA could reduce the transformation frequency, it is unlikely that L. pneumophila has a mechanism to recognize specific sequences prior to uptake of DNA. Rather, the specificity of transformation of DNA seems to be related to the frequency of recombination. Therefore, the inability to detect transformation after addition of pUC4K or pBC-K plasmid DNA is probably due to an inability of heterologous DNA to recombine into the L. pneumophila chromosome.
Transformation of L. pneumophila with gspA, ppa, and asd loci.
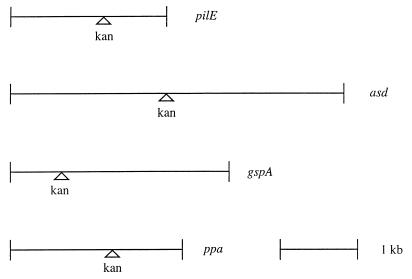
Whether the acquisition of DNA by L. pneumophila was specific only for the pilEL locus was addressed by attempting transformation of L. pneumophila after addition of exogenous plasmid DNA harboring various L. pneumophila loci: gspA, ppa, and asd. Synthesis of a global stress protein (GspA) can be induced under various in vitro stress conditions as well as during intracellular infection within macrophages (1, 2, 4). The ppa locus encodes a pyrophosphatase that is also induced during intracellular infection (2). Production of aspartate-β-semialdehyde dehydrogenase (Asd) is required for peptidoglycan biosynthesis in L. pneumophila (25). All of these loci were interrupted by a Kanr marker and introduced into L. pneumophila on a pBOC20-based vector. The relative position of the Kanr marker in each cloned DNA fragment is shown in Fig. 3. Transformation of AA100 was examined for each plasmid and compared to transformation after addition of pBJ115, a pBOC20-based plasmid containing the pilEL locus interrupted by a Kanr insertion. The pilEL, gspA, ppa, and asd loci from the plasmids were all able to be recovered by L. pneumophila (Fig. 1). In contrast, the vector pBOC20 was unable to transfer a detectable phenotype to AA100. Therefore, transformation of L. pneumophila may occur after addition of a variety of L. pneumophila loci on plasmid DNA. The frequency of transformation was variable between different experiments (up to 10-fold), but transformation ability was consistent in all experiments. The relative amount of L. pneumophila chromosomal DNA on each transforming plasmid did not correlate to the transformation frequency. The differences between the transformation frequencies of the plasmids were not significant except for comparison of pBJ114 and pBJ115 with pJA2A-K2 (unpaired t test, P < 0.05).
FIG. 3.
Position of the Kanr marker in cloned DNA fragments. The relative position of the marker is shown for pBJ114 and pBJ115 (pilE), pOHBOC1-K (asd), pGS-K (gspA), and pJA2A-K2 (ppa). A 1-kb fragment is shown for scale.
All of the loci examined contained at least 2.0 kb of chromosomal DNA which had been disrupted by the insertion of the Kanr marker. The ability of L. pneumophila to be transformed by plasmid DNA without a resistance marker disrupting the L. pneumophila-derived DNA was also assessed. Two different plasmids containing the asd locus were tested for the ability to transform L. pneumophila AA400. AA400 was generated from strain AA100 by the insertion of a Kanr marker into asd and is auxotrophic, requiring addition of DAP for growth (25). The transformation frequency after addition of plasmids containing the uninterrupted asd locus to AA400 (regenerating the wild-type phenotype) was determined. pOHBOC1 contains ∼4.3 kb of chromosomal DNA, while pOHBOC2 contains ∼1.4 kb, including 209 bp upstream and 209 bp downstream of the open reading frame of asd. Although AA400 was transformed at a frequency of 8.2 × 10−7 after addition of pOHBOC1 DNA, transformation was not detected after addition of pOHBOC2 or pBOC20 (Fig. 1). Thus, transformation of L. pneumophila occurred independently of the presence of the Kanr marker on the plasmid. However, the inability of AA400 to be transformed after addition of pOHBOC2 DNA may be due to a requirement for the presence of a minimum amount of DNA for recombination.
Dependence of competence upon culture conditions.
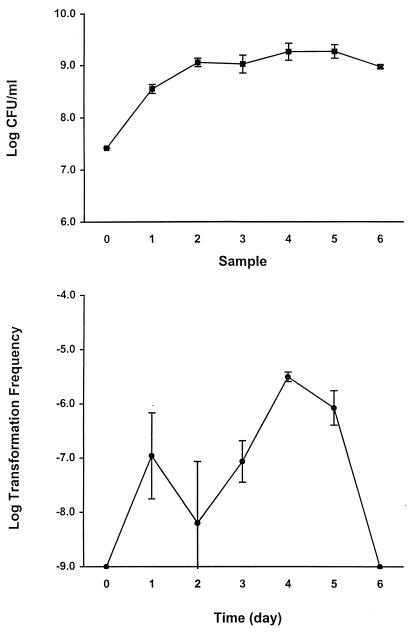
The frequency of transformation of L. pneumophila AA100 to Nalr after addition of isogenic BS200 chromosomal DNA was examined at different phases of growth. The transformation frequency of AA100 increased over time when the bacteria were grown under conditions required for pilus expression (37°C, broth culture, without aeration). Transformation was undetectable when DNA was added to the culture at day 0 (transformants were sampled after a 2-day incubation with the DNA [see Materials and Methods]) and peaked at 4 days after inoculation of cultures (Fig. 4, bottom panel). Transformation was undetectable when DNA was added to cultures grown for 6 days (Fig. 4, bottom panel). Bacterial viability did not decrease significantly during the duration of the experiment (Fig. 4, top panel).
FIG. 4.
Induction of competence in L. pneumophila. (Top) Viable counts before addition of DNA. Sample numbers correspond to samples taken at the time points indicated in the bottom panel. (Bottom) L. pneumophila BS200 chromosomal DNA (400 μg/ml) was added to cultures at the time points indicated, and the cultures were allowed to incubate for an additional 2 days before transformation frequency was determined. No transformants were detected after addition of DNA at days 0 and 6. The limit of detection for transformation frequency was 10−9. Error bars indicate standard deviations.
To determine if the transformation frequency could be altered by changes in culture conditions, transformation of L. pneumophila AA100 was examined after addition of pBJ114 DNA under various culture conditions. The transformation frequency was measured after bacterial culture at different temperatures to determine if competence was affected by temperature. Transformation was apparent after incubation of the bacteria at 37, 30, and 24°C (Table 2). No consistent difference in the transformation frequencies measured at different temperatures was observed. Also, the presence of 5% CO2 during culture at 37°C had no effect (Table 2). The ability of L. pneumophila to become competent in an aqueous environment, as might be relevant in the environment, was measured. Bacteria were grown on solid medium and resuspended in double-distilled sterile water for 4 days at 37 or 24°C, and pBJ114 DNA was added. However, no transformation was detected (Table 2). Therefore, as determined with the culture conditions tested, L. pneumophila is transformation competent when grown at 24 to 37°C in BYE broth without aeration.
TABLE 2.
Effect of culture conditions on transformation frequency
| Plasmid | Culture medium | Temp (°C) | Other culture condition | Transformation frequencya |
|---|---|---|---|---|
| pBC-K | BYE | 37 | <10−9 | |
| pBJ114 | BYE | 37 | 3.9 × 10−7 ± 1.3 × 10−7 | |
| pBC-K | BYE | 30 | <10−9 | |
| pBJ114 | BYE | 30 | 9.5 × 10−6 ± 2.0 × 10−6 | |
| pBC-K | BYE | 24 | <10−9 | |
| pBJ114 | BYE | 24 | 1.6 × 10−7 ± 1.4 × 10−8 | |
| pBC-K | BYE | 37 | Aeration | <10−9 |
| pBJ114 | BYE | 37 | Aeration | <10−9 |
| pBC-K | BYE | 37 | CO2 | <10−9 |
| pBC-K | BCYE | 37 | CO2 | <10−9 |
| pBJ114 | BCYE | 37 | <10−9 | |
| pBC-K | Water | 37 | <10−9 | |
| pBJ114 | Water | 37 | <10−9 | |
| pBC-K | Water | 24 | <10−9 | |
| pBJ114 | Water | 24 | <10−9 |
Transformation frequency was measured after addition of 40 μg of plasmid DNA per ml.
Because recognition sequence-dependent transformation in N. gonorrhoeae requires factors involved in type IV pilus biogenesis (18), the correlation between type IV pilus expression and competence in L. pneumophila was examined. The ability of L. pneumophila to be transformed after addition of chromosomal DNA was originally determined for bacteria grown under conditions which allow expression of type IV pili (50). Therefore, transformation of L. pneumophila was measured after growth of bacteria under conditions where no type IV pili are expressed. Expression of type IV pili by L. pneumophila was not evident as determined by electron microscopic examination after culture of the bacteria in BYE broth with aeration at 37°C or on BCYE agar at 37°C (data not shown). No transformation of L. pneumophila was detected after addition of pBJ114 when the cultures were aerated or when L. pneumophila was grown on solid medium (Table 2). Therefore, the transformation-competent phenotype and expression of type IV pili are induced under similar culture conditions.
A pilEL mutant is not competent for DNA transformation.
The gene products required for type IV pilus expression in N. gonorrhoeae are essential for uptake sequence-dependent competence resulting in DNA transformation (18). Since type IV pilus expression in L. pneumophila and natural transformation competence are induced under similar growth conditions, the requirement for type IV pilus expression in L. pneumophila for transformation competence was examined. In contrast to wild-type strain AA100, the pilEL mutant BS100 is unable to express type IV pili (50). Therefore, the requirement of L. pneumophila transformation for the pilEL locus was determined with BS100 and compared to transformation of AA100.
In contrast to AA100, the pilEL mutant BS100 was unable to be transformed after addition of chromosomal DNA from strain BS200 (transformation frequency of >10−9). To confirm that type IV pili were required for competence, the ability of the wild-type pilEL locus to restore competence to BS100 was determined. Cosmid pCC3 contains the pilEL locus and restores expression of type IV pili to strain BS100 (50). The cosmid also restored transformation frequency of BS100 to wild-type levels (1.9 × 10−6 ± 7.1 × 10−8). Since pCC3 contains a large fragment of chromosomal DNA from L. pneumophila, restoration of competence to BS100 was determined with pBJ120, a plasmid that contains only one complete open reading frame, pilEL. Introduction of pBJ120 into BS100 minimally restored transformation frequency (7.6 × 10−8 ± 1.0 × 10−7). Neither the pTLP6 nor the pBC vector was able to restore competence to BS100 (transformation frequency of <10−9). However, BS100 harboring pBJ120 does not express type IV pili (50). We cannot exclude the possibility that low levels of PilEL on the surface of L. pneumophila may result in both a minimal transformation frequency and reduced numbers of type IV pili, which would be difficult to detect by electron microscopic examination, particularly since only 10% of wild-type bacteria express the type IV pili (50). Nevertheless, the type IV pilin gene is sufficient to restore a minimal level of competence for DNA transformation.
Prevalence of natural competence for transformation in legionellae.
The prevalence of the pilEL locus in L. pneumophila serogroups and in various Legionella species has been recently determined by Southern analysis (50). The locus was present in all 13 L. pneumophila serogroups (analyzed under high-stringency conditions) and showed variable hybridization for 16 Legionella species examined (analyzed under low-stringency conditions). The natural transformation competence of representatives of the L. pneumophila serogroups and Legionella species was determined after addition of plasmid DNA (pOHBOC1-K) containing a Kanr marker inserted within asd obtained from L. pneumophila AA100. The asd locus was chosen because the gene was likely to be present in all species, since none of the Legionella species tested were auxotrophic prior to transformation. This was further confirmed by DNA hybridization with asd as a probe. In situ hybridizations of all tested serogroups and strains showed hybridization to all 13 serogroups (data not shown). Four of six Legionella species tested hybridized to asd as well (L. wadsworthii and L. santicrucis did not hybridize).
As expected, transformation was detected for L. pneumophila AA100 (serogroup 1). However, even though all serogroups hybridized with a probe containing the pilEL locus in high-stringency Southern hybridizations, transformation was detected only for L. pneumophila serogroup 5, at a frequency of 1.4 × 10−6 ± 4.2 × 10−7. All other Legionella species tested were not transformable whether they hybridized to the type IV pilin gene or not (transformation frequency, <10−9). The inability of these Legionella species to be transformed after addition of the asd-containing plasmid suggests that the pilEL locus is not functional. However, the chromosomal DNA from L. pneumophila AA100 may be a barrier to homologous recombination in different serogroups or species if the asd loci are not similar enough to allow homologous recombination. Alternatively, DNA uptake or recombination by other serogroups or species may be hindered by alternative restriction systems not encountered in L. pneumophila AA100.
DISCUSSION
This is the first description of natural competence for transformation in L. pneumophila. The ability of L. pneumophila to become competent for DNA transformation is dependent on growth conditions and is induced under conditions similar to those for expression of type IV pili on the bacterial surface. Disruption of the pilEL locus of L. pneumophila results in loss of type IV pili and also abolishes induction of competence. Restoration of both expression of pili and transformation competence is accomplished by reintroduction of the wild-type pilEL locus on a cosmid. Since type IV pili of L. pneumophila are also involved in adherence to protozoan and mammalian cells, the pili have been designated CAP, for competence- and adherence-associated pili.
PilEL is similar to type IV pilin proteins found in N. gonorrhoeae, P. aeruginosa, and Moraxella bovis, among others (50). However, only in N. gonorrhoeae has a connection between the type IV pilus structure and natural competence for DNA transformation been observed (18). While B. subtilis and H. influenzae both maintain DNA uptake mechanisms with components that resemble proteins found in association with type IV pilin systems (13, 30, 53), neither has been shown to produce any type IV pilus structure. In addition, many pathogenic species, such as P. aeruginosa, produce type IV pili (51) but have not found to be naturally competent. Therefore, L. pneumophila is the second example of a bacterium which requires a type IV pilin locus for both pilus expression and natural competence for DNA transformation.
In N. gonorrhoeae, all of the gene products required for type IV pilus biogenesis are also required for DNA uptake (18). These include PilE, the major pilus structural component (35), and PilC, an adhesin (43) which has been localized to the tip of the pilus (44) and also to the outer membrane (18). Interestingly, transformation requires the presence of PilE and PilC but not necessarily the assembled pilus structure (18). A similar phenomenon may be present in L. pneumophila. Reintroduction of only the pilEL open reading frame into the pilEL mutant consistently restored competence, but at a minimal level compared to complementation with a cosmid harboring the locus. The pilEL gene alone did not restore expression of type IV pili, suggesting perhaps reduced levels of PilEL at the bacterial surface. Incomplete complementation may be due to inefficient transcription of pilEL from the plasmid or may represent the requirement for additional loci that were disrupted by a polar effect in the pilEL mutant. Regardless, the pilEL gene alone is sufficient for restoration of minimal levels of transformation, and the pilEL locus is required for both type IV pilus expression and transformation competence.
Acquisition of DNA fragments by bacteria can be dependent upon particular sequences found within the fragment or can be independent of these sequences. In N. gonorrhoeae, for example, type IV pilus-dependent uptake is species specific and requires the presence of a 10-bp sequence frequently found as part of transcription terminators within gonococcal chromosomal DNA (16, 23). However, transformation can also occur independently of such sequences in the absence of pili (12). The exact function of such sequences during DNA uptake is unknown. However, other mechanisms for DNA uptake appear to be independent of particular sequences, as is the case in S. pneumoniae (33), B. subtilis (33), and A. calcoaceticus (32, 38). Since L. pneumophila-derived DNA on a plasmid, vector DNA, L. pneumophila chromosomal DNA, and E. coli chromosomal DNA were capable of reducing the transformation frequency of a plasmid containing L. pneumophila DNA, the presence of an uptake mechanism specific for L. pneumophila sequences is unlikely. In this respect, L. pneumophila would be different from N. gonorrhoeae in the requirement for recognition sequence-specific DNA uptake during type IV pilin-dependent transformation.
However, it is unknown if uptake of vector DNA without the presence of L. pneumophila DNA occurs. We have been unable to differentiate between a lack of DNA uptake and DNA degradation after uptake because of the low transformation frequency and high levels of background during attempted radiolabelled-DNA uptake experiments (data not shown). Our failure to detect transformation of vector DNA may therefore be due either to failure of L. pneumophila to internalize the DNA or to degradation of DNA after uptake. Since vector DNA and E. coli chromosomal DNA block transformation, it is most likely that vector DNA is taken up and degraded.
In other bacterial transformation systems, transforming DNA is taken up as single-stranded DNA fragments and thus requires recombination onto the bacterial chromosome or recircularization of DNA into a plasmid (33). In L. pneumophila, there is a barrier to transformation for plasmid DNA that does not contain L. pneumophila chromosomal DNA. This barrier is not due to a general failure to maintain plasmid DNA, since electroporated plasmids are maintained in L. pneumophila. However, Southern hybridization analysis confirmed that transformation of a plasmid containing L. pneumophila DNA is a result of homologous recombination (of L. pneumophila DNA but not the vector) into the L. pneumophila chromosome. It is probable that plasmid DNA taken up by the bacteria is unable to recircularize and replicate autonomously and therefore that homologous recombination must occur for transformation to be detected.
Transformation of L. pneumophila after addition of plasmid DNA was dependent on culture conditions which correlated with growth conditions used during previous characterization of CAP pili (50). Temperature-regulated pilus expression has previously been described for L. pneumophila (31). Pili were found to be differentially expressed at 30°C but not at 37°C when grown on solid media. Expression correlated with transcription of pilBCD, whose products are homologues of P. aeruginosa PilB, PilC, and PilD (31), which are required for type II protein secretion and pilus biogenesis (52). As reported here, growth of L. pneumophila at 30°C in broth did not result in a consistent difference in transformation frequency when compared to growth at 24 or 37°C. However, as there was variability between experiments (up to 10-fold), and because type IV pilus expression is low (10% of that for wild-type bacteria), it is not possible to evaluate slight differences in transformation frequency that may have occurred due to temperature regulation of pilus expression.
It is possible that the variability seen during transformation experiments could be caused by nuclease activity present in the culture medium during introduction of DNA into L. pneumophila or by different levels of shearing of DNA in individual DNA preparations. Two different chromosomal DNA isolation procedures were used to prepare DNA. However, variability between experiments was seen regardless of the isolation procedure used. Because of the observed variability, the inconsistent differences between transformation efficiencies were difficult to interpret. However, despite the variability in frequency seen, the ability of L. pneumophila to become competent for transformation after addition of exogenous DNA and the dependence of transformation upon the pilEL locus were very consistent for all experiments.
The discovery that L. pneumophila is naturally competent for DNA transformation will be beneficial in genetic manipulations of the bacteria. However, the consequences of the ability of L. pneumophila to acquire genetic elements by natural competence for DNA transformation are unknown. The ability to incorporate DNA into the bacterial chromosome from the extracellular environment is an important mechanism for horizontal exchange of genetic information. Considering the ubiquitous presence of L. pneumophila in the environment, natural competence for DNA transformation may allow incorporation of genes into the bacterial chromosome which could increase environmental fitness or increase the ecological niches available to the microorganism.
ACKNOWLEDGMENTS
We thank Omar Harb and Lian-Young Gao for critical analysis of the manuscript.
B. J. Stone was supported by National Research Service Award CA09509. Y. Abu Kwaik is supported by NIH grant AI38410.
REFERENCES
- 1.Abu Kwaik Y. Fatal attraction of mammalian cells to Legionella pneumophila. Mol Microbiol. 1998;30:689–695. doi: 10.1046/j.1365-2958.1998.01092.x. [DOI] [PubMed] [Google Scholar]
- 2.Abu Kwaik Y. Induced expression of the Legionella pneumophila gene encoding a 20-kilodalton protein during infection. Infect Immun. 1998;66:203–212. doi: 10.1128/iai.66.1.203-212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik Y, Eisenstein B I, Engleberg N C. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect Immun. 1993;61:1320–1329. doi: 10.1128/iai.61.4.1320-1329.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu Kwaik Y, Engleberg N C. Cloning and molecular characterization of a Legionella pneumophila gene induced by intracellular infection and by various in vitro stress stimuli. Mol Microbiol. 1994;13:243–251. doi: 10.1111/j.1365-2958.1994.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 5.Abu Kwaik Y, Gao L-Y, Stone B J, Venkataraman C, Harb O S. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol. 1998;64:3127–3133. doi: 10.1128/aem.64.9.3127-3133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abu Kwaik Y, Pederson L L. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 7.Arroyo J, Hurley M C, Wolf M, McClain M S, Eisenstein B I, Engleberg N C. Shuttle mutagenesis of Legionella pneumophila: identification of a gene associated with host cell cytopathogenicity. Infect Immun. 1994;62:4075–4080. doi: 10.1128/iai.62.9.4075-4080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avery O T, McLeod C M, McCarthy M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnhart B J, Herriott R M. Penetration of deoxyribonucleic acid into Hemophilus influenzae. Biochim Biophys Acta. 1963;76:25–39. [PubMed] [Google Scholar]
- 10.Bellinger-Kawahara C, Horwitz M A. Complement component C3 fixes selectively to the major outer membrane protein (MOMP) of Legionella pneumophila and mediates phagocytosis of liposome-MOMP complexes by human monocytes. J Exp Med. 1990;172:1201–1210. doi: 10.1084/jem.172.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswas G D, Sox T, Blackman E, Sparling P F. Factors affecting genetic transformation of Neisseria gonorrhoeae. J Bacteriol. 1977;129:983–992. doi: 10.1128/jb.129.2.983-992.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyle-Vavra S, Seifert H S. Uptake-sequence-independent DNA transformation exists in Neisseria gonorrhoeae. Microbiology. 1996;142:2839–2845. doi: 10.1099/13500872-142-10-2839. [DOI] [PubMed] [Google Scholar]
- 13.Breitling R, Dubnau D. A pilin-like membrane protein is essential for DNA binding by competent Bacillus subtilis. J Bacteriol. 1990;172:1499–1508. doi: 10.1128/jb.172.3.1499-1508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catlin B W, Cunningham L S. Transforming activities and base contents of deoxyribonucleate preparations from various Neisseriae. J Gen Microbiol. 1961;26:303–312. doi: 10.1099/00221287-26-2-303. [DOI] [PubMed] [Google Scholar]
- 15.Donnenberg M S, Giron J A, Nataro J P, Kaper J B. A plasmid encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 16.Elkins C, Thomas C E, Seifert H S, Sparling P F. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J Bacteriol. 1991;173:3911–3913. doi: 10.1128/jb.173.12.3911-3913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faast R, Ogierman M A, Stroeher U H, Manning P A. Nucleotide sequence of the structural gene, tcpA, for a major pilin subunit of Vibrio cholerae. Gene. 1989;85:227–231. doi: 10.1016/0378-1119(89)90486-1. [DOI] [PubMed] [Google Scholar]
- 18.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer T F. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene. 1997;192:125–134. doi: 10.1016/s0378-1119(97)00038-3. [DOI] [PubMed] [Google Scholar]
- 19.Gao L, Harb O S, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao L-Y, Harb O S, Abu Kwaik Y. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect Immun. 1998;66:883–892. doi: 10.1128/iai.66.3.883-892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, L.-Y., B. J. Stone, J. K. Brieland, and Y. Abu Kwaik. Different fates of Legionella pneumophila pmi and mil mutants within macrophages and epithelial cells. Microb. Pathog., in press. [DOI] [PubMed]
- 22.Gibson F C, III, Tzianabos A O, Rodgers F C. Adherence of Legionella pneumophila to U-937 cells, guinea-pig alveolar macrophages, and MRC-5 cells by a novel, complement-independent binding mechanism. Can J Microbiol. 1994;40:865–872. doi: 10.1139/m94-137. [DOI] [PubMed] [Google Scholar]
- 23.Goodman S D, Scocca J J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 25.Harb O S, Abu Kwaik Y. Identification of the aspartate-β-semialdehyde dehydrogenase gene of Legionella pneumophila and characterization of a null mutant. Infect Immun. 1998;66:1898–1903. doi: 10.1128/iai.66.5.1898-1903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harb O S, Venkataraman C, Haack B J, Gao L-Y, Abu Kwaik Y. Heterogeniety in the attachment and uptake mechanisms of the Legionnaires’ disease bacterium, Legionella pneumophila, by protozoan hosts. Appl Environ Microbiol. 1998;64:126–132. doi: 10.1128/aem.64.1.126-132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 28.Johnson K, Parker M L, Lory S. Nucleotide sequence and transcriptional initiation site of two Pseudomonas aeruginosa pilin genes. J Biol Chem. 1986;261:15703–15708. [PubMed] [Google Scholar]
- 29.Juni E, Janik A. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larson T G, Goodgal S H. Sequence and transcriptional regulation of com101A, a locus required for genetic transformation in Haemophilus influenzae. J Bacteriol. 1991;173:4683–4691. doi: 10.1128/jb.173.15.4683-4691.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liles M R, Viswanathan V K, Cianciotto N P. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilin biosynthesis and type II secretion. Infect Immun. 1998;66:1776–1782. doi: 10.1128/iai.66.4.1776-1782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenz M G, Reipschlager K, Wackernagel W. Plasmid transformation of naturally competent Acinetobacter calcoaceticus in non-sterile soil extract and groundwater. Arch Microbiol. 1992;157:355–360. doi: 10.1007/BF00248681. [DOI] [PubMed] [Google Scholar]
- 33.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClain M S, Hurley M C, Brieland J K, Engleberg N C. The Legionella pneumophila hel locus encodes intracellularly induced homologs of heavy-metal ion transporters of Alcaligenes spp. Infect Immun. 1996;64:1532–1540. doi: 10.1128/iai.64.5.1532-1540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer T F, Billyard E, Haas R, Storzbach S, So M. Pilus genes of Neisseria gonorrhoeae: chromosomal organization and DNA sequence. Proc Natl Acad Sci USA. 1984;81:6110–6114. doi: 10.1073/pnas.81.19.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mody C H, Paine III R, Shahrabadi M S, Simon R H, Pearlman E, Eisenstein B I, Toews G B. Legionella pneumophila replicates within rat alveolar epithelial cells. J Infect Dis. 1993;167:1138–1145. doi: 10.1093/infdis/167.5.1138. [DOI] [PubMed] [Google Scholar]
- 37.Nash T W, Libby D M, Horwitz M A. Interaction between the Legionnaires’ disease bacterium (Legionella pneumophila) and human alveolar macrophages. J Clin Invest. 1984;74:771–782. doi: 10.1172/JCI111493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmen R, Vosman B, Buijsman P, Breek C K D, Hellingwerf K J. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J Gen Microbiol. 1993;139:295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 39.Potts W J, Saunders R J. Nucleotide sequence of the structural gene for class I pilin from Neisseria meningitidis: homologies with the pilE locus of Neisseria gonorrhoeae. Mol Microbiol. 1988;2:647–653. doi: 10.1111/j.1365-2958.1988.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 40.Rodgers F G, Gibson I F C. Opsonin-independent adherence and intracellular development of Legionella pneumophila within U-937 cells. Can J Microbiol. 1993;39:718–722. doi: 10.1139/m93-103. [DOI] [PubMed] [Google Scholar]
- 41.Rodgers F G, Greaves P W, Macrae A D. Flagella and fimbriae on legionella organisms. Lancet. 1979;ii:753–4. doi: 10.1016/s0140-6736(79)90691-3. [DOI] [PubMed] [Google Scholar]
- 42.Rodgers F G, Greaves P W, Macrae A D, Lewis M J. Electron microscopic evidence of flagella and pili on Legionella pneumophila. J Clin Pathol. 1980;33:1184–1188. doi: 10.1136/jcp.33.12.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudel T, v. Putten J P M, Gibbs C P, Haas R, Meyer T F. Interaction of two variable proteins (PilE and PilC) required for pilus-mediated adherence of Neisseria gonorrhoeae to human epithelial cells. Mol Microbiol. 1992;22:3439–3450. doi: 10.1111/j.1365-2958.1992.tb02211.x. [DOI] [PubMed] [Google Scholar]
- 44.Rudel T, Scheuerpflug I, Meyer T F. Neisseria PilC protein identified as a type-4 pilus tip-located adhesin. Nature. 1995;373:357–359. doi: 10.1038/373357a0. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Shaw C E, Taylor R K. Vibrio cholerae 0395 tcpA pilin gene sequence and comparison of predicted protein structural features to those of type 4 pilins. Infect Immun. 1990;58:3042–3049. doi: 10.1128/iai.58.9.3042-3049.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sisco K L, Smith H O. Sequence-specific DNA uptake in Haemophilus transformation. Proc Natl Acad Sci USA. 1979;76:972–976. doi: 10.1073/pnas.76.2.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith O H, Danner D B, Deich R A. Genetic transformation. Annu Rev Biochem. 1981;50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- 49.Sparling P F. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966;92:1364–1370. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stone B J, Abu Kwaik Y A. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strom M S, Lory S. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J Bacteriol. 1986;165:367–372. doi: 10.1128/jb.165.2.367-372.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 53.Tomb J F, El-Hajj H, Smith H O. Nucleotide sequence of a cluster of genes involved in transformation of Haemophilus influenzae Rd. Gene. 1991;104:1–10. doi: 10.1016/0378-1119(91)90457-m. [DOI] [PubMed] [Google Scholar]
- 54.Young F E, Spizizen J. Incorporation of deoxyribonucleic acid in the Bacillus subtilis transformation system. J Bacteriol. 1963;86:392–400. doi: 10.1128/jb.86.3.392-400.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]