Abstract
The alginate lyase-encoding gene (algL) of Azotobacter chroococcum was localized to a 3.1-kb EcoRI DNA fragment that revealed an open reading frame of 1,116 bp. This open reading frame encodes a protein of 42.98 kDa, in agreement with the value previously reported by us for this protein. The deduced protein has a potential N-terminal signal peptide that is consistent with its proposed periplasmic location. The analysis of the deduced amino acid sequence indicated that the gene sequence has a high homology (90% identity) to the Azotobacter vinelandii gene sequence, which has very recently been deposited in the GenBank database, and that it has 64% identity to the Pseudomonas aeruginosa gene sequence but that it has rather low homology (15 to 22% identity) to the gene sequences encoding alginate lyase in other bacteria. The A. chroococcum AlgL protein was overproduced in Escherichia coli and purified to electrophoretic homogeneity in a two-step chromatography procedure on hydroxyapatite and phenyl-Sepharose. The kinetic and molecular parameters of the recombinant alginate lyase are similar to those found for the native enzyme.
Alginates are linear polysaccharides composed of (1,4)-linked β-d-mannuronic acid and its C-5 epimer, α-l-guluronic acid. These uronic acids are arranged in block structures which may be homopolymeric (polymannuronic acid or polyguluronic acid) or heteropolymeric random sequence (15). The proportion and arrangement of the block structures vary greatly in alginates from different sources and determine the physical properties of the polymer, particularly the ability to form gels in the presence of divalent cations (21). Alginates are synthesized as cell wall components by brown seaweeds and as exopolysaccharides by two families of heterotrophic bacteria, Pseudomonadaceae and Azotobacteriaceae. Marine alga alginate is used widely in the food, pharmaceutical, textile, and oil industries. Bacterial alginate is an acetylated polymer of d-mannuronic and l-guluronic acids, and evidence has been presented for the location of O-acetyl groups at the 2 and/or 3 position of d-mannuronosyl residues (41). In their pioneering work, Lawson and Stacey (26) described the existence of two capsular polysaccharides in Azotobacter chroococcum, and, more recently, one of these exocellular polysaccharides was identified as an alginate (9). In cystic fibrosis patients, Pseudomonas aeruginosa produces alginate, which facilitates the attachment of the bacterium to tracheal mucins. The exopolysaccharide protects the microorganism from phagocytes and prevents antibiotic uptake. Consequently, it is a major pathogenic factor in these patients (3).
Alginates are degraded by a group of enzymes that catalyze the β-elimination of the 4-O-linked glycosidic bond, with formation of unsaturated uronic acid-containing oligosaccharides (6). Several of the bacteria that synthesize alginate-like polysaccharides also produce alginate lyases, but they cannot use the polymers as the sole carbon and energy source. Typically, alginate lyases have an absolute specificity for either d-mannuronic or l-guluronic acid at the nonreducing side of the bond to be cleaved but no limitation on the uronic acids at the reducing side (5). Although alginate lyase may prove useful in the elucidation of alginate molecular structure, its degrading action on alginate represents a drawback in the bioproduction of the polymer by fermentation. In studies of P. aeruginosa, it is necessary to look for inhibitors of alginate synthesis with potential use as therapeutic agents in cystic fibrosis patients. Therefore, research on this enzyme is important from different points of view.
In this paper, we describe the cloning of the algL gene encoding the alginate lyase from A. chroococcum and the purification and characterization of its product. We also report the conditions under which the enzyme can be overproduced in Escherichia coli and easily purified to electrophoretic homogeneity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
A. chroococcum ATCC 4412 was grown at 30°C on nitrogen-free Burk’s medium supplemented with 0.5% (wt/vol) sucrose as the sole energy and carbon source.
E. coli DH5α (Bethesda Research Laboratories) was used for all plasmid constructions, E. coli MC1061 (30) was used for gene library construction, and E. coli BL-21 (DE3) (40) was used as the host for pAPET-2 to produce alginate lyase. These strains were grown on Luria-Bertani (LB) and M9 minimal media containing ampicillin (100 μg/ml) as described by Sambrook et al. (37).
P. aeruginosa 8830, a gift from A. M. Chakrabarty (University of Illinois, Chicago), was grown as described by May and Chakrabarty (29) to isolate alginic acid, which was used as substrate for A. chroococcum alginate lyase.
Plasmid pRL500 (12) was used to construct the partial A. chroococcum gene library, and pBluescript II SK(+) (Stratagene) was used as cloning vector. Plasmid pET-3a, used for the expression of recombinant proteins, was from Novagen. pCL8, containing alginate lyase gene (algL) from P. aeruginosa (4), was a gift from A. M. Chakrabarty. Plasmids pAPL4.7 and pAPET-2, containing the algL gene from A. chroococcum, were constructed for this work.
DNA manipulation and Southern blot hybridization.
Total DNA from A. chroococcum was isolated as described by Ausubel et al. (1). All DNA manipulations and E. coli transformations were performed by standard procedures (37). DNA fragments were purified from agarose gels by using the GeneClean kit (Bio 101, Inc.). For Southern hybridization, DNA was digested and fragments were electrophoresed on 0.7% agarose gels with the Tris-borate-EDTA buffer system (37). DNA was transferred to Z-probe membranes (Bio-Rad) under vacuum, and Southern blot hybridizations were performed as described by Ausubel et al. (1). DNA probes were 32P labelled with a DNA-labelling kit (-dCTP) (Pharmacia) and [α-32P]dCTP (Amersham).
Cloning of the algL gene.
Genomic DNA from A. chroococuum was completely digested with EcoRI and fractionated on a 0.7% agarose gel. DNA fragments of approximately 2.0 to 4.0 kb were purified and ligated into the EcoRI site of pRL500. E. coli MC1061 was transformed with the ligation mixture, and transformants were grown on LB agar plates containing ampicillin. Transformants were screened for the presence of the algL gene by colony hybridization blotting with a 1.8-kb XbaI-EcoRI fragment from plasmid pCL8 as a probe.
DNA sequencing and nucleotide sequence analysis.
DNA fragments containing the algL gene were subcloned into pBluescript II SK(+) and sequenced by the dideoxy chain termination method (38) with M13 universal and reverse oligonucleotides or synthetic oligonucleotides as primers. Sequencing reactions were carried out with Sequenase version 2.0 (US Biochemical Corp.). Nested unidirectional deletions were generated with the double-stranded nested-deletion kit (Pharmacia LKB). Both strands of DNA were sequenced.
Computer sequence analysis was carried out with the Genetics Computer Group software package (10). Amino acid sequences were compared with the FASTA program, and alignments were produced with the PileUp program and default parameters (33).
Expression of AlgL and purification of alginate lyase.
To express AlgL protein, a PCR with plasmid template pAPL4.7 and primers designed to introduce the NdeI (5′-TATTCATATGAAGACCAGACTTGCCC-3′) and BamHI (5′-CTTCGGGATCCCTGCGGAATACCAG-3′) restriction sites encompassing algL was performed by using a final volume of 50 μl that contained 1.5 ng of DNA from plasmid pAPL4.7, 0.2 mM each deoxynucleoside triphosphate, 50 pmol of each oligonucleotide, and 2.5 U of Taq polymerase and 1× buffer (Boehringer).
An expected PCR product of 0.2 kb containing BamHI and NdeI sites was amplified. It was digested with NdeI and BamHI, ethanol precipitated, and ligated into a similarly digested and phosphatase-treated pET-3a expression plasmid. Possible constructs were confirmed by restriction digestion and sequencing.
To create pAPET-2, a 1.8-kb BstXI-EcoRV fragment from pAPL4.7 (containing the algL gene) was isolated and cloned in pET-3a (containing the PCR product) previously digested with BamHI, refilled by a Klenow reaction, and finally digested with BstXI. This plasmid contained the whole A. chroococcum algL gene under the control of the T7/lac promoter; in consequence, its expression was inducible by isopropyl-β-d-thiogalactopyranoside (IPTG).
E. coli BL-21 (DE3) cells were transformed with plasmid pAPET-2. Transformation mixtures were used to directly inoculate 2 liters of M9 minimal medium containing 100 μg of ampicillin per ml. The culture was incubated at 30°C to an absorbance at 550 nm (A550) of 0.5; at this time, T7 polymerase was induced by adding IPTG at a final concentration of 1 mM, and cells were incubated for another 3 h at 30°C. Then cells were collected by centrifugation (8,000 × g for 5 min), and the periplasmic fraction was prepared as described by Efterkhar and Schiller (11). The periplasmic proteins were loaded onto a hydroxyapatite column (1.8 by 14 cm) equilibrated with 20 mM potassium phosphate buffer (pH 7.5). The adsorbed alginate lyase activity was eluted with a linear gradient of 0.1 to 1.0 M potassium phosphate buffer (pH 7.5). Most active fractions were pooled and loaded onto a phenyl-Sepharose column (1 by 8 cm) equilibrated with 25 mM Tris-HCl buffer (pH 8.2) containing 1 M NaCl. The alginate lyase activity was eluted within the void volume of this column. Most active fractions were combined and used as the purified AlgL enzyme.
Alginate lyase assay.
As the substrate for the enzyme assay, alginic acid from either Macrocystis pyrifera (60% mannuronate; Sigma), A. chroococcum, or P. aeruginosa was used. Alginic acid from A. chroococcum was prepared as described by Jarman et al. (22), and that from P. aeruginosa was prepared as described by May and Chakrabarty (29). When needed, the substrate from P. aeruginosa was deacetylated before being used as substrate (16). Alginate lyase activity was quantitatively measured by the thiobarbituric acid method (42). Enzyme preparations (5 to 15 μg of total protein) in 50 mM Tris-HCl buffer (pH 7.5)–0.2 M MgCl2 containing substrate (0.1 mg) were incubated at 30°C for 15 min. One unit of enzyme activity is defined as the amount of enzyme required to generate 1 μmol of β-formylpyruvate per min. The protein concentration was measured by the method of Bradford (8) with bovine serum albumin (Sigma) as the standard.
Alginate lyase plate assay.
Alginate lyase-producing E. coli strains were identified by being grown at 37°C on LB plates containing 1% agarose, alginate from the seaweed M. pyrifera (2 mg/ml), and 100 μg of ampicillin per ml. The plates were stained by flooding with 10% (wt/vol) cetylpyridinium chloride, and clear zones of depolymerization on a white background were observed, indicating lyase activity (17).
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was done by the method of Laemmli (24). Enzymatic activity after SDS-PAGE and subsequent renaturation were visualized exactly as described previously (34). All PAGE runs were performed at room temperature with a Bio-Rad mini Protean slab gel apparatus.
Nucleotide sequence accession number.
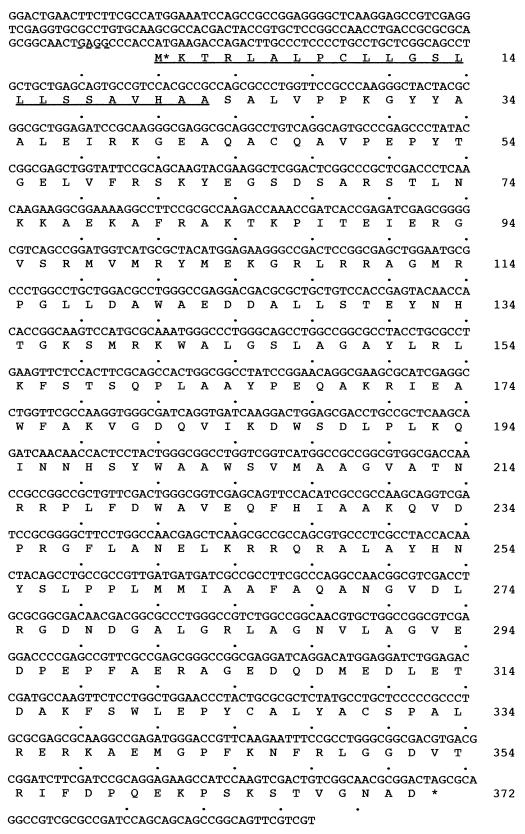
The nucleotide sequence of the A. chroococcum algL gene (see Fig. 3) has been deposited in the DDBJ, EMBL, and GenBank DNA databases under accession no. AJ223605.
FIG. 3.
Nucleotide sequence of A. chroococcum algL and deduced amino acid sequence of its product. The deduced amino acid sequence of AlgL is shown in single-letter code below the nucleotide sequence of algL. The possible Shine-Dalgarno sequence is indicated by a dotted underline. The initial methionine is indicated by an asterisk, and the possible signal peptide is indicated by a solid underline.
RESULTS AND DISCUSSION
Cloning of the A. chroococcum algL gene.
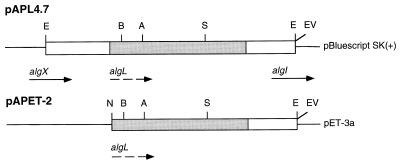
To clone algL, total genomic DNA from A. chroococcum was digested and subjected to Southern blotting with, as the probe, a 1.8-kb XbaI-EcoRI fragment from plasmid pCL8, containing the algL gene from P. aeruginosa (4). A 3.2-kb EcoRI fragment hybridized with this probe (data not shown). Total A. chroococcum DNA was then digested with EcoRI, and fragments of approximately 2.0 to 4.0 kb were purified and ligated to pRL500 to construct a partial A. chroococcum gene library. Transformants in E. coli MC1061 were screened for the presence of the algL gene by using the same probe (see Materials and Methods). A positive clone was analyzed. This clone contained a 3.1-kb EcoRI insert, which was cloned into pBluescript to generate plasmid pAPL4.7 for further analysis (Fig. 1).
FIG. 1.
Restriction map of pAPL4.7 and pAPET-2 containing the A. chroococcum algL gene. Thin lines and open bars correspond to vector plasmid DNA and cloned DNA fragments, respectively. The coding region is represented by solid bars. A broken arrow indicates the direction of the transcription of algL. E, EcoRI; A, ApaI; B, BstXI, S, SacI; N, NdeI; EV, EcoRV. Potential algX and algI genes upstream and downstream of algL gene are indicated by solid arrow underlines.
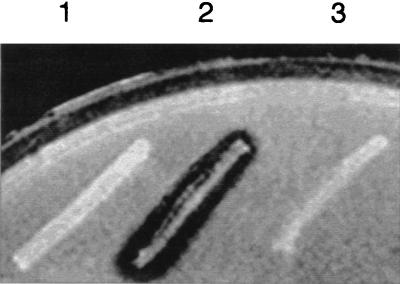
To determine whether this DNA fragment encoded the alginate lyase enzyme, E. coli DH5α cells were transformed with pAPL4.7 and tested for alginate lyase activity on LB plates containing alginate. A clear zone of depolymerization was observed around cells containing pAPL4.7. Untransformed E. coli DH5α did not synthesize an endogenous activity that was able to degrade alginate, and DH5α cells containing the pBluescript vector showed no clear zones (Fig. 2). Therefore, the 3.1-kb EcoRI fragment harbors the alginate lyase enzyme from A. chroococcum.
FIG. 2.
Plate assay for alginate lyase activity. Cells were restreaked on LB plates containing alginate and assayed for alginate lyase activity (see Materials and Methods). 1, untransformed E. coli DH5α cells; 2, E. coli DH5α cells containing pAPL4.7 plasmid; 3, E. coli cells containing pBluescript SK(+) plasmid.
Nucleotide sequence of the algL gene.
To determine if the 3.1-kb fragment contains the bonafide algL gene from A. chroococcum, nested deletions were generated from pAPL4.7 with exonuclease III and a number of the resulting clones were checked for alginate lyase activity to locate the algL gene in the pAPL4.7 plasmid and were used for sequencing. The complete nucleotide sequence of algL gene and its translation of the open reading frame into a 372-residue amino acid sequence are shown in Fig. 3. The coding region ends with a TAG stop codon and it encodes a polypeptide with a calculated molecular mass of 42.98 kDa. As reported previously, the molecular mass of the alginate lyase from A. chroococcum, determined by activity staining after SDS-PAGE and subsequent renaturation, is 43 kDa (34) and the molecular mass estimated by gel filtration is about 46 kDa (35). A potential Shine-Dalgarno (GAGG) sequence lies just 7 nucleotides upstream from the start site. In the region of DNA upstream of the ATG codon, sequences related to the −35 and −10 consensus promoter regions were not identified. The G+C content of the nucleotide sequence is 67.2%, in good agreement with the value found for the Azotobacter genes, which is within 65 to 68% (2).
When sequences contained in plasmid pAPL4.7 flanking the algL open reading frame were sequenced and compared with other nucleotide sequences deposited in the data banks, it was observed that these sequences showed homologies to those of P. aeruginosa algX and algI genes, which are constituents of the alginate biosynthesis operon (14, 31). These data suggest that the A. chroococcum algL gene is probably located in an operon and has, as flanking genes, the algX gene at its 5′ end and the algI gene at its 3′ end. The existence of an alginate biosynthesis operon has also been described for A. vinelandii (13). The sequence of the algXLIVFA operon from A. vinelandii (as appeared in the GenBank database) indicates that the alginate biosynthesis operon in this organism has the same structure as the one described in the present report for A. chroococcum. The flanking sequences of the A. chroococcum algL gene show a high homology to the recently reported sequence of the algXLIVFA operon from A. vinelandii.
Deduced amino acid sequence of AlgL.
The deduced protein sequence of AlgL was compared with entries in the GenBank and SWISS-PROT databases. Multiple alignment of several alginate lyase sequences indicates that A. chroococcum AlgL is more similar to P. aeruginosa AlgL (63.6% identity) than to K. pneumoniae AlgL (16.3% identity), P. alginovora ΔlgL (16.3% identity), or Photobacterium ΔlgL (18.8%). As mentioned above, the A. vinelandii AlgL deduced amino acid sequence (GenBank) is very similar (90% identity) to that from A. chroococcum.
The presence at the N-terminal region of a signal peptide of 20 to 30 amino acids, rich in hydrophobic residues, had been described for several alginate lyases (4, 27, 28, 39). This signal peptide has not been experimentally investigated in A. chroococcum, but the hydrophobicity profile deduced from the algL gene shows a highly hydrophobic region of 25 amino acids at the amino-terminal end. A potential cleavage site appears between amino acid residues Ala22 and Ala23. This zone has high homology to the amino acid sequence of the signal peptide described for P. aeruginosa. These data suggest the existence of a signal peptide in A. chroococcum alginate lyase and the maturing of this protein. The mature protein would then begin at Ala23. Processing and export through the inner membrane are consistent with the periplasmic location of AlgL in A. chroococcum (35) and its purification from the periplasmic fraction from E. coli.
Purification and characterization of recombinant alginate lyase.
The algL gene was expressed from the bacteriophage T7 polymerase promoter of pET-3a expression vector. The final construct, called pAPET-2, was transformed in E. coli BL21 (DE3), and the AlgL enzyme was produced following induction of T7 polymerase. The best conditions for the overexpression of the enzyme were direct culture inoculation from the transformation mixture, 30°C, initiation of induction by adding 1 mM IPTG at an A550 of 0.5, and an induction time of 3 h. These conditions prevented the overexpressed protein from aggregation into inclusion bodies.
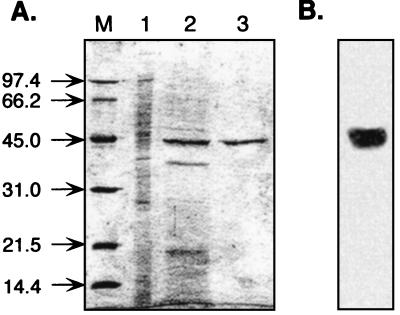
The gene product of algL was purified 10.4-fold from the periplasm fraction of E. coli BL21 (DE3)(pAPET-2), with a recovery of 6% by hydroxyapatite chromatography and phenyl-Sepharose column chromatography (Table 1). The final preparation gave a single band on SDS-PAGE with a molecular mass of approximately 43 kDa (Fig. 4). This value is in good agreement with the value estimated from the deduced amino acid sequence of AlgL and with the value determined by activity staining in SDS-PAGE for the native AlgL from A. chroococcum (34).
TABLE 1.
Purification of recombinant alginate lyase from A. chroococcum ATCC 4412a
| Step | Total amt of protein (mg) | Total activity (U) | Sp act (U/mg of protein) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Periplasmic fraction | 69.35 | 641.7 | 9.25 | 100 | 1 |
| Hydroxyapatite | 5.6 | 331.8 | 59.25 | 51.7 | 6.4 |
| Phenyl-Sepharose | 0.36 | 33.8 | 93.88 | 5.26 | 10.14 |
The enzyme was purified from 5 g (fresh mass) of E. coli BL-21 (DE3) containing plasmid pAPET-2.
FIG. 4.
SDS-PAGE analysis of the recombinant A. chroococcum AlgL protein. (A) Gel stained with Coomassie brilliant blue R-250; (B) alginate lyase activity detected on a gel containing 1% alginic acid. Lanes: M, molecular mass markers; 1, periplasmic fraction (10 μg); 2, hydroxyapatite column AlgL fraction (10 μg); 3, phenyl-Sepharose AlgL fraction (10 μg).
In agreement with other reports, which have described the partial purification of AlgL from both A. chroococcum and A. vinelandii (23), attempts to purify the native protein to homogeneity were unsuccessful even though a variety of purification steps were used. In this case, the recovery of partially purified enzyme (0.06 mg/liter of culture) was also lower than the obtained with the overexpressed alginate lyase (0.36 mg/liter).
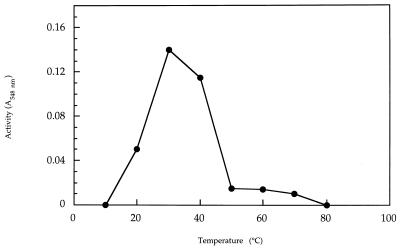
To characterize the purified enzyme, it was found that the optimum temperature for the recombinant enzyme assay is 30°C at pH 7.5 and that alginate lyase was inactive when analyzed at 70°C (Fig. 5). These values are rather similar to those for the native enzyme (data not shown).
FIG. 5.
Optimum temperature for the recombinant alginate lyase assay. Aliquots (25 μl) of an enzyme preparation (1.5 mU/μl) were analyzed for activity at the indicated temperatures, and the values found after a 30-min assay were spectrographically recorded at 548 nm by the thiobarbituric acid method.
The enzyme activity was measured in the presence of cations at 100 mM. Like the native protein (35), the activity of recombinant AlgL increased in the presence of Na+ and K+; the combined action of these cations and Mg2+ increased the activity 100-fold with respect to the control without added cations (Table 2). When these monovalent and divalent cations were used at concentrations in the range from 1 to 50 mM, the increase in alginate lyase activity was not significant. Other cations, like Ca2+, Co2+, Mn2+, and Zn2+ or the presence of EDTA does not affect the enzymatic activity. This enhancing effect of Mg2+ on alginate lyase activity has been reported for the enzyme from Klebsiella aerogenes (25), P. alginovora (7), and a marine bacterium that has not been identified (36). In all these cases, the presence of Mg2+ is stimulative but is not essential for the activity. There are other situations, however, where the cations are strictly required. Thus, Bacillus circulans JBH2 alginate lyase requires Mg2+ (18), and a Pseudomonas sp., P. aeruginosa, and Littorina sp. alginate lyase depends on Ca2+ (16). The enzyme from A. chroococcum 4A1M is activated by Ca2+ and inhibited strongly by Hg2+ (19). It should be mentioned that the enzyme from A. chroococcum 4A1M has a molecular mass of 23 kDa by SDS-PAGE and 24 kDa by gel filtration. Therefore, its molecular mass is roughly half that of the enzyme found in, and isolated by us from, A. chroococcum ATCC 4412. Furthermore, the enzyme from strain 4A1M showed maximum activity at 60°C whereas the one used in the present work has maximum activity at 30°C.
TABLE 2.
Effect of different monovalent and divalent cations on alginate lyase activity assaya
| Cation added | Alginate lyase activity (A548) |
|---|---|
| None | 0.010 |
| K+ | 0.173 |
| Na+ | 0.014 |
| Mg2+ | 0.017 |
| K+ plus Mg2+ | 0.927 |
| Mg2+ plus Na+ | 0.018 |
| K+ plus Mg2+ plus Na+ | 1.059 |
Aliquots of a purified A. chroococcum alginate lyase preparation (30 mU) were assayed for enzyme activity in the presence of the indicated cations at 100 mM (final concentration) each. Other experimental conditions were as in the thiobarbituric acid assay, except that 10 mM Tris-HCl buffer (pH 7.5) was used.
The Km value for the recombinant A. chroococcum alginate lyase was found to be 0.08 mM for uronic acids, which is in the range from 0.1 to 0.5 mM reported for other alginate lyases (16). From a Lineweaver-Burk double-reciprocal plot, a Vmax value of 0.183 mM min−1 was calculated for the enzyme.
To study the substrate specificity, the recombinant alginate lyase was used to degrade a series of alginates and the activity was assayed. AlgL was 17-fold more active against alginate from the seaweed M. pyriferia than against that from P. aeruginosa. The enzymatic activity against the A. chroococcum alginate was lower than with both above-mentioned substrates. Alginate from P. aeruginosa was 37% acetylated, but that from M. pyriferia was completely deacetylated. Therefore, it is likely that the acetylation of the bacterial alginate confers some protection against depolymerization by alginase. In favor of this conclusion is the fact that after chemical deacetylation of the Pseudomonas alginate, A. chroococcum AlgL increased its activity around 10-fold.
Finally, the possible roles of alginate lyases in some environments and several potential applications for both the oligosaccharides derived from the action of alginase and the enzyme itself have recently been discussed (41). Along these lines, Murata et al. (32) reported that they obtained an alginate lyase in large quantity and suggested its possible value as a therapeutic agent for the treatment of cystic fibrosis patients infected with mucoid P. aeruginosa. In fact, the use of alginate lyases to degrade the alginate in the lungs of cystic fibrosis patients, allowing a major diffusion of antibiotic, has been recently reported (20). The alginate lyase from A. chroococcum could be used as therapeutic agent because, as mentioned above, it is able to degrade the alginate produced by P. aeruginosa.
ACKNOWLEDGMENTS
This work was supported by financial help from Plan Andaluz de Investigación and Consejo Superior de Investigaciones Científicas (Spain). A. Pascual was supported by a fellowship from the Spanish Ministry of Education.
We thank F. J. Cejudo and J. de la Cruz for critical reading of the manuscript.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1989. [Google Scholar]
- 2.Becking J H. The family Azotobacteraceae. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The prokaryotes. Berlin, Germany: Springer-Verlag KG; 1981. pp. 794–817. [Google Scholar]
- 3.Boyd A, Chakrabarty A M. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl Environ Microbiol. 1994;60:2355–2359. doi: 10.1128/aem.60.7.2355-2359.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd A, Ghosh M, May T B, Shinaberger D, Koegh R, Chakrabarty A M. Sequence of the algL gene of Pseudomonas aeruginosa and purification of its alginate lyase product. Gene. 1993;131:1–8. doi: 10.1016/0378-1119(93)90662-m. [DOI] [PubMed] [Google Scholar]
- 5.Boyd J, Turvey J R. Isolation of a poly-α-l-guluronate lyase from Klebsiella aerogenes. Carbohydr Res. 1977;57:163–171. doi: 10.1016/s0008-6215(00)81928-x. [DOI] [PubMed] [Google Scholar]
- 6.Boyd J, Turvey J R. Structural studies of alginic acid using a bacterial poly-α-l-guluronate lyase. Carbohydr Res. 1978;66:187–194. [Google Scholar]
- 7.Boyen C, Bertheau Y, Barbeyront T, Kloare G B. Preparation of guluronate lyase from Pseudomonas alginovora for protoplast isolation in Laminaria. Enzyme Microb Technol. 1990;12:885–890. [Google Scholar]
- 8.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Cote G L, Krull L H. Characterization of the exocellular polysaccharides from Azotobacter chroococcum. Carbohydr Res. 1988;181:143–152. [Google Scholar]
- 10.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Efterkhar F, Schiller N L. Partial purification and characterization of a manuronan-specific alginate lyase from Pseudomonas aeruginosa. Curr Microbiol. 1994;29:37–42. [Google Scholar]
- 12.Elhai J, Wolk C. A versatile class of positive-selection vectors based on the nonviability of palindrome containing plasmid that allows cloning into long polylinker. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 13.Fialho A M, Zielinski N A, Fett W F, Chakrabarty A M, Berry A. Distribution of alginate gene sequences in the Pseudomonas rRNA homology group I-Azomonas-Azotobacter lineage of superfamily B procaryotes. Appl Environ Microbiol. 1990;56:436–443. doi: 10.1128/aem.56.2.436-443.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin M J, Ohman D E. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J Bacteriol. 1996;178:2186–2195. doi: 10.1128/jb.178.8.2186-2195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gacesa P. Alginates. Carbohydr Polym. 1988;8:161–182. [Google Scholar]
- 16.Gacesa P. Enzymic degration of alginates. Int J Biochem. 1992;24:545–552. doi: 10.1016/0020-711x(92)90325-u. [DOI] [PubMed] [Google Scholar]
- 17.Gacesa P, Wusteman F S. Plate assay for simultaneous detection of alginate lyase and determination of substrate specificity. Appl Environ Microbiol. 1990;56:2265–2267. doi: 10.1128/aem.56.7.2265-2267.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen J B, Doubet R S, Ram J. Alginase enzyme production by Bacillus circulans. Appl Environ Microbiol. 1984;47:704–709. doi: 10.1128/aem.47.4.704-709.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haraguchi K, Kodama T. Purification and properties of poly(β-d-mannuronate) lyase from Azotobacter chroococcum. Appl Microbiol Biotechnol. 1996;44:576–581. [Google Scholar]
- 20.Hatch R A, Schiller N L. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:974–977. doi: 10.1128/aac.42.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haug A, Larsen B, Baardseth E. Comparison of the constitution of alginates from differents sources. Proc Int Seaweed Symp. 1969;6:443–451. [Google Scholar]
- 22.Jarman T R, Deavin L, Scolombe S, Righelato R C. Investigation of the effect of environmental conditions on the rate of exopolysaccharide synthesis in Azotobacter vinelandii. J Gen Microbiol. 1978;107:59–64. [Google Scholar]
- 23.Kennedy L, McDowell K, Sutherland I W. Alginases from Azotobacter species. J Gen Microbiol. 1992;138:2465–2471. [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lange B, Wingender J, Winkler U K. Isolation and characterization of an alginate lyase from Klebsiella aerogenes. Arch Microbiol. 1989;152:302–308. doi: 10.1007/BF00409667. [DOI] [PubMed] [Google Scholar]
- 26.Lawson G L, Stacey M. Immunopolysaccharides. I. Preliminary studies of a polysaccharide from Azotobacter chroococcum containing an uronic acid. J Chem Soc. 1954;1954:1925–1931. [Google Scholar]
- 27.Maki H, Mori A, Fujiyama K, Kinoshita S, Yoshida T. Cloning, sequence analysis and expression in Escherichia coli of a gene encoding an alginate lyase form Pseudomonas sp. OS-ALG-9. J Gen Microbiol. 1993;139:987–993. doi: 10.1099/00221287-139-5-987. [DOI] [PubMed] [Google Scholar]
- 28.Malissard M, Duez C, Guinand M, Vacheron M-J, Michel G, Marty N, Joris B, Thamm T, Ghuysen J-M. Sequence of a gene encoding a (polyManA) alginate lyase active on Pseudomonas aeruginosa alginate. FEMS Microbiol Lett. 1993;110:101–106. doi: 10.1111/j.1574-6968.1993.tb06302.x. [DOI] [PubMed] [Google Scholar]
- 29.May T B, Chakrabarty A M. Isolation and assay of Pseudomonas aeruginosa alginate. Methods Enzymol. 1994;235:295–304. doi: 10.1016/0076-6879(94)35148-1. [DOI] [PubMed] [Google Scholar]
- 30.Meissner P S, Sisk W P, Berman M L. Bacteriophage 1 cloning system for the construction of directional cDNA libraries. Proc Natl Acad Sci USA. 1987;84:4171–4175. doi: 10.1073/pnas.84.12.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monday S R, Schiller N L. Alginate synthesis in Pseudomonas aeruginosa: the role of AlgL (alginate lyase) and AlgX. J Bacteriol. 1996;178:625–632. doi: 10.1128/jb.178.3.625-632.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murata K, Inose T, Hisano T, Abe S, Yonemoto Y, Yamashita T, Takagi M, Sakaguchi K, Kimura A, Imanaka T. Bacterial alginate lyase: enzymology, genetics and application. J Ferment Bioeng. 1993;76:427–437. [Google Scholar]
- 33.Pearson W, Lipman D. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peciña A, Paneque A. Detection of alginate lyase by activity staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequent renaturation. Anal Biochem. 1994;217:124–127. doi: 10.1006/abio.1994.1092. [DOI] [PubMed] [Google Scholar]
- 35.Peciña A. Ph.D. thesis. Seville, Spain: University of Seville; 1997. [Google Scholar]
- 36.Romeo A, Preston J F. Purification and structural properties of an extracellular (1→4)-β-d-mannuronan specific alginate lyase from a marine bacterium. Biochemistry. 1986;25:8385–8391. [Google Scholar]
- 37.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiller N L, Monday S R, Boyd C M, Keen N T, Ohman D E. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J Bacteriol. 1993;175:4780–4789. doi: 10.1128/jb.175.15.4780-4789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of clones genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 41.Sutherland I W. Polysaccharide lyases. FEMS Microbiol Lett. 1995;16:323–347. doi: 10.1111/j.1574-6976.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 42.Weissbach A, Hurwitz J. The formation of 2-keto-3-deoxy heptonic acid in extracts of Escherichia coli B. I and II. J Biol Chem. 1958;234:705–712. [PubMed] [Google Scholar]