Abstract
Endometriosis is a chronic, debilitating, gynecologic condition with a non-specific clinical presentation. Globally, patients can experience diagnostic delays of ~6 to 12 years, which significantly hinders adequate management and places a significant financial burden on patients and the healthcare system. Through artificial intelligence (AI), it is possible to create models that can extract data patterns to act as inputs for developing interventions with predictive and diagnostic accuracies that are superior to conventional methods and current tools used in standards of care. This literature review explored the use of AI methods to address different clinical problems in endometriosis. Approximately 1309 unique records were found across four databases; among those, 36 studies met the inclusion criteria. Studies were eligible if they involved an AI approach or model to explore endometriosis pathology, diagnostics, prediction, or management and if they reported evaluation metrics (sensitivity and specificity) after validating their models. Only articles accessible in English were included in this review. Logistic regression was the most popular machine learning method, followed by decision tree algorithms, random forest, and support vector machines. Approximately 44.4% (n = 16) of the studies analyzed the predictive capabilities of AI approaches in patients with endometriosis, while 47.2% (n = 17) explored diagnostic capabilities, and 8.33% (n = 3) used AI to improve disease understanding. Models were built using different data types, including biomarkers, clinical variables, metabolite spectra, genetic variables, imaging data, mixed methods, and lesion characteristics. Regardless of the AI-based endometriosis application (either diagnostic or predictive), pooled sensitivities ranged from 81.7 to 96.7%, and pooled specificities ranged between 70.7 and 91.6%. Overall, AI models displayed good diagnostic and predictive capacity in detecting endometriosis using simple classification scenarios (i.e., differentiating between cases and controls), showing promising directions for AI in assessing endometriosis in the near future. This timely review highlighted an emerging area of interest in endometriosis and AI. It also provided recommendations for future research in this field to improve the reproducibility of results and comparability between models, and further test the capacity of these models to enhance diagnosis, prediction, and management in endometriosis patients.
Subject terms: Diagnostic markers, Predictive markers, Reproductive disorders
Introduction
Endometriosis is a chronic, gynecologic condition1 estimated to affect 190 million women worldwide2. This benign, but often debilitating condition is thought to impact ~10% of women based on extrapolations of pelvic pain and subfertility in the general population3 and of those that are symptomatic, the prevalence is thought to be 30% to 50%4. True prevalence rates are difficult to estimate because this condition is often underreported, undiagnosed or misdiagnosed1. In Canada, the national societal burden of endometriosis is estimated at CAD $1.8 billion annually based on treatment costs, caregiver costs, quality of life and work absenteeism5. Endometriosis poses a large economic and disease burden on society and the precise scope of the problem remains unknown.
Endometriosis is characterized by extrauterine growth of endometrial-like tissue in areas of the pelvis (i.e., ovaries), bowels, bladder, and peritoneum6. These growths are rarely found in the thoracic region, and other organ systems7,8. Endometriosis has three predominant phenotypes: superficial endometriosis, endometriomas and deep endometriosis (DE)8,9. There are many staging systems for endometriosis, including the American Society for Reproductive Medicine classification system: stage I (minimal), stage II (mild), stage III (moderate), and stage IV (severe)10,11. However, given the complexity of this disease, it is difficult to universally stage and characterize under the present systems. Significant research has been done in recent years in attempts to elucidate the pathogenesis of this disease and many etiological factors are currently being explored including immune-mediated, inflammatory, genetic and environmental components12,13.
The signs and symptoms of this disease are non-specific and can vary in severity, creating clinical heterogeneity, which adds to the diagnostic difficulty associated with this disease8. Patients can present with a range of symptomatology depending on the type of endometriosis, location of implants, stage, and severity including but not limited to dysmenorrhea, dyspareunia, abdominal pain, chronic pelvic pain, menorrhagia, bowel symptoms, urinary symptoms, and subfertility or infertility8. Due to the combination of non-specific symptoms, a long differential list, lack of provider awareness, unnecessary investigations, and a lack of non-invasive diagnostic tools, many patients experience significant delays in receiving an endometriosis diagnosis1,14–16. The current literature has documented diagnostic delays of up to 6 to 12 years globally before patients receive a definitive diagnosis and adequate management1,17,18. Currently, the gold standard diagnostic procedure for endometriosis remains laparoscopic visualization of lesions followed by histologic confirmation of ectopic endometriotic implants8, a costly and invasive process that requires a skilled clinician. Transvaginal ultrasonography is a commonly used clinical technique in endometriosis screening and diagnosis, given its non-invasive nature and widespread accessibility8.
In the past 5 years, the emergence of artificial intelligence (AI) has spread rapidly into healthcare; it has demonstrated marked potential in disease diagnostics, treatments, and a higher-level analysis of large biomedical datasets19,20. With the increase in digitization in healthcare, AI presents novel opportunities to decrease the amount of time required for diagnosis and to streamline care in many settings19. Machine learning (ML) is a subset of AI and includes common methods such as logistic regression with the use of training and test sets and support vector machines (SVMs)19. Currently, AI has been used to analyze multi-omics, clinical, behavioral/wellness, environmental and research and developmental data19, and it has been applied to decision-making, patient self-management, triage, understanding disease mechanisms, and drug discovery21,22. However, AI methods require an expert’s oversight to help inform the model’s development since clinical problems are often complex and multifaceted19. Additionally, the privacy and the security of patient data remain a consideration when introducing new technology into healthcare; thus researchers should be aware of any risks associated with AI models19.
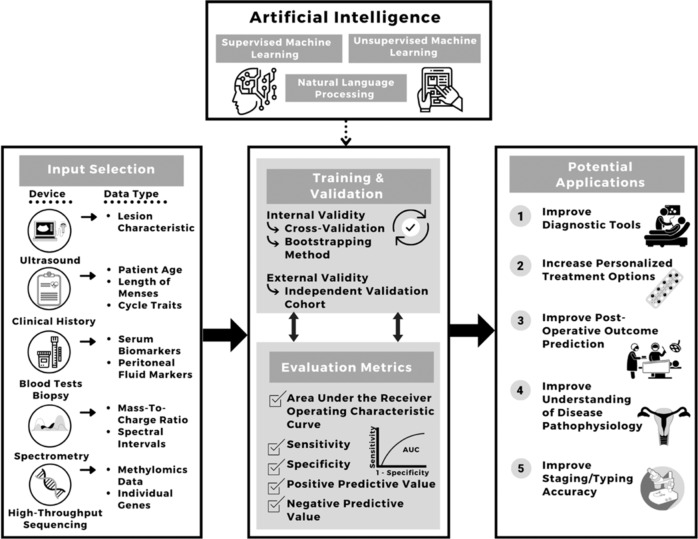
From fetal heart monitoring to reproductive medicine, AI technologies have been used in the field of obstetrics and gynecology and have demonstrated the potential to significantly aid in prediction of outcomes22–25. Given the diversity of its use in the clinical context, there is great potential to apply AI to the complex challenges presented by endometriosis and improve non-invasive diagnostics to reduce the delays and human error associated with diagnosis22. However, clinicians face significant challenges in the field of AI applications including a widespread lack of understanding about different AI methods and the competencies and limitations of AI technologies21. This review examines the different ways AI methods have been applied to solve pressing issues in endometriosis diagnostics, prediction, and research as shown in Fig. 1. By providing a thorough understanding of the different models and their application to clinical problems, and by analyzing their strengths and limitations, recommendations will be provided to help future researchers adequately develop AI models to advance the field of endometriosis.
Fig. 1. Potential area of use for artificial intelligence applications in endometriosis.
This figure was created by B.S. and M.E.
Results
Study selection
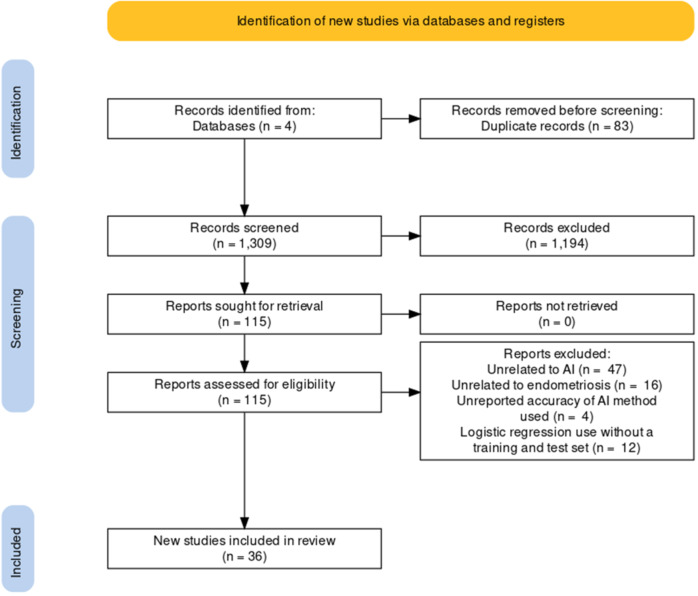
A total of 1309 titles were identified by searching the PubMed, Medline-OVID, EMBASE, and CINAHL database, and 115 full-texts were eligible for screening after studies were excluded during the title and abstract-screening stages. Of these, 79 papers were excluded in the final review based on our exclusion criteria and 36 studies were included in the final review (Fig. 2). A summary of the eligible studies and extracted study characteristics is shown in Table 1. The majority of studies were predominantly retrospective designs (n = 20) using data from large clinical databases and registries and some prospective designs (n = 16); no randomized studies were included. Samples sizes ranged from modest numbers of 26 patients with endometriosis26 to 1396 symptomatic patients27, with the average sample size being 245 individuals for studies exploring diagnosis and prediction in endometriosis.
Fig. 2. Workflow of the study.
Flowchart of study identification, inclusion, and exclusion criteria.
Table 1.
Description of the studies.
| Year | Author [ref.] | Study design | Intervention | Purpose | Objective | Sample size | AI accuracy for best model |
|---|---|---|---|---|---|---|---|
| 2022 | Bendifallah et al.50 | Retrospective | Logistic Regression, Random Forest, Decision Tree, eXtreme Gradient Boosting, Voting Classifier (soft/hard) | Prediction | Predict likelihood of endometriosis based on 16 essential clinical and symptom-based features related to patient history, demographics, endometriosis phenotype and treatment | 1126 endometriosis patients, 608 controls |
SE = 93% SP = 92% |
| 2022 | Bendifallah et al.35 | Prospective | Logistic Regression, Random Forest eXtreme Gradient Boosting, AdaBoost | Diagnosis | Diagnosis of endometriosis using a blood-based mRNA diagnostic signature | 200 plasma samples (153 cases, 47 controls) |
SE = 96.8% SP = 100% |
| 2021 | Maicus et al.61 | Prospective | Resnet (2 + 1)D | Diagnosis | Classification of the state of the Pouch of Douglas using the sliding sign test on ultrasound | 749 transvaginal ultrasound videos (414 training set, 139 validation set, 196 test set) |
SE = 88.6% SP = 90% |
| 2021 | Guerriero et al.59 | Retrospective | K-Nearest Neighbor, Naïve Bayes, Neural Networks, SVM, Decision Tree, Random Forest, Logistic Regression | Prediction | Detection of endometriotic bowel involvement in rectosigmoid deep endometriosis | 333 patients |
SE = 72% SP = 73% |
| 2021 | Li et al.52 | Retrospective | Deep Machine Learning Algorithm (NNET) | Diagnosis | Diagnosis of endometriosis based on genes | 213 patients |
SE = 100% SP = 61.1% |
| 2020 | Matta et al.30 | Retrospective Case–Control | Logistic Regression, ANN, SVM, Adaptive Boosting, PLSDA | Research | Identify biomarkers of internal exposure in adipose tissue most associated with endometriosis | 99 women (44 controls, 55 cases) |
SE = NR SP = NR |
| 2020 | Akter et al.53 | Retrospective | New Ensemble Machine Learning Classifier (GenomeForest) | Diagnosis | Classifying endometriosis versus control patients using RNAse and enrichment-based DNA-methylation datasets | 38 single-end RNA-sequence samples, 80 MBD-sequence DNA-methylation samples |
Transcriptomics Data SE = 93.8% SP = 100% Methylomics Data SE = 92.9% SP = 88.6% |
| 2020 | Perrotta et al.54 | Prospective Observational Cross-Sectional Pilot | Random Forest-Based Machine Learning Classification Analysis | Diagnosis | Diagnosis of endometriosis using gut and/or vaginal microbiome profiles | 59 women (24 controls, 35 endometriosis patients) |
SE = NR SP = NR |
| 2020 | Guo et al.58 | Retrospective Cohort | Logistic Regression | Prediction | Predict any-stage and stage 3/4 endometriosis before surgery in infertile women | 1016 patients (443 without endometriosis, 377 patients with stage 1/2 endometriosis, 196 patients with stage 3/4 endometriosis) |
SE = NR SP = NR |
| 2021 | Vesale et al.45 | Retrospective | Logistic Regression | Prediction | Predict likelihood of voiding dysfunction after surgery for deep endometriosis | 789 patients |
SE = NR SP = NR |
| 2019 | Benoit et al.46 | Retrospective | Logistic Regression | Prediction | Predict likelihood of a live birth after surgery followed by ART for patients with endometriosis-related infertility | 297 women |
SE = NR SP = NR |
| 2019 | Lee et al.29 | Retrospective | Recommendation System | Research | Identify diseases associated with endometriosis | 1,730,562 controls, 11,273 cases |
SE = NR SP = NR |
| 2019 | Braga et al.36 | Prospective Case–Control | PLSDA | Diagnosis | Develop an adjuvant tool for diagnosis of grades 3 and 4 endometriosis in infertile patients | 50 endometriosis serum samples, 50 control samples |
SE = NR SP = NR |
| 2019 | Chattot et al.57 | Prospective Observational | Logistic Regression | Prediction | Predict rectosigmoid involvement in endometriosis using preoperative score | 119 women undergoing surgery for endometriosis |
SE = NR SP = NR |
| 2019 | Knific et al.31 | Retrospective | Decision Tree, Linear Model, K-Nearest Neighbor, Random Forest | Diagnosis | Diagnosis of endometriosis based on plasma levels of proteins and patients’ clinical data | 210 patients |
SE = 40% SP = 65% |
| 2019 | Parlatan et al.37 | Retrospective | K-Nearest Neighbor, SVM, PCA | Diagnosis | Diagnosis of endometriosis using non-invasive Raman spectroscopy-based classification model | 94 serum samples (49 endometriosis, 45 controls) |
SE = 89.7% SP = 80.5% |
| 2019 | Akter et al.55 | Retrospective | Decision Tree, PLSDA, SVM, Random Forest | Diagnosis | Classify endometriosis versus control biopsy samples using transcriptomics or methylomics data | 38 samples in transcriptomics dataset, 77 samples in methylomics dataset |
Transcriptomics Data SE = 81.3% SP = 95.5% Methylomics Data SE = 76.2% SP = 80% |
| 2018 | Bouaziz et al.28 | Retrospective | NLP | Research | Using NLP to extract data by text mining of the endometriosis-related genes in the PubMed database | 724 genes retrieved |
SE = NR SP = NR |
| 2017 | Dominguez et al.33 | Prospective Case–Control | SVM | Diagnosis | Diagnosis of endometriosis using lipidomic profiling of endometrial fluid in patients with ovarian endometriosis | 12 endometriosis, 23 controls |
SE = 58.3% SP = 100% |
| 2016 | Ghazi et al.38 | Prospective Cohort | PLSDA, Multi-Layer Feed Forward ANN, QDA | Prediction | Determine classifier metabolites for early prediction risk of disease | 31 infertile women with endometriosis, 15 controls |
SE = NR SP = NR |
| 2015 | Reid et al.60 | Prospective Observational | Logistic Regression | Prediction | Use mathematical ultrasound models to determine whether a combination of transvaginal sonography markers could improve prediction of Pouch of Douglas obliteration | 189 women with suspected endometriosis |
Model 1 SE = 88% SP = 97% Model 2 SE = 88% SP = 99% |
| 2014 | Lafay Pillet et al.47 | Prospective | Logistic Regression | Diagnosis | Diagnose DE before surgery for patients operated on for endometriomas | 164 patients with DIE, 162 with no DIE |
SE = 51% SP = 94% |
| 2014 | Tamaresis et al.56 | Retrospective | Margin Tree Classification | Diagnosis | Detect and stage pelvic endometriosis using genomic data from endometrium | 148 endometrial samples |
SE = NR SP = NR |
| 2014 | Wang et al.39 | Prospective Case–Control | Genetic Algorithm, Decision Tree Algorithm, Quick Classifier Algorithm | Diagnosis | Diagnosis of endometriosis and stage using peptide profiling | 122 patients |
SE = 90.9% SP = 92.9% |
| 2013 | Wang et al.51 | Retrospective | Decision Tree | Prediction | Predict medical care decision rules for patients with recurrent pelvic cyst after surgical interventions | 178 case records |
SE = NR SP = NR |
| 2012 | Ballester et al.48 | Prospective Longitudinal Study | Logistic Regression | Prediction | Prediction of clinical pregnancy rate in patients with endometriosis | 142 infertile patients with DIE |
SE = 66.7% SP = 95.7% |
| 2012 | Fassbender et al.40 | Retrospective | LSSVM | Diagnosis | Diagnosis of endometriosis undetectable by ultrasonography | 254 plasma samples (89 controls, 165 endometriosis patients) |
SE = 88% SP = 84% |
| 2012 | Fassbender et al.41 | Retrospective | LSSVM | Diagnosis | Diagnosis of endometriosis through mRNA expression profiles in luteal phase endometrium biopsies | 49 endometrial biopsies |
SE = 91% SP = 80% |
| 2012 | Vodolazkaia et al.34 | Retrospective Cohort | Logistic Regression, LSSVM | Diagnosis | Diagnosis of endometriosis in symptomatic patients without U/S evidence of endometriosis | 121 controls, 232 endometriosis patients |
SE = 81% SP = 81% |
| 2012 | Dutta et al.42 | Prospective | PLSDA | Prediction | Identification of predictive biomarkers in serum for early diagnosis of endometriosis in a minimally invasive manner | 22 endometriosis, 23 controls |
SE = 81.8% SP = 91.3% |
| 2012 | Nnoaham et al.27 | Prospective Observational | Logistic Regression | Prediction | Predict any-stage endometriosis and stage 3 and 4 disease with a symptom-based model | 1396 symptomatic women |
SE = 82.6% SP = 75.8% |
| 2010 | Wang et al.26 | Retrospective | ANN | Prediction | Screening for biomarkers of eutopic endometrium in endometriosis patients | 26 patients |
SE = 91.7% SP = 90.9% |
| 2009 | Wolfler et al.43 | Prospective Exploratory Cohort | Genetic Algorithm | Prediction | Predict endometriosis before laparoscopy using patterns of serum proteins in symptomatic patients | 91 symptomatic patients |
SE = 81.3% SP = 60.3% |
| 2009 | Stegmann et al.62 | Prospective Cohort | Logistic Regression | Prediction | Prediction of lesions that have high probability of containing histologically-confirmed endometriosis | 114 women with complete data on 487 lesions |
SE = 88.4% SP = 24.6% |
| 2008 | Wang et al.44 | Retrospective | ANN | Diagnosis | Diagnostic model to correctly detect endometriosis and no endometriosis in serum samples using potential biomarkers of endometriosis | 66 serum samples |
SE = 91.7% SP = 90% |
| 2005 | Chapron et al.49 | Prospective | Logistic Regression | Prediction | Predict presence of posterior deep endometriosis among women with chronic pelvic pain symptoms | 134 women scheduled for laparoscopy for chronic pelvic pain symptoms |
SE = 68.6% SP = 77.1% |
NR not reported, PLSDA partial least squares discriminant analysis, QDA quadratic discriminant analysis, SVMs support vector machines, ANNs artificial neural networks, LSSVMs least squares support vector machines, PCA principal component analysis, NLP natural language processing, DE deep endometriosis, U/S ultrasound, miRNAs microRNAs, ART assisted reproductive technology, RNA ribonucleic acid, DNA deoxyribonucleic acid, MBD methyl binding domain, SE sensitivity, SP specificity.
Study characteristics
In the field of endometriosis, AI utilization spanned three overarching categories: predicting outcomes in endometriosis populations, building diagnostic models, and improving research efficacy. Most interventions were developed to assist with prediction of endometriosis in patients. However, the type, stage and specific characteristics of endometriosis that these interventions predicted, differed among the studies, depending on the research question generated by the authors. Approximately 44.4% (n = 16) of the studies analyzed the predictive capabilities of AI approaches in patients with endometriosis, while 47.2% (n = 17) explored diagnostic capabilities. The predictive capabilities differed between studies but included many aims such as predicting fertility therapy success in endometriosis patients, the likelihood of endometriosis versus other pelvic pain pathologies, predicting the presence of DE, and many more as seen in Table 1. Only 8.33% (n = 3) of the studies used AI technologies to advance the understanding of disease pathophysiology28–30. The AI methods that were used included: logistic regression, K-nearest neighbor, Naïve Bayes, random forest, decision tree, SVMs, neural networks, classification tree analysis, genetic algorithm, least squares support vector machines (LSSVMs), partial least squares discriminant analysis (PLSDA), margin tree classification, quick classifier algorithm, quadratic discriminant analysis (QDA), natural language processing (NLP), principle component analysis (PCA), adaptive boosting, eXtreme gradient boosting, voting classifier (hard/soft), deep learning and new ensemble ML classifiers. However, logistic regression (n = 15) was the AI intervention that was most frequently used to build predictive and diagnostic models.
The types of inputs used in different AI models varied among the studies. Four studies used biomarkers as the specific inputs for their final predictive model, but the types of biomarkers differed including: angiogenic factors, cytokines, serum microRNAs signatures, and other metabolite biomarkers. Some studies also used metabolite spectra as inputs for their AI models (n = 10) however, there was significant diversity between the type of spectrometry method (i.e., Raman spectrometry versus hydrogen nuclear magnetic resonance [1H-NMR] Carr-Purcell-Meiboom-Gill [CPMG] spectrometry) and the specific mass-dependent velocity (m/z, mass divided by charge number) peak ranges that were used among the studies. Other studies also used genetic variables such as large transcriptomics datasets (n = 5) and clinical factors (n = 6) as inputs for their final models. The clinical factors that were used in different models demonstrated some similarity with age, history of pelvic surgery, dysmenorrhea, and pelvic pain being commonly used variables. However, many studies used different combinations, thresholds and classifiers for these variables in their models. For instance, various combinations of severe dysmenorrhea, primary dysmenorrhea, and secondary dysmenorrhea were used in different ML models.
Although the AI approaches were heterogenous, most models generally achieved sensitivity and specificity above 85%, as demonstrated in Table 1. All of the studies (n = 33) used a validation process to train and validate AI models with various methods of cross-validation (i.e., bootstrapping method, leave-one-out cross-validation, etc.) or by implementing a validation/test cohort not used in the initial training set. Table 1 also reports on sensitivity and specificity for the models.
Given the heterogeneity in the purpose of the AI intervention, type and stage of endometriosis being examined, type of AI methodology used, and evaluation metrics, the included studies were grouped into six categories based on the inputs used to create the AI models. These categories are discussed in detail below.
Diagnostic or predictive models for endometriosis using biomarkers
Four different studies31–35 examined the use of biomarkers as inputs to create diagnostic or predictive AI models in endometriosis populations. As seen in Table 2, the type of biomarkers used differed among the studies. Knific et al.31 was the only study that used protein ratios while others used metabolites33, miRNAs35 and other biomarkers34. Knific et al.31 and Bendifallah et al.35 were the only studies in this category to use the random-forest method to develop a diagnostic model for endometriosis and the accuracy of Knific et al.’s31 model was reported to be 59%31 —the lowest accuracy for all the models in this category—while the clinical accuracy of Bendifallah et al.’s35 model was significantly higher with a sensitivity and specificity of 96.8 and 100%. One study used LSSVMs34 and the accuracy of this method was deemed to be 79% with a sensitivity and specificity of 82% and 75%, respectively. One study also used SVMs to develop a diagnostic model for endometriosis using lipidomic profiling of endometrial fluid in patients with ovarian endometriosis33. The accuracy of this method was reported to be 85.7% with a sensitivity and specificity of 58.3% and 100%, respectively. It should be noted that among the four studies that were examined, there were no commonalities in the specific biomarker inputs used; thus, it is difficult to compare the accuracy of each AI model given the differences in the inputs used. The pooled SE and SP for each study’s most accurate model were 85.6% and 85%, respectively33–35.
Table 2.
Diagnostic and predictive moels built using biomarkers.
| AI methods used | Authors [ref.] | Stage of endometriosis | Type of endometriosis | Sample size | Inputs used | Method accuracy |
|---|---|---|---|---|---|---|
| Random Forest | Bendifallah et al.35 | rASRM Class I–II and Class III–IV | Not specified | 200 patients (153 endometriosis, 47 controls) | 86 miRNAs composing a diagnostic blood signature |
SE = 96.8% SP = 100% |
| Knific et al.31 | All four stages of endometriosisa | Not specified | 210 patients (116 endometriosis, 94 controls) | Proteins ratios for the following: CTACK/MCP-3, MCP-3/CTACK, CCL11/I-309, X6Ckine/MCP-1, CTACK/SCYB16, Gro-alpha/CTACK |
SE = NR SP = NR |
|
| Logistic Regression | Bendifallah et al.35 | rASRM Class I–II and Class III–IV | Not specified | 200 patients (153 endometriosis, 47 controls) | 86 miRNAs composing a diagnostic blood signature |
SE = 96.8% SP = 100% |
| Vodolazkaia et al.34 | Not specified | U/S negative endometriosis | 353 EDTA samples (232 endometriosis, 121 controls) | VEGF, Annexin V, CA-125, glycodelin, sICAM-1 |
SE = 82% SP = 75% |
|
| eXtreme Gradient Boost | Bendifallah et al.35 | rASRM Class I–II and Class III–IV | Not specified | 200 patients (153 endometriosis, 47 controls) | 86 miRNAs composing a diagnostic blood signature |
SE = 90.3% SP = 100% |
| AdaBoost | Bendifallah et al.35 | rASRM Class I–II and Class III–IV | Not specified | 200 patients (153 endometriosis, 47 controls) | 86 miRNAs composing a diagnostic blood signature |
SE = 96.8% SP = 100% |
| Support Vector Machines | Dominguez et al.33 | Not specified | Ovarian endometriosis | 35 patients (12 endometriosis, 23 controls) | 123 differentially expressed metabolites in endometrial fluid |
SE = 58.3% SP = 100% |
| Least Squares Support Vector Machines | Vodolazkaia et al.34 | Not specified | U/S negative endometriosis | 353 EDTA samples (232 endometriosis, 121 controls) | VEGF, Annexin V, CA-125, sICAM-1 |
SE = 82% SP = 75% |
rASRM revised American Society of Reproductive Medicine, NR not reported, U/S ultrasound, CTACK cutaneous T cell-attracting chemokine, MCP-3 monocyte chemotactic protein 3, CCL-11 C-C motif chemokine ligand 11, I-309 chemokine ligand 1, X6Ckine C-C motif chemokine 21, MCP-1 monocyte chemoattractant protein 1, SCYB16 chemokine ligand 16, Gro-alpha growth regulated oncogene-alpha, VEGF vascular endothelial growth factor, CA-125 cancer antigen 125, sICAM-1 soluble intercellular adhesion molecule-1, SE sensitivity, SP specificity.
aMinimal, mild, moderate and severe stages of endometriosis were included.
Diagnostic or predictive models for endometriosis using protein spectra
Ten studies26,36–44 used various metabolite spectra as their primary inputs to develop diagnostic and predictive models in endometriosis populations. In this specific problem formulation, it is important to note the methodology that is used. The most popular method to determine metabolite spectra for model development was surface-enhanced laser desorption/ionization time-of-flight mass spectrometry, which was used by four studies26,41,43,44. The pooled SE for the models with highest accuracy in each study was 91.7%, while the pooled SP was 81.1%26,37–44. Table 3 presents the other methods of spectrometry and spectroscopy that were used to determine the metabolite spectra of interest for the model inputs.
Table 3.
Diagnostic and predictive models built using protein spectra.
| AI methods used | Authors [ref.] | Spectrometry or spectroscopy method | Stage of endometriosis | Type of endometriosis | Sample size | Inputs used | Method accuracy |
|---|---|---|---|---|---|---|---|
| Support Vector Machines | Parlatan et al.37 | Raman Spectroscopy | All four stages of endometriosisa | Not specified | 94 serum samples (49 endometriosis, 45 controls) | 790–1729 cm−1 spectral interval |
SE = 87.5% SP = 100% |
| k-nearest neighbor (weighted) | Parlatan et al.37 | Raman Spectroscopy | All four stages of endometriosisa | Not specified | 94 serum samples (49 endometriosis, 45 controls) | 790–1729 cm−1 spectral interval |
SE 100% SP = 100% |
| Partial least squares discriminant analysis (PLSDA) | Braga et al.36 | Mass Spectrometry | Stage 3 and 4 | Not specified | 100 patients (50 endometriosis, 50 controls) | Positive ionization m/z = 758.7234, 786.7585, 758.7155, 782.7239, 369.4541; negative ionization m/z = 279.3316, 215.1182, 255.3261, 281.3487, 283.36375 |
SE = NR SP = NR |
| Dutta et al.42 | 1H-NMR Spectroscopy | Stage 1 and 2 | Not specified | 45 patients (22 endometriosis, 23 controls) | TSP, lipoproteins (LDL and VLDL), unsaturated lipid, creatinine, L-Arginine, glucoerophosphatidylcholine, D-glucose, ornithine, citrate, L-lysine, tyrosine, L-histidine, L-phenylalanine, formate, choline, L-threonine, acetate, L-glutamine, succinate, acetone, adipic acid, L-isoleucine, alanine, L-aspartate, 3-hydroxybutyric acid, propylene glycol, valine, leucine, creatine, pyruvate, lactate, 2-hydroxybutyrate |
SE = 81.8% SP = 91.3% |
|
| Quadratic discriminant analysis | Ghazi et al.38 | Nuclear magnetic resonance spectroscopy | Stage 2 and 3 | Not specified | 45 patients (31 endometriosis, 15 controls) | Chemical shift for all spectra between 0 to 5.5ppm |
SE = NR SP = NR |
| Genetic algorithm | Wang et al.39 | Liquid chromatography tandem mass spectrometry | All four stages of endometriosisa | Not specified | 122 patients (60 endometriosis, 62 without endometriosis) | m/z = 1433.9, 1599.4, 2085.6, 6798, 3217.2 |
SE = 90.9% SP = 92.9% |
| Wolfler et al.43 | Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry | Not specified | Not specified | 91 symptomatic patients | Mass peaks between 2000 and 20000 Da |
SE = 55.6% SP = 64.9% |
|
| Decision tree algorithm | Wang et al.39 | Liquid chromatography tandem mass spectrometry | All four stages of endometriosisa | Not specified | 122 patients (60 endometriosis, 62 without endometriosis) | 36 differentially expressed peptide spectra |
SE = 90% SP = 80.6% |
| Wolfler et al.43 | Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry | Not specified | Not specified | 91 symptomatic patients | Mass peaks between 2000 and 20000 Da |
SE = 92.7% SP = 62.8% |
|
| Quick classifier algorithm | Wang et al.39 | Liquid chromatography tandem mass spectrometry | All four stages of endometriosisa | Not specified | 122 patients (60 endometriosis, 62 without endometriosis) | 36 differentially expressed peptide spectra |
SE = 73.3% SP = 77.4% |
| Least squares support vector machines | Fassbender et al.40 | Matrix-assisted laser desorption ionization time-of-flight mass spectrometry | Stage 1/2, stage 3/4 | U/S negative endometriosis | 254 plasma samples (165 endometriosis, 89 without endometriosis) | Minimal to mild endometriosis m/z = 4898, 5715, 8328, 9926, 14.698; moderate to severe endometriosis m/z = 3192, 4519, 2189, 4373, 7457; ultrasonography-negative endometriosis m/z = 2.058, 2456, 3.883, 14.694, 42.065 |
Minimal to mild endometriosis: SE = 75% SP = 86% Moderate to severe endometriosis: SE = 98% SP = 81% Ultrasonography-negative endometriosis: SE = 88% SP = 84% |
| Fassbender et al.41 | Proteomic surface-enhanced laser desorption ionization time-of-flight mass spectrometry | All four stages of endometriosisa | Not specified | 49 endometrial biopsies (31 endometriosis, 18 without endometriosis) | m/z = 2072, 2973, 3623, 3680, 21113 |
SE = 91% SP = 80% |
|
| Artificial neural networks | Ghazi et al.38 | Nuclear magnetic resonance spectroscopy | Stage 2 and 3 | Not specified | 45 patients (31 endometriosis, 15 controls) | Chemical shift for all spectra between 0 and 5.5ppm |
SE = 50% SP = 17% |
| Wang et al.26 | Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry | All four stages of endometriosisa | Not specified | 39 patients (26 endometriosis, 13 controls) | m/z = 6898, 5891, 5385, 6448, 5425 |
SE = 91.7% SP = 90.9% |
|
| Wang et al.44 | Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry | All four stages of endometriosisa | Not specified | 66 serum samples (36 endometriosis, 30 controls) | m/z = 8142, 5640, 5847, 8940, 3269 |
SE = 91.7% SP = 90% |
NR not reported, m/z mass-to-charge ratio, ppm parts per million, Da Dalton, TSP thrombospondin, VLDL very-low-density lipoprotein, LDL low-density lipoprotein, 1H-NMR hydrogen-1 nuclear magnetic resonance, U/S ultrasound, SE sensitivity, SP specificity.
aMinimal, mild, moderate and severe stages of endometriosis were included.
Among the studies in this category, artificial neural networks (ANNs) were the most popular method used in three of the models26,38,44. However, although these three studies used the same type of AI intervention, the inputs varied greatly between them. Two studies used PLSDA to compute their final models36,42, albeit using different methodologies (mass spectroscopy36 and 1H-NMR spectrophotometer42). While the inputs also varied between both models, they both had a similar correct classification rates of 84%36 and 86.67%42. Further studies between similar inputs are needed to determine if PLSDA is an appropriate AI intervention to compute diagnostic and predictive models in endometriosis populations.
Diagnostic or predictive models for endometriosis using clinical variables and symptoms
Six studies45–50 grouped in this category strongly preferred using logistic regression; two studies50,51 used decision tree methods to build a model and one study50 also used random forest, eXtreme gradient boosting and voting classifier (soft/hard) ML algorithms as shown in Table 4. Interestingly many studies in this category examined predictive and diagnostic model capabilities in patients with some form of deep endometriosis (n = 5). The pooled SE for the models with highest accuracy in each study was 81.7% while the pooled SP was 91.6%47–50. Specific inputs into each model varied as seen in previous categories with Bendifallah et al.50 using the largest number of clinical features for their models. However, there were some commonalities in the types of inputs that were used in each model. Patient age was the most frequently used input (n = 5) in diagnostic and predictive models using clinical variables. Given that endometriosis most commonly presents in reproductive-aged women, it is not surprising that age is the most frequent input in a diagnostic/predictive AI model. Other significant inputs included the presence or severity of dysmenorrhea, presence or severity of dyspareunia, visual analogic scale for dyspareunia, infertility, and previous surgery for endometriosis or pelvic surgery. Among the studies that did report SE and SP metrics, the SE values ranged from 51% to 95% and SP values ranged from 77.1 to 95.7%47–50.
Table 4.
Diagnostic and predictive models built using clinical variables and symptoms.
| AI methods used | Authors [ref.] | Stage of endometriosis | Type of endometriosis | Sample size | Inputs used | Method accuracy |
|---|---|---|---|---|---|---|
| Logistic Regression | Bendifallah et al.50 | Not specified | Ovarian, superficial or deep endometriosis | Training set (1126 patients), validation set (100 patients) | Mother/daughter history of endometriosis, history of surgery for endometriosis, age, BMI, dysmenorrhea/VAS of dysmenorrhea, abdominal pain outside menstruation, pain suggesting of sciatica, pain during sexual intercourse, lower back pain outside menstruation, painful defecation, urinary pain during menstruation, right shoulder pain near or during menstruation, blood in the stools during menstruation, blood in urine during menstruation, absenteeism duration in the last 6 months, number of non-hormonal pain treatments used |
SE = 95% SP = 81% |
| Vesale et al.45 | Not specified | Deep endometriosis with colorectal involvement | Training set (789 patients), validation set (333 patients) | Age, type of colorectal management, colpectomy and parametrectomy |
SE = NR SP = NR |
|
| Benoit et al.46 | All four stages of endometriosisa | Not specified | 297 patients who underwent ART after surgery for endometriosis-associated infertility | Age, duration of infertility, number of ICSI-IVF cycles, ovarian reserve, rAFS score |
SE = NR SP = NR |
|
| Lafay Pillet et al.47 | Not specified | Deep endometriosis in patients with ovarian endometrioma | 326 patients (164 with DE lesions associated with endometrioma, 162 patients with no associated DE lesions) | VAS of gastrointestinal symptoms ≥5 or of deep dyspareunia >5, duration of pain greater than 24 months, severe dysmenorrhea (defined as the prescription of the OCP for the treatment of a primary dysmenorrhea or the worsening of a secondary dysmenorrhea), primary or secondary infertility |
SE = 51% SP = 94% |
|
| Ballester et al.48 | Not specified | Deep endometriosis | training set: 94 patients who underwent ICSI-IVF, validation set: 48 consecutive patients | Patient’s age, presence of DIE, AMH serum level >1 ng/ml, number of ICS-IVF cycles |
SE = 66.7% SP = 95.7% |
|
| Chapron et al.49 | Not specified | Posterior deep endometriosis | 134 patients (51 with posterior DE, 83 with other disorders) | Painful defecation during menses, VAS for dyspareunia > or =8, previous surgery for endometriosis, pain other than non-cyclic |
SE = 68.6% SP = 77.1% |
|
| Decision Tree | Bendifallah et al.50 | Not specified | Ovarian, superficial or deep endometriosis | Training set (1126 patients), validation set (100 patients) | See above. |
SE = 91% SP = 66% |
| Wang et al.51 | Not specified | Ovarian endometriomas | 178 case records | Patients’ basic information (age, number of pregnancies, number of births, number of miscarriages, past histories, menstruation periods, regularity of menstruations, periods of menstrual flow, severity of dysmenorrhea, urges to defecate, dyspareunia, whether other pains exist and other concomitant histories); clinical test values (endometrioma counts, sizes of endometriomas, follicle counts, CA125 blood values, sizes of uteruses, level of ovarian adhesions and contents of endometriomas); treatment-related information (medication prior to surgery, medication following surgery, route of drug administration, surgical method, surgical routine, UGA method, UGA site, UGA with irrigation and medication used) |
SE = NR SP = NR |
|
| Random Forest | Bendifallah et al.50 | Not specified | Ovarian, superficial or deep endometriosis | Training set (1126 patients), validation set (100 patients) | See above. |
SE = 92% SP = 92% |
| eXtreme Gradient Boosting | Bendifallah et al.50 | Not specified | Ovarian, superficial or deep endometriosis | Training set (1126 patients), validation set (100 patients) | See above. |
SE = 93% SP = 92% |
| Voting Classifier (soft/hard) | Bendifallah et al.50 | Not specified | Ovarian, superficial or deep endometriosis | Training set (1126 patients), validation set (100 patients) | See above. |
Voting Classifier Soft SE = 93% SP = 88% Voting Classifier Hard SE = 91% SP = 92% |
NR not reported, DE deep endometriosis, ICSI-IVF intracytoplasmic sperm injection in vitro fertilization, rAFS revised American Fertility Society, OCP oral contraceptive pill, VAS visual analogic scale, BMI body mass index, CA-125 cancer antigen 125, UGA ultrasound-guided aspiration, SE sensitivity, SP specificity.
aMinimal, mild, moderate and severe stages of endometriosis were included.
Diagnostic or predictive models for endometriosis using genetic variables
Models that were built using genetic variables as their primary inputs used a significantly larger number of inputs than any of the other six input categories referenced in this review. Only five studies52–56 used genetic variables to build their predictive and diagnostic models, however, the type of input varied between individual gene candidates52,56, large protein-coding gene datasets from transcriptomics and methylomics data53,55, and 16S rRNA gene amplicon data54. The AI methods used in this category included: deep ML algorithm, decision tree, GenomeForest (a new ensemble ML classifier), random-forest-based ML classification analysis, PLSDA, SVM, random forest, and margin tree classification. The pooled SE for the models with highest accuracy in each study was 96.7%, while the pooled SP was 70.7%52,53,55.
Two studies compared the use of large transcriptomics and methylomics datasets to build different AI models that were compared with each other53,55. As seen in Table 5, regardless of which AI method was used, the models built using the transcriptomics dataset outperformed the models built with the methylomics dataset, albeit marginally. Akter53 used GenomeForest, a novel ensemble technique based on chromosomal partitioning, to classify endometriosis and control samples using both transcriptomics and methylomics datasets. The authors concluded that this new classifier could help identify candidate biomarkers for endometriosis; they further demonstrated that three different ML models (GenomeForest, decision tree, and Biosigner) independently identified NOTCH3 as candidate gene with differential expression in the endometriosis samples53,55. ML methods may be of particular use when analyzing very large genomic datasets to help identify candidate genes that have altered expression in endometriosis patients versus control samples.
Table 5.
Diagnostic and predictive models built using genetic variables.
| AI methods used | Authors [ref.] | Stage of endometriosis | Type of endometriosis | Sample size | Inputs used | Method accuracy |
|---|---|---|---|---|---|---|
| Deep Machine Learning Algorithm | Li et al.52 | All four stages of endometriosisa | Not specified | 213 patients (142 endometriosis, 71 controls) | SCAF11, KIF3A, KRAS, MDM2 |
SE = 100% SP = 61.1% |
| GenomeForest | Akter et al.53 | All four stages of endometriosisa | Not specified | Transcriptomics dataset: 16 endometriosis, 22 controls; methylomics dataset: 44 endometriosis, 36 controls | Genes in transcriptomics data and genomic regions in methylated data. 11 687 protein-coding genes (14 154 genes total) |
For transcriptomics data: SE = 93.8% SP = 100% For methylomics data: SE = 92.9% SP = 88.6% |
| Random-Forest-based Machine Learning Classification Analysis | Perrotta et al.54 | All four stages of endometriosisa | Not specified | 59 patients (35 endometriosis, 24 controls) | Operational taxonomic unit and community state types in vaginal microbiome |
SE = NR SP = NR |
| Decision Tree | Akter et al.55 | All four stages of endometriosisa | Not specified | Transcriptomics dataset: 38 samples (16 endometriosis, 22 controls); methylomics dataset: 77 samples (42 endometriosis, 35 controls) | Transcriptomics: 14 154 genes; methylomics: 2 577 382 methylated regions |
For transcriptomics: SE = 81.3% SP = 95.5% For methylomics: SE = 76.2% SP = 80% |
| Partial Least Squares Discrimination Analysis | Akter et al.55 | All four stages of endometriosisa | Not specified | Transcriptomics dataset: 38 samples (16 endometriosis, 22 controls); methylomics dataset: 77 samples (42 endometriosis, 35 controls) | Transcriptomics: 14 154 genes; methylomics: 2 577 382 methylated regions |
For transcriptomics: SE = 86.4% SP = 56.3% For methylomics: SE = 60% SP = 76.2% |
| Support Vector Machines | Akter et al.55 | All four stages of endometriosisa | Not specified | Transcriptomics dataset: 38 samples (16 endometriosis, 22 controls); methylomics dataset: 77 samples (42 endometriosis, 35 controls) | Transcriptomics: 14 154 genes; methylomics: 2 577 382 methylated regions |
For transcriptomics: SE = 63.6% SP = 43.8% For methylomics: SE = 40% SP = 61.9% |
| Random Forest | Akter et al.55 | All four stages of endometriosisa | Not specified | Transcriptomics dataset: 38 samples (16 endometriosis, 22 controls); methylomics dataset: 77 samples (42 endometriosis, 35 controls) | Transcriptomics: 14 154 genes; methylomics: 2 577 382 methylated regions |
For transcriptomics: SE = 45.5% SP = 43.8% For methylomics: SE = 31.4% SP = 52.4% |
| Margin Tree Classification | Tamaresis et al.56 | All four stages of endometriosisa | Not specified | 148 endometrial samples (77 endometriosis, 37 without endometriosis but other uterine/pelvic pathology, 34 controls) | FOSB, FOS, EGR1, JUNB, MTSS1L, CTSW, TGFB1, SOC3, IL32, FKBP8, ISYNA1, CCL3, GNLY, MAP3K11, C1QA, NOTCH3, CYR61, NPTXR, FBN1, PNRC2, ITGA6, DHFR, SLC39A6, MYO10, HSP90B1, SMC3, PKP4, PALLD, DIO2 |
SE = NR SP = NR |
NR not reported, SCAF11 SR-related CTD-associated factor 11, KIF3A kinesin family member 3A, KRAS Kirsten rat sarcoma viral oncogene homolog, MDM2 mouse double minute 2 homolog, FOSB Fbj murine osteosarcoma oncogene B, EGR1 early growth response 1, JUNB JunB proto-oncogene, MTSS1L metastasis suppressor 1-like, CTSW cathepsin W, TGFB1 transforming growth factor beta 1, SOC3 suppressor of cytokine signaling 3, IL32 interleukin 32, FKBP8 FKBP prolyl isomerase 8, ISYNA1 inositol-3-phosphate synthase 1, CCL3 chemokine ligand 3, GNLY granulysin, MAP3K11 mitogen-activated protein kinase kinase kinase 11, C1QA complement C1q A chain, NOTCH3 notch receptor 3, CYR61 cysteine-rich angiogenic inducer 61, NPTXR neuronal pentraxin receptor, FBN1 fibrillin 1, PNRC2 protein rich nuclear receptor coactivator 2, ITGA6 integrin subunit alpha 6, DHFR dihydrofolate reductase, SLC39A6 Dolutegravir carrier family 39 member 6, MYO10 myosin X, HSP90B1 heat shock protein 90 beta family member 1, SMC3 structural maintenance of chromosomes 3, PKP4 plakophillin 4, PALLD Palladin, cytoskeletal associated protein, DIO2 iodothyronine deiodinase 2, SE sensitivity, SP specificity.
aMinimal, mild, moderate and severe stages of endometriosis were included.
Diagnostic or predictive models for endometriosis using mixed variables
Three studies27,57,58 used mixed variable types to create predictive or diagnostic models for endometriosis as shown in Table 6. All three studies used logistic regression as the methodology to construct models and the sample sizes ranged from 119 patients57 to 1396 patients27. Inputs included clinical variables collected from patient medical history, physical exam findings, ultrasonography evidence, and MRI visualization. It should be noted that Chattot et al.57 had the smallest sample size. The study with the largest sample size27 reported a SE and SP of 82.6% and 75.8%, respectively. The accuracy for studies in this category was relatively consistent compared to other categories with similar SE and SP.
Table 6.
Diagnostic and predictive models built using mixed variables.
| AI methods used | Authors [ref.] | Stage of endometriosis | Type of endometriosis | Sample size | Inputs used | Evaluation Metric |
|---|---|---|---|---|---|---|
| Logistic Regression | Guo et al.58 | All stages of endometriosis and stage 3/4 endometriosis | NR | 1016 infertile patients | for any-stage endometriosis nomogram: BMI, Cycle length, parity, palpable nodularity, endometrioma diagnosed on TVS, tubal pathology; for stage 3–4 endometriosis nomogram: pain, palpable nodularity, endometrioma diagnosed on TVS |
SE = NR SP = NR |
| Logistic Regression | Chattot et al.57 | Not specified | NR | 119 patients (47 endometriosis with rectosigmoid involvement, 72 endometriosis without rectosigmoid involvement) | Palpation of a posterior nodule on digital examination, UBESS score of 3 on ultrasonography, rectosigmoid involvement in endometriosis infiltration on MRI, presence of blood in the stools during menstruation |
SE = NR SP = NR |
| Logistic Regression | Nnoaham et al.27 | Stage 3 and 4 endometriosis | NR | 1396 symptomatic women | Ultrasound evidence, menstrual dyschezia, ethnicity, history of benign ovarian cysts |
SE = 82.6% SP = 75.8% |
NR not reported, BMI body mass index, TVS transvaginal ultrasound, UBESS ultrasound-based endometriosis staging system, MRI magnetic resonance imaging, SE sensitivity, SP specificity.
Diagnostic or predictive models for endometriosis using imaging
Only three studies59–61 explored the use of imaging variables as their primary inputs for their AI models as seen in Table 7. Guerriero59 built models specifically for rectosigmoid endometriosis and compared the accuracy of the different AI methods using the same inputs for each model. This specific study allows one to draw conclusions about the accuracy of different methodologies in developing predictive models to increase suspicion for rectosigmoid endometriosis. The Naïve Bayes and SVM approaches produced the models with the highest accuracy (75%) in this study and K-nearest neighbor produced the lowest accuracy (69%). SVM also produced the highest SE at 84% while Naïve Bayes and decision tree showed the highest SP (77%). The pooled SE for the models with highest accuracy in each study was 88% while the pooled SP was 89.7%59–61.
Table 7.
Diagnostic and predictive models built using imaging.
| Authors [ref.] | Stage of endometriosis | Type of endometriosis | Sample size | Inputs used | AI methods used | Method accuracy |
|---|---|---|---|---|---|---|
| Maicus et al.61 | NR | Endometriosis with POD obliteration | 749 sliding sign transvaginal ultrasound videos | Presence of sliding sign on transvaginal U/S | Resnet (2 + 1)D |
SE = 89% SP = 90% |
| Guerriero et al.59 | NR | Rectosigmoid endometriosis | 106 patients with U/S diagnosis of rectosigmoid endometriosis | Age; presence of U/S signs of uterine adenomyosis; presence of an endometrioma; adhesions of the ovary to the uterus; presence of “kissing ovaries”; absence of sliding sign | K-nearest Neighbor |
SE = 66% SP = 71% |
| Guerriero et al.59 | NR | Rectosigmoid endometriosis | 106 patients with U/S diagnosis of rectosigmoid endometriosis | Age; presence of U/S signs of uterine adenomyosis; presence of an endometrioma; adhesions of the ovary to the uterus; presence of “kissing ovaries”; absence of sliding sign | Naive Bayes |
SE = 72% SP = 77% |
| Guerriero et al.59 | NR | Rectosigmoid endometriosis | 106 patients with U/S diagnosis of rectosigmoid endometriosis | Age; presence of U/S signs of uterine adenomyosis; presence of an endometrioma; adhesions of the ovary to the uterus; presence of “kissing ovaries”; absence of sliding sign | Neural Networks |
SE = 72% SP = 73% |
| Guerriero et al.59 | NR | Rectosigmoid endometriosis | 106 patients with U/S diagnosis of rectosigmoid endometriosis | Age; presence of U/S signs of uterine adenomyosis; presence of an endometrioma; adhesions of the ovary to the uterus; presence of “kissing ovaries”; absence of sliding sign | Support Vector Machine |
SE = 84% SP = 71% |
| Guerriero et al.59 | NR | Rectosigmoid endometriosis | 106 patients with U/S diagnosis of rectosigmoid endometriosis | Age; presence of U/S signs of uterine adenomyosis; presence of an endometrioma; adhesions of the ovary to the uterus; presence of “kissing ovaries”; absence of sliding sign | Decision Tree |
SE = 66% SP = 77% |
| Guerriero et al.59 | NR | Rectosigmoid endometriosis | 106 patients with U/S diagnosis of rectosigmoid endometriosis | Age; presence of U/S signs of uterine adenomyosis; presence of an endometrioma; adhesions of the ovary to the uterus; presence of “kissing ovaries”; absence of sliding sign | Random Forest |
SE = 66% SP = 72% |
| Guerriero et al.59 | NR | Rectosigmoid endometriosis | 106 patients with U/S diagnosis of rectosigmoid endometriosis | Age; presence of U/S signs of uterine adenomyosis; presence of an endometrioma; adhesions of the ovary to the uterus; presence of “kissing ovaries”; absence of sliding sign | Logistic Regression |
SE = 72% SP = 73% |
| Reid et al.60 | NR | NR | 189 women (100 training set, 89 test set) with suspected endometriosis | POD 1 model: posterior compartment deep endometriosis, right ovarian fixation, negative “sliding sign”; POD 2 model: unilateral ovarian fixation, unilateral endometrioma, negative “sliding sign” | Logistic Regression |
POD 1: SE = 88% SP = 97% POD 2: SE = 88% SP = 99% |
U/S ultrasound, POD pouch of Douglas, NR not reported, SE sensitivity, SP specificity.
Reid et al.60 also produced two logistic regression models using different imaging variables; the accuracy of both models was higher than the logistic regression model produced by Guerriero et al.59 indicating that perhaps the inputs for Reid’s model60 played a role in the higher accuracy, SE and SP. All three studies in this category explored “sliding sign” on transvaginal ultrasound as an important features in their models.
Maicus et al.61 was the only study to use a deep learning model called Resnet (2 + 1)D to classify the state of the pouch of Douglas with regards to adhesions indicative of endometriosis in patients. Their model was trained, internally validated, and externally tested on a dataset to evaluate the sliding sign on ultrasound, demonstrating an accuracy of 88.8%.
Discussion
In the field of endometriosis, AI interventions have proven to be heterogenous in terms of their purpose, methodology, input selection and accuracy. Given the wide range of problems that exist in the field of endometriosis diagnosis, prediction and research, it is not surprising that models were built to tackle many different problem formulations. This study performed a thorough scoping review on the literature intersecting endometriosis and AI, and it provides a timely understanding of AI technology in the field of endometriosis. A meta-analysis of the data was not possible due to the diverse nature of studies included in this scoping review. Our study identified six major categories of model inputs that were used to build AI interventions in addition to three studies that used AI methods to improve research techniques28–30 and one study that only used lesion characteristics to build a predictive model62. Of the six major input categories, biomarkers, clinical variables, genetic variables and metabolite spectra were the most frequently used input types for building diagnostic and predictive AI models.
AI interventions that were built using biomarker inputs included diagnostic and predictive models for ultrasound-negative endometriosis34, and ovarian endometriomas33. Biomarker inputs for these models included plasma biomarkers collected in all phases of the menstrual cycle34, lipidomic profiling of endometrial fluid33, and serum miRNA markers35. AI interventions built using metabolite spectra as their primary input included detecting endometriosis in serum samples43,44, screening for biomarkers in eutopic endometrium26, diagnosing ultrasound-negative endometriosis40, diagnosing endometriosis using messenger RNA expression in endometrium biopsies41, identifying predictive serum biomarkers42, diagnosing and staging endometriosis using peptide profiling39, determining classifier metabolites for early prediction risk38, and diagnosing stage 3 and stage 4 endometriosis in infertile patients36. Studies that used genetic variables to build AI interventions included classifying endometriosis using RNAse and enrichment-based DNA-methylation datasets53, diagnosing endometriosis using gut and/or vaginal microbiome profiles54, using transcriptomics or methylomics to classify endometriosis55, and staging pelvic endometriosis using genomic data56. Some studies also used clinical signs and symptoms collected when obtaining a patient’s medical history as well as other clinical variables to build models. These AI interventions included predicting the presence of posterior deep endometriosis in patients with chronic pelvic pain symptoms49, predicting pregnancy rates in patients with endometriosis48, predicting medical care decision rules for patients with recurrent endometriomas51, diagnosing DE pre-operatively for patients with endometriomas47 and differentiating between patients with and without endometriosis50.
Our scoping review was able to evaluate the current literature and map out the field of study to demonstrate that AI applications in endometriosis look promising for improving diagnostics, research efficacy and outcome prediction in this patient population. Pooled SE ranged between 81.7 and 96.7% and pooled SP ranged between 70.7 and 91.6%. Our review included a range of heterogenous study designs, large retrospective analyses, various ML interventions and diverse research questions in the field of endometriosis. This is a timely review providing clinicians and computer scientists with an extensive understanding of AI applications in endometriosis. Clinical decision-making by humans is often prone to errors, biases and heuristics63. However, this review shows strong promise for AI’s ability to mitigate these human errors and provide superior outcome prediction with high SE and SP. Although many of the studies included in this review relied on a human component for data analysis/collection and determining feature extraction, AI technologies (especially when using standardized and validated models) may present the potential to reduce diagnostic error that can result from individual practicing biases and clinical heuristics. Future studies with human comparators are required to determine this. This review also demonstrated how AI can be used to improve research efficacy particularly through the use of natural language processing28 and identification of potential biomarkers30 and diseases29 associated with endometriosis pathophysiology. Lastly, this scoping review adds to future recommendations for research in this field and supports the need for standardized guidelines for ML applications in medicine.
Approximately 44.4% (n = 16) of AI interventions were predictive models meant to predict various outcomes in patients with endometriosis or undifferentiated symptomatic patients. Models were built to predict the presence of posterior DE in patients with chronic pelvic pain49, the clinical pregnancy rate in patients with endometriosis48, and many other outcomes in this patient population. However, many of these studies were conducted retrospectively and they did not adequately compare the AI’s ability to outperform existing decision tools and clinical diagnostics. Additionally, none of the studies involved a human comparator (since many models were trained and validated on retrospectively diagnosed patient datasets) and thus make it difficult to comment on AI’s superiority as a tool clinicians can use for predictive modeling.
The type and stage of endometriosis varied among the included studies; thus, the AI approaches to prediction and diagnosis also differed. This makes it difficult to compare AI models used in the studies. Many studies lacked detailed information on the methods used to verify patients with endometriosis with regards to a reference standard, while others cited gold standard laparoscopic visualization with subsequent histopathologic confirmation as the modality of diagnosis. Additionally, the heterogeneity of the study designs, input data used, and AI interventions, made it difficult to compare the accuracy and efficacy of the different models. Many studies lacked transparent descriptions of their modeling making it difficult to critique methodology and determine if the right AI model was being used to predict the outcome in question.
Applying AI to assess endometriosis is relatively new, and most AI methods used are still relatively simple. Various data types continue to be explored; however, each data type was utilized exclusively up to date. As can be seen from the tables, the use of protein spectra continues to be perhaps the most common approach, but generally only with small sample sizes. In the future, the increasing adoption of AI in assessing endometriosis will also likely play an essential role in women’s healthcare.
Our recommendations, based on this review and challenges of employing AI, are as follows:
The types and stages of endometriosis included in the study sample need to be clearly defined, and models should specify what type/stage of endometriosis they are built to predict, classify or diagnose.
The gold standard (a reference where we compare the AI model against) has to be defined and justified to assess reliability.
The evaluation metric (e.g., sensitivity and specificity) needs to be tested and reported clearly.
Transparent descriptions of the used AI model is needed for reproducibility.
Applying multiple AI models to determine the most accurate one for specific outcomes and diagnostic goals.
A large sample size with a diverse age group used is required for achieving generalizability.
Training and testing phases need to be clearly explained, specifically stating whether cross-validation or holdout is implemented; and
Logistic regression models incorporating a training and test/validation cohort would be more effective in establishing external validation of the model; and
Studies using retrospective analyses of large clinical datasets to build models should attempt to validate their models in prospective controlled clinical trials. Controlled clinical trials are required to determine whether AI can outperform human decision-making and remove any potential biases. Although internal validation samples are essential to test a model’s performance, these models should also be tested through prospective controlled trials to ensure that they are generalizable in a clinical context and that their performance is not limited to an artificial set of parameters.
Of the 36 studies included in this review, 50% were published in the last 5 years, indicating that there is recent and rapidly growing interest in AI applications to improve diagnostic, predictive and research capabilities for a complex disease such as endometriosis. Further research should be conducted using human comparators and should include comparisons with existing scoring systems and diagnostic tools to determine AI’s superiority for predictive and diagnostic modeling in endometriosis. These AI algorithms should also be externally validated or tested through prospective controlled trials to ensure that they contribute to advancing real-world clinical practice and diagnostics. This review was able to identify this interest in AI and highlight the benefits and shortcomings of AI interventions to improve future models for endometriosis.
Methods
Study guidelines
Given the heterogeneity and breadth of research in this field, a scoping review was performed to summarize the use of AI applications in endometriosis research, diagnostics, and prediction to help identify gaps in knowledge and address broad research questions64. The guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses for Scoping Review (PRISMA-ScR)65 and Arksey and O’Malley’s recommendations for scoping review methodology66 were followed. A prior review protocol was drafted using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Protocols67 for internal use amongst the research team but it was not externally published or registered prospectively.
Search strategy and study eligibility
The PubMed, Medline-OVID, EMBASE, and CINAHL databases were searched sequentially from January 2000 to March 2022 for all English-language papers using the following search strategy (adapted for each database): [(Endometriosis) OR (Endometrioma)] AND [(AI) OR (ML) OR (Prediction Model) OR (Classification)]. Gray literature was not included in this scoping review in attempt to only include peer-reviewed studies. This timeframe was chosen to reflect advances in AI technologies and applications in medicine. The scope of the search was not restricted to a particular type or stage of endometriosis. The search for this scoping review was completed in March 2022.
Inclusion and exclusion criteria
The following inclusion criteria were used to determine study eligibility for this review: (1) the study involved assessing an AI approach or model to advance prediction, diagnosis, management or disease understanding in the field of endometriosis; (2) the study reported a quantitative metric on the accuracy/performance of the AI method; (3) the study was conducted using humans; (4) the article was accessible in English; and (5) the study used a validation method to test its model. Studies were excluded if: (1) they were not conducted using humans; (2) did not assess or evaluate an AI approach or model; (3) did not pertain to the field of endometriosis; and (4) developed a logistic regression model without the use of a training and test/validation set. One reviewer (BS) conducted the literature search and two reviewers (BS and ME) screened the titles, abstracts and full-texts independently for potentially eligible studies. Reference lists of eligible studies were also hand-searched but no additional studies were included on this basis.
Study selection and data extraction
One author (B.S.) conducted the literature search, and two authors (B.S. and M.E.) independently screened the titles and abstracts for potentially eligible studies. Each potential study for inclusion underwent full-text screening and was assessed to extract study-specific information and data; Table 1 presents a summary of the title, lead author, publication year, study design, AI intervention, purpose/aim, sample size, type of inputs used in the AI method, specific inputs in the final model, evaluation metrics used and AI accuracy. Two reviewers (B.S. and M.E.) independently conducted a full-text screening and extracted information from potentially eligible studies. They then cross-checked the identified studies to determine eligibility through discussion and used consensus to resolve discrepancies. The information collated in the initial evidence table was used to aggregate data and determine the main themes of use for AI in endometriosis in the currently published literature. Where studies explored more than one AI model, the model with the highest accuracy was assessed and included in the review.
Pooled evaluation metric
Pooled sensitivities and specificities were calculated for studies within the same input category. The following formula68 was used to combine means across different studies where SE or SP is the pooled mean for sensitivity or specificity, as follows:
| 1 |
where, for example, N1 is the number of participants in study 1 and X1 is the value of the reported sensitivity or specificity in study 1.
Author contributions
B.S., M.E., C.M., C.A., P.Y., and M.B. conceived the study. B.S. and M.E. developed the search strategy and made the inclusion decisions and the quality assessment. C.M., C.A., P.Y., and M.B. provided methodological and clinical expertise. B.S. and M.E. wrote the draft of the paper. B.S. and M.E. created all figures. All authors approved final paper.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Brintha Sivajohan, Mohamed Elgendi.
References
- 1.Nnoaham KE, et al. Impact of endometriosis on quality of life and work productivity: a multicenter study across ten countries. Fertil. Steril. 2011;96:366. doi: 10.1016/j.fertnstert.2011.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zondervan KT, Becker CM, Missmer SA. Endometriosis. N. Engl. J. Med. 2020;382:1244–1256. doi: 10.1056/NEJMra1810764. [DOI] [PubMed] [Google Scholar]
- 3.Shafrir AL, et al. Risk for and consequences of endometriosis: a critical epidemiologic review. Best. Pract. Res. Clin. Obstet. Gynaecol. 2018;51:1–15. doi: 10.1016/j.bpobgyn.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Barbieri RL. Etiology and epidemiology of endometriosis. Am. J. Obstet. Gynecol. 1990;162:565–567. doi: 10.1016/0002-9378(90)90430-F. [DOI] [PubMed] [Google Scholar]
- 5.Levy AR, et al. Economic burden of surgically confirmed endometriosis in Canada. J. Obstet. Gynaecol. Can. 2011;33:830–837. doi: 10.1016/S1701-2163(16)34986-6. [DOI] [PubMed] [Google Scholar]
- 6.Practice bulletin no. 114: Management of endometriosis. Obst. Gynecol.116, 223–236 (2010). [DOI] [PubMed]
- 7.Johnson NP, et al. World Endometriosis Society consensus on the classification of endometriosis. Hum. Reprod. 2017;32:315–324. doi: 10.1093/humrep/dew293. [DOI] [PubMed] [Google Scholar]
- 8.Zondervan KT, et al. Endometriosis. Nat. Rev. Dis. Prim. 2018;4:9. doi: 10.1038/s41572-018-0008-5. [DOI] [PubMed] [Google Scholar]
- 9.International working group of AAGL, ESGE, ESHRE and WES et al. An international terminology for endometriosis. J. Minim. Invasive Gynecol.28, 1849–1859 (2021). [DOI] [PubMed]
- 10.Canis M, et al. Revised American Society for Reproductive Medicine classification of endometriosis. Fertil. Steril. 1997;67:817–821. doi: 10.1016/S0015-0282(97)81391-X. [DOI] [PubMed] [Google Scholar]
- 11.Gruppo Italiano per lo Studio dell’Endometriosi. Relationship between stage, site and morphological characteristics of pelvic endometriosis and pain. Hum. Reprod.16, 2668–2671 (2011). [DOI] [PubMed]
- 12.Zondervan KT, Cardon LR, Kennedy SH. The genetic basis of endometriosis. Curr. Opin. Obstet. Gynecol. 2001;13:309–314. doi: 10.1097/00001703-200106000-00011. [DOI] [PubMed] [Google Scholar]
- 13.Mihalyi A, et al. Role of immunologic and inflammatory factors in the development of endometriosis: indications for treatment strategies. Clin. Pract. 2005;2:623. [Google Scholar]
- 14.Gao X, et al. Economic burden of endometriosis. Fertil. Steril. 2006;86:1561–1572. doi: 10.1016/j.fertnstert.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy S, et al. ESHRE guideline for the diagnosis and treatment of endometriosis. Hum. Reprod. 2005;20:2698–2704. doi: 10.1093/humrep/dei135. [DOI] [PubMed] [Google Scholar]
- 16.Chiaffarino F, et al. Endometriosis and irritable bowel syndrome: a systematic review and meta-analysis. Arch. Gynecol. Obstet. 2021;303:17–25. doi: 10.1007/s00404-020-05797-8. [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaki S, et al. Relationship between delay of surgical diagnosis and severity of disease in patients with symptomatic deep infiltrating endometriosis. Fertil. Steril. 2006;86:1314–1316. doi: 10.1016/j.fertnstert.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 18.Prast J, et al. Costs of endometriosis in Austria: a survey of direct and indirect costs. Arch. Gynaecol. 2013;288:569–576. doi: 10.1007/s00404-013-2793-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang F, Preininger A. AI in health: state of the art, challenges, and future directions. Yearb. Med. Inform. 2019;28:016–026. doi: 10.1055/s-0039-1677908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang R, et al. Artificial intelligence in reproductive medicine. Reproduction. 2019;158:R139–R154. doi: 10.1530/REP-18-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M, Decary M. Artificial intelligence in healthcare: An essential guide for health leaders. Healthc. Manag. Forum. 2020;33:10–18. doi: 10.1177/0840470419873123. [DOI] [PubMed] [Google Scholar]
- 22.Yoldemir T. Artificial intelligence and women’s health. Climacteric. 2020;23:1–2. doi: 10.1080/13697137.2019.1682804. [DOI] [PubMed] [Google Scholar]
- 23.Siristatidis C, Pouliakis A. Artificial Intelligence in IVF: a need. Syst. Biol. Reprod. Med. 2011;57:179–185. doi: 10.3109/19396368.2011.558607. [DOI] [PubMed] [Google Scholar]
- 24.Lutomski, J. E., Meaney, S., Greene, R. A., Ryan, A. C. & Devane, D. Expert systems for fetal assessment in labour. Cochrane Database Syst. Rev.410.1002/14651858.CD010708 (2015).
- 25.Elgendi, M., Allaire, C., Williams, C., Bedaiwy, M. A. & Yong, P. J. Machine learning revealed new correlates of chronic pelvic pain in women. Front. Digit. Health2, 600604 (2020). [DOI] [PMC free article] [PubMed]
- 26.Wang L, et al. Identification biomarkers of eutopic endometrium in endometriosis using artificial neural networks and protein fingerprinting. Fertil. Steril. 2010;93:2460–2462. doi: 10.1016/j.fertnstert.2009.08.061. [DOI] [PubMed] [Google Scholar]
- 27.Nnoaham KE, Hummelshoj L, Kennedy SH, Jenkinson C, Zondervan KT. Developing symptom-based predictive models of endometriosis as a clinical screening tool: Results from a multicenter study. Fertil. Steril. 2012;98:692–701. doi: 10.1016/j.fertnstert.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouaziz, J. et al. How artificial intelligence can improve our understanding of the genes associated with endometriosis: natural language processing of the pubmed database. BioMed Res. Int.10.1155/2018/6217812 (2018). [DOI] [PMC free article] [PubMed]
- 29.Lee JH, Kwon SY, Chang J, Yuk JS. Machine learning approach to find the relation between endometriosis, benign breast disease, cystitis and non-toxic goiter. Sci. Rep. 2019;9:1–7. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matta K, et al. Associations between persistent organic pollutants and endometriosis: A multipollutant assessment using machine learning algorithms. Environ. Pollut. 2020;260:114066. doi: 10.1016/j.envpol.2020.114066. [DOI] [PubMed] [Google Scholar]
- 31.Knific T, et al. Multiplex analysis of 40 cytokines do not allow separation between endometriosis patients and controls. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-52899-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cosar E, et al. Serum microRNAs as diagnostic markers of endometriosis: a comprehensive array-based analysis. Fertil. Steril. 2016;106:402–409. doi: 10.1016/j.fertnstert.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 33.Domínguez F, et al. Lipidomic profiling of endometrial fluid in women with ovarian endometriosis. Biol. Reprod. 2017;96:772–779. doi: 10.1093/biolre/iox014. [DOI] [PubMed] [Google Scholar]
- 34.Vodolazkaia A, et al. Evaluation of a panel of 28 biomarkers for the non-invasive diagnosis of endometriosis. Hum. Reprod. 2012;27:2698–2711. doi: 10.1093/humrep/des234. [DOI] [PubMed] [Google Scholar]
- 35.Bendifallah S, et al. MicroRNome analysis generates a blood-based signature for endometriosis. Sci. Rep. 2022;12:4051. doi: 10.1038/s41598-022-07771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braga DPAF, et al. Metabolomic profile as a noninvasive adjunct tool for the diagnosis of grades III and IV endometriosis-related infertility. Mol. Reprod. Dev. 2019;86:1044–1052. doi: 10.1002/mrd.23221. [DOI] [PubMed] [Google Scholar]
- 37.Parlatan, U. et al. Raman spectroscopy as a non-invasive diagnostic technique for endometriosis. Sci. Rep.9, 1–7. 10.1038/s41598-019-56308-y (2019). [DOI] [PMC free article] [PubMed]
- 38.Ghazi N, et al. 1H NMR-based metabolomics approaches as non-invasive tools for diagnosis of endometriosis. Int J. Reprod. BioMed. 2016;14:1–8. [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Liu HY, Shi HH, Lang JH, Sun W. Urine peptide patterns for non-invasive diagnosis of endometriosis: a preliminary prospective study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2014;177:23–28. doi: 10.1016/j.ejogrb.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Fassbender A, et al. Proteomics analysis of plasma for early diagnosis of endometriosis. Obstet. Gynecol. 2012;119:276–285. doi: 10.1097/AOG.0b013e31823fda8d. [DOI] [PubMed] [Google Scholar]
- 41.Fassbender A, et al. Combined mRNA microarray and proteomic analysis of eutopic endometrium of women with and without endometriosis. Hum. Reprod. 2012;27:2020–2029. doi: 10.1093/humrep/des127. [DOI] [PubMed] [Google Scholar]
- 42.Dutta M, et al. A metabonomics approach as a means for identification of potential biomarkers for early diagnosis of endometriosis. Mol. Biosyst. 2012;8:3281–3287. doi: 10.1039/c2mb25353d. [DOI] [PubMed] [Google Scholar]
- 43.Wölfler MM, et al. Mass spectrometry and serum pattern profiling for analyzing the individual risk for endometriosis: promising insights? Fertil. Steril. 2009;91:2331–2337. doi: 10.1016/j.fertnstert.2008.03.064. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Zheng W, Mu L, Zhang SZ. Identifying biomarkers of endometriosis using serum protein fingerprinting and artificial neural networks. Int. J. Gynecol. Obstet. 2008;101:253–258. doi: 10.1016/j.ijgo.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 45.Vesale E, et al. Predictive approach in managing voiding dysfunction after surgery for deep endometriosis: a personalized nomogram. Int. Urogynecol. J. 2021;32:1205–1212. doi: 10.1007/s00192-020-04428-9. [DOI] [PubMed] [Google Scholar]
- 46.Benoit L, et al. Predicting the likelihood of a live birth for women with endometriosis-related infertility. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019;242:56–62. doi: 10.1016/j.ejogrb.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 47.Lafay Pillet MC, et al. A clinical score can predict associated deep infiltrating endometriosis before surgery for an endometrioma. Hum. Reprod. 2014;29:1666–1676. doi: 10.1093/humrep/deu128. [DOI] [PubMed] [Google Scholar]
- 48.Ballester M, et al. Nomogram to predict pregnancy rate after ICSI-IVF cycle in patients with endometriosis. Hum. Reprod. 2012;27:451–456. doi: 10.1093/humrep/der392. [DOI] [PubMed] [Google Scholar]
- 49.Chapron C, et al. Presurgical diagnosis of posterior deep infiltrating endometriosis based on a standardized questionnaire. Hum. Reprod. 2005;20:507–513. doi: 10.1093/humrep/deh627. [DOI] [PubMed] [Google Scholar]
- 50.Bendifallah S, et al. Machine learning algorithms as new screening approach for patients with endometriosis. Sci. Rep. 2022;12:639. doi: 10.1038/s41598-021-04637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang YF, et al. Mining medical data: A case study of endometriosis. J. Med. Syst. 2013;37:9899. doi: 10.1007/s10916-012-9899-y. [DOI] [PubMed] [Google Scholar]
- 52.Li B, Wang S, Duan H, Wang Y, Guo Z. Discovery of gene module acting on ubiquitin-mediated proteolysis pathway by co-expression network analysis for endometriosis. Reprod. BioMed. Online. 2021;42:429–441. doi: 10.1016/j.rbmo.2020.10.005. [DOI] [PubMed] [Google Scholar]
- 53.Akter, S. et al. GenomeForest: an ensemble machine learning classifier for endometriosis. AMIA Summits Transl. Sci. Proc. 33–42. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7233069/ (2020). [PMC free article] [PubMed]
- 54.Perrotta AR, et al. The vaginal microbiome as a tool to predict rASRM stage of disease in endometriosis: a pilot study. Reprod. Sci. 2020;27:1064–1073. doi: 10.1007/s43032-019-00113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akter, S. et al. Machine learning classifiers for endometriosis using transcriptomics and methylomics data. Front. Genet.10, 1–17. 10.3389/fgene.2019.00766 (2019). [DOI] [PMC free article] [PubMed]
- 56.Tamaresis JS, et al. Molecular classification of endometriosis and disease stage using high-dimensional genomic data. Endocrinology. 2014;155:4986–4999. doi: 10.1210/en.2014-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chattot, C. et al. ENDORECT: a preoperative score to accurately predict rectosigmoid involvement in patients with endometriosis. Hum. Reprod. Open2, 10.1093/hropen/hoz007 (2019). [DOI] [PMC free article] [PubMed]
- 58.Guo Z, Feng P, Chen X, Tang R, Yu Q. Developing preoperative nomograms to predict any-stage and stage III-IV endometriosis in infertile women. Front. Med. 2020;7:695. doi: 10.3389/fmed.2020.570483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guerriero S, et al. Artificial intelligence (AI) in the detection of rectosigmoid deep endometriosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2021;261:29–33. doi: 10.1016/j.ejogrb.2021.04.012. [DOI] [PubMed] [Google Scholar]
- 60.Reid S, Lu C, Condous G. Can we improve the prediction of pouch of Douglas obliteration in women with suspected endometriosis using ultrasound-based models? A multicenter prospective observational study. Acta Obstet. Gynecol. Scand. 2015;94:1297–1306. doi: 10.1111/aogs.12779. [DOI] [PubMed] [Google Scholar]
- 61.Maicas G, et al. Deep learning to diagnose pouch of Douglas obliteration with ultrasound sliding sign. Reprod. Fertil. 2021;2:236–243. doi: 10.1530/RAF-21-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stegmann BJ, et al. A logistic model for the prediction of endometriosis. Fertil. Steril. 2009;91:51–55. doi: 10.1016/j.fertnstert.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beam AL, Kohane IS. Translating artificial intelligence into clinical care. JAMA. 2016;316:2368–2369. doi: 10.1001/jama.2016.17217. [DOI] [PubMed] [Google Scholar]
- 64.Peters MD, et al. Guidance for conducting systematic scoping reviews. JBI Evid. Implant. 2015;13:141–146. doi: 10.1097/XEB.0000000000000050. [DOI] [PubMed] [Google Scholar]
- 65.Tricco AC, et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): checklist and explanation. Ann. Intern. Med. 2018;169:467–473. doi: 10.7326/M18-0850. [DOI] [PubMed] [Google Scholar]
- 66.Arksey H, O’malley L. Scoping studies: towards a methodological framework. Int. J. Soc. Res. Methodol. 2005;8:19–32. doi: 10.1080/1364557032000119616. [DOI] [Google Scholar]
- 67.Moher D, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015;4:1–9. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bird K, et al. Assessment of hypertension using clinical electrocardiogram features: a first-ever review. Front. Med. 2020;7:583331. doi: 10.3389/fmed.2020.583331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper.