Abstract
Neoantigen provides a promising breakthrough in tumor immunotherapy, although only a subset of patients responds well due to the quality of their immune response. However, few biomarkers have been reported to measure the quality of immune response to neoantigens and to predict prognosis of patients with multiple myeloma (MM). Here, we first developed a neoantigen-prediction pipeline starting from outcomes of somatic mutations and gene-expression profiles. Given the expression of some specific marker genes, the human leukocyte antigen (HLA)-I score and the cytolytic score were evaluated respectively to reflect HLA-I molecular expression and CD8+ T/natural killer (NK) cell abundance. According to the process of the immune response to neoantigens, we comprehensively took neoantigen load, cytolytic score, and HLA-I score to construct a neoantigen immune response score (NAIRscore), in which the HLA-I score presented a hazard ratio (HR) of less than 1, while the cytolytic score and neoantigen load presented a HR of greater than 1. Meanwhile, NAIRscore presented a competitive advantage to stratify MM samples. Especially, those exhibiting high NAIRscore correlated with an increased overall survival (OS), echoing the underlying molecular signatures of lower driver-gene mutations and down-regulated immune response. Notably, an online tool based on this study is provided to identify neoantigens and predict OS.
Keywords: neoantigen, neoantigen prediction pipeline, neoantigen immune response score, NAIRscore, quality of immune response to neoantigens, multiple myeloma, overall-survival prediction
Graphical abstract

To date, few biomarkers were reported to measure the quality of immune response to neoantigens and to predict prognoses of patients with MM. Jian et al. first provided a user-friendly online tool to evaluate neoantigen immune response score (NAIRscore), in which NAIRscore showed competitive advantage to relate with an increased OS.
Introduction
Multiple myeloma (MM) is the second most frequent hematological malignancy as characterized by the clonal expansion of malignant plasma cells within bone marrow, affecting >30,000 individuals per year in the United States.1,2 In recent decades, many explorations and advances in therapeutics have prolonged the survival for most patients with MM, but MM remains incurable and prone to relapse in the vast majority of cases.3, 4, 5, 6 The treatment of MM remains challenging, especially for relapsed/refractory patients. Due to clinical and pathological heterogeneity, response and survival in MM treatment vary to prognostic risk factors, such as disease stage at diagnosis and cytogenetic abnormalities.7,8 Therefore, correlating prognostic factors with biological function could enable the development of novel and targeted therapeutic classes.
In recent years, cancer immunotherapy with neoantigen vaccines has been considered a promising strategy for stimulating the body’s own immune system to eliminate tumor cells; however, only small subsets of patients within large cohorts favorably respond to the treatment due to the quality of their immune response.9,10 Neoantigens are a class of short peptides with aberrant expression in tumor cells, which result from somatic mutations and are used as tumor-specific antigens. Therein, neoantigens can bind to human leukocyte antigen (HLA) molecules and be presented on the surface, thereby being recognized as non-self by T cell receptors (TCRs) to elicit a T cell-mediated anti-tumor immune response.11,12 Previously, our team developed a customized proteogenomic workflow for neoantigen prediction and selection, i.e., ProGeo-neo,13 and we recently updated this tool (v.2.0) for expanding the source of candidate neoantigens.14 Also, we issued a manually curated database of human tumor neoantigen peptides, i.e., dbPepNeo,15 which helps screen immunogenic neoantigens from candidates. This database was updated recently to v.2.0.16 Besides, we have developed additional tools for identification of immunogenic neoantigens and for the subsequent design of candidate peptide vaccines.17, 18, 19
Several clinical trials have shown that personalized neoantigen vaccines have the ability to induce CD8+ and CD4+ T cell responses and improve the overall survival of patients with melanoma and glioblastoma, highlighting the considerable potential of neoantigen used in immunotherapy.20,21 In addition, neoantigen load was correlated with somatic mutations, and both were reported to relate to prolonged survival with checkpoint inhibitors in melanoma and lung cancer.22,23 High tumor mutational burden (H-TMB) has received FDA approval for selecting suitable patients for immunotherapy;24 however, little has been reported with regard to low-TMB cancers, such as MM. Studies have shown that the quality of immune response to neoantigens may act as a better biomarker for immunotherapy.10,25
Here, based on the 516 paired outcomes (i.e., Maf and FPKM files) of 478 patients with MM downloaded from the MMRF-CoMMpass project, we developed a neoantigen-prediction pipeline to identify neoantigens in each tumor. Then, to represent the quality of immune response to neoantigens, we took the neoantigen load, the cytolytic score, and the HLA-I score to construct a neoantigen immune response score (NAIRscore). A high NAIRscore was found to be associated with an increased overall survival. For ease of use by clinicians, based on this study, we have established a friendly web-based tool with URL at http://www.biostatistics.online/MMPrgognosis/.
Result
Higher mutation burden and neoantigen load in recurrent patients compared with primary patients
In this study, based on the MMRF-CommPass project on the GDC data portal, we downloaded 1,092 simple nucleotide variation files (i.e., Maf) and 859 transcriptomic profiles (i.e., FPKM), respectively. These files were derived from whole-exon sequencing (WES) and RNA sequencing (RNA-seq) of CD138+ cells from patients with myeloma, respectively. As shown in Figure S1, after sample selection, a total of 516 matched Maf files and FPKM files derived from 478 patients with MM were retained, including 462 primary samples and 54 recurrent samples (Table 1). In particular, paired primary and recurrent samples from 32 patients with MM were simultaneously obtained.
Table 1.
Clinical characteristics of 516 samples used in this study
| Primary (n = 462) | Recurrent (n = 54) | |
|---|---|---|
| Age (years) | ||
| Mean | 65.2 | 68.1 |
| SD | 10.5 | 10.4 |
| Gender (%) | ||
| Male | 283 (61.3) | 26 (48.1) |
| Female | 179 (38.7) | 28 (51.9) |
| Race (%) | ||
| White | 462 (100) | 54 (100) |
| ISS stage (%) | ||
| I | 152 (32.9) | 14 (25.9) |
| II | 154 (33.3) | 17 (31.5) |
| III | 140 (30.3) | 22 (40.7) |
| NA | 16 (3.5) | 1 (1.9) |
SD, standard deviation; ISS, International Staging System; NA, not available.
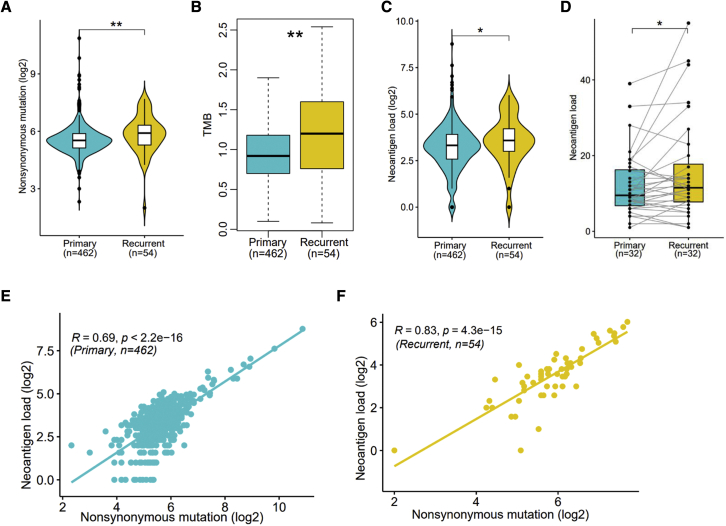
We first carried out analysis of somatic mutations in 516 Maf files using R package maftools.26 In 462 primary samples, a total of 27,953 nonsynonymous variations were detected, ranging from 5 to 1,864 variations per sample, with a median of 46 variations (Figure S2A). Similarly, in 54 recurrent samples, a total of 3,815 nonsynonymous variations were identified, ranging from 4 to 207 variations per sample, with a median of 60 variations (Figure S2B). Comparatively, more nonsynonymous mutations were found in recurrent samples (Figure 1A). And, in recurrent samples, significantly higher TMB was observed as well (Figure 1B). When compared with the 33 TCGA tumors cohorts by function “tcgaCompare” of R package maftools,26 MM expresses as a low-TMB tumor (Figure S3).
Figure 1.
Comparison of characteristics between recurrent and primary samples
(A–C) Comparison of nonsynonymous mutation (A), TMB (B), and neoantigen load (C) between primary and recurrent samples. (D) Comparison of neoantigen load between primary and paired recurrent samples. (E and F) The Pearson’s correlation between nonsynonymous mutations and neoantigen load in primary (E) and recurrent (F) samples. Significance was determined by Wilcoxon test. ∗p < 0.05, ∗∗p < 0.01.
As shown in Figure S1, a workflow for predicting neoantigens starting with the Maf file and the FPKM file was established through referring to the published neoantigen-prediction pipeline.27 As the nonsynonymous variations in chromosomes may lead to the changes in amino acid sequences of mutated genes in various forms, including single amino acid variation (SAAV), insertion or deletion of amino acids (indelAA), and frameshift mutation of amino acids (FSAA). Based on the mutated genes and their altered amino acids, the 8–11 amino acid short peptides containing those changed amino acid(s) were computationally cut from the reference amino acid sequence of mutated genes, and then their binding to HLA type was separately predicted for each patient. HLA-A·02:01 was applied to all patients with MM, as this HLA type is most prevalent among White people in the United States, according to the Allele Frequency Net Database (AFND).28
Statistically, a total of 6,329 neoantigens were identified in 462 primary samples, ranging between 1 and 437 per sample, with a median of 10. In 54 recurrent samples, a total of 887 neoantigens were identified, ranging between 1 and 65 per sample, with a median of 12. Here, we directly defined the number of neoantigens as neoantigen load in each sample. Comparatively, a significantly higher neoantigen load in recurrent samples was observed when compared with the primary samples (Figure 1C). Also, a higher neoantigen load was found when comparing those recurrent samples with their paired primary samples (Figure 1D). Furthermore, when we compared those samples diagnosed at different International Staging System (ISS) stages, no significance was observed on neoantigen load in primary and recurrent samples (Figures S4A and S4B), suggesting that the production of neoantigens has no significant correlation with ISS stage.
In addition, in both primary (Figure 1E) and recurrent (Figure 1F) samples, a significant positive correlation was discovered between the nonsynonymous mutations and neoantigen load. Additional nonsynonymous mutations may be responsible for relapse in recurrent patients. At the same time, in recurrent samples, more nonsynonymous mutations resulted in a relatively higher TMB, but this may also lead to production of more neoantigens. Consistent with the top mutated genes (Figures S2A and S2B), we found that the neoantigens’ peptides were predominantly derived from immunoglobulin heavy- and light-chain genes IRF4 and TP53.
A high NAIRscore was related to an increased overall survival
Neoantigens are considered a class of tumor-specific antigens that can bind to HLA moleculars and present on the surface of tumor cells, thereby being recognized specifically by T cells.12 Since only HLA-I neoantigens were predicted in this study, we took HLA-I molecular expression and CD8+ T cell abundance into consideration. In light of the expression level of some specific marker genes according to a prior study,29 the HLA-I score and the cytolytic score can be evaluated to represent HLA-I molecular expression and CD8+ T cell abundance in each sample, respectively. Marker genes B2M, HLA-A, HLA-B, and HLA-C were used to evaluate the HLA-I score, while marker genes GZMA, GZMH, PRF1, GNLY, and GZMM were used to calculate the cytolytic score.
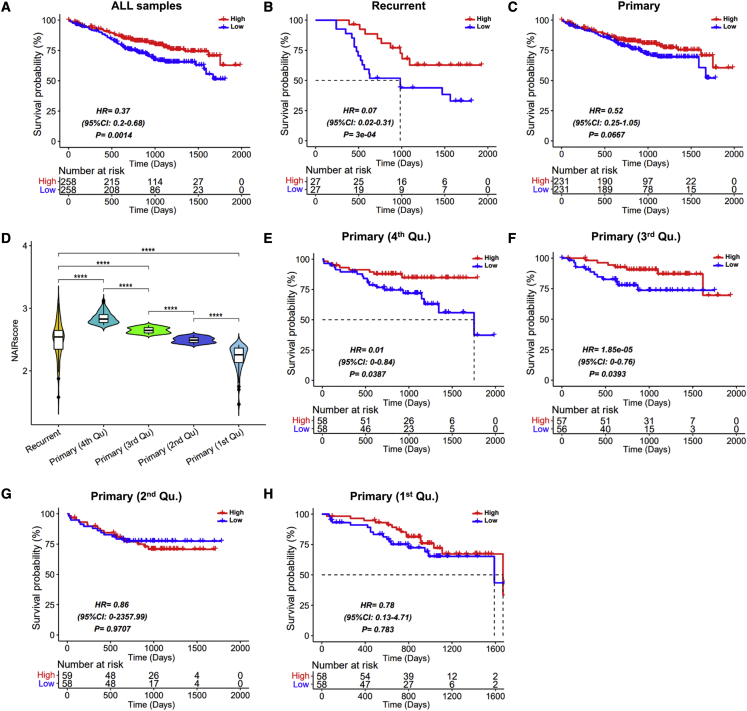
Given the process of immune response to neoantigens, we used the cytolytic score, the HLA-I score, and neoantigen load to construct a NAIRscore to measure the quality of immune response to tumor neoantigens by performing a multivariable Cox proportional hazards regression model. As shown in Figure 2A, we observed that the median of NAIRscore significantly stratified samples into high- and low-NAIRscore groups, and the high-NAIRscore group was associated with an increased overall survival (OS) (hazard ratio [HR] = 0.37; log rank test, p = 0.0014). Due to the significant difference in neoantigen load between primary and recurrent samples above, the NAIRscore was separately applied for sample stratification. With one accord, high NAIRscores were observed to relate to an increased OS in both recurrent (Figure 2B; HR = 0.07, p = 3e-04) and primary samples (Figure 2C; HR = 0.52, p = 0.0667), although there was no significance in primary samples.
Figure 2.
Prognostic analysis in different subsets of patients with MM
The median of NAIRscore in each prognostic model was used to divide samples into high- and low-NAIRscore groups. (A–C) Prognostic analysis in all samples (A), recurrent samples (B), and primary samples (C). (D) Comparison of NAIRscore between the recurrent samples and the four quartiles of primary samples. (E–H) Prognostic analysis in the fourth quartile (Qu.) (E), third Qu. (F), second Qu. (G), and first Qu. (H) in primary samples.
To further investigate the association between NAIRscore and OS in primary samples, we divided those primary samples into four quartiles based on the rank of the NAIRscore (Figure 2D). Interestingly, those primary samples in the third and fourth quartiles can both be divided significantly into high- and low-NAIRscore groups by the median NAIRscore, respectively (Figures 2E and 2F), and the high-NAIRscore group exhibited better OS. In contrast, those primary samples in the first and second quartiles cannot both be significantly stratified by the median of their NAIRscore (Figures 2G and 2H). However, when those primary samples in the first and second quartiles were further differentiated between high- and low-NAIRscore groups by the optimal p value, respectively, we also observed that the high-NAIRscore group exhibited better outcomes (Figures S5A and S4B). Indeed, the quality of immune response to tumor antigens can play a pivotal role in survival.10 In addition, we also considered that the NAIRscore has no significant correlation with the ISS stage, as the comparisons presented in Figures S4C and S4D reveal.
NAIRscore as a biomarker has a competitive advantage
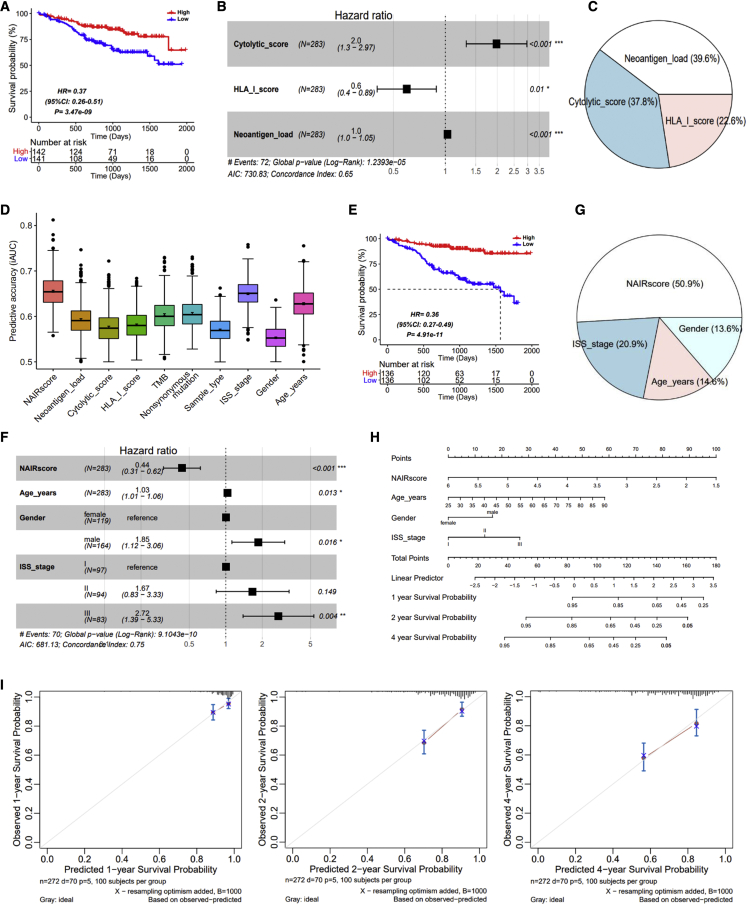
Considering the observation of significant stratification in recurrent samples and in the third and fourth quartiles of those primary samples above, we merged these samples to re-construct NAIRscore by re-computing a multivariable Cox regression model. Expectedly, these samples were significantly stratified into high- and low-NAIRscore groups, and the high-NAIRscore group was found to be associated with increased OS (Figure 3A). Consequently, in the subgroups of MM samples, i.e., ISS stage, gender, and age, NAIRscore can be used as an effective biomarker for sample stratification, and a high NAIRscore was associated with an increased OS (Figures S6A–S6C). In addition, in this fitted Cox model, a competitive concordance index was measured, and the three features were all significant (Figure 3B). Therein, an HLA-I score with an HR of less than 1 indicated that the higher expression of the HLA-I molecular expression, the more conducive to the patients’ OS. In contrast, a cytolytic score and neoantigen load with an HR of greater than 1 indicated that the higher value of cytolytic score and neoantigen load, the worse the patients’ OS. Meanwhile, the relative contribution of each variable in this model was estimated (Figure 3C), suggesting that the three variables all critically contribute to construct NAIRscore.
Figure 3.
Comparison of NAIRscore and other characteristics in a subset of samples, including the recurrent samples and the fourth Qu. and third Qu. primary samples
(A) Prognostic analysis based on NAIRscore. (B) The forest plot based on cytolytic score, HLA-I score, and neoantigen load. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. (C) The relative contribution of cytolytic score, HLA-I score, and neoantigen load in prognostic model. (D) Comparison of predictive accuracy on between NAIRscore and other features. (E) Prognostic analysis based on NAIRscore, age, gender, and ISS stage. (F) The forest plot based on NAIRscore, age, gender, and ISS stage. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. (G) The relative contribution of NAIRscore, age, gender, and ISS stage in prognostic model. (H) Nomogram for predicting the 1-, 2-, and 4-year OS probabilities. (I) Calibration curves for 1-, 2-, and 4-year OS.
Furthermore, by performing a 1,000× bootstrap resampling to calculate the integrated area under the receiver operating characteristic (ROC) curve (iAUC), we further compared the predictive accuracy of NAIRscore with other factors. As shown in Figure 3D, the iAUCs of NAIRscore and ISS stage were comparable, and both were superior to the other features. Interestingly, neoantigen load, TMB, and nonsynonymous mutation were found to yield the same iAUC, suggesting that they had similar predictive accuracy, possibly due to their correlation.
Moreover, combining NAIRscore with three clinical features, i.e., ISS type, gender, and age, we further carried out a prognostic analysis and fitted the SCORE. Uniformly, when the median of SCORE were used to divide those samples into high- and low-SCORE groups, much smaller p values were observed (Figure 3E). In the forest plot shown in Figure 3F, we observed a larger concordance index, indicating that the addition of clinical features improved the predictive power. Meanwhile, as shown in Figure 3G, NAIRscore was found to make a greater contribution to this model, followed by ISS stage, age, and gender. In addition, a nomogram was constructed for clinicians, illustrating that a quantitative probability can be evaluated in predicting 1-, 2-, and 4-year OS for an individual (Figure 3H). The calibration curves of the nomogram in predicting 1-, 2-, and 4-year survival rates were also validated (Figure 3I), suggesting that a good predictive robustness can be achieved using this nomogram. Also, the NAIRscore was observed to contribute the most risk points when compared with others, indicating that the NAIRscore was a more important feature.
Comparison of molecular characteristics between high- and low-NAIRscore groups
To explore the underlying molecular characteristics of different OSs between samples with high and low NAIRscores, we separately compared their somatic mutations and differentially expressed genes (DEGs). Primarily, driver genes in high- and low-NAIRscore groups were identified (Figures 4A and 4B). Meanwhile, genes NRAS, KRAS, and BRAF were considered as drivers in both high- and low-NAIRscore groups. In the low-NAIRscore group, however, we identified more driver genes, especially genes KRAS, TP53, and IRF4 with higher mutational frequency (Figure 4C), which may explain in part the poor prognosis in the low-NAIRscore group. Compared with wild-type samples, samples with KRAS mutant (Figure 4D) and samples with TP53 mutant (Figure 4E) all exhibited a decreased survival. Indeed, KRAS mutation was reported to relate to tumor-promoting inflammation and immune modulation, leading to immune escape in the tumor microenvironment.30,31 The loss or mutation of TP53 resulted in immune evasion by affecting the activity of myeloid and T cells and modulating the immune response during cancer development.32 The transcription factor IRF4 is required during immune response, including lymphocyte activation and the generation of immunoglobulin-secreting plasma cells.33
Figure 4.
Molecular characteristics between high- and low-NAIRscore groups
(A and B) The driver genes identified in samples of high- (A) and low-NAIRscore group (B). (C) The mutational waterfall of driver genes in samples of high- and low-NAIRscore groups. (D) Prognostic analysis in samples with KRAS mutant. (E) Prognostic analysis in samples with TP53 mutant. (F) The up- and down-regulated genes between high- and low-NAIRscore groups. (G) The GO enrichment analysis of the down-regulated genes, including biological process (BP), cellular component (CC), and molecular function (MF).
Moreover, the DEGs between high and lowNAIRscores were investigated as well. As shown in Figure 4F, we identified 14 up-regulated genes and 171 down-regulated genes in total (Table S1). In particular, genes GZMA, GZMH, GNLY, and PRF1 marked to the cytolytic score were down-regulated in the high-NAIRscore group. Therefore, lower cytolytic scores were evaluated in the high-NAIRscore group, which corresponded to the cytolytic score with an HR greater than 1 (Figure 3B). Further, these down-regulated DEGs in the high-NAIRscore group were applied for GO enrichment analysis (Table S2). Consistently, several significant biology processes (BPs) with regard to immune response were enriched (Figure 4G), such as T cell activation, immune-response-activating cell-surface receptor signaling pathway, antigen-receptor-mediated signaling pathway, and regulation of T cell activation. The cell component (CC) and molecular function (MF) also were enriched, including TCR complex, antigen binding, TCR binding, and major histocompatibility complex (MHC) class I protein binding. More specifically, these down-regulated immune response induced by neoantigens may lead to better survival in the high-NAIRscore group.
A web-based tool to identify neoantigens and predict OS for patients with MM
Based on the neoantigen-predictive pipeline proposed above and the prognostic model constructed by 516 patients with MM, we developed a web-based tool to identify neoantigens and predict the OS for each patient with MM (http://www.biostatistics.online/MMPrgognosis/). In this tool, users need to provide two text files from the same patient with MM patient, i.e., a Maf file to reflect somatic mutations in tumor and an FPKM file to represent expression level of genes in tumor. After uploading the two files, those neoantigens in the tumor can be identified within several minutes, and users can directly download results for further exploration, such as personalized neoantigen vaccines. In the meantime, users also can obtain the predictive OS table and risk curve.
In this tool, the scoring system of NAIRscore was fitted as follow: (−1 × 0.3222 × cytolytic score) + [−1 × (−0.2699) × HLA-I score] + (−1 × 0.0021 × neoantigen load), and the median NAIRscore in this study (2.5686) was used to divide patients into high- and low-NAIRscore groups. The patients with a NAIRscore greater than 2.5686 would exhibit a high OS rate. The OS rate at different time points could be calculated and presented in the table and curve. Moreover, if three clinical features, i.e., ISS stage, age, and gender, were provided by users, the scoring system would be further upgraded as follow: (−1 × 0.7009 × NAIRscore) + (−1 × 0.0316 × age) + [−1 × (0female | 0.3634male)] + [−1 × (0ISS-I | 0.9343ISS-II | 1.4124ISS-III)]. Similarly, the median SCORE (−1.3509) was used to stratify samples and predict OS. Importantly, this tool is easy to operate and is user friendly for clinicians, which is very helpful for the generalization and application of our models.
Discussion
In this study, an effective neoantigen-prediction pipeline starting from Maf and FPKM profiles was proposed and integrated into a web-based tool, in which neoantigens resulted from somatic mutations can be identified within minutes. It can serve as an alternative when raw sequencing data is not available. Here, consistent with a prior report,34 we found that larger mutational burden and neoantigens load in recurrent MM samples compared with the primary MM samples. This indicated that the larger mutational burden may be the cause of relapse in recurrent patients, as well as more neoantigen production.
According to the process of immune response to tumor antigens, we treated neoantigen load, the HLA-I score, and the cytolytic score as features to construct the NAIRscore, representing the quality of immune response to tumor antigens in samples. Meanwhile, we observed that neoantigen load and the cytolytic score have HRs of greater than 1, and the HLA-I score has an HR of less than 1. Consistently, neoantigen load has been reported as an HR of greater than 1, and high somatic mutation and neoantigen load were correlated with decreased progression-free survival in MM.35 These suggested that a higher cytolytic score resulting from neoantigen load did not yield a better prognosis in MM, although a high neoantigen load and tumor-infiltrating CD8+ T cells have been reported to imply a better survival in some solid tumors, such as melanoma and neuroblastoma.36,37 This might indicate a difference between low- and high-TMB tumors.
In addition, patients whose samples had a high NAIRscore were found to be significantly associated with a better OS, indicating that this is consistent with previous research that a high quality of immune response to tumor antigens was beneficial to prognosis.10 In particular, we found that the NAIRscore was able to significantly stratify those MM samples with a high NAIRscore. That suggests that a comprehensive consideration of immune response processes may better serve as a biomarker, rather than any one individually, i.e., neoantigen load, tumor-infiltrating CD8+ T cells, and HLA molecular expression. Indeed, in the high-NAIRscore group, we observed fewer driver genes, such as KRAS, TP53, and IRF4, and a down-regulated BP of immune response induced by neoantigens, such as immune-response-activating cell-surface receptor signaling pathway and antigen-receptor-mediated signaling pathway. Of course, in the future, NAIRscore needs to be validated in more MM samples, including taking monoclonal gammopathy of undetermined significance (MGUS) and smoldering MM (SMM) into account. Especially, NAIRscore needs to be verified and compared in more tumors, including high- and low-TMB tumors.
As we have known, H-TMB has received FDA approvals for selecting patients for promising immune checkpoint inhibitor (ICI) treatment. However, compared with TMB, neoantigen load was observed to be of equal advantage, while NAIRscore presented a greater advantage (Figure 3D). TMB refers to the number of somatic mutations per megabase of interrogated genomic sequence, while neoantigens denote those immunogenic short peptides originated from somatic mutations. To some extent, TMB and neoantigen load are all positively correlated with somatic mutations in tumor. Meanwhile, emerging research showed that the quality of the immune response is more important in tumors,10 which may explain why NAIRscore exhibits a better advantage.
In summary, we constructed NAIRscore for the first time to evaluate the quality of immune response to tumor neoantigens, in which a high NAIRscore was found to be an effective biomarker to relate with increased OS. A friendly web-based tool was implemented to identify neoantigens and predict prognosis for patients with MM.
Materials and methods
Sample collection and selection
In this study, open sequencing files of CD138+ cells from patients with MM were downloaded from the MMRF-CoMMpass project (dbGaP study accession: phs000748) through the GDC data portal (https://portal.gdc.cancer.gov/projects/MMRF-COMMPASS), including WES and RNA-seq (Figure S1). We obtained 1,092 simple nucleotide variation files (Maf format) derived from WES and 859 transcriptomic profiles (FPKM) derived from RNA-seq. Here, we only retained the matched WES and RNA-seq samples with clinical information. Further, we selected those files sequenced from bone marrow samples from White patients with MM. Eventually, a total of 516 matched Maf files and FPKM files derived from 478 patients with MM were retained, including 462 primary samples and 54 recurrent samples. Therein, the paired primary and recurrent samples from 32 patients with MM were obtained simultaneously.
Sequencing and statistical analysis
As shown in GDC, the tumor cells (CD138+ cells sorted from bone marrow) and normal cells (whole blood cells from peripheral blood) from matched samples were evaluated by WES. Based on the workflow of aliquot ensemble somatic variant merging and masking and human reference genome (GRCh38), the simple nucleotide variations of each tumor sample were separately filtered and saved in Maf format. On the R (v.4.0.3) platform, the mutated classifications and mutated genes in each Maf file were filtered and analyzed by R package maftools (v.2.6.05),26 the driver genes in different groups were implemented by the oncodrive function with mutations of greater than 8 and adjusted p values of less than 0.05.
In addition, the tumor cells were evaluated by RNA-seq, and the merged FPKM expression matrix was downloaded and used in this study. The FPKM matrix was applied for filtering of candidate neoantigens and evaluation of the cytolytic score and the HLA-I score.29 The FPKM matrix was used to screen DEGs between groups by performing Wilcoxon rank-sum test and Benjamini-Hochberg (BH) adjustment, in which we set the cutoff at the absolute value of log2, a fold change of greater than 1, and an adjusted p value of less than 0.05.
Neoantigen-prediction pipeline
Referring to the published neoantigen-prediction pipeline,27 as shown in Figure S1, we constructed a similar workflow for a neoantigen-prediction pipeline starting from Maf file and FPKM file. In this workflow, the mutated genes and their altered amino acids were first filtered from each Maf file. We downloaded the amino acid sequences of all human genes from UniprotKB. Then, based on those mutated genes and their altered amino acids, we can select those short peptides containing the mutated amino acids from their reference amino acid sequences. Only HLA-I neoantigens consisting of 8–11 amino acids were considered. Subsequently, the binding of these short peptides to HLA type was predicted by netMHCpan4.0,38 and we screened those short peptides with %Rank of less than 2. Eventually, the expression level of genes mapped by those screening short peptides in the paired RNA-seq files was applied for selection, and we selected those peptides with FPKM of greater than 2.
Somatic mutations in chromosomes may lead to the changes in amino acid sequences of mutated genes in various forms, including SAAV, indelAA, or FSAA. For SAAV, we first found the mutant site in the amino acid sequence of the reference gene and replaced it with the mutant amino acid. Then, we can theoretically cut the sequence into 8–11 amino acid short peptides containing the mutated position. Similarly, for indelAA, we found the mutant site in the amino acid sequence of the reference gene and inserted or deleted the mutant amino acid(s). Also, those 8–11 amino acid short peptides containing the mutant amino acid(s) can be cut from the sequence. For FSAA, all mutant sites in the amino acid sequence of the reference gene were first identified, and the sequence containing these mutation sites was cut into 8–11 peptides.
Due to lack of sequence files (e.g., fastq), we cannot accurately predict HLA types for each patient with MM. In this study, as only White patients with MM were selected, according to the AFND,28 we found that HLA-A·02:01 is the major HLA type among White people in the United States. Therefore, those short peptides were applied to predict the binding to HLA-A·02:01 by using netMHCpan4.0.
Evaluation of cytolytic score and HLA-I score
A previous report29 showed that the cytolytic score and the HLA-I score in hematologic tumors can be evaluated by the geometric mean of expression of several specific marker genes as follows:
in which i denotes the i-th marker genes. Accordingly, the marker genes GZMA, GZMH, PRF1, GNLY, and GZMM were used to calculate the cytolytic score, which is positively related to the cytolytic activity of CD8+ T/natural killer (NK) cells. The marker genes B2M, HLA-A, HLA-B, and HLA-C was used to detect the HLA-I score, which is significantly associated with the presentation of HLA-I neoantigens.
NAIRscore construction and prognostic analysis
In this study, we directly treated the number of neoantigens as neoantigen load in each sample. Taking neoantigen load, the cytolytic score, and the HLA-I score as features, a multivariable Cox proportional hazards regression model was constructed using the R package survival (v.3.2-13) and survminer (v.0.4.9), in which the coefficient (weight) of each feature can be generated in view of the importance of this feature. Meanwhile, NAIRscore can be constructed as follows:
in which i denotes the i-th feature, and Feature and Coef represent its value and coefficient in the fitted model, respectively. The NAIRscore was designed to reflect the quality of immune response to neoantigens in each sample. We applied the median of NAIRscore to stratify samples into high- and low-NAIRscore groups.
Further, when combining NAIRscore and three clinical features, i.e., ISS stage, age, and gender, the scoring system was further upgraded as follows:
in which i denotes the i-th feature. When the continuous features (i.e., NAIRscore, age) were applied, Feature and Coef represent its value and coefficient, respectively. When the feature was gender, “female” was treated as reference, and then “male” was applied by its Coef. When the feature was ISS stage, “ISS-I” was treated as reference, and then “ISS-II” and “ISS-III” were individually applied by their Coef.
The Kaplan-Meier survival curve was implemented by R package survminer (v.0.4.9), and the log rank test was used to compare the OS between high- and low-NAIRscore groups. The nomogram and calibration plots were visualized by using the R package rms (v.6.2-0).
To compare the predictive accuracy of the different variables or models, the iAUC with 100,00× bootstrap resampling was performed and analyzed. Referring to two previous studies,39,40 the relative contribution of each variable in the model was estimated by the proportion of overall χ2 from Harrell’s rms R package (v.6.2-0). The R package compareC (v.1.3.2) was used to compare if the difference in two correlated overall concordance indices were statistically significant.
Establishment of online web tool
An easy-to-use web tool starting from the paired Maf and FPKM files was developed, which can allow users to identify neoantigens in tumor and predict a patient’s OS. To implement an interaction interface and improve user experience, the front-end web framework was built by Bootstrap and Jquery, and the back-end was implemented by PHP.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31900102 and 31870829), China Postdoctoral Science Foundation (2019M652792), Shanghai Municipal Health Commission Collaborative Innovation Cluster Project (2019CXJQ02), and Natural Science Foundation of Hunan Province (2021JJ40995). We appreciate the High Performance Computing Center of Central South University. The authors would like to acknowledge Dr. Michael Liebman for his critical reading and editing of this manuscript.
Author contributions
X.J. and L.X. conceived and designed this study. X.J. carried out data collection and analysis and wrote the draft manuscript. L.X. revised the manuscript. L.X. contributed to building the web-based tool. W.Z., J.Z., and Y.W. contributed to data analysis and interpretation. The authors read and approved the final manuscript.
Declaration of interest
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.07.006.
Contributor Information
Xingxing Jian, Email: jianxingxing@foxmail.com.
Lu Xie, Email: xielu@sibpt.com.
Supplemental information
Significance was determined by Wilcoxon test, and P values were adjusted by BH method.
Those highlighted GO terms were presented in Figure 4G.
Data availability
All data used in this study were downloaded from project Multiple Myeloma CoMMpass Study (project ID: MMRF-COMMPASS; dbGaP study accession: phs000748) through the GDC data portal (https://portal.gdc.cancer.gov/projects/MMRF-COMMPASS).
References
- 1.Kuehl W.M., Bergsagel P.L. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J. Clin. Invest. 2012;122:3456–3463. doi: 10.1172/JCI61188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Roy M., Liang L., Xiao X., Peng Y., Luo Y., Zhou W., Zhang J., Qiu L., Zhang S., Liu F., et al. Lycorine downregulates HMGB1 to inhibit autophagy and enhances bortezomib activity in multiple myeloma. Theranostics. 2016;6:2209–2224. doi: 10.7150/thno.15584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia J., He Y., Meng B., Chen S., Zhang J., Wu X., Zhu Y., Shen Y., Feng X., Guan Y., et al. NEK2 induces autophagy-mediated bortezomib resistance by stabilizing Beclin-1 in multiple myeloma. Mol. Oncol. 2020;14:763–778. doi: 10.1002/1878-0261.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jian X., Zhu Y., Ouyang J., Wang Y., Lei Q., Xia J., Guan Y., Zhang J., Guo J., He Y., et al. Alterations of gut microbiome accelerate multiple myeloma progression by increasing the relative abundances of nitrogen-recycling bacteria. Microbiome. 2020;8:74. doi: 10.1186/s40168-020-00854-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng X., Guo J., An G., Wu Y., Liu Z., Meng B., He N., Zhao X., Chen S., Zhu Y., et al. Genetic aberrations and interaction of NEK2 and TP53 accelerate aggressiveness of multiple myeloma. Adv. Sci. 2022;9:e2104491. doi: 10.1002/advs.202104491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palumbo A., Avet-Loiseau H., Oliva S., Lokhorst H.M., Goldschmidt H., Rosinol L., Richardson P., Caltagirone S., Lahuerta J.J., Facon T., et al. Revised international staging system for multiple myeloma: a report from international myeloma working group. J. Clin. Oncol. 2015;33:2863–2869. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greipp P.R., San Miguel J., Durie B.G.M., Crowley J.J., Barlogie B., Bladé J., Boccadoro M., Child J.A., Avet-Loiseau H., Harousseau J.L., et al. International staging system for multiple myeloma. J. Clin. Oncol. 2005;23:3412–3420. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 9.Sambi M., Bagheri L., Szewczuk M.R. Current challenges in cancer immunotherapy: multimodal approaches to improve efficacy and patient response rates. J. Oncol. 2019;2019:e4508794. doi: 10.1155/2019/4508794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balachandran V.P., Łuksza M., Zhao J.N., Makarov V., Moral J.A., Remark R., Herbst B., Askan G., Bhanot U., Senbabaoglu Y., et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Lu M., Qin Y., Gao W., Tao L., Su W., Zhong J. Neoantigen: a new breakthrough in tumor immunotherapy. Front. Immunol. 2021;12:672356. doi: 10.3389/fimmu.2021.672356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng M., Mo Y., Wang Y., Wu P., Zhang Y., Xiong F., Guo C., Wu X., Li Y., Li X., et al. Neoantigen vaccine: an emerging tumor immunotherapy. Mol. Cancer. 2019;18:128. doi: 10.1186/s12943-019-1055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y., Wang G., Tan X., Ouyang J., Zhang M., Song X., Liu Q., Leng Q., Chen L., Xie L. ProGeo-neo: a customized proteogenomic workflow for neoantigen prediction and selection. BMC Med. Genomics. 2020;13:52. doi: 10.1186/s12920-020-0683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C., Zhang Y., Jian X., Tan X., Lu M., Ouyang J., Liu Z., Li Y., Xu L., Chen L., et al. ProGeo-Neo v2.0: a one-stop software for neoantigen prediction and filtering based on the proteogenomics strategy. Genes. 2022;13:783. doi: 10.3390/genes13050783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan X., Li D., Huang P., Jian X., Wan H., Wang G., Li Y., Ouyang J., Lin Y., Xie L. dbPepNeo: a manually curated database for human tumor neoantigen peptides. Database. 2020;2020:baaa004. doi: 10.1093/database/baaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu M., Xu L., Jian X., Tan X., Zhao J., Liu Z., Zhang Y., Liu C., Chen L., Lin Y., Xie L. dbPepNeo2.0: a database for human tumor neoantigen peptides from mass spectrometry and TCR recognition. Front. Immunol. 2022;13:855976. doi: 10.3389/fimmu.2022.855976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G., Li Y., Xie L. Advances in the prediction of antigenic peptides in personalized tumor neoantigen vaccine. Prog. Biochem. Biophys. 2019;46:441–448. [Google Scholar]
- 18.Zhao J., Tan X., Jian X., Wang G., Xie L. Bioinformatics prediction of candidate epitopes for peptide vaccines based on pathogen antigenic protein sequences. Life Sci. Res. 2021;25:363–371. [Google Scholar]
- 19.Wang G., Wan H., Jian X., Li Y., Ouyang J., Tan X., Zhao Y., Lin Y., Xie L. INeo-Epp: a novel T-cell HLA class-I immunogenicity or neoantigenic epitope prediction method based on sequence-related amino acid features. BioMed Res. Int. 2020;2020:5798356. doi: 10.1155/2020/5798356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ott P.A., Hu Z., Keskin D.B., Shukla S.A., Sun J., Bozym D.J., Zhang W., Luoma A., Giobbie-Hurder A., Peter L., et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keskin D.B., Anandappa A.J., Sun J., Tirosh I., Mathewson N.D., Li S., Oliveira G., Giobbie-Hurder A., Felt K., Gjini E., et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L., Sucker A., Hillen U., Foppen M.H.G., Goldinger S.M., et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S., et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcus L., Fashoyin-Aje L.A., Donoghue M., Yuan M., Rodriguez L., Gallagher P.S., Philip R., Ghosh S., Theoret M.R., Beaver J.A., et al. FDA approval summary: pembrolizumab for the treatment of tumor mutational burden-high solid tumors. Clin. Cancer Res. 2021;27:4685–4689. doi: 10.1158/1078-0432.CCR-21-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aurisicchio L., Pallocca M., Ciliberto G., Palombo F. The perfect personalized cancer therapy: cancer vaccines against neoantigens. J. Exp. Clin. Cancer Res. 2018;37:86. doi: 10.1186/s13046-018-0751-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hundal J., Carreno B.M., Petti A.A., Linette G.P., Griffith O.L., Mardis E.R., Griffith M. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8:11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Galarza F.F., McCabe A., Santos E.J.M.D., Jones J., Takeshita L., Ortega-Rivera N.D., Cid-Pavon G.M.D., Ramsbottom K., Ghattaoraya G., Alfirevic A., et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48:D783–D788. doi: 10.1093/nar/gkz1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dufva O., Pölönen P., Brück O., Keränen M.A.I., Klievink J., Mehtonen J., Huuhtanen J., Kumar A., Malani D., Siitonen S., et al. Immunogenomic landscape of hematological malignancies. Cancer Cell. 2020;38:424–428. doi: 10.1016/j.ccell.2020.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Hamarsheh S., Groß O., Brummer T., Zeiser R. Immune modulatory effects of oncogenic KRAS in cancer. Nat. Commun. 2020;11:5439. doi: 10.1038/s41467-020-19288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ischenko I., D'Amico S., Rao M., Li J., Hayman M.J., Powers S., Petrenko O., Reich N.C. KRAS drives immune evasion in a genetic model of pancreatic cancer. Nat. Commun. 2021;12:1482. doi: 10.1038/s41467-021-21736-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blagih J., Buck M.D., Vousden K.H. p53, cancer and the immune response. J. Cell. Sci. 2020;133:jcs237453. doi: 10.1242/jcs.237453. [DOI] [PubMed] [Google Scholar]
- 33.Shaffer A.L., Emre N.C.T., Lamy L., Ngo V.N., Wright G., Xiao W., Powell J., Dave S., Yu X., Zhao H., et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perumal D., Imai N., Laganà A., Finnigan J., Melnekoff D., Leshchenko V.V., Solovyov A., Madduri D., Chari A., Cho H.J., et al. Mutation-derived neoantigen-specific T-cell responses in multiple myeloma. Clin. Cancer Res. 2020;26:450–464. doi: 10.1158/1078-0432.CCR-19-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller A., Asmann Y., Cattaneo L., Braggio E., Keats J., Auclair D., Lonial S., MMRF CoMMpass Network. Russell S.J., Stewart A.K. High somatic mutation and neoantigen burden are correlated with decreased progression-free survival in multiple myeloma. Blood Cancer J. 2017;7:e612. doi: 10.1038/bcj.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spranger S., Spaapen R.M., Zha Y., Williams J., Meng Y., Ha T.T., Gajewski T.F. Up-regulation of PD-L1, Ido, and T(regs) in the melanoma tumor microenvironment is driven by CD8+ T cells. Sci. Transl. Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bao R., Spranger S., Hernandez K., Zha Y., Pytel P., Luke J.J., Gajewski T.F., Volchenboum S.L., Cohn S.L., Desai A.V. Immunogenomic determinants of tumor microenvironment correlate with superior survival in high-risk neuroblastoma. J. Immunother. Cancer. 2021;9:e002417. doi: 10.1136/jitc-2021-002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan 4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunother. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pagès F., Mlecnik B., Marliot F., Bindea G., Ou F.-S., Bifulco C., Lugli A., Zlobec I., Rau T.T., Berger M.D., et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 40.Ren J., Xu L., Zhou S., Ouyang J., You W., Sheng N., Yan L., Peng D., Xie L., Wang Z. Clinicopathological features combined with immune infiltration could well distinguish outcomes in stage II and stage III colorectal cancer: a retrospective study. Front. Oncol. 2021;11:776997. doi: 10.3389/fonc.2021.776997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Significance was determined by Wilcoxon test, and P values were adjusted by BH method.
Those highlighted GO terms were presented in Figure 4G.
Data Availability Statement
All data used in this study were downloaded from project Multiple Myeloma CoMMpass Study (project ID: MMRF-COMMPASS; dbGaP study accession: phs000748) through the GDC data portal (https://portal.gdc.cancer.gov/projects/MMRF-COMMPASS).