Abstract
Exosomes has attracted tremendous research interests as they are emerging as a new paradigm of liquid biopsy. Although the concentration of exosomes in blood is relatively abundant, there still exists various vesicle-like nanoparticles, such as microvesicles, apoptotic bodies. It's an urgent need to isolate and enrich exosomes from the complex contaminants in biofluid samples. Moreover, the expressing level of exosomal biomarkers varies a lot, which make the sensitive molecular detection of exosomes in high demand. Unfortunately, the efficient isolation and sensitive molecular quantification of exosomes is still a major obstacle hindering the further development and clinical application of exosome-based liquid biopsy. Nanomaterials, with unique physiochemical properties, have been widely used in biosensing and analysis aspects, thus they are thought as powerful tools for effective purification and molecular analysis of exosomes. In this review, we summarized the most recent progresses in nanomaterials assisted exosome isolation and analysis towards liquid biopsy. On the one hand, nanomaterials can be used as capture substrates to afford large binding area and specific affinity to exosomes. Meanwhile, nanomaterials can also be served as promising signal transducers and amplifiers for molecular detection of exosomes. Furthermore, we also pointed out several potential and promising research directions in nanomaterials assisted exosome analysis. It's envisioned that this review will give the audience a complete outline of nanomaterials in exosome study, and further promote the intersection of nanotechnology and bio-analysis.
Keywords: Liquid biopsy, Exosomes, Nanomaterials, Exosome isolation, Molecular detection
Graphical abstract
Nomenclature
Abbreviations
- CTCs
Circulating tumor cells
- ctDNA
Circulating tumor DNA
- EVs
Extracellular vesicle
- PEG
Polyethylene glycol
- COF
Covalent-organic framework
- MMP
Matrix Metalloproteinase
- EG
Endoplasmic reticulum
- PDAC
Pancreatic ductal adenocarcinoma
- NCs
Normal controls
- DLD
Deterministic lateral displacement
- 3D
Three-dimensional
- PDMS
Polydimethylsiloxane
- QDs
Quantum dots
- PS
Phosphatidylserine
- CDs
Carbon dots
- GO
Graphene oxide
- ssDNAs
Single-stranded DNA
- SERS
Surface-enhanced Raman spectroscopy
- GSSNTs
Gold-silver-silver core-shell-shell nanotrepangs
- TiN
Titanium nitride
- ZIC-HILIC
Zwitterionic hydrophilic interaction liquid chromatography
- MOFs
Metal-Organic Frameworks
- GBM
Glioblastoma
- FcNHSH
N-(2-mercaptoethyl) ferrocene carboxamide
- AuNPFe2O3NC
Gold-loaded ferric oxide nanocubes
- APTES
3-Aminopropyltriethoxysilane
- PDA
Polydopamine
- ELISA
Enzyme-linked immunosorbent assay
- GDH
Glucose dehydrogenase
- ZIF-8
Zeolitic imidazolate framework-8
- HCR
Hybridization chain reaction
- ExoADM
DNA molecular nanomachine
- BBA
10-benzyl-2-amino-acridone
- ECL
Electrochemiluminescence
- LSPR
Localized surface plasmon resonance
- GCDs
Gold-carbon quantum dots
- NPs
Nanoparticles
- SWCNTs
Single-walled carbon nanotubes
- MIO
Au-coated TiO2 macroporous inverse opal
- SPR
Surface plasmon resonance
- LDI-MS
Matrix-assisted laser desorption/ionization mass spectrometry
- PMO
Periodic mesoporous organosilica
1. Introduction
With the rapid development of precision medicine and individualized treatment, tissue biopsy has been broadly used in the diagnosis and prognosis of cancer [1,2]. However, tissue biopsy is suffering from the restricted methods for sample acquisition, poor accessibility to deep tumor tissue, intra-tumor heterogeneity, and difficulties in characterizing multiple tumor sites [3,4]. To overcome these shortcomings, many studies focusing on molecular analysis in liquid bio-fluids have emerged and gradually replaced traditional tissue biopsy. In liquid biopsy, circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and tumor-derived exosomes carrying tumor-specific information are released from tumor tissue into blood and other body fluids, which make tumor biopsy samples easily to be obtained in a non-invasive way [[5], [6], [7], [8]]. Meanwhile, through a series of morphological and molecular measurements as well as genetic characterizations, liquid biopsy can be used to look for new early diagnostic markers and therapeutic targets, monitor progression and prognosis of diseases [[9], [10], [11]]. Therefore, liquid biopsy has been considered a revolutionary technology in precision medicine, which will offer an attractive alternative for cancer diagnosis and treatment.
Among all the analysis targets in liquid biopsy, exosomes are rapidly emerging as a new paradigm in the past decade [[12], [13], [14], [15]]. Exosomes are small extracellular vesicle (EVs) with diameters between 30 and 150 nm and secreted by all cell types [16,17]. They are released by fusion of multivesicular endosomes with the plasma membrane, carry a cargo of proteins, nucleic acids and lipids from their parental cells [18,19]. Release of exosomes has been found to increase significantly in many cellular processes, including the most neoplastic cells and occurs continuously at all stages of tumor development [20]. Therefore, accumulation of the tumor-derived exosomes in blood and malignant effusions has been widely reported [21,22]. Growing evidences have shown that tumor derived exosomes carry characteristic protein and miRNA markers in various cancer types and the expression level of these molecules is correlated with tumor progression [[23], [24], [25]]. These exosomal markers may constitute a “cancer signature” to facilitate early detection and monitoring of disease progression in a non-invasive manner [26,27]. Meanwhile, exosomes and their cargos could be new targets for effective cancer therapy if their components and functions in regulating tumor origination and progression can be revealed [28,29]. To facilitate early detection and progression monitoring of tumors in a noninvasive manner, clear understanding on the biogenesis and functions of exosomes is desirable. Therefore, to deep insight into the tumor-derived exosomes, efficient isolation and molecular detection of exosomes are absolutely necessary.
Although the concentration of exosomes in blood is relative abundant, reach to ≥109 vesicles/mL, they still confront many contaminant vesicles, such as microvesicles, apoptotic bodies and so on. Recently, numerous efforts have been made toward the isolation of exosomes, including ultracentrifugation, polymer-based precipitation, affinity-based capture, and filtration. Currently the most commonly used method for exosome purification is ultracentrifugation, a method that includes differential centrifugation steps reaching speeds of up to 100 000×g [30,31]. Nevertheless, it always turns out to be inefficient with regards to low exosome yield (5–25% recovery rate) and involves a cumbersome procedure [32]. Polymer-based precipitation method, which relies on the formation of polymer network under specific conditions to entwines all components present in the sample and cause a decrease in solubility, always suffer from the interference of potential non-exosome contaminants and the polyethylene glycol (PEG) contained in samples are incompatible with the down-stream molecular analysis [33]. Affinity-related methods are widely available in form of various ready-to-use kits and affords highly selective separation of exosomes using specific antibodies, yet they are not applicable for large sample volumes and the isolated vesicles may lose the functional activity [34,35]. Besides, with regards to the recently developed filtration, exosomes are prone to adhere to the filtration membranes, thereby resulting in sample loss [36] Also, since the additional force is applied to pass the analyzed liquid through the membranes, the exosomes can potentially be deformed or damaged [37,38]. Therefore, the currently available methods for the isolation of exosomes are far from maturity and these technical challenges severely impede extensive biological and clinical studies of exosomes.
To overcome the current challenges and limitations of conventional methods, nanomaterials with unique physiochemical properties are emerging as powerful tools for effective purification and analysis of exosomes. The small size of nanomaterials enhances interaction with biological components at the micro-scale, resulting in good biocompatibility and biological activity [39,40]. In the past decades, the significant advances of nanotechnology have made it possible to controllably prepare nanomaterials, which not only exhibit unique interfacial, mechanical, optical and electromagnetic properties compared to macroscopic materials, but also enable them to perform a variety of functional adjustments to the in vivo microenvironment and micro-tissue [41,42]. Because of the excellent characteristics, nanomaterials have been widely used in biomedical fields in recent years, which provides a new way to explore the clinical utility of exosomes. Exosomes can be isolated effectively by nanomaterial through label-free methods, such as affinitive binding [43]. Nanomaterials functionalized devices can enahnce the isolation efficiency and performance than traditional approach through various synergistic interactions [44]. Meanwhile, benefiting from the variable chemical composition, morphology and physicochemical characteristics, nanomaterials have been served as various biosensors for exosome analysis with high sensitivity, good specificity, and low cost. Thus, nanomaterial provides a promising alterative for exosome isolation and molecular detection, leading to further application of exosome-based liquid biopsy.
In this review, we first provide an overview of the most recent research progresses (2017–2022) in nanomaterials assisted exosome isolation and analysis towards liquid biopsy. Different from the previous work, our review summarizes the most recent developments in designing nanomaterials assisted isolation techniques, which mainly cover phosphate groups directed metal oxides and nanocomposite as captures, as well as nano modified interface enhanced exosomes isolation. Meanwhile, nanomaterials assisted electrochemical probes, fluorescence probes etc. for exosomes analysis were also introduced. The future research perspectives and challenges in nanomaterials-based exosomes isolation and analysis in liquid biopsy are discussed.
2. Nanomaterials assisted exosome isolation
Although a lot of reviews about nanomaterials-based exosomes assay have been published, most of them only focused on the detection of exosomes. Actually, isolation and analysis of exosomes are two very important parts in exosomes assay. The isolation of exosomes from body fluids is an essential step for any exosome related scientific research and application. Highly efficient and specific exosomes isolation not only solve all the aforementioned problems of traditional exosomes isolation approaches, but also can realize accurate profiling of exosomes molecules and functions. Nanotechnology used in exosomes provides a facile, low cost, but highly efficient approach for exosomes isolation, which is of great value to facilitate the downstream analysis of exosomes in liquid biopsy. Nanomaterials mainly play two roles in exosomes isolation: as a capturer for exosomes isolation or as a nanointerface to improve the isolation efficiency of exosomes.
2.1. Nanomaterials based capturers for exosomes isolation
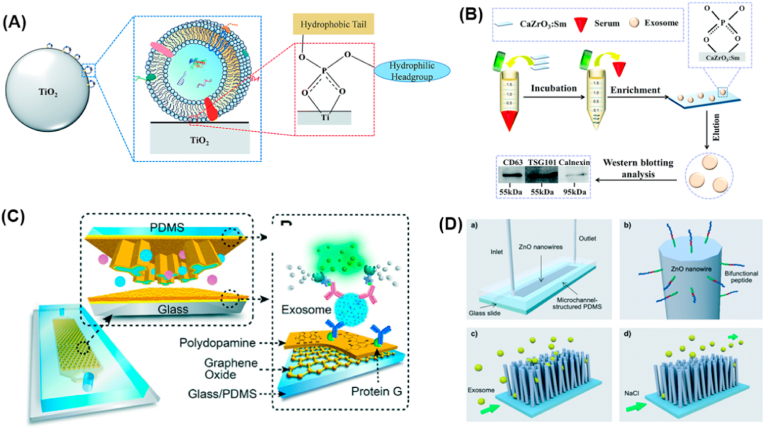
Some metal oxides, such as TiO2 and ZrO2, are widely used for highly selective enrichment of phosphorylated peptides via reversible and specifical binding with phosphate groups [[45], [46], [47]]. Based on this, such kind of metal oxides have been developed for effective exosome isolation. As shown in Fig. 1a, Gao and co-workers developed a novel strategy for facile serum exosome isolation based on specific interactions between phospholipid bilayers and TiO2 [43]. Benefiting from its simplicity and highly affinitive binding ability between TiO2 and exosomes, improved isolation efficiency, reduced nonspecific adsorption and shorter sample processing time were achieved. In addition, Sm-doped CaZrO3 nanosheets were applied for facile human serum exosome isolation by making the best of the high affinity of CaZrO3 with the phosphate groups on the lipid bilayer of exosomes (Fig. 1b) [48]. Compared with commercially available TiO2, Sm-doped CaZrO3 achieved a higher enrichment efficiency for target proteins CD63 and TSG101 than the endoplasmic reticulum (ER) marker Calnexin. Moreover, CaZrO3: Sm was stable and could be reused at least three times with satisfactory isolation efficiency which greatly reduce the cost.
Fig. 1.
Nanomaterials based exosomes isolation. (A) Mechanism of TiO2-based exosomes isolation [43]. (B) Schematic illustrations of the Sm-doped CaZrO3 nanosheets based exosomes isolation [48]. (C) Schematic of a single-channel PDMS/glass device, with the exploded-view highlighting the coated PDMS chip containing an array of Y-shaped microposts [57]. (D) Schematic illustrations of the peptide–nanowire interface within a microfluidic channel for capture and release of tumor-derived exosomes [58].
To achieve much higher efficiency of exosomes isolation, metal oxides functionalized hybrids were proposed. For instance, hydrophilic bimetallic magnetic MOF composite Fe3O4@UiO-66-NH2@PA-Ti4+ was synthesized and used for rapid capture of exosomes [49]. The presence of bimetals in the magnetic nanocomposites not only enhanced the chelating effect of the particles with the phospholipid bilayer, but also increased the hydrophilicity of the material to reduce non-specific adsorption of contaminated proteins, thereby improving the capturing efficiency of exosomes with low contamination. Due to the dynamic characteristic of urine, fast and efficient exosome isolation method is desirable. Thus, Fe3O4@TiO2-CD63 aptamer was proposed for rapid urine-derived exosomes isolation [50]. With the synergistic interactions of TiO2 with phosphate groups as well as aptamers with specific exosome proteins, 92.6% exosomes with intact structure were captured within 10 min and 999 proteins were detected through LC-MS/MS.
In general, most of designed hybrids can integrate multi-functions into one platform and gain satisfactory results. Therefore, some hybrids involved in exosomes isolation, detection and even molecules profiling have been developed and obtained ideal sensitivity and selectivity. In 2021, Li et al. reported a dual-functional Fe3O4@SiO2@TiO2 platform composed of reversible conjunction and “off-on” signal responses [51]. Taking advantages of the high affinity binding of TiO2 with phosphate groups on the lipid bilayer of exosomes and magnetic response property of Fe3O4, exosomes could be isolated within 20 min with a capture efficiency of 91.5% by Fe3O4@SiO2@TiO2 particles. In addition, a hairpin-like PSMA aptasensors was constructed with improved selectivity to detect tumor exosomes with a detection limit of 5 × 102 particles/μL in solution. To further characterize the molecules of exosomes, hybrids assisted exosomes isolation coupled with following analysis methods were also developed. Pang et al. introduced Fe3O4@TiO2 nanoparticles to enrich exosomes through the binding of TiO2 shell and hydrophilic phosphate head of the exosome phospholipids [52]. The captured exosomes can be isolated by magnet, and the isolation process can be finished within 5 min with a capture efficiency of 96.5%. And then anti-PD-L1 antibody modified Au@Ag@MBA SERS tags were added to label the exosomal PD-L1 for quantification with a LOD of 1 PD-L1+ exosome/μL by using 4 μL clinical serum sample. In addition, they also reported an in situ exosomal microRNA determination platform by target-triggered SERS and Fe3O4@TiO2 [53]. SERS tag-encapsulated exosomes can be enriched by Fe3O4@TiO2 and then concentrated by external magnet to enhance the Raman intensity. Thus, the combination of the hot-spot assemblies and the magnet concentration led to a dual increase in the Raman signal. Based on this platform, pancreatic ductal adenocarcinoma (PDAC) patients can be recognized from normal controls (NCs) with an accuracy of 99.6%.
2.2. Nanointerface enhanced exosomes isolation
Owing to the nanoscale of exosome, it is very important use the isolation material in the same scale. Nanomaterials assembled into various dimensions have shown great advantages in the applications of biomedical fields [54,55]. In recent years, nanomaterials assembled nanointerface with controlled structures and functions has been used for the isolation of exosomes. For example, deterministic lateral displacement (DLD) pillar arrays were developed for the separation of exosomes by using size-based exosome displacement [56]. DLD arrays with uniform gap sizes ranging from 25 to 235 nm was produced by silicon via double-stage lithographic process and e-beam lithography. Compared with conventional approaches for exosomes isolation, the sample volumes were small and exosomes labelling was not required. Moreover, the DLD technology also realized rapid and non-destructive exosomes sorting in a continuous flow with a single-exosome resolution. Such kind of progress not only explored the potential for on-chip exosomes separation and diagnostics, but also facilitated the manufacturable and scalable nanodevices for chip-based liquid biopsies.
Microfluidic devices are emerging as powerful technologies for efficient isolation of exosomes due to their outstanding ability in sample isolation and analysis [59]. During the past few years, numberless microfluidic devices with different structure have been developed for the isolation exosomes, such as Y shape, W shape etc. [59,60] However, the existing microfluidic devices do not have the desired nanoscale interactions with exosomes, resulting in low on-chip capture efficiency of exosomes. Besides, integrating all the steps for sample pretreatment, exosomes isolation and analysis into a single device is the ultimate goal of exosomes based liquid biopsy. Obviously, the traditional microfluidic technology could not integrate all the operations onto a single chip. Gradually, nanomaterials are used to modify the microfluidic device interface for exosomes isolation and downstream analysis, which is capable to improve mass transfer and reaction characteristics, reduce boundary effects. Due to the similar nanoscale of nanointerface with exosomes, not only efficient exosomes isolation was achieved, but also the sensitivity and selectivity of exosomes analysis in liquid biopsy have been improved significantly. For example, Zhang and co-workers developed a nano-IMEX microfluidic platform for exosome capture [57]. As demonstrated in Fig. 1c, the surface of the channel and Y-shaped microposts was coated with a nanostructured GO/PDA film by a layer-by-layer coating method to increase the surface area and antibody immobilization density. This design markedly expedited the PDA deposition kinetics which enhanced the anti-fouling property and provided numerous reactive sites for the covalent coupling of protein G to immobilize CD81 monoclonal antibodies in an oriented fashion. Such a sandwich exosome ELISA assisted with enzymatic signal amplification strategy afforded a detection limit as low as 80 aM with three marker cocktails. In 2018, Huang's group designed a ZnO nanowires coated three-dimensional (3D) polydimethylsiloxane (PDMS) scaffold chip device for effective immunocapture of exosomes [61]. The coated ZnO nanowire array increased the surface area for exosomes specific antibody immobilization as well as created size exclusion-like effect for exosomes retaining. Combining the fluid flow with chaotic or vortex feature of scaffold chip, efficient exosomes capture at a high flow rate can be achieved. Similarly, Tan group also introduced a ZnO nanowire modified microfluidic chip for exosomes isolation [62]. Unlike the design of Huang, they fabricated the herringbone microfluidic chip by maskless photolithography and ZnO nanowire arrays were anchored on the herringbone structure by chemical bathing. Such a design reduced the flow rates, resulting in the mixing and contact of exosomes with ZnO nanowire increased significantly. This easy-to-operate and low-cost nanointerface offered highly efficient exosomes capture, but the isolation method without specificity may lead to low purity in liquid biopsy application.
Exosome-based diagnostics and therapeutics have drawn great attentions in clinical application. Great efforts have been made to release the high purity of captured exosomes with complete structure and components. Suwatthanarak et al. reported multifunctional peptide-functionalized ZnO nanowires within a microfluidic chip for the isolation and release of exosomes [58]. The peptide consists of an exosome-binding site, a linker, and a ZnO-binding site. As shown in Fig. 1d, exosomes can be captured by the nanointerface specifically through the specific interaction between CD9 of exosome and P238 peptide on the nanointerface. As a result of multifunctional peptide modification, the proposed nanointerface significantly improved the exosome isolation efficiency by the synergistic interactions. Moreover, the captured exosome could be released from the nanointerface with neutral salt, a non-damaging condition to both nanowires and exosomes. Thus, exosomes can be trapped and released efficiently. Considering the precise diagnosis and treatment are very important in exosomes based liquid biopsy, a covalent chemistry-based hepatocellular carcinoma-specific exosomes purification and release chip was developed by Sun and co-workers [63]. Through the synergistic interactions of covalent binding, multimarker antibody cocktails, nanostructured substrates, and microfluidic chaotic mixers originated from the multi-functions of the chip, hepatocellular carcinoma derived exosomes can be captured efficiently and specifically. More importantly, the click chemistry-mediated specific exosomes capture can be released form the nanointerface by breaking the embedded disulfide bond. Such a versatile nanointerface provided an alternative foundation for developing nanomaterials modified microfluidic platforms for exosome-based preclinical study of specific diseases.
Apart from the specificity and efficiency of exosomes isolation increasing greatly, nanomaterials modified microchip interface also facilitates the integration of exosomes isolation and analysis into one platform. Zhang et al. developed a 3D-nanopatterned microfluidic chip with herringbone structure for ultrasensitive detection of tumor derived-exosomes [44]. Silica colloids or silica nanorods were used to fabricate the 3D-nanopatterned microfluidic chip through colloidal self-assembly strategy. The unique structure of chip facilitated the mass transfer of exosomes in microscale, improved the binding efficiency and speed, and decreased the near-surface hydrodynamic resistance. Combining with sandwich exosome enzyme-linked immunosorbent assay method, some potential markers of ovarian cancer can be detected from low levels of exosomes in plasma. Although the 3D-nanopatterned microfluidic chip shows its great potential in liquid biopsy for cancer diagnosis, cancer monitoring is also crucial to clinical implementation of precision medicine. To solve the current challenges in the monitoring of tumor progression and metastasis, the same group reported a 3D nanoengineering device fabricated by high-resolution colloidal inkjet printing method for molecules and functions analysis of exosomes [64]. Compared with the existing chips, boundary effects, fundamental limits in mass transfer, and surface reaction can be overcome by the 3D nanoengineering device, resulting in immense improvement of sensitivity for exosomes analysis. Distinct from conventional approaches, the 3D nanoengineering device integrates molecular and functional phenotyping of MMP14 for accurate cancer classification and tumor metastasis monitoring. Therefore, the developed nanointerface offered a useful liquid biopsy tool for longitudinal surveillance of tumor evolution in patients, and increased cancer management and precision medicine.
3. Nanomaterials enhanced exosomes analysis
Exosomes are thought to participate in tumor initiation, progression, and metastasis, which have been regarded as a promising biomarker for non-invasive cancer diagnosis and prognosis in liquid biopsy [[65], [66], [67], [68]]. To thoroughly reveal the clinical significance of circulating exosomes, a series of strategies have been developed for sensitive and multiplexed detection of exosomal biomarkers, such as proteins and nucleic acids. Conventional approaches including western blotting, enzyme-linked immunosorbent assay ELISA, and mass spectrometry etc. have been widely used for molecular analysis of exosomes [[69], [70], [71], [72]]. Nevertheless, these methods always turn out to be sample and time-consuming, as well as involve cumbersome procedures. Therefore, there is growing need for sensitive and reliable methods to probe tumor-derived exosomes rapidly and specifically, yet with modest requirements for sample volumes. Because of the intrinsic merits of biological compatibility, excellent physicochemical features and unique catalytic ability, numerous researchers attempt to establish novel nanomaterials based nanoprobes for exosomes analysis with improve high sensitivity and selectivity. To meet the increasing demands of ultrasensitive detection in liquid biopsy, nanomaterials have been integrated with various detectors as powerful nanoprobes for exosomes analysis. The advanced exosome analysis nanoprobes typically require only trace amounts of exosomes and short analysis time, relying on different detectors, such as electrochemistry, fluorescence etc.
3.1. Nanomaterials based electrochemical probes for exosomes analysis
Electrochemical sensors have been recognized as excellent approaches for bioanalysis mainly due to their advantages of convenient operation, rapid response, fast analysis, lost cost, sensitive recognition, easy miniaturization and portability, as well as high selectivity and simplicity [[73], [74], [75]]. Electrochemical analysis is achieved by monitoring the change of electrical conductivity of electrode. The signal is generated as a consequence of an electrochemical interaction between the electrode surface and the analyte [76]. Thus, effective electron transfer and direct attachment of biomolecules is in favor of improving the sensitivity of electrochemical detection. Benefiting from the interesting properties of nanomaterials, nanomaterial based-electrochemical sensor may provide a promising platform for exosomes detection. Nanomaterials, such as noble metals, quantum dots (QDs), metallic oxides, polymeric biomaterials and carbon-based materials have been used in electrochemical sensors, mainly act as electrode substrate to improve electron transfer efficiency or functional tags for signal amplification [[77], [78], [79]].
3.1.1. Nanomaterials act as electrode substrate to enhance exosomes analysis
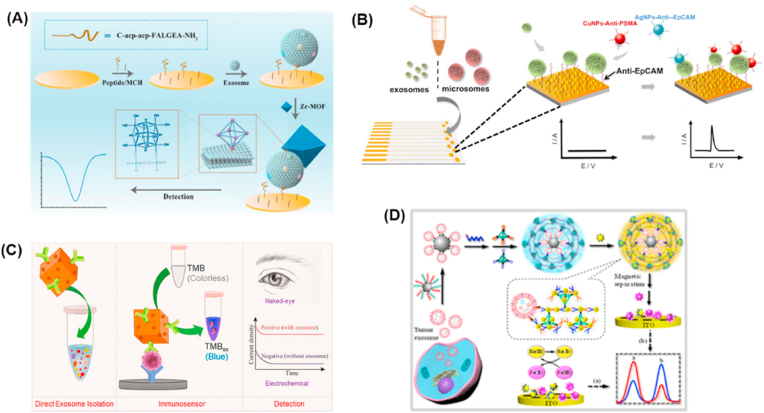
Due to the inherent redox properties of certain nanomaterials, electrochemical analysis can be achieved by directly monitoring the change of electrode conductivity caused by the change of electrical signal induced by the target. Such nanomaterial used in electrochemical sensor can provide real-time signal response, resulting in simple and rapid detection. In the past few decades, this type of sensor has attracted broad interests and offered distinctive advantages in food, environmental, medical, and other fields. Of course, such electrochemical sensors have also been successfully applied to the detection of exosomes. For example, Zr-Based Metal-Organic Frameworks (MOFs) based electrochemical sensor was developed for Glioblastoma (GBM)-derived exosomes analysis [80]. The specific design of the MOFs ensured the electrode surface to generate a high electrochemical signal. As shown in Fig. 2a, the captured exosomes can be directly quantified by monitoring the electrochemical signal inside Zr-MOF without extra recognition and amplification elements, while the sensor demonstrated very high sensitivity and selectivity. GBM-derived exosomes with detection range from 9.5 × 103 to 1.9 × 107 particles/μL and detection limit of 7.83 × 103 particles/μL was demonstrated. Taking advantage of the excellent conductivity of gold nanoparticles (Au NPs) and good supramolecular recognition ability of cucurbituril [7] towards ferrocene, a host-guest interaction based electrochemical sensor was reported for exosomes analysis with much lower detection limit [81]. Herein, cucurbit was self-assembled on the Au NPs composed electrodes, and then the captured exosomes by CD63 aptamer linked ferrocene was released from the electrode, thereby leading to the decrease of electrochemical signal. The host-guest based electrochemical sensor enables sensitive and selective electrochemical detection of exosomes with the detection limit of 482 particles/μL. To realize more sensitive exosomes analysis, Zhang and co-workers combined the g-C3N4 conjugated polydopamine coated Galinstan nanoprobes and multivalent PAIVIAM-AuNPs electrode interface for exosomes detection [82]. Benefiting from the excellent features of the Galinstan NPs in inhibiting the g-C3N4 passivation during the electrochemical reduction process and accelerating electron transfer, the antibody modified g-C3N4@Galinstan-PDA acted as both capturers of exosomes and signal probes for exosomes analysis. The work showed excellent performances in exosomes analysis with a detection limit of 31 particles/μL. In 2020, Kashefi-Kheyrabadi et al. realized ultra-low detection limit of 17 particles/μL over a wide dynamic range (1 × 102 to 1 × 109) particles/μL by using a detachable microfluidic device implemented with the electrochemical aptasensor [83]. The captured exosomes were determined by MoS2 nanosheets, chitosan, and graphene nanoplatelets fabricated gold electrode. Furthermore, Gao et al. developed a closed gold wireless nanopore electrode for single exosomes phenotyping, which solved the heterogeneity issue of exosomes analysis efficiently [84].
Fig. 2.
Nanomaterials enhanced electrochemical analysis of exosomes. (A) The principle of the electrochemical biosensor for the detection of GBM-derived exosomes [80]. (B) Schematic representation of the two-step isolation and analysis of exosomes and microsomes [86]. (C) Schematic representation of the Au–NPFe2O3NC based assay for direct exosome isolation and detection from cell culture media [90]. (D) Schematic illustration of the ratiometric immobilization-free electrochemical sensing system for tumor exosome detection [91].
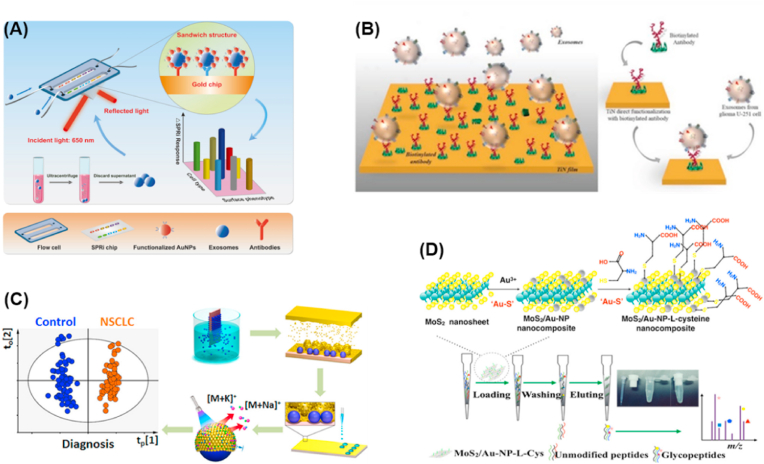
To obtain deep insight into the molecules and functions of exosomes, nanomaterials based electrochemical sensors were further used for exosomal proteins analysis. In 2018, a low cost and single-used gold screen-printed electrode was used for the research of lung cancer phosphoproteins with a sensitivity down to 15 ng/μL [85]. To search for much more cancer related biomarkers, Kelley's group reported a chip-based approach for electrochemical analysis of exosomal proteins [86]. As shown in Fig. 2b, gold and copper were chosen as probes for electrochemical readout due to their direct electro-oxidation of metal nanoparticles result in the potentials fall in the potential window of the gold electrodes, and the electrochemical signals were well separated, which allowed multi-protein detection at the same time. Combining the specific capture ability and excellent electrochemical features of the developed sensor chip, surface markers associated with PCa directly from minimally invasive serum samples were found and significant increase in the levels of EpCAM and PSMA expressed on exosomes was also observed. The work provides a successful nanomaterials-based electrochemical sensor for protein marker detection from tumor-derived exosomes and shows great potential in cancer diagnosis at early stage. To realize simultaneous determination of tumor exosomal proteins, Zhang's group developed magneto-mediated electrochemical sensor based on host-guest recognition for exosomal proteins analysis [87]. Stable complexes formed on screen-printed carbon electrode through the host-guest recognition between graphene oxide-cucurbit [7] and N-(2-mercaptoethyl) ferrocene carboxamide (FcNHSH), which effectively prevented the modification of exosomes on the electrode. In this way, four tumors derived-exosomal proteins were sensitively detected by the oxidation current signal of FcNHSH. The developed nanomaterials based electrochemical sensor shows great potential in clinical detection, early diagnosis and prognosis of breast cancer.
The selectively encapsulated microRNA by exosomes plays an important role in cell-to-cell communication, which have become reliable biomarker of disease progression. Therefore, sensitive detection of exosomal microRNAs will allow for non-invasive detection in liquid biopsy. Recently, multi covalent attachment p19 based electrochemical sensor was developed by Ghazizadeh et al. for exosomal microRNA analysis [88]. Due to the electrochemical induced reaction between [Fe(CN)6]−3/−4 and positive charged biomarkers, captured exosomes could covalently bind onto the electrode and make the sensor stable. The developed nanomaterials based electrochemical sensor showed high specificity and as low as 1 a.M. detection limit of exosomal miR-21. To overcome the limitations of unique structure and heterogeneity of exosomes, a highly integrated electrochemical platform was developed for accurate molecular profiling of tumorous exosomes [89]. Taking the advantage of self-assembly of ZIF-8 on DNA-modified electrodes, the proposed integrated strategy allowed low-abundant exosomes detection. The detection limit reached as low as ∼250 vesicles in 10 μL plasma sample. Furthermore, exosomal protein and RNA molecules can be simultaneously profiled with 100% sensitivity and specificity in a single sensor chip. The integrated electrochemical platform provides a significant tool for profiling tumorous exosome information in clinical cohorts.
3.1.2. Nanomaterials act as signal amplification element of electrochemical probes to enhance exosomes analysis
Apart from electrode substrates, nanomaterials can also act as nanoelectrocatalysts, nanocarriers and electroactive tags for signal amplification in electrochemical detection due to the inherent redox and electrocatalytic properties of the materials. Different signal amplification strategies have been proposed in electrochemical sensors, such as metal NPs, MOFs, and DNA-based amplification assays. For example, Boriachek and co-workers designed a multifunctional gold-loaded ferric oxide nanocubes (Au–NPFe2O3NC) for electrochemical detection of exosomes [90]. As shown in Fig. 2c, Au–NPFe2O3NC was employed for the following enzyme-linked immunosorbent assay (ELISA)-based sensing protocol due to the inherent peroxidase-like activity after accomplished exosomes isolation and transferred to screen-printed electrode. The captured exosomes were analyzed by naked-eye observation along with UV–visible and electrochemical sensor. Such a low-cost approach demonstrated good performance in the analysis of placental cell-derived exosomes, which enabled to detect 103 particles/mL with a relative standard deviation of <5.5%.
As a new type of material, MOFs used in electrochemical assay not only provide much more active sites for reactions but also served as nanocarriers to encapsulate other nanoparticles to increase the catalytic activity. For example, MOFs with dual functions was presented for sensitive exosomes analysis [92]. Herein, glucose dehydrogenase (GDH) and anodic enzyme were encapsulated by zeolitic imidazolate framework-8 (ZIF-8) to form GDH@ZIF-8 composites, in which the stability and catalytic activity of GDH improved significantly. On the other hand, another zirconium MOFs (UiO-66-NH2) loaded with electroactive molecules (K3[Fe(CN)6]) acted as nano-enrichment carriers to improve the capability of the cathode to transfer electrons from the anode. Benefiting from the synergistic effects of the two MOFs composites, the sensitivity of the as-proposed biosensor was further improved with 300 particle/mL detection limit. To realize much lower detection limit, a novel COFs-based nanoprobe named HRP-pSC4-AuNPs@COFs was designed and fabricated for colorectal cancer-derived exosomes detection [93]. Excellent conductivity of Au improved the response of electrochemical and a large amount of HRP loaded COFs endowed the composites with high catalytic activity. The COFs-based nanoprobe highlighted excellent analytical performance for CRC-derived exosomes in the linear range from 5 × 102 to 107 particles/μL with a detection limit down to 160 particles/μL.
Due to the predictability and programmability, DNA nanotechnology has become the most popular technology for signal amplification. In general, the sensors were formed by controllable novel nanoscale structures, which were constructed by using the specific molecular properties of DNA. In 2020, Yang et al. developed a dual-aptamer recognition system and hyperbranched DNA superstructure signal amplification strategy for direct quantification of tumor derived exosomes [91]. As shown in Fig. 2d, the sandwich-like complex was generated by activating DNA tetrahedron-based hyperbranched hybridization chain reaction after cholesterol-modified DNA probe was anchored on the captured exosomes. And then numerous Ru(NH3)63+ (Ru(III)) were immobilized on the sandwich complex resulting in the redox reaction between [Fe(CN)6]3– (Fe(III)) and Ru(II) was significantly prevented. In this way, obvious enhancement of IFe(III)/IRu(III) value was achieved. Consequently, highly reliable and accurate exosomes analysis was obtained. The developed signal amplification strategy also displayed good feasibility for clinical sample analysis. Benefiting from their large surface area and excellent biocompatibility, Au NPs have widely used as DNA carrier for signal amplification. In 2019, Huang et al. developed a hemin/G-quadruplex-assisted signal amplification approach for electrochemical analysis of gastric cancer exosomes [94]. Significant improvement of electrochemical signal on gold electrode was generated through the hemin/G-quadruplex DNAzyme induced H2O2 reduction and signal amplification produced by RCA reaction. This method allowed sensitive detection of exosomes in the range of 4.8 × 103 to 4.8 × 106 particles/μL with a detection limit of 9.54 × 102 mL−1. Similarly, An and co-workers developed a DNA hybridization chain reaction (HCR) for signal amplification and click chemistry-based electrochemical sensor for sensitive exosomes analysis with a detection limit of 96 particles/μL, which demonstrated great potential for exosome analysis in clinical samples and application in cancer diagnosis [95].
Not only sensitive quantification of exosomes can be achieved based on the nanomaterials induced signal amplification in electrochemical sensor, but also rapid and accurate profiling the exosomal molecules can be realized. For example, Jin et al. reported a bioinspired exosome-activated DNA molecular nanomachine (ExoADM) with multivalent cyclic amplification for the detection of exosomal markers [96]. Relying on the high specificity of aptamer-exosome recognition and cyclic amplification ability of ExoADM, the dynamic expression level changes of ExoPD-L1 and ExoCD63 were tracked simultaneously induced by signaling molecules. Moreover, they found the expression levels of the two proteins on exosomes could well differentiate cancer patients from normal individuals. In 2021, Park et al. reported an integrated magneto-electrochemical device for rapid characterization of cancer-derived exosomes [97]. Both exosomal proteins and microRNA can be profiled simultaneously by the integrated device, which effectively extended the clinical utility of exosomes for cancer diagnosis, recurrence monitoring and prognosis. MicroRNAs in tumor-derived exosomes have attracted rapidly growing interest due to their potential in cancer diagnostic and prognostic applications. Zhang et al. reported DNA walkers based ratiometric electrochemical biosensor for attomolar detection of exosomal miR-21 [98]. Moreover, Guo et al. developed a HCR based electrochemical sensor for sensitive detection of exosomal miR-122 [99]. Herein, the gold electrode was modified with hairpin DNA, and electroactive [Ru(NH3)6]3+ was used as an electrochemical signal reporter for signal amplification, resulting in significant increase of electrochemical signal. The proposed electrochemical assay also provided attomolar level detection of exosomal miR-122 with a linear range with 9 orders of magnitude in different tumor-derived exosomes.
3.2. Nanomaterials assisted photoelectrochemical probes for exosomes analysis
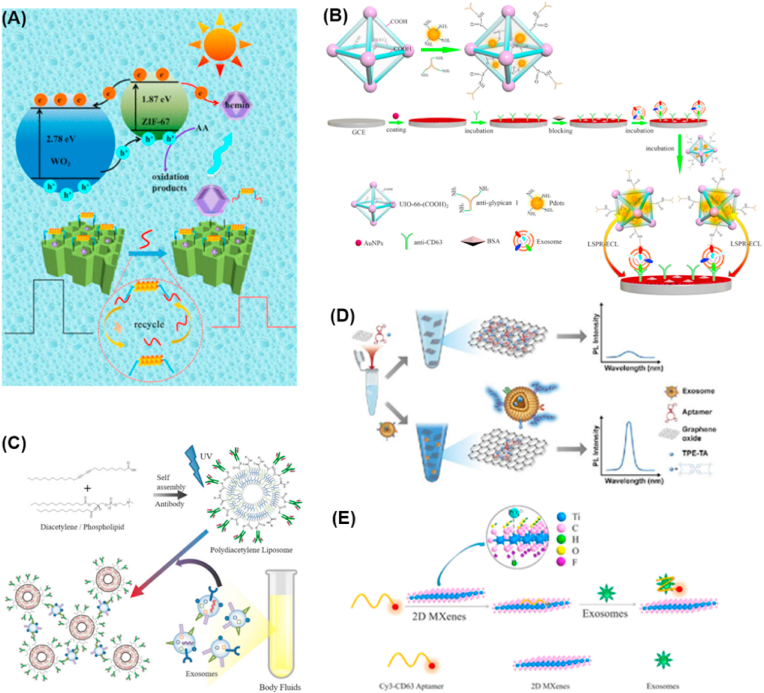
To further improve the sensitivity of exosomes analysis in liquid biopsy, photoelectrochemical sensors have become a rapid developing technology to replace the electrochemical assay [100,101]. In general, the specific recognition between photoelectrochemically active nanomaterials and target molecules under illumination will cause the corresponding change of electrical signal. Different from electrochemical assay, the excitation light from the detection signal is separate. Thus, photoelectrochemical assay not only inherits the advantages of electrochemical sensor, but also has many unique advantages, including excellent detectivity, stability and sensitivity due to its low background signal and high signal-to noise ratio [102,103]. Chen's group designed a 10-benzyl-2-amino-acridone (BAA) based multifunctional signal probe, which acted as a dual-modal aptasensor for photoelectrochemical detection of exosomes [104]. Interestingly, the released Cu2+ for exosome capture probe could inhibit the visible-light-induced oxidase mimic activity and photoelectrochemical activity of BAA simultaneously. Because of the changes of absorbance and photocurrent intensities are directly proportional to the concentration of exosomes, thereby exosomes can be quantified accurately with 1.38 × 103 particles/μL detection limit. In 2021, Pei et al. developed a NiO/BiOI/Au NP/CdSe composite based cathodic photoelectrochemical sensor for exosomes detection. Taking advantages of the synergistic effects of NiO/BiOI/Au NP/CdSe composite to increase photocurrent signal, the cathodic photoelectrochemical sensor showed good selectivity and sensitivity with 1.2 × 102 particles/μL detection limit in complex exosomes sample [105]. Furthermore, the photoelectrochemical biosensors was also used for the quantitation of exosomal microRNA. For example, Pang et al. constructed a TiO2 nanosilks (NSs)@MoS2 QDs based photoelectrochemical sensor and realized sensitive microRNA analysis with as low as 5 fg/mL detection limit [106]. Recently, Wang and co-workers presented a target miRNA-powered λ-exonuclease induced amplification strategy for exosomal RNA detection [107]. As shown in Fig. 3a, the photoelectrode was fabricated by MOFs-decorated WO3 nanoflakes. Meanwhile, both signal quencher and electronic mediator can be acted by the programmed release nanodevice with efficient target exosomal miRNA-responsive release profiles of hemin, leading to the quench of photocurrent. Due to the smart integration of these units to one device, the developed photoelectrochemical sensor yielded a highly sensitive assay for the detection of exosomal miRNAs with the detection limit as low as 0.5 fM.
Fig. 3.
(A) Schematic illustration for the detection mechanism of the MOFs-decorated WO3 nanoflakes based photoelectrochemical biosensing platform [107]. (B) Principle of the localized surface plasmon resonance between Au NPs and polymer dots based electrochemiluminescence immunosensor for pancreatic exosome detection [114]. Nanomaterials enhanced fluorescent analysis of exosomes (C–D). (C) Schematic illustration of the fabrication of polydiacetylene liposome immunosensor for exosome detection [116]. (D) Schematic illustration of the facile “turn-on” fluorescence aptasensor for exosome surface proteins profiling [117]. (E) Cy3-CD63 aptamer was mixed with MXenes aqueous solution and then added exosomes for the downstream analysis [118].
3.3. Nanomaterials assisted electrochemiluminescence probes for exosomes analysis
Electrochemiluminescence (ECL) is the combination of electrochemistry and chemiluminescence [108,109]. As a powerful analytical method, ECL sensor has been widely applied in the field of biosensing due to the remarkable advantages of high sensitivity, wide detection range, good reproducibility and selectivity, strong anti-interference capability, as well as simple equipment and operation [110]. To meet the ultra-sensitive demands of exosomes detection, various nanomaterials were loaded with luminophores to boost the efficiency of ECL and improve assay sensitivity. As QDs are facile in synthesis and modification, as well as possess stable optical and electrochemical properties, they have become attractive ECL luminophores. For example, Feng and co-workers developed a homogeneous sensing system for exosomes analysis by using CdS QDs as ECL emitters [111]. The developed ECL sensor demonstrated high sensitivity and selectivity, as well as wide detection range in complex biological samples through the enzymatic recycling binding of target amplification. To improve the sensitivity of ECL sensor, a large amount of Ru(bpy)32+ were immobilized on silica NPs by Li et al. used as an amplification strategy for exosomes assay [112]. The Ru@SiO2 NPs based ECL signal amplification displayed a wide calibration range of 3.22 × 10−4 - 156 μg/mL with a 2.73 × 10−4 μg/mL detection limit for exosomes. In the same year, Chen group synthesized a g-C3N4 nanosheet loaded with luminol capped Au NPs (Lum-AuNPs@g-C3N4) nanocomposite and used as signal nanoprobe for the analysis of phosphatidylserine (PS)-positive exosomes, a potential biomarker for early diagnosis of ovarian malignancy [113]. Herein, the g-C3N4 nanosheets with large surface area acted as a catalyst to catalyze the co-reactant H2O2 decomposition, resulting in the ECL signal amplification of luminol-H2O2 system. Thus, the ECL biosensor realized highly sensitive PS-positive exosomes assay and enabled accurate quantification of ovarian tumor-derived exosomes in complex sample. This year, Xiong et al. constructed a localized surface plasmon resonance (LSPR) between Au NPs and polymer dots based ECL immunosensor for exosomes analysis [114]. As shown in Fig. 3b, the excited hot electrons were transferred to conduction band of polymer dots after the hot electrons of Au NPs were photoexcited to surface plasmon states by ECL emission of polymer dots, resulting in significant improvement of ECL efficiency of polymer dots. The ECL immunosensor exhibited linear responses in linear range of 1.0 × 103 to 1.0 × 106 exosomes/mL with a detection limit of 400 exosomes/mL. Recently, nanomaterials based ECL have been used in downstream analysis of exosomes. Adhikari et al. developed a label-free ECL nanoimmunosensor for CD63 protein detection by using carbon nanochips/iron oxide/nafion-nanocomposite modified mesoporous carbon interface [115]. The ECL immunosensor not only showed notable stability and reproducibility, but also displayed a wide linear range and very low detection limit in profiling of the target protein.
3.4. Nanomaterials assisted fluorescence probes for exosomes analysis
Due to the high sensitivity and versatility, simplicity, fast signaling speed, good tolerance to interference, as well as nondestructive way of analyzing or tracking targets, fuorescence-based assays have become the most popular technique applied in liquid biopsy [119]. Recently, the rapid developments of fluorescent nanomaterials, such as QDs, upconversion NPs, and semiconducting polymer dots, have make them gradually replaced conventional fluorophores as the signaling units in fluorescence-based exosomes assay. Benefiting from the controllable optical characteristics of nanomaterials, there is a diverse probe selection to improve the detection throughput of exosomes assay. Moreover, nanomaterials can act as solid support for the target recognition to simplify fluorescence-based assays. Interestingly, nanomaterials can also be used to eliminate interference in fluorescence assay. Therefore, nanomaterials used in fluorescence biosensors can be used as direct or stimuli-responsive signaling units to improve detection flexibility and performance.
3.4.1. Nanomaterials act as direct signal units of fluorescence to enhance exosomes analysis
QDs have been widely used in fluorescence-based assays due to their brightness and tunable electronic and optical properties. For example, Yuan and co-workers fabricated blinking silicon QDs and used to single molecule localization imaging of exosomes [120]. However, one of the obstacles for direct application of QDs in exosome analysis is that their surface property may not be compatible with the salt concentration of PBS buffers, which will likely cause aggregation or even dissolution. Moreover, exosomes are often overwhelmed with other vesicles that could nonspecifically adsorb onto the QDs. To overcome these limitations of QDs in exosomes assay, surface modifications have been used to change the surface properties and reduce nonspecific binding of QDs. Bian's group developed a one-step quantification platform for exosomes detection by using aptamer functionalized QDs [121]. The designed nanoprobe was comprised of recognition aptamer and fluorescent materials. The specific capture of exosome with strong anti-interference ability and fluorescent amplification effect was achieved, thereby enabled robust quantification of exosomes in complex matrices.
As a new class of QDs, carbon dots (CDs) not only exhibit excellent fluorescence property due to quantum confinement but also possess good biocompatible, which effectively avoids the potential toxicity of QDs in liquid biopsy application [[122], [123], [124]]. In general, CDs are mixed with metal ions and fluorescent dyes to achieve ratiometric detection, so as to improve the capability of the fluorescence-based assays. In 2018, Jiang et al. synthesized gold-carbon quantum dots (GCDs) for fluorescence imaging of exosomes [125]. Through immuno-reactions, cancer-derived exosomes were labeled by GCDs and thus facilitated fluorescent imaging of exosomes. In the same year, DNA-labeled CDs and acridone derivate were used as ratiometric fluorescent bioprobes for the detection of exosomal microRNA [126]. By combining the self-referencing capability of ratiometric fluorescenc between 5,7-dinitro-2-sulfo-acridone and CDs, as well as the strand displacement reaction-based target catalysis signal amplification strategy, highly sensitive and stable exosomal miRNA-21 assay with as low as 3.0 fM detection limit could be realized. Moreover, the introduction of strand displacement reaction in the strategy also promoted the remarkable improvement of detection selectivity in complex sample and realized the monitoring of the dynamic change of exosomal miRNA-21 simultaneously. Therefore, the proposed ratiometric fluorescent bioprobe showed great potential to distinguish nontumorigenic exosomes and cancer exosomes in liquid biopsy.
Apart from QDs and CDs, fluorescent silica NPs that prepared from doping organic fluorophores with colloidal silica particles have been developed [127,128]. Because the selection of particle sizes is wide, size control during NPs synthesis is less strict, surface functionalization is easy, as well as excellent biocompatibility and water solubility, fluorescent silica NPs are particularly promising in fluorescence-based assays. For example, Yuan and co-workers developed blinking probe for exosomes imaging by using ultrasmall silica nanospheres as the scaffolds [120]. Silica NPs were synthetized by hydrolysis reaction of tetraethylorthosilicate and further modified by APTES. The reactive amines functionalized silica NPs was further attached by organic dyes. The obtained fluorescent silica nanospheres hold sustainable and excellent fluorescence switching behavior, allowed long-term super-resolution imaging of exosomes with high localization precision. Moreover, the fluorescent silica nanospheres still maintained their intrinsic fluorescence behavior in cell culture medium, which confirmed their great potential as biocompatible exosomes imaging agents in liquid biopsy.
Compared with the aforementioned inorganic nanomaterials, fluorescent polymeric NPs prepared from doping with fluorophores or intrinsically fluorescent characteristics provide some advantages, such as exceptional brightness, remarkable stability, easy-controlled surface properties [[129], [130], [131]]. Therefore, fluorescent polymeric NPs have attracted great attentions in fluorescence-based exosomes assays. As shown in Fig. 3c, Kim et al. fabricated a polydiacetylene based liposomal biosensor for exosomes detection [116]. Taking advantages of its unique fluorescent properties derived from the eneyne-conjugated backbone of polydiacetylene, sensitive exosomes assay was achieved with a detection limit of 3 × 108 particles/mL. Such a low detection concentration can be used in clinical applications. In 2022, Hua and co-workers designed and constructed a multisite-targeting polymer based fluorescence sensing for exosomes recognition and target tumor lesions [132]. The prepared fluorescent polymeric NPs could bind with exosomes without any interference through Schiff segment, which showed strong selectivity and sensitivity in in-vivo and in-vitro exosomes recognition.
3.4.2. Nanomaterials act as stimuli-responsive signaling units of fluorescence to enhance exosomes analysis
To improve the sensitivity and flexibility of exosomes assay design, a variety of novel fluorescent nanomaterials with thoughtful design have been developed as energy acceptors to quench the fluorescence of diverse dye molecules [133,134]. Typical nanomaterials employed in such systems for exosomes assay are 2D nanomaterials such as graphene oxide and MoS2. For example, Li and co-workers developed a GO-based sensor for exosomal proteins profiling [117]. As shown in Fig. 3d, fluorescence was quenched when TPE-TAs/aptamer complex was absorbed by GO in the absence of tumor-derived exosomes. In contrast, the specifically recognized aptamer will preferentially bind with their targets when exosomes are introduced. Therefore, the TPE-TAs/aptamer complexes break away from GO surface, resulting in a “turn-on” fluorescence signal. Not only sensitive exosomes detection with a detection limit of 3.43 × 105 particles/μL was achieved, but also exosomal proteins profiling with high sensitivity could be applied. The GO based sensor provided a promising approach to profile tumor-derived exosomal proteins for the early diagnosis in liquid biopsy. In 2022, Liao and co-workers reported an aptamer/GO fluorescence resonance energy transfer system based selective “turn-on” fluorescence igniting in sandwich mode for exosome labeling [135]. Integrating GO and aptamer into one fluorescent aptasensor, which showed outstanding performance for the detection of exosomes. Qualitative and quantitative analysis of exosomes with low background interference was achieved. Exosomal microRNAs are ideal biomarkers for the early diagnosis and prognosis of lung cancer. Recently, MoS2 nanosheets have also been used as the energy acceptors for fluorogenic quantitative detection of exosomal microRNAs [136]. MoS2 nanosheets adsorbed onto the dye labeled hairpin probes and exhibited excellent quench ability. The MoS2 nanosheets based fluorescence sensor provided a rapid, simple, and highly specific quantitative approach for exosomal miRNA.
In recent years, 2D transition-metal carbides and carbonitrides materials named as MXenes have attracted great attentions and offered distinctive advantages in fluorescence-based assays. For instance, Zhang and co-workers constructed a self-standard ratiometric fluorescence resonance energy transfer nanoprobe for sensitive detection of exosomes based on Cy3 labeled CD63 aptamer/Ti3C2 MXenes nanocomplex [118]. As illustrated in Fig. 3e, the Cy3 labeled CD63 aptamer can be selectively adsorbed by the Ti3C2 MXene nanosheets through hydrogen bond and metal chelate interaction between phosphate groups of aptamers and Ti2+ of MXenes, hence the fluorescence signal of Cy3 labeled CD63 aptamer was quenched quickly due to the fluorescence resonance energy transfer between MXene and Cy3. With the addition of exosomes, the fluorescence signal of Cy3 recovered rapidly. Meanwhile, the fluorescence signal of MXenes could be used as a standard reference owing to its little change in the whole process. As a result, a MXene based self-standard turn-on fluorescence resonance energy transfer sensing platform was constructed for sensitive exosomes detection with 1.4 × 103 particles/mL detection limit. What's more, the fluorescence sensor can also be applied in the identification of exosomal biomarkers. The proposed biosensing platform not only offered a universal strategy for exosomes, but also provided a new way for multiple biomarkers detection.
3.5. Nanomaterials assisted colorimetric probes for exosomes analysis
Colorimetric biosensors for exosomes detection have attracted considerable attentions due to their convenience and simplification [137,138]. Typically, the signal changes of colorimetric biosensors can be easily and instantly observed with naked eye through color change, which are very suitable for point-of-care diagnosis and on-site analysis [139]. However, the analytical performance and application of conventional colorimetric biosensors in harsh environments are seriously restrained as natural enzymes are involved in the process of color change, the well-known complex biological catalysts are temperature sensitive, high cost, and low operational stability. With the rapid development of nanotechnology, nanomaterials-based colorimetric exosomes assay methods have been demonstrated to be promising alternatives to conventional colorimetric biosensors [140,141]. Not only the drawbacks of natural enzyme-based sensing can be overcome, but also the selectivity and sensitivity of exosomes assay can be improved by the nanomaterials based colorimetric biosensors. Based on these advantageous, a variety of nanomaterials based colorimetric biosensors for exosomes detection have been developed. In principle, nanomaterials employed in colorimetric biosensors mainly paly two roles: colorimetric substrates and signal transduction.
3.5.1. Nanomaterials act as colorimetric substrates of colorimetry to enhance exosomes analysis
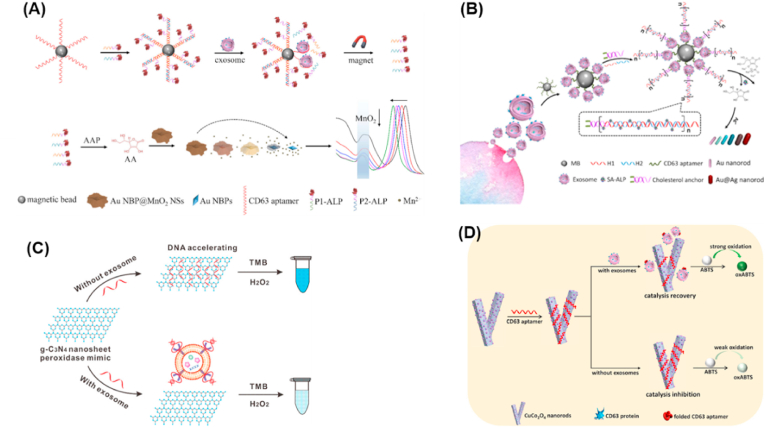
Owing to their extraordinary scattering and optical absorption properties, noble metal nanoparticles, such as Au and Ag NPs have been introduced as colorimetric substrates with localized surface plasmon resonance absorption. As early as 2017, Tan's group constructed an aptamer/Au NPs based biosensor for colorimetric detection of exosomal proteins through the assembly of Au NPs with a panel of aptamers targeting ubiquitous or putative exosome surface proteins [142]. The weak and non-specific binding equilibrium between Au NPs and aptamers is broken in the presence of exosomes. Instead, the strong and specific binding between aptamer and the exosome surface protein takes place, leading to a rapid displacement of aptamers from the exosomes surface and consequent aggregation of Au NPs. The aggregation of Au NPs can induce interparticle plasmon coupling, resulting in the LSPR shift, and a visible color change from red to blue, which can be used as a colorimetric signal. Depending on the specific exosome surface protein-aptamer interaction, multiple proteins on different cancer-derived exosomes can be identified simultaneously, which opened the door to better understanding of cancer development. In the following year, Au NPs have also been used by Liu et al. as colorimetric substrate for exosomes and their biomarkers assay [143]. To further improve the performance of nanomaterials based colorimetric biosensors, noble metal NPs with various modifications have been widely applied in designing colorimetric biosensors. As shown in Fig. 4a, Zhang and co-workers designed gold nanobipyramid@MnO2 nanosheets as plasmonic nanoparticle etching substrates for colorimetric biosensor-based exosome detection [144]. The inherent enhanced sensitivity in plasmonic sensing of gold NPs and further modification with poly(4-styrenesulfonic acid) ensured the favorable sensitivity and stability for exosome assay. Thus, the proposed gold nanobipyramid@MnO2 nanosheets based colorimetric biosensor exhibited satisfactory sensitivity down to 1.35 × 102 particles/μL for exosomes. Moreover, this method also showed good accuracy in the analysis of human serum samples and great potential for exosome-based tumor diagnosis.
Fig. 4.
Nanomaterials enhanced colorimetric analysis of exosomes. (A) Schematic illustration of the plasmonic colorimetry for exosome detection via competitive reaction and etching of Au NBP@MnO2 NSs [144]. (B) Schematic illustration of the mechanism for multicolor visual detection of exosomes based on HCR and enzyme-catalyzed metallization of Au NRs [145]. (C) Illustration of DNA aptamer accelerating the intrinsic peroxidase-like activity of g-C3N4 NSs for the detection of exosomes [146]. (D) Schematic representation of the mechanism for label-free detection of exosomes based on CD63 aptamer inhibiting oxidase activity of CuCo2O4 nanorods [147].
The aggregation of nanoparticles can induce interparticle plasmon coupling leading to the shift of localized surface plasmon resonance (LSPR), which can also be induced by non-aggregation of nanomaterials. Typically, the color change of such kind of colorimetric sensor is derived from the growth of plasmonic nanoparticles on enzymes and nanostructured templates induced by LSPR shift. In this way, the above group continued reporting a plasmonic colorimetric assay method for exosomes detection [145]. As illustrated in Fig. 4b, the Au@Ag nanorod was obtained by the enzyme modulated growth of Ag NPs on Au nanostars. In detail, the alkaline phosphatase boosted the ascorbic acid generation to reduce the deposition of silver shells on the surface of Au nanostars, giving rise to a LSPR blue shift. Therefore, the color changes of solutions and absorption blue shift of Au nanostars indicated the concentration of exosomes, which realized sensitive exosomes detection with detection limit as low as 9 × 103 particles/μL by naked eyes. Most importantly, clinical samples analysis also demonstrated satisfactory results.
3.5.2. Nanomaterials act as signal transduction of colorimetry to enhance exosomes analysis
Since Fe3O4 NPs were used as nanozymes, more and more nanomaterials with enzyme-mimicking characteristics have been designed and applied in colorimetric assay [148,149]. Nanozymes can be served as enzyme mimics to be signal-transduction tools and produce distinct color changes in catalytic reaction. Compared to conventional natural enzymes, nanozymes show attractive advantages of low cost, improved stability and ease of storage, which accelerate their practical applications in liquid biopsy [150,151]. To date, peroxidase-mimicking nanomaterials have been widely used in colorimetric sensor for exosomes assay. Benefiting from the excellent affinity of g-C3N4 NSs to single-stranded DNA (ssDNAs) and promising capability of DNA in improving the peroxidase-like activity of nanozymes, Wang et al. developed an exosome sensing platform by coupling g-C3N4 NSs with ssDNAs [146]. They demonstrated that ssDNA adsorbed on g-C3N4 NSs could accelerate the intrinsic peroxidase-like activity of the nanosheets through the aromatic stacking and electrostatic interactions between ssDNAs and substrate. As shown in Fig. 4c, CD63, the surface marker of exosome could competitively bind with ssDNAs aptamer, which prevented the improvement of peroxidase-like activity and permitted sensitive colorimetric assay of exosomes. The method could recognize differential expression and patients of exosomal CD63, which highlighted the great value of the ssDNA-g-C3N4 NSs in clinical diagnosis using liquid biopsy. Similarly, Chen's group also developed a colorimetric aptasensor for exosomes assay based on DNA-capped single-walled carbon nanotubes (SWCNTs) [152]. The developed aptasensor achieved a detection of limit of 5.2 × 105 particles/μL. Recently, nanozymes based colorimetric sensors have been used to profile exosomal proteins. Di et al. reported a nanozyme-assisted immunosorbent assay based on the installation of peroxidase-like Exo@Au nanozymes onto the phospholipid membranes of exosomes enabled sensitive detection of exosomal proteins for rapid cancer diagnosis [153]. Such progress revealed tumor-derived exosomal proteins can serve as promising biomarkers for cancer diagnosis in a cooperative detection mode.
Most of the peroxidase-mimicking nanomaterial-based colorimetric assays require H2O2 as an oxidant, which might lead to a harmful impact on exosomes, while there is no impact on the oxidase-mimicking nanomaterials involved in catalytic reaction. For example, Zhang et al. developed a label-free method for exosome detection based on the target-responsive controllability of oxidase-like activity of Cu/Co bimetallic MOFs (CuCo2O4 nanorods) [147]. As shown in Fig. 4d, CuCo2O4 nanorods could directly catalyze the oxidation of substrates with molecular oxygen as the electron acceptor instead of volatile H2O2, which was more stable, simple and eco-friendly. The negatively charged CD63 aptamers absorbed by CuCo2O4 nanorods through electrostatic interaction, resulting in the inhibition of oxidase-like activity. In contrast, the oxidase-like activity would restore in the presence of exosomes because of CD63 aptamers released from the CuCo2O4 nanorods by virtue of CD63 aptamer-exosome recognition. The established colorimetric method not only realized sensitive exosomes detection over a range of 5.6 × 104 to 8.9 × 105 particles/μL with a detection limit of 4.5 × 103 particles/μL, but also greatly avoided the interference of nonspecific adsorption, thereby ensured the reliability of the method.
3.6. Nanomaterials assisted surface-enhanced Raman spectroscopy probes for exosomes analysis
Raman spectroscopy biosensors have been used for various bioanalytical studies because of their remarkable enhanced analytical signals, including single molecule-level sensitivity and tolerance to quenching [154,155]. However, the signal intensity of Raman scattering is very low to be distinguished. To overcome this problem, several nanomaterials or metal nanostructure have been employed as signal enhancers to enhance the electric field by surface treatment. Benefiting from the excellent detectability and specificity, nanomaterials assisted SERS sensors have been applied to the label free detection of exosomes. Nanomaterials used in SERS sensors mainly act as SERS substrate or SERS-active nanotags to enhance the signal intensity.
3.6.1. Nanomaterials act as SERS substrate to enhance exosomes analysis
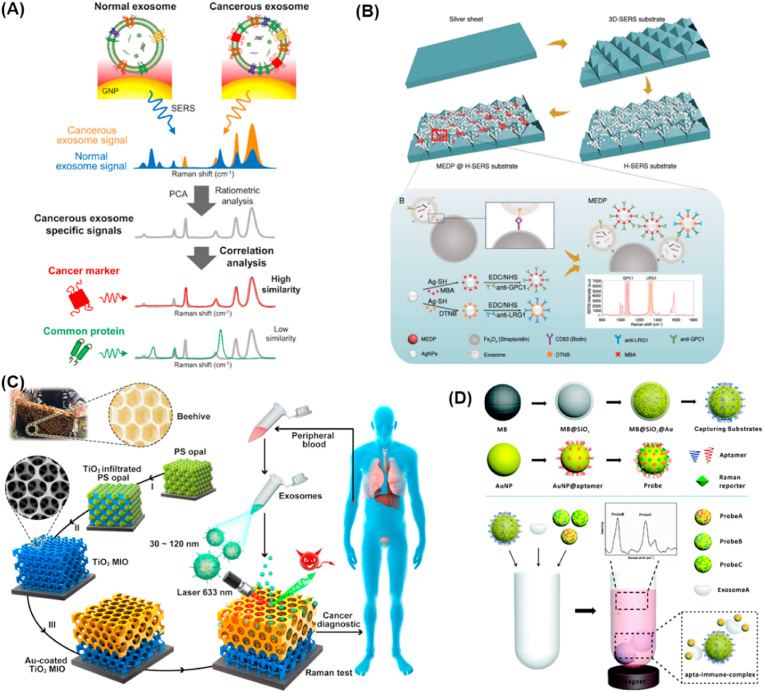
Label free SERS assay of exosomes is mainly based on the use of nanosized or roughened SERS substrate to enhance the low Raman signal of exosomes particles. Metal nanomaterials such as Ag, Au have become the most widely used SERS substrate due to their unique optical and electrical properties, small size, as well as large specific surface area. For example, silver NPs were used directly by Rojalin et al. as inexpensive, biocompatible, and label free plasmonic substrates for SERS based liquid biopsy diagnostics [156]. Gold NPs-coated plate was used by Shin and co-workers for SERS signals collection of exosomes derived from normal and lung cancer cell lines [157]. Combing with deep learning algorithm, the proposed strategy predicted the presence and progression of lung cancer. The same group also fabricated an aggregated gold NPs substrate to form nanogaps for SERS based correlation between protein markers and cancerous exosomes [158]. As shown in Fig. 5a, a strong electromagnetic field can be formed in the nanogaps between aggregated NPs nanoparticles. To prevent the deformation of signal through damage to the salt formation and exosome, the SERS spectra of exosomes was obtained in a liquid state. In addition, the gold NPs based SERS substrate was modified with cysteamine to induce the electrostatic adsorption of exosomes. Thus, exosomes could be captured by the cationic amino groups of the cysteamine. In this way, the correlation of non-small cell lung cancer cell-derived exosomes and potential protein markers was demonstrated.
Fig. 5.
Nanomaterials enhanced Raman analysis of exosomes. (A) Detection of unique lung cancer cell-derived exosomes and comparison to the profiles of their potential surface protein markers [158]. (B) The synthesis process of the H-SERS substrate and illustration of the construction of exosome capture system, the fabrication of SERS detection probes, and the SERS detection of exosomes [159]. (C) Detection process and design inspiration of the Au-coated TiO2 MIO SERS probe [160]. (D) The principle of the SERS-based simultaneous multiple detection method of exosomes [161].
To provide much more hot spots between closely spaced metal NPs leading to intense SERS enhancement, various structures of SERS substrate were constructed from gold NPs for exosomes assay. In 2019, a uniform plasmonic head-flocked gold nanopillar substrate was developed by Lee et al. for highly specific and ultrasensitive detection of exosomal miRNAs [162]. The specific structure of the proposed SERS substrate created multiple hotspots, resulting in the enhancement of local plasmonic fields. Therefore, the SERS sensor highlighted extremely low detection limit without any amplification process, multiplex sensing capability, wide dynamic range from 1 a.m. to 100 nm, as well as sound miRNA recovery in serum sample. What's more, exosomal miRNA expression patterns in breast cancer subtype can be recognized by the SERS sensor, which provided a promising tool for early cancer detection and monitoring for cancer recurrence after treatment or resection. In 2021, Kang and co-workers developed a close-packed gold octahedra array as a SERS sensing platform for the quantitative determination of exosomal miRNAs [163]. Because of Au octahedron in the array was evenly located on its triangular surface, such kind of orientation produced numerous hot spots and greatly improved the assay uniformity and sensitivity. The SERS sensor demonstrated a broad linear range from 10 aM to 10 nM and a low detection limit of 5.3 aM without any amplification strategy. This year, a hierarchical SERS substrate (H-SERS substrate) was constructed by Li et al. for SERS based exosomes detection [159]. As demonstrated in Fig. 5b, the hierarchical SERS substrate can provide much more hot spots due to the uniform layer of Au NPs and ordered pyramid array. The H-SERS sensor not only achieved sensitive detection of exosomes with a detection limit of 15 particles/μL, but also improved the detection rate of pancreatic cancer by combing the LRG1-positive exosomes and GPC1-positive exosomes. All these superiorities indicated that the proposed H-SERS sensor is of great promise to facilitate the early diagnosis of pancreatic cancer. Inspired by the unique SPR properties of gold NPs assembled in triangular pyramid DNA, Zhang and co-workers prepared a similar structure and served as a novel Raman probe for the determination of exosomes [164]. Due to strong electromagnetic hot spots appeared at the junctions between AuNPs, significant enhancement of the SERS signals could be achieved. Under optimal conditions, sensitive and selective detection of exosomes could be realized with 1.1 × 102 particles/μL. Meanwhile, exosome extracted from breast cancer patients and healthy individuals also could be distinguished by the strategy.
The construction of composites based on metal NPs is also an effective strategy to improve the Raman detection signal. For example, Fraire and co-workers developed Au@Ag NPs as SERS substrate for label-free detection of individual exosome [165]. Compared with previous strategies, the core-shell plasmonic Au@Ag NPs as SERS substrate showed higher near-field enhancements, thereby resulting in the signal-to-noise ratio of the SERS spectra improved obviously. Individual vesicles derived from B16F10 melanoma cells and red blood cells could be successfully discriminated with an unprecedented sensitivity and specificity. Moreover, the Au@Ag NPs based SERS sensor also reduced the acquisition time because of the higher near field enhancement, which provided a label free platform for high-throughput identification of single exosomes. Yan and co-workers designed a plasmonic hybrid SERS substrate made of graphene overlaid on engineered periodic Au-pyramid nanostructure for label-free identification of exosomes [166]. Benefiting from the synergistic interaction of electromagnetic enhancement mechanism though plasmonic Au-pyramid nanostructure and chemical enhancement mechanism through the hybrid, obvious enhancement of SERS signal could be enhanced by several orders of magnitudes. The established SERS sensor realized the assay of single exosomes from different source. Similarly, Ray's group also developed a mixed-dimensional heterostructure SERS platform by using 2D graphene oxide and 0D plasmonic gold nanostar for exosomes detection [167]. Through light-matter and matter-matter interactions, 4 orders of magnitude of Raman signal enhancement was achieved. The proposed mixed-dimensional heterostructure SERS sensor realized trace level identification of exosomes with a detection limit of 3.8 × 102 exosomes/mL for TNBC-derived exosomes and 4.4 × 102 exosomes/mL for HER2(+) breast cancer-derived exosomes. Moreover, this sensor also showed great potential in the identification of cancer biomarkers. Inspired by the concept of beehives, Au-coated TiO2 macroporous inverse opal (MIO) structure was designed by Dong et al. and used as SERS substrate for label free assay of exosomes (Fig. 5c) [160]. Taking advantage of the prominent “slow light effect”, the Raman signals of exosomes enhanced significantly. Different from the current liquid biopsy techniques, the MID based SERS sensor for exosomes assay was noninvasive and time-saving, which made the vitro cancer monitoring and diagnostics as simple as the common diseases.
3.6.2. Nanomaterials act as SERS-active nanotags to enhance exosomes analysis
Owing to the unique LSPR characteristic, Ag and Au NPs have been used as SERS-active nanotags to increase the weak signal of the common dyes in exosomes assay. For example, SERS-active exosome was developed by Chen et al. and used for the monitoring of intracellular trafficking processes of exosomes [168]. The SERS-active exosome was prepared by modifying Au nanoparticles with Raman reporter groups, and then conjugated to an exosome. Herein, Au NPs acted as SERS-active generators, while exosome was used as a vehicle for loading and delivery of Au NPs into cells. By monitoring the SERS signals of exosomes, the potential pathways involved in the internalization of exosomes were investigated. Gold nanorods coated with QSY21 Raman reporters were proposed by Kwizera and co-workers for SERS based molecular detection and analysis of exosomes [169]. The SERS effects were facilitated by the high electromagnetic fields at the ends of gold nanorods. HER2 and EpCAM biomarkers on exosomes in plasma from HER2-positive breast cancer patients were identified by the developed SERS-active nanotags. To improve the specificity of exosomes, some antibody or aptamer modified SERS-active nanotags have been reported for the detection of exosomes through the formation of aptamer-exosomes-antibody sandwich-type immunocomplexes. Wang and co-workers developed a microfluidic Raman biochip for exosomes analysis based on EpCAM-functionalized Raman-active polymeric nanomaterials [170]. The biochip showed rapid analysis of exosome samples within 1 h and low detection limit with 1.6 × 102 particles/mL with 20 μL exosomes samples. Gold NPs decorated with a Raman reporter and specific aptamer were designed by Wang et al. for the screening and multiple detection of tumor-derived exosome [161]. Three kinds of SERS probes were synthesized by using different types of SERS reporters and aptamers for simultaneous multiple detection. As demonstrated in Fig. 5d, only specific SERS probes could recognize the target exosomes and formed a sandwich-type apta-immunocomplex the magnetic substrates when one kind of target exosome was added to the mixture of SERS probes and capturing substrates. And then the SERS signals of supernatant could be detected once the complexes were precipitated by a magnet. With the addition of exosomes, the signal intensity of the SERS probe become weaker, while the signals intensity of other non-specific SERS probes kept the same. Thus, SERS signal with a decreased intensity indicated the existence of the target exosome. Similarly, gold nanoparticles functionalized with Raman reporters and detection antibodies based SERS nanotag was developed by Wang et al. for sensitive direct detection of serum exosomes [171]. Unique Raman signals corresponding to the molecular structures of Raman reporters could be generated form the SERS nanotags under laser excitation. Not only the expression profiles of protein biomarkers were multiplexed, the developed SERS sensor also showed significantly higher signal intensities of protein biomarkers. The hot spots generated in the metal NPs-metal NPs junctions because of the plasmon coupling effect could improve the Raman signal intensity. In view of this, Pang et al. developed anti-PD-L1 antibody modified Au@Ag@MBA SERS tags for label free quantification of exosomal PD-L1 [52]. Coupling with Fe3O4@TiO2 isolation, the strategy realized accurate and rapid quantification of exosomal PD-L1 by using only 4 μL clinic serum sample and within 40 min. Meanwhile, the exosomal PD-L1 levels from different clinical samples could be discriminated clearly based on the SERS signal analysis. In the same year, Ning and co-workers also developed gold-silver-silver core-shell-shell nanotrepangs (GSSNTs) based SERS-active nanotags and achieved multiple and sensitive SERS detection of cancer-derived exosomes [172].
3.7. Nanomaterials assisted surface plasmon resonance probes for exosomes analysis
Surface plasmon resonance (SPR) is a label-free, real-time analysis technique that can detect molecular interactions on the surface of a gold layer by monitoring the changes in its refractive index [173,174]. SPR is extremely sensitive to the biological binding events occurring within 200 nm (wave depth) of the gold layer. Such distance closely matches the dimension of exosomes [175]. Therefore, SPR-based biosensors are perfectly suited for the study of exosomes. Recently, a various of label free SPR sensors for exosomes assay have been reported.
Gold NPs are the commonly used signal amplification probe in SPR biosensors. Wang et al. reported a SPR aptasensor for direct quantification of cancerous exosomes with dual Au NP-assisted signal amplification [176]. Exosomes were captured by the aptamers functionalized Au chip, and then labeled by aptamer/T30 linked Au NPs on the Au film. Owing to the electronic coupling between Au film and Au NPs, detection of exosomes in low concentration could be achieved. Fan et al. developed an antibody-modified SPR chip for highly sensitive detection of non-small cell lung cancer derived exosomes [177]. As shown in Fig. 6a, antibody-functionalized Au NPs are injected into the flow chamber of chip to form sandwich structures of antibodies/exosome/Au NPs hybrid, resulting in amplification of SPR signal. The detection limit of the SPR-based biosensing is 104 particles/μL. For increasing the specificity, Mao and co-workers proposed a simple and ultra-sensitive SPR platform for the detection of PD-L1 exosomes based on graphene modified gold chip using multifunctional peptide as recognition supermolecule [178]. Not only sensitive detection of PD-L1 exosomes without prior labelling could be achieved based on the direct SPR assay, but also the exosomes could be quantified accurately. Apart from aforementioned SPR biosensors, nanoplasmonic biosensors that employ nanoscale topographies are a new category of SPR based biosensors. Liu and co-workers developed an intensity-modulated, compacted SPR biosensor by using conventional SPR sensing mechanism and did not require fabrication of any nanostructures [179]. With lung cancer as the disease model, the compact SPR biosensor demonstrated much higher sensitivity than traditional ELISA approach.
Fig. 6.
Nanomaterials assisted SPR analysis of exosomes. (A) Working principle of plasmonic AuNPs-enhanced SPRi biosensor for multiplex analysis of NSCLC-derived exosomes via different recognition sites [177]. (B) Schematic illustration of TiN functionalized by biotinylated anti-CD63 antibody for the detection of U251 GMs-derived exosomes. Nanomaterials assisted mass spectrometry analysis of exosomes [183]. (C) Scheme of preparation and application of the plasmonic chip for LDI MS based metabolic analysis [184]. (D) Synthesis and application of MoS2/Au-NP−L-cysteine in exosomal glycopeptide enrichment [185].
As an excellent alternative plasmonic supporting material compared to silver and gold NPs, titanium nitride (TiN) showed tunable plasmonic properties in visible and near-infrared spectra [180,181]. Moreover, TiN is a nanomaterial with excellent biocompatibility and chemical stability [182]. Benefiting from the unique features of TiN, Qiu and co-workers developed a biotinylated antibody-functionalized TiN (BAF-TiN) based SPR biosensor for label free detection of glioma-derived exosomes [183]. As illustrated in Fig. 6b, the biotinylated anti-EGFRvIII antibody was immobilized on the TiN surface through high affinity biotin-TiN interaction. The obtained substrate could be used as a reporter in the SPR biosensor to ensure the excellent sensitivity and selectivity toward GMs-derived exosomes. Compared with traditional gold NPs based SPR biosensors, the developed BAF-TiN biosensor exhibited satisfied sensitivity and selectivity in the detection of glioma-derived exosomes. The detection limit for epidermal growth factor receptor variant-III and CD63 were 2.75 × 10−3 μg/mL and 4.29 × 10−3 μg/mL, respectively. The BAF-TiN biosensor showed great potential in the detection of cancer biomarkers and monitoring the progression of malignant glioma.
3.8. Nanomaterials assisted mass spectrometry assay of exosomes
Matrix-assisted laser desorption/ionization mass spectrometry (LDI-MS) has shown great potential in large-scale clinical use due to its fast detection speed, low sample consumption, mass measurement for molecular identification, and high sensitivity with low costs. Notably, the performance of LDI-MS is normally determined by the property of matrix. To improve the detection ability of LDI-MS and overcome the shortcomings of traditional matrix, a few nanomaterials with excellent light absorption and energy transfer properties have been developed as matrix of LDI-MS for exosomes assay. For example, Sun and co-workers developed a plasmonic gold chips as a new matrix for direct LDI-MS detection of small metabolites in exosomes. As shown in Fig. 6c, the gold chips were prepared from gold nanoshells on the surface through a three-step process including controlled particle synthesis, dip-coating, and gold sputtering for mass production. Benefiting from the specific nanogaps and nanocrevices of gold shells on-chip, the small metabolite molecules could be trapped selectively and transfer the laser energy, thereby sensitive, selective, and multiplex metabolic quantification was achieved by LDI MS using 500 nL of exosomes. Moreover, in vitro metabolic diagnosis of lung cancer patients using exosomes was demonstrated by the developed strategy for the first time [184]. Similarly, titania was used by Chen et al. as the matrix of LDI-MS for urinary exosome metabolic analysis [186]. Due to the excellent electron transferability, the mix-crystal titania highlighted superior sensitivity, selectivity and reproducibility for metabolic patterns of exosomes. The titania-assisted intact exosome mass spectrometry assay revealed the clinical value of exosome metabolic analysis, and provided a strategy for exosome-based diagnosis at metabolomic level toward large-scale clinical use.
In light of the significance of exosomes in cancer diagnosis and treatment, it is important to understand the components and functions of exosomes. Growing evidence has shown that the components and functions of exosomes are highly related to their carried proteins. Therefore, comprehensive characterization of exosomal proteins will facilitate the understanding of their functions and identification of more exosome-based disease markers. As a powerful tool for proteome analysis, mass spectrometry has been widely in most of biological samples, such as cell, exosomes [187,188]. To improve the mass spectrometry identification efficiency of low-abundant proteins, some nanomaterials have been used as extractors or nanoreactors for exosomal proteins characterization. For example, Zhang's group designed a novel MoS2/Au-NP-l-cysteine nanocomposite for exosomal glycopeptide enrichment [185]. As shown in Fig. 6d, MoS2 nanosheets with large surface area were used as zwitterionic hydrophilic interaction liquid chromatography (ZIC-HILIC) materials for immobilizing hydrophilic l-cysteine with low steric hindrance between the materials and glycopeptides. When the MoS2/Au-NP-l-cysteine nanocomposites were applied for the analysis of complex samples, 775 glycoproteins were identified from 50 μg of proteins extracted from HeLa cell exosomes. Another effective way to improve the low-abundant protein identification is to enhance the efficiency of protein enzymolysis. In view of this, a versatile periodic mesoporous organosilica (PMO) nanomaterials was used by Fang et al. for the analysis of exosomal membrane proteins [189]. Benefiting from the amphiphilic porous structure of PMO, the lysed exosomal membrane proteins were enriched by PMO directly through hydrobic-hydrobic interaction, besides an extractor for exosomal proteins, the inherent pores of PMO could be served as a nanoreactor for in situ proteolytic digestion of the exosomal proteins. As a result, highly efficient identification of exosomal membrane proteins was achieved. Such progress provided a valuable tool for the study of exosomes markers and their functions, thereby resulting in the development of exosomes in liquid biopsy.
4. Conclusion and outlook
Exosomes are rapidly emerging as a new paradigm of liquid biopsy, efficient isolation and sensitive analysis of exosomes are crucial for promoting their application in clinical settings. Benefiting from the variable chemical compositions and morphologis, as well as excellent physicochemical characteristics, nanomaterials have been widely used in exosomes research in recent years. Nanomaterials used in exosomes provides a facile, low cost, but highly efficient and sensitive approach for exosomes isolation and analysis, which is of great value to facilitate the downstream analysis of exosomes in liquid biopsy. Despite significant progress have been made in nanomaterials assisted exosome isolation and analysis, there are still some challenges to be addressed which will hinder their clinical application in liquid biopsy due to the nature of exosomes. Therefore, there is still much room for the development of nanomaterial assisted exosomes isolation and analysis to realize the vision of exosomes based clinical diagnosis and therapeutic values in liquid biopsy.
One interesting direction for novel nanomaterials application in exosomes deserving more future research efforts is integrating specific isolation and downstream molecular analysis of exosomes into one platform. As the multifunctional design in nanomaterials assisted assays, such nanomaterial permit exosomes isolation and analysis in one nanodevice, requiring no additional assay platforms. With proper design, the specificity and efficiency of exosomes capture can be greatly increased and the interference particles, such as microvesicle, proteins be dramatically reduced, enabling quick and sensitive detection of exosomes. Another attractive feature of such designs, the cost and detection time can be can be greatly decreased, thereby facilitates the simultaneous detection of a great deal of clinical samples. Such progress will provide a significant tool for the research of exosomes with important biological significance and produce revolutionary development of exosomes based clinical diagnosis and treatment of tumors. Ensuring accurate and reliable exosomes assay is another difficulty in nanomaterials assisted exosomes isolation and analysis. Due to the interdisciplinary characteristics of nanomaterials coupled with different testing instruments, inconsistent results of exosomes detection from different research group or operation protocols are very common. One promising approach to solve these problems is develop a feasible and practical standardization procedure, including biological samples collection, exosomes isolation from biopsy and analysis operation.
Credit Author Statement
Xiaoni Fang: Conceptualization; Writing – original draft; Yuqing Wang: Formal analysis; Resources; Shurong Wang: Resources; Baohong Liu: Conceptualization; Validation; Supervision; Funding acquisition
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors would like to acknowledge financial support from the National Natural Science Foundation of China (Grant 21934001, 22004019) and Scientific Research Project of Shanghai Municipal Health Commission (Grant 20204Y0113).
Contributor Information
Xiaoni Fang, Email: xnfang@fudan.edu.cn.
Baohong Liu, Email: bhliu@fudan.edu.cn.
References
- 1.Rivellese F., Surace A.E.A., Goldmann K., Sciacca E., Çubuk C., Giorli G., John C.R., Nerviani A., Fossati-Jimack L., Thorborn G., Ahmed M., Prediletto E., Church S.E., Hudson B.M., Warren S.E., McKeigue P.M., Humby F., Bombardieri M., Barnes M.R., Lewis M.J., Pitzalis C., Rivellese F., Giorli G., Nerviani A., Fossati-Jimack L., Thorborn G., Humby F., Bombardieri M., Lewis M.J., Durez P., Buch M.H., Rizvi H., Mahto A., Montecucco C., Lauwerys B., Ng N., Ho P., Romão V.C., da Fonseca J.E.C., Verschueren P., Kelly S., Sainaghi P.P., Gendi N., Dasgupta B., Cauli A., Reynolds P., Cañete J.D., Ramirez J., Celis R., Moots R., Taylor P.C., Edwards C.J., Isaacs J., Sasieni P., Choy E., Thompson C., Bugatti S., Bellan M., Congia M., Holroyd C., Pratt A., White L., Warren L., Peel J., Hands R., Hadfield G., Pitzalis C. R.R.A.c.g. the, Rituximab versus tocilizumab in rheumatoid arthritis: synovial biopsy-based biomarker analysis of the phase 4 R4RA randomized trial. Nat. Med. 2022 doi: 10.1038/s41591-022-01789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astolfi M., Péant B., Lateef M.A., Rousset N., Kendall-Dupont J., Carmona E., Monet F., Saad F., Provencher D., Mes-Masson A.M., Gervais T. Micro-dissected tumor tissues on chip: an ex vivo method for drug testing and personalized therapy. Lab Chip. 2016;16(2):312–325. doi: 10.1039/c5lc01108f. [DOI] [PubMed] [Google Scholar]
- 3.Parikh A.R., Leshchiner I., Elagina L., Goyal L., Levovitz C., Siravegna G., Livitz D., Rhrissorrakrai K., Martin E.E., Van Seventer E.E., Hanna M., Slowik K., Utro F., Pinto C.J., Wong A., Danysh B.P., de la Cruz F.F., Fetter I.J., Nadres B., Shahzade H.A., Allen J.N., Blaszkowsky L.S., Clark J.W., Giantonio B., Murphy J.E., Nipp R.D., Roeland E., Ryan D.P., Weekes C.D., Kwak E.L., Faris J.E., Wo J.Y., Aguet F., Dey-Guha I., Hazar-Rethinam M., Dias-Santagata D., Ting D.T., Zhu A.X., Hong T.S., Golub T.R., Iafrate A.J., Adalsteinsson V.A., Bardelli A., Parida L., Juric D., Getz G., Corcoran R.B. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019;25(9):1415–1421. doi: 10.1038/s41591-019-0561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoshino A., Kim H.S., Bojmar L., Gyan K.E., Cioffi M., Hernandez J., Zambirinis C.P., Rodrigues G., Molina H., Heissel S., Mark M.T., Steiner L., Benito-Martin A., Lucotti S., Di Giannatale A., Offer K., Nakajima M., Williams C., Nogués L., Pelissier Vatter F.A., Hashimoto A., Davies A.E., Freitas D., Kenific C.M., Ararso Y., Buehring W., Lauritzen P., Ogitani Y., Sugiura K., Takahashi N., Alečković M., Bailey K.A., Jolissant J.S., Wang H., Harris A., Schaeffer L.M., García-Santos G., Posner Z., Balachandran V.P., Khakoo Y., Raju G.P., Scherz A., Sagi I., Scherz-Shouval R., Yarden Y., Oren M., Malladi M., Petriccione M., De Braganca K.C., Donzelli M., Fischer C., Vitolano S., Wright G.P., Ganshaw L., Marrano M., Ahmed A., DeStefano J., Danzer E., Roehrl M.H.A., Lacayo N.J., Vincent T.C., Weiser M.R., Brady M.S., Meyers P.A., Wexler L.H., Ambati S.R., Chou A.J., Slotkin E.K., Modak S., Roberts S.S., Basu E.M., Diolaiti D., Krantz B.A., Cardoso F., Simpson A.L., Berger M., Rudin C.M., Simeone D.M., Jain M., Ghajar C.M., Batra S.K., Stanger B.Z., Bui J., Brown K.A., Rajasekhar V.K., Healey J.H., de Sousa M., Kramer K., Sheth S., Baisch J., Pascual V., Heaton T.E., La Quaglia M.P., Pisapia D.J., Schwartz R., Zhang H., Liu Y., Shukla A., Blavier L., DeClerck Y.A., LaBarge M., Bissell M.J., Caffrey T.C., Grandgenett P.M., Hollingsworth M.A., Bromberg J., Costa-Silva B., Peinado H., Kang Y., Garcia B.A., O'Reilly E.M., Kelsen D., Trippett T.M., Jones D.R., Matei I.R., Jarnagin W.R., Lyden D. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182(4):1044–1061. doi: 10.1016/j.cell.2020.07.009. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmirotta R., Lovero D., Cafforio P., Felici C., Mannavola F., Pellè E., Quaresmini D., Tucci M., Silvestris F. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Therap. Adv. Med. Oncol. 2018;10 doi: 10.1177/1758835918794630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig N., Whiteside T.L., Reichert T.E. Challenges in exosome isolation and analysis in Health and disease. Int. J. Mol. Sci. 2019;20(19) doi: 10.3390/ijms20194684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ignatiadis M., Sledge G.W., Jeffrey S.S. Liquid biopsy enters the clinic — implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021;18(5):297–312. doi: 10.1038/s41571-020-00457-x. [DOI] [PubMed] [Google Scholar]
- 8.Poudineh M., Sargent E.H., Pantel K., Kelley S.O. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat. Biomed. Eng. 2018;2(2):72–84. doi: 10.1038/s41551-018-0190-5. [DOI] [PubMed] [Google Scholar]
- 9.Vaidyanathan R., Soon R.H., Zhang P., Jiang K., Lim C.T. Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab Chip. 2019;19(1):11–34. doi: 10.1039/c8lc00684a. [DOI] [PubMed] [Google Scholar]
- 10.Siravegna G., Mussolin B., Venesio T., Marsoni S., Seoane J., Dive C., Papadopoulos N., Kopetz S., Corcoran R.B., Siu L.L., Bardelli A. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019;30(10):1580–1590. doi: 10.1093/annonc/mdz227. [DOI] [PubMed] [Google Scholar]
- 11.Ye Q., Ling S., Zheng S., Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol. Cancer. 2019;18(1):114. doi: 10.1186/s12943-019-1043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng W., Dean D.C., Hornicek F.J., Shi H., Duan Z. Exosomes promote pre-metastatic niche formation in ovarian cancer. Mol. Cancer. 2019;18(1):124. doi: 10.1186/s12943-019-1049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castillo J., Bernard V., San Lucas F.A., Allenson K., Capello M., Kim D.U., Gascoyne P., Mulu F.C., Stephens B.M., Huang J., Wang H., Momin A.A., Jacamo R.O., Katz M., Wolff R., Javle M., Varadhachary G., Wistuba I.I., Hanash S., Maitra A., Alvarez H. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann. Oncol. 2018;29(1):223–229. doi: 10.1093/annonc/mdx542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halvaei S., Daryani S., Eslami-S Z., Samadi T., Jafarbeik-Iravani N., Bakhshayesh T.O., Majidzadeh-A K., Esmaeili R. Exosomes in cancer liquid biopsy: a focus on breast cancer. Mol. Ther. Nucleic Acids. 2018;10:131–141. doi: 10.1016/j.omtn.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alimirzaie S., Bagherzadeh M., Akbari M.R. Liquid biopsy in breast cancer: a comprehensive review. Clin. Genet. 2019;95(6):643–660. doi: 10.1111/cge.13514. [DOI] [PubMed] [Google Scholar]
- 16.Tămaș F., Bălașa R., Manu D., Gyorki G., Chinezu R., Tămaș C., Bălașa A. The importance of small extracellular vesicles in the cerebral metastatic process. Int. J. Mol. Sci. 2022;23(3) doi: 10.3390/ijms23031449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., Fissell W.H., Patton J.G., Rome L.H., Burnette D.T., Coffey R.J. Reassessment of exosome composition. Cell. 2019;177(2):428–445. doi: 10.1016/j.cell.2019.02.029. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mastoridis S., Bertolino G.M., Whitehouse G., Dazzi F., Sanchez-Fueyo A., Martinez-Llordella M. Multiparametric analysis of circulating exosomes and other small extracellular vesicles by advanced imaging flow cytometry. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Sousa K.P., Potriquet J., Mulvenna J., Sotillo J., Groves P.L., Loukas A., Apte S.H., Doolan D.L. Proteomic identification of the contents of small extracellular vesicles from in vivo Plasmodium yoelii infection. Int. J. Parasitol. 2022;52(1):35–45. doi: 10.1016/j.ijpara.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Lee K., Fraser K., Ghaddar B., Yang K., Kim E., Balaj L., Chiocca E.A., Breakefield X.O., Lee H., Weissleder R. Multiplexed profiling of single extracellular vesicles. ACS Nano. 2018;12(1):494–503. doi: 10.1021/acsnano.7b07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolè L., Cappello F., Cappellesso R., VandenBussche C.J., Fassina A. MicroRNA profiling in serous cavity specimens: diagnostic challenges and new opportunities. Cancer Cytopathology. 2019;127(8):493–500. doi: 10.1002/cncy.22143. [DOI] [PubMed] [Google Scholar]
- 22.Qu X., Li Q., Yang J., Zhao H., Wang F., Zhang F., Zhang S., Zhang H., Wang R., Wang Q., Wang Q., Li G., Peng X., Zhou X., Hao Y., Zhu J., Xiao W. Double-stranded DNA in exosomes of malignant pleural effusions as a novel DNA source for EGFR mutation detection in lung adenocarcinoma. Front. Oncol. 2019;9 doi: 10.3389/fonc.2019.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boussadia Z., Lamberti J., Mattei F., Pizzi E., Puglisi R., Zanetti C., Pasquini L., Fratini F., Fantozzi L., Felicetti F., Fecchi K., Raggi C., Sanchez M., D'Atri S., Carè A., Sargiacomo M., Parolini I. Acidic microenvironment plays a key role in human melanoma progression through a sustained exosome mediated transfer of clinically relevant metastatic molecules. J. Exp. Clin. Cancer Res. 2018;37(1):245. doi: 10.1186/s13046-018-0915-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X., Ying X., Wang X., Wu X., Zhu Q., Wang X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 2017;38(1):522–528. doi: 10.3892/or.2017.5697. [DOI] [PubMed] [Google Scholar]
- 25.Luo C., Xin H., Zhou Z., Hu Z., Sun R., Yao N., Sun Q., Borjigin U., Wu X., Fan J., Huang X., Zhou S., Zhou J. Hepatology; 2022. Tumor-derived Exosomes Induce Immunosuppressive Macrophages to Foster Intrahepatic Cholangiocarcinoma Progression. n/a(n/a. [DOI] [PubMed] [Google Scholar]
- 26.Kharaziha P., Ceder S., Li Q., Panaretakis T. Tumor cell-derived exosomes: a message in a bottle. Biochim. Biophys. Acta Rev. Canc. 2012;1826(1):103–111. doi: 10.1016/j.bbcan.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 27.Avgeris M., Panoutsopoulou K., Papadimitriou M.-A., Scorilas A. Circulating exosomal miRNAs: clinical significance in human cancers. Expert Rev. Mol. Diagn. 2019;19(11):979–995. doi: 10.1080/14737159.2019.1673732. [DOI] [PubMed] [Google Scholar]
- 28.Choi H., Yim H., Park C., Ahn S.-H., Ahn Y., Lee A., Yang H., Choi C. Targeted delivery of exosomes armed with anti-cancer therapeutics. Membranes. 2022;12(1) doi: 10.3390/membranes12010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morishita M., Takahashi Y., Nishikawa M., Takakura Y. Pharmacokinetics of exosomes—An important factor for elucidating the biological roles of exosomes and for the development of exosome-based therapeutics. J. Pharmaceut. Sci. 2017;106(9):2265–2269. doi: 10.1016/j.xphs.2017.02.030. [DOI] [PubMed] [Google Scholar]
- 30.Yamada T., Inoshima Y., Matsuda T., Ishiguro N. Comparison of methods for isolating exosomes from bovine milk. J. Vet. Med. Sci. 2012;74(11):1523–1525. doi: 10.1292/jvms.12-0032. [DOI] [PubMed] [Google Scholar]
- 31.Liga A., Vliegenthart A.D.B., Oosthuyzen W., Dear J.W., Kersaudy-Kerhoas M. Exosome isolation: a microfluidic road-map. Lab Chip. 2015;15(11):2388–2394. doi: 10.1039/c5lc00240k. [DOI] [PubMed] [Google Scholar]
- 32.Lamparski H.G., Metha-Damani A., Yao J.-Y., Patel S., Hsu D.-H., Ruegg C., Le Pecq J.-B. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods. 2002;270(2):211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- 33.Rider M.A., Hurwitz S.N., Meckes D.G., ExtraPEG A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 2016;6(1) doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C., Skog J., Hsu C.-H., Lessard R.T., Balaj L., Wurdinger T., Carter B.S., Breakefield X.O., Toner M., Irimia D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010;10(4):505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakai W., Yoshida T., Diez D., Miyatake Y., Nishibu T., Imawaka N., Naruse K., Sadamura Y., Hanayama R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016;6(1) doi: 10.1038/srep33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z., Wu H.-j., Fine D., Schmulen J., Hu Y., Godin B., Zhang J.X.J., Liu X. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip. 2013;13(15):2879–2882. doi: 10.1039/c3lc41343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu M., Ouyang Y., Wang Z., Zhang R., Huang P.-H., Chen C., Li H., Li P., Quinn D., Dao M., Suresh S., Sadovsky Y., Huang Tony J. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA. 2017;114(40):10584–10589. doi: 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P., Kaslan M., Lee S.H., Yao J., Gao Z. Vol. 7. 2017. Progress in Exosome Isolation Techniques, Theranostics; pp. 789–804. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roach K.A., Stefaniak A.B., Roberts J.R. Metal nanomaterials: immune effects and implications of physicochemical properties on sensitization, elicitation, and exacerbation of allergic disease. J. Immunot. 2019;16(1):87–124. doi: 10.1080/1547691X.2019.1605553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han Z., Peng C., Yi J., Zhang D., Xiang X., Peng X., Su B., Liu B., Shen Y., Qiao L. Highly efficient exosome purification from human plasma by tangential flow filtration based microfluidic chip. Sensor. Actuator. B Chem. 2021;333 [Google Scholar]
- 41.Yao S., Ren P., Song R., Liu Y., Huang Q., Dong J., O'Connor B.T., Zhu Y. Nanomaterial-enabled flexible and stretchable sensing systems: processing, integration, and applications. Adv. Mater. 2020;32(15) doi: 10.1002/adma.201902343. [DOI] [PubMed] [Google Scholar]
- 42.Chan C.K.W., Zhang L., Cheng C.K., Yang H., Huang Y., Tian X.Y., Choi C.H.J. Recent advances in managing atherosclerosis via nanomedicine. Small. 2018;14(4) doi: 10.1002/smll.201702793. [DOI] [PubMed] [Google Scholar]
- 43.Gao F., Jiao F., Xia C., Zhao Y., Ying W., Xie Y., Guan X., Tao M., Zhang Y., Qin W., Qian X. A novel strategy for facile serum exosome isolation based on specific interactions between phospholipid bilayers and TiO2. Chem. Sci. 2019;10(6):1579–1588. doi: 10.1039/c8sc04197k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang P., Zhou X., He M., Shang Y., Tetlow A.L., Godwin A.K., Zeng Y. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 2019;3(6):438–451. doi: 10.1038/s41551-019-0356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsen M.R., Thingholm T.E., Jensen O.N., Roepstorff P., Jørgensen T.J.D. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics. 2005;4(7):873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Li Y., Leng T., Lin H., Deng C., Xu X., Yao N., Yang P., Zhang X. Preparation of Fe3O4@ZrO2 Core−Shell microspheres as affinity probes for selective enrichment and direct determination of phosphopeptides using matrix-assisted laser desorption ionization mass spectrometry. J. Proteome Res. 2007;6(11):4498–4510. doi: 10.1021/pr070167s. [DOI] [PubMed] [Google Scholar]
- 47.Kweon H.K., Håkansson K. Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal. Chem. 2006;78(6):1743–1749. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 48.Geng H., Wu G., Li C., Song J., Chen P., Cai Q. Preparation of Sm-doped CaZrO3 nanosheets for facile human serum exosome isolation. New J. Chem. 2021;45(26):11719–11726. [Google Scholar]
- 49.Zhang C., Pan Y., Zhao Y., Wang P., Zhang L., Zhang W. Design and application of hydrophilic bimetallic metal-organic framework magnetic nanoparticles for rapid capture of exosomes. Anal. Chim. Acta. 2021;1186 doi: 10.1016/j.aca.2021.339099. [DOI] [PubMed] [Google Scholar]
- 50.Zhang N., Sun N., Deng C. Rapid isolation and proteome analysis of urinary exosome based on double interactions of Fe3O4@TiO2-DNA aptamer. Talanta. 2021;221 doi: 10.1016/j.talanta.2020.121571. [DOI] [PubMed] [Google Scholar]
- 51.Li Q., Wang Y., Ling L., Qiao L., Chen H., Ding C., Yu S. Rapid and specific detection nanoplatform of serum exosomes for prostate cancer diagnosis. Microchim. Acta. 2021;188(8):283. doi: 10.1007/s00604-021-04934-7. [DOI] [PubMed] [Google Scholar]
- 52.Pang Y., Shi J., Yang X., Wang C., Sun Z., Xiao R. Personalized detection of circling exosomal PD-L1 based on Fe3O4@TiO2 isolation and SERS immunoassay. Biosens. Bioelectron. 2020;148 doi: 10.1016/j.bios.2019.111800. [DOI] [PubMed] [Google Scholar]
- 53.Jiang S., Li Q., Wang C., Pang Y., Sun Z., Xiao R. In situ exosomal MicroRNA determination by target-triggered SERS and Fe3O4@TiO2-based exosome accumulation. ACS Sens. 2021;6(3):852–862. doi: 10.1021/acssensors.0c01900. [DOI] [PubMed] [Google Scholar]
- 54.Xu J.-W., Yao K., Xu Z.-K. Nanomaterials with a photothermal effect for antibacterial activities: an overview. Nanoscale. 2019;11(18):8680–8691. doi: 10.1039/c9nr01833f. [DOI] [PubMed] [Google Scholar]
- 55.Kim S., Kim J.H., Lee J.S., Park C.B. Beta-sheet-forming, self-assembled peptide nanomaterials towards optical, energy, and healthcare applications. Small. 2015;11(30):3623–3640. doi: 10.1002/smll.201500169. [DOI] [PubMed] [Google Scholar]
- 56.Wunsch B.H., Smith J.T., Gifford S.M., Wang C., Brink M., Bruce R.L., Austin R.H., Stolovitzky G., Astier Y. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat. Nanotechnol. 2016;11(11):936–940. doi: 10.1038/nnano.2016.134. [DOI] [PubMed] [Google Scholar]
- 57.Zhang P., He M., Zeng Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip. 2016;16(16):3033–3042. doi: 10.1039/c6lc00279j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suwatthanarak T., Thiodorus I.A., Tanaka M., Shimada T., Takeshita D., Yasui T., Baba Y., Okochi M. Microfluidic-based capture and release of cancer-derived exosomes via peptide–nanowire hybrid interface. Lab Chip. 2021;21(3):597–607. doi: 10.1039/d0lc00899k. [DOI] [PubMed] [Google Scholar]
- 59.Li X., Popel A.S., Karniadakis G.E. Blood–plasma separation in Y-shaped bifurcating microfluidic channels: a dissipative particle dynamics simulation study. Phys. Biol. 2012;9(2) doi: 10.1088/1478-3975/9/2/026010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Capretto L., Cheng W., Hill M., Zhang X. In: Microfluidics: Technologies and Applications. Lin B., editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 2011. Micromixing within microfluidic devices; pp. 27–68. [Google Scholar]
- 61.Chen Z., Cheng S.-B., Cao P., Qiu Q.-F., Chen Y., Xie M., Xu Y., Huang W.-H. Detection of exosomes by ZnO nanowires coated three-dimensional scaffold chip device. Biosens. Bioelectron. 2018;122:211–216. doi: 10.1016/j.bios.2018.09.033. [DOI] [PubMed] [Google Scholar]
- 62.Sooriyaarachchi D., Maharubin S., Tan G.Z. ZnO nanowire-anchored microfluidic device with herringbone structure fabricated by maskless photolithography. Biomed. Eng. Comput. Biol. 2020;11 doi: 10.1177/1179597220941431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun N., Lee Y.-T., Zhang R.Y., Kao R., Teng P.-C., Yang Y., Yang P., Wang J.J., Smalley M., Chen P.-J., Kim M., Chou S.-J., Bao L., Wang J., Zhang X., Qi D., Palomique J., Nissen N., Han S.-H.B., Sadeghi S., Finn R.S., Saab S., Busuttil R.W., Markovic D., Elashoff D., Yu H.-h., Li H., Heaney A.P., Posadas E., You S., Yang J.D., Pei R., Agopian V.G., Tseng H.-R., Zhu Y. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat. Commun. 2020;11(1):4489. doi: 10.1038/s41467-020-18311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang P., Wu X., Gardashova G., Yang Y., Zhang Y., Xu L., Zeng Y. Molecular and functional extracellular vesicle analysis using nanopatterned microchips monitors tumor progression and metastasis. Sci. Transl. Med. 2020;12(547) doi: 10.1126/scitranslmed.aaz2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Di Meo A., Bartlett J., Cheng Y., Pasic M.D., Yousef G.M. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol. Cancer. 2017;16(1):80. doi: 10.1186/s12943-017-0644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin L.-Y., Yang L., Zeng Q., Wang L., Chen M.-L., Zhao Z.-H., Ye G.-D., Luo Q.-C., Lv P.-Y., Guo Q.-W., Li B.-A., Cai J.-C., Cai W.-Y. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol. Cancer. 2018;17(1):84. doi: 10.1186/s12943-018-0834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujita K., Nonomura N. Urinary biomarkers of prostate cancer. Int. J. Urol. 2018;25(9):770–779. doi: 10.1111/iju.13734. [DOI] [PubMed] [Google Scholar]
- 68.Liu Q., Yu Z., Yuan S., Xie W., Li C., Hu Z., Xiang Y., Wu N., Wu L., Bai L., Li Y. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. 2016;8(8) doi: 10.18632/oncotarget.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ren J., Ding L., Zhang D., Shi G., Xu Q., Shen S., Wang Y., Wang T., Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X., Wang S., Wang H., Cao J., Huang X., Chen Z., Xu P., Sun G., Xu J., Lv J., Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer. 2019;18(1):20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doyle L.M., Wang M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8(7) doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Purushothaman A. In: The Extracellular Matrix: Methods and Protocols. Vigetti D., Theocharis A.D., editors. Springer New York; New York, NY: 2019. Exosomes from cell culture-conditioned medium: isolation by ultracentrifugation and characterization; pp. 233–244. [DOI] [PubMed] [Google Scholar]
- 73.Karimi-Maleh H., Karimi F., Alizadeh M., Sanati A.L. Electrochemical sensors, a bright future in the fabrication of portable kits in analytical systems. Chem. Rec. 2020;20(7):682–692. doi: 10.1002/tcr.201900092. [DOI] [PubMed] [Google Scholar]
- 74.Maduraiveeran G., Sasidharan M., Ganesan V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens. Bioelectron. 2018;103:113–129. doi: 10.1016/j.bios.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 75.Hwang D.-W., Lee S., Seo M., Chung T.D. Recent advances in electrochemical non-enzymatic glucose sensors – a review. Anal. Chim. Acta. 2018;1033:1–34. doi: 10.1016/j.aca.2018.05.051. [DOI] [PubMed] [Google Scholar]
- 76.Felix F.S., Angnes L. Electrochemical immunosensors – a powerful tool for analytical applications. Biosens. Bioelectron. 2018;102:470–478. doi: 10.1016/j.bios.2017.11.029. [DOI] [PubMed] [Google Scholar]
- 77.Karimi-Maleh H., Fakude C.T., Mabuba N., Peleyeju G.M., Arotiba O.A. The determination of 2-phenylphenol in the presence of 4-chlorophenol using nano-Fe3O4/ionic liquid paste electrode as an electrochemical sensor. J. Colloid Interface Sci. 2019;554:603–610. doi: 10.1016/j.jcis.2019.07.047. [DOI] [PubMed] [Google Scholar]
- 78.Xie F., Cao X., Qu F., Asiri A.M., Sun X. Cobalt nitride nanowire array as an efficient electrochemical sensor for glucose and H2O2 detection. Sensor. Actuator. B Chem. 2018;255:1254–1261. [Google Scholar]
- 79.Cesewski E., Johnson B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020;159 doi: 10.1016/j.bios.2020.112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun Z., Wang L., Wu S., Pan Y., Dong Y., Zhu S., Yang J., Yin Y., Li G. An electrochemical biosensor designed by using Zr-based metal–organic frameworks for the detection of glioblastoma-derived exosomes with practical application. Anal. Chem. 2020;92(5):3819–3826. doi: 10.1021/acs.analchem.9b05241. [DOI] [PubMed] [Google Scholar]
- 81.Liu Q., Yue X., Li Y., Wu F., Meng M., Yin Y., Xi R. A novel electrochemical aptasensor for exosomes determination and release based on specific host-guest interactions between cucurbit [7]uril and ferrocene. Talanta. 2021;232 doi: 10.1016/j.talanta.2021.122451. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y., Wang F., Zhang H., Wang H., Liu Y. Multivalency interface and g-C3N4 coated liquid metal nanoprobe signal amplification for sensitive electrogenerated chemiluminescence detection of exosomes and their surface proteins. Anal. Chem. 2019;91(18):12100–12107. doi: 10.1021/acs.analchem.9b03427. [DOI] [PubMed] [Google Scholar]
- 83.Kashefi-Kheyrabadi L., Kim J., Chakravarty S., Park S., Gwak H., Kim S.-I., Mohammadniaei M., Lee M.-H., Hyun K.-A., Jung H.-I. Detachable microfluidic device implemented with electrochemical aptasensor (DeMEA) for sequential analysis of cancerous exosomes. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112622. [DOI] [PubMed] [Google Scholar]
- 84.Au - Gao R., Au - Cui L.-F., Au - Ruan L.-Q., Au - Ying Y.-L., Au - Long Y.-T. A closed-type wireless nanopore electrode for analyzing single nanoparticles. JoVE. 2019;145 doi: 10.3791/59003. [DOI] [PubMed] [Google Scholar]
- 85.Ahmed M., Carrascosa L.G., Wuethrich A., Mainwaring P., Trau M. An exosomal- and interfacial-biosensing based strategy for remote monitoring of aberrantly phosphorylated proteins in lung cancer cells. Biomater. Sci. 2018;6(9):2336–2341. doi: 10.1039/c8bm00629f. [DOI] [PubMed] [Google Scholar]
- 86.Zhou Y.-G., Mohamadi R.M., Poudineh M., Kermanshah L., Ahmed S., Safaei T.S., Stojcic J., Nam R.K., Sargent E.H., Kelley S.O. Interrogating circulating microsomes and exosomes using metal nanoparticles. Small. 2016;12(6):727–732. doi: 10.1002/smll.201502365. [DOI] [PubMed] [Google Scholar]
- 87.An Y., Li R., Zhang F., He P. Magneto-mediated electrochemical sensor for simultaneous analysis of breast cancer exosomal proteins. Anal. Chem. 2020;92(7):5404–5410. doi: 10.1021/acs.analchem.0c00106. [DOI] [PubMed] [Google Scholar]
- 88.Ghazizadeh E., Naseri Z., Jaafari M.R., Forozandeh-Moghadam M., Hosseinkhani S. A fires novel report of exosomal electrochemical sensor for sensing micro RNAs by using multi covalent attachment p19 with high sensitivity. Biosens. Bioelectron. 2018;113:74–81. doi: 10.1016/j.bios.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 89.Wang F., Gui Y., Liu W., Li C., Yang Y. Precise molecular profiling of circulating exosomes using a metal–organic framework-based sensing interface and an enzyme-based electrochemical logic platform. Anal. Chem. 2022;94(2):875–883. doi: 10.1021/acs.analchem.1c03644. [DOI] [PubMed] [Google Scholar]
- 90.Boriachek K., Masud M.K., Palma C., Phan H.-P., Yamauchi Y., Hossain M.S.A., Nguyen N.-T., Salomon C., Shiddiky M.J.A. Avoiding pre-isolation step in exosome analysis: direct isolation and sensitive detection of exosomes using gold-loaded nanoporous ferric oxide nanozymes. Anal. Chem. 2019;91(6):3827–3834. doi: 10.1021/acs.analchem.8b03619. [DOI] [PubMed] [Google Scholar]
- 91.Yang L., Yin X., An B., Li F. Precise capture and direct quantification of tumor exosomes via a highly efficient dual-aptamer recognition-assisted ratiometric immobilization-free electrochemical strategy. Anal. Chem. 2021;93(3):1709–1716. doi: 10.1021/acs.analchem.0c04308. [DOI] [PubMed] [Google Scholar]
- 92.Gu C., Bai L., Pu L., Gai P., Li F. Highly sensitive and stable self-powered biosensing for exosomes based on dual metal-organic frameworks nanocarriers. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112907. [DOI] [PubMed] [Google Scholar]
- 93.Wang M., Pan Y., Wu S., Sun Z., Wang L., Yang J., Yin Y., Li G. Detection of colorectal cancer-derived exosomes based on covalent organic frameworks. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112638. [DOI] [PubMed] [Google Scholar]
- 94.Huang R., He L., Xia Y., Xu H., Liu C., Xie H., Wang S., Peng L., Liu Y., Liu Y., He N., Li Z. A sensitive aptasensor based on a hemin/G-quadruplex-assisted signal amplification strategy for electrochemical detection of gastric cancer exosomes. Small. 2019;15(19) doi: 10.1002/smll.201900735. [DOI] [PubMed] [Google Scholar]
- 95.An Y., Jin T., Zhu Y., Zhang F., He P. An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry. Biosens. Bioelectron. 2019;142 doi: 10.1016/j.bios.2019.111503. [DOI] [PubMed] [Google Scholar]
- 96.Jin D., Peng X.-X., Qin Y., Wu P., Lu H., Wang L., Huang J., Li Y., Zhang Y., Zhang G.-J., Yang F. Multivalence-actuated DNA nanomachines enable bicolor exosomal phenotyping and PD-L1-guided therapy monitoring. Anal. Chem. 2020;92(14):9877–9886. doi: 10.1021/acs.analchem.0c01387. [DOI] [PubMed] [Google Scholar]
- 97.Park J., Park J.S., Huang C.-H., Jo A., Cook K., Wang R., Lin H.-Y., Van Deun J., Li H., Min J., Wang L., Yoon G., Carter B.S., Balaj L., Choi G.-S., Castro C.M., Weissleder R., Lee H. An integrated magneto-electrochemical device for the rapid profiling of tumour extracellular vesicles from blood plasma. Nat. Biomed. Eng. 2021;5(7):678–689. doi: 10.1038/s41551-021-00752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J., Wang L.-L., Hou M.-F., Xia Y.-K., He W.-H., Yan A., Weng Y.-P., Zeng L.-P., Chen J.-H. A ratiometric electrochemical biosensor for the exosomal microRNAs detection based on bipedal DNA walkers propelled by locked nucleic acid modified toehold mediate strand displacement reaction. Biosens. Bioelectron. 2018;102:33–40. doi: 10.1016/j.bios.2017.10.050. [DOI] [PubMed] [Google Scholar]
- 99.Guo Q., Yu Y., Zhang H., Cai C., Shen Q. Electrochemical sensing of exosomal MicroRNA based on hybridization chain reaction signal amplification with reduced false-positive signals. Anal. Chem. 2020;92(7):5302–5310. doi: 10.1021/acs.analchem.9b05849. [DOI] [PubMed] [Google Scholar]
- 100.Viter R., Kunene K., Genys P., Jevdokimovs D., Erts D., Sutka A., Bisetty K., Viksna A., Ramanaviciene A., Ramanavicius A. Photoelectrochemical bisphenol S sensor based on ZnO-nanoroads modified by molecularly imprinted polypyrrole. Macromol. Chem. Phys. 2020;221(2) [Google Scholar]
- 101.Yan P., Jiang D., Tian Y., Xu L., Qian J., Li H., Xia J., Li H. A sensitive signal-on photoelectrochemical sensor for tetracycline determination using visible-light-driven flower-like CN/BiOBr composites. Biosens. Bioelectron. 2018;111:74–81. doi: 10.1016/j.bios.2018.03.054. [DOI] [PubMed] [Google Scholar]
- 102.Wang H., Zhang B., Tang Y., Wang C., Zhao F., Zeng B. Recent advances in bismuth oxyhalide-based functional materials for photoelectrochemical sensing. TrAC, Trends Anal. Chem. 2020;131 [Google Scholar]
- 103.Wang J., Liu Z. Recent advances in two-dimensional layered materials for photoelectrochemical sensing. TrAC, Trends Anal. Chem. 2020;133 [Google Scholar]
- 104.Xia Y., Chen T., Chen W., Chen G., Xu L., Zhang L., Zhang X., Sun W., Lan J., Lin X., Chen J. A dual-modal aptasensor based on a multifunctional acridone derivate for exosomes detection. Anal. Chim. Acta. 2022;1191 doi: 10.1016/j.aca.2021.339279. [DOI] [PubMed] [Google Scholar]
- 105.Pei Y., Ge Y., Zhang X., Li Y. Cathodic photoelectrochemical aptasensor based on NiO/BiOI/Au NP composite sensitized with CdSe for determination of exosomes. Microchim. Acta. 2021;188(2):51. doi: 10.1007/s00604-021-04716-1. [DOI] [PubMed] [Google Scholar]
- 106.Pang X., Zhang X., Gao K., Wan S., Cui C., Li L., Si H., Tang B., Tan W. Visible light-driven self-powered device based on a straddling nano-heterojunction and bio-application for the quantitation of exosomal RNA. ACS Nano. 2019;13(2):1817–1827. doi: 10.1021/acsnano.8b07944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Y., Yang M., Shi H., Ge S., Wang X., Yu J. Photoelectrochemical detection of exosomal miRNAs by combining target-programmed controllable signal quenching engineering. Anal. Chem. 2022;94(7):3082–3090. doi: 10.1021/acs.analchem.1c04086. [DOI] [PubMed] [Google Scholar]
- 108.Moghaddam M.R., Carrara S., Hogan C.F. Multi-colour bipolar electrochemiluminescence for heavy metal ion detection. Chem. Commun. 2019;55(8):1024–1027. doi: 10.1039/c8cc08472f. [DOI] [PubMed] [Google Scholar]
- 109.Maar R.R., Zhang R., Stephens D.G., Ding Z., Gilroy J.B. Near-infrared photoluminescence and electrochemiluminescence from a remarkably simple boron difluoride formazanate dye. Angew. Chem. Int. Ed. 2019;58(4):1052–1056. doi: 10.1002/anie.201811144. [DOI] [PubMed] [Google Scholar]
- 110.Zhao Y., Bouffier L., Xu G., Loget G., Sojic N. Electrochemiluminescence with semiconductor (nano)materials. Chem. Sci. 2022;13(9):2528–2550. doi: 10.1039/d1sc06987j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo Y., Cao Q., Zhao C., Feng Q. Stimuli-responsive DNA microcapsules for homogeneous electrochemiluminescence sensing of tumor exosomes. Sensor. Actuator. B Chem. 2021;329 [Google Scholar]
- 112.Li R., An Y., Jin T., Zhang F., He P. Detection of MUC1 protein on tumor cells and their derived exosomes for breast cancer surveillance with an electrochemiluminescence aptasensor. J. Electroanal. Chem. 2021;882 [Google Scholar]
- 113.Liu X., Wang Q., Chen J., Chen X., Yang W. Ultrasensitive electrochemiluminescence biosensor for the detection of tumor exosomes based on peptide recognition and luminol-AuNPs@g-C3N4 nanoprobe signal amplification. Talanta. 2021;221 doi: 10.1016/j.talanta.2020.121379. [DOI] [PubMed] [Google Scholar]
- 114.Xiong H., Huang Z., Lin Q., Yang B., Yan F., Liu B., Chen H., Kong J. Surface plasmon coupling electrochemiluminescence immunosensor based on polymer dots and AuNPs for ultrasensitive detection of pancreatic cancer exosomes. Anal. Chem. 2022;94(2):837–846. doi: 10.1021/acs.analchem.1c03535. [DOI] [PubMed] [Google Scholar]
- 115.Adhikari J., Rizwan M., Dennany L., Ahmed M.U. Electrochemiluminescence nanoimmunosensor for CD63 protein using a carbon nanochips/iron oxide/nafion-nanocomposite modified mesoporous carbon interface. Measurement. 2021;170 [Google Scholar]
- 116.Kim C., Lee K. Polydiacetylene (PDA) liposome-based immunosensor for the detection of exosomes. Biomacromolecules. 2019;20(9):3392–3398. doi: 10.1021/acs.biomac.9b00641. [DOI] [PubMed] [Google Scholar]
- 117.Li B., Liu C., Pan W., Shen J., Guo J., Luo T., Feng J., Situ B., An T., Zhang Y., Zheng L. Facile fluorescent aptasensor using aggregation-induced emission luminogens for exosomal proteins profiling towards liquid biopsy. Biosens. Bioelectron. 2020;168 doi: 10.1016/j.bios.2020.112520. [DOI] [PubMed] [Google Scholar]
- 118.Zhang Q., Wang F., Zhang H., Zhang Y., Liu M., Liu Y. Universal Ti3C2 MXenes based self-standard ratiometric fluorescence resonance energy transfer platform for highly sensitive detection of exosomes. Anal. Chem. 2018;90(21):12737–12744. doi: 10.1021/acs.analchem.8b03083. [DOI] [PubMed] [Google Scholar]
- 119.Fang X., Zheng Y., Duan Y., Liu Y., Zhong W. Recent advances in design of fluorescence-based assays for high-throughput screening. Anal. Chem. 2019;91(1):482–504. doi: 10.1021/acs.analchem.8b05303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yuan J., Zong S., Liu Y., Wang Z., Cui Y. Ultrasmall silica nanospheres based blinking nanoprobes for optical super resolution imaging. Opt. Mater. 2021;112 [Google Scholar]
- 121.Wu M., Chen Z., Xie Q., Xiao B., Zhou G., Chen G., Bian Z. One-step quantification of salivary exosomes based on combined aptamer recognition and quantum dot signal amplification. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112733. [DOI] [PubMed] [Google Scholar]
- 122.Li M., Chen T., Gooding J.J., Liu J. Review of carbon and graphene quantum dots for sensing. ACS Sens. 2019;4(7):1732–1748. doi: 10.1021/acssensors.9b00514. [DOI] [PubMed] [Google Scholar]
- 123.Hu C., Li M., Qiu J., Sun Y.-P. Design and fabrication of carbon dots for energy conversion and storage. Chem. Soc. Rev. 2019;48(8):2315–2337. doi: 10.1039/c8cs00750k. [DOI] [PubMed] [Google Scholar]
- 124.Xia C., Zhu S., Feng T., Yang M., Yang B. Evolution and synthesis of carbon dots: from carbon dots to carbonized polymer dots. Adv. Sci. 2019;6(23) doi: 10.1002/advs.201901316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jiang X., Zong S., Chen C., Zhang Y., Wang Z., Cui Y. Gold–carbon dots for the intracellular imaging of cancer-derived exosomes. Nanotechnology. 2018;29(17) doi: 10.1088/1361-6528/aaaf14. [DOI] [PubMed] [Google Scholar]
- 126.Xia Y., Wang L., Li J., Chen X., Lan J., Yan A., Lei Y., Yang S., Yang H., Chen J. A ratiometric fluorescent bioprobe based on carbon dots and acridone derivate for signal amplification detection exosomal microRNA. Anal. Chem. 2018;90(15):8969–8976. doi: 10.1021/acs.analchem.8b01143. [DOI] [PubMed] [Google Scholar]
- 127.Mochizuki C., Nakamura J., Nakamura M. Photostable and biocompatible luminescent thiol-terminated organosilica nanoparticles with embedded Au(I)–Thiolate complexes for fluorescent microscopic imaging. ACS Appl. Nano Mater. 2021;4(12):13305–13318. [Google Scholar]
- 128.Cursi L., Vercellino S., McCafferty M.M., Sheridan E., Petseva V., Adumeau L., Dawson K.A. Multifunctional superparamagnetic nanoparticles with a fluorescent silica shell for the in vitro study of bio-nano interactions at the subcellular scale. Nanoscale. 2021;13(38):16324–16338. doi: 10.1039/d1nr04582b. [DOI] [PubMed] [Google Scholar]
- 129.Khalin I., Heimburger D., Melnychuk N., Collot M., Groschup B., Hellal F., Reisch A., Plesnila N., Klymchenko A.S. Ultrabright fluorescent polymeric nanoparticles with a stealth pluronic shell for live tracking in the mouse brain. ACS Nano. 2020;14(8):9755–9770. doi: 10.1021/acsnano.0c01505. [DOI] [PubMed] [Google Scholar]
- 130.Ong S.Y., Zhang C., Dong X., Yao S.Q. Recent advances in polymeric nanoparticles for enhanced fluorescence and photoacoustic imaging. Angew. Chem. Int. Ed. 2021;60(33):17797–17809. doi: 10.1002/anie.202101964. [DOI] [PubMed] [Google Scholar]
- 131.Cai H., Dai X., Wang X., Tan P., Gu L., Luo Q., Zheng X., Li Z., Zhu H., Zhang H., Gu Z., Gong Q., Luo K. A nanostrategy for efficient imaging-guided antitumor therapy through a stimuli-responsive branched polymeric prodrug. Adv. Sci. 2020;7(6) doi: 10.1002/advs.201903243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hua Q., Jin Y., Wei G., Wang W., Wang L., Yin Y., Yang J., Gu Y., Ni C. Design and development of novel fluorescence sensing material for exosome recognition. Colloids Surf. B Biointerfaces. 2022;214 doi: 10.1016/j.colsurfb.2022.112421. [DOI] [PubMed] [Google Scholar]
- 133.Glembockyte V., Grabenhorst L., Trofymchuk K., Tinnefeld P. DNA origami nanoantennas for fluorescence enhancement. Accounts Chem. Res. 2021;54(17):3338–3348. doi: 10.1021/acs.accounts.1c00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu X., Zhang H., Song Z., Guo L., Fu F., Wu Y. A ratiometric nanoprobe for biosensing based on green fluorescent graphitic carbon nitride nanosheets as an internal reference and quenching platform. Biosens. Bioelectron. 2019;129:118–123. doi: 10.1016/j.bios.2019.01.032. [DOI] [PubMed] [Google Scholar]
- 135.Liao Z., Peng J., Chen S., Zhang P., Chen H., Feng D., Zhang T., Ye K., Deng Y., Dong Y., Geng L. Sensitive fluorescent sensor for the fuzzy exosomes in serum based on the exosome imprinted polymer sandwiched with aggregation induced emission. Sensor. Actuator. B Chem. 2022;358 [Google Scholar]
- 136.Gao Z., Yuan H., Mao Y., Ding L., Effah C.Y., He S., He L., Liu L.-e., Yu S., Wang Y., Wang J., Tian Y., Yu F., Guo H., Miao L., Qu L., Wu Y. In situ detection of plasma exosomal microRNA for lung cancer diagnosis using duplex-specific nuclease and MoS2 nanosheets. Analyst. 2021;146(6):1924–1931. doi: 10.1039/d0an02193h. [DOI] [PubMed] [Google Scholar]
- 137.Su S., Sun Q., Gu X., Xu Y., Shen J., Zhu D., Chao J., Fan C., Wang L. Two-dimensional nanomaterials for biosensing applications. TrAC, Trends Anal. Chem. 2019;119 [Google Scholar]
- 138.Yang T., Luo Z., Tian Y., Qian C., Duan Y. Design strategies of AuNPs-based nucleic acid colorimetric biosensors. TrAC, Trends Anal. Chem. 2020;124 [Google Scholar]
- 139.Wang L., Lin J. Recent advances on magnetic nanobead based biosensors: from separation to detection. TrAC, Trends Anal. Chem. 2020;128 [Google Scholar]
- 140.Hassan E.M., DeRosa M.C. Recent advances in cancer early detection and diagnosis: role of nucleic acid based aptasensors. TrAC, Trends Anal. Chem. 2020;124 [Google Scholar]
- 141.Wang J., Huang X., Xie J., Han Y., Huang Y., Zhang H. Exosomal analysis: advances in biosensor technology. Clin. Chim. Acta. 2021;518:142–150. doi: 10.1016/j.cca.2021.03.026. [DOI] [PubMed] [Google Scholar]
- 142.Jiang Y., Shi M., Liu Y., Wan S., Cui C., Zhang L., Tan W. Aptamer/AuNP biosensor for colorimetric profiling of exosomal proteins. Angew. Chem. Int. Ed. 2017;56(39):11916–11920. doi: 10.1002/anie.201703807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu W., Li J., Wu Y., Xing S., Lai Y., Zhang G. Target-induced proximity ligation triggers recombinase polymerase amplification and transcription-mediated amplification to detect tumor-derived exosomes in nasopharyngeal carcinoma with high sensitivity. Biosens. Bioelectron. 2018;102:204–210. doi: 10.1016/j.bios.2017.11.033. [DOI] [PubMed] [Google Scholar]
- 144.Zhang Y., Jiao J., Wei Y., Wang D., Yang C., Xu Z. Plasmonic colorimetric biosensor for sensitive exosome detection via enzyme-induced etching of gold nanobipyramid@MnO2 nanosheet nanostructures. Anal. Chem. 2020;92(22):15244–15252. doi: 10.1021/acs.analchem.0c04136. [DOI] [PubMed] [Google Scholar]
- 145.Zhang Y., Wang D., Yue S., Lu Y., Yang C., Fang J., Xu Z. Sensitive multicolor visual detection of exosomes via dual signal amplification strategy of enzyme-catalyzed metallization of Au nanorods and hybridization chain reaction. ACS Sens. 2019;4(12):3210–3218. doi: 10.1021/acssensors.9b01644. [DOI] [PubMed] [Google Scholar]
- 146.Wang Y.-M., Liu J.-W., Adkins G.B., Shen W., Trinh M.P., Duan L.-Y., Jiang J.-H., Zhong W. Enhancement of the intrinsic peroxidase-like activity of graphitic carbon nitride nanosheets by ssDNAs and its application for detection of exosomes. Anal. Chem. 2017;89(22):12327–12333. doi: 10.1021/acs.analchem.7b03335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang Y., Su Q., Song D., Fan J., Xu Z. Label-free detection of exosomes based on ssDNA-modulated oxidase-mimicking activity of CuCo2O4 nanorods. Anal. Chim. Acta. 2021;1145:9–16. doi: 10.1016/j.aca.2020.12.018. [DOI] [PubMed] [Google Scholar]
- 148.Xiao J., Hai L., Li Y., Li H., Gong M., Wang Z., Tang Z., Deng L., He D. An ultrasmall Fe3O4-decorated polydopamine hybrid nanozyme enables continuous conversion of oxygen into toxic hydroxyl radical via GSH-depleted cascade redox reactions for intensive wound disinfection. Small. 2022;18(9) doi: 10.1002/smll.202105465. [DOI] [PubMed] [Google Scholar]
- 149.Kim M.S., Kweon S.H., Cho S., An S.S.A., Kim M.I., Doh J., Lee J. Pt-decorated magnetic nanozymes for facile and sensitive point-of-care bioassay. ACS Appl. Mater. Interfaces. 2017;9(40):35133–35140. doi: 10.1021/acsami.7b12326. [DOI] [PubMed] [Google Scholar]
- 150.Jiao L., Yan H., Wu Y., Gu W., Zhu C., Du D., Lin Y. When nanozymes meet single-atom catalysis. Angew. Chem. Int. Ed. 2020;59(7):2565–2576. doi: 10.1002/anie.201905645. [DOI] [PubMed] [Google Scholar]
- 151.Ma Q., Liu Y., Zhu H., Zhang L., Liao X. Nanozymes in tumor theranostics. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.666017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xia Y., Liu M., Wang L., Yan A., He W., Chen M., Lan J., Xu J., Guan L., Chen J. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 2017;92:8–15. doi: 10.1016/j.bios.2017.01.063. [DOI] [PubMed] [Google Scholar]
- 153.Di H., Mi Z., Sun Y., Liu X., Liu X., Li A., Jiang Y., Gao H., Rong P., Liu D. Nanozyme-assisted sensitive profiling of exosomal proteins for rapid cancer diagnosis. Theranostics. 2020;10(20):9303–9314. doi: 10.7150/thno.46568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sarycheva A., Gogotsi Y. Raman spectroscopy analysis of the structure and surface chemistry of Ti3C2Tx MXene. Chem. Mater. 2020;32(8):3480–3488. [Google Scholar]
- 155.Jones R.R., Hooper D.C., Zhang L., Wolverson D., Valev V.K. Raman techniques: fundamentals and frontiers. Nanoscale Res. Lett. 2019;14(1):231. doi: 10.1186/s11671-019-3039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rojalin T., Koster H.J., Liu J., Mizenko R.R., Tran D., Wachsmann-Hogiu S., Carney R.P. Hybrid nanoplasmonic porous biomaterial scaffold for liquid biopsy diagnostics using extracellular vesicles. ACS Sens. 2020;5(9):2820–2833. doi: 10.1021/acssensors.0c00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shin H., Oh S., Hong S., Kang M., Kang D., Ji Y.-g., Choi B.H., Kang K.-W., Jeong H., Park Y., Hong S., Kim H.K., Choi Y. Early-stage lung cancer diagnosis by deep learning-based spectroscopic analysis of circulating exosomes. ACS Nano. 2020;14(5):5435–5444. doi: 10.1021/acsnano.9b09119. [DOI] [PubMed] [Google Scholar]
- 158.Shin H., Jeong H., Park J., Hong S., Choi Y. Correlation between cancerous exosomes and protein markers based on surface-enhanced Raman spectroscopy (SERS) and principal component analysis (PCA) ACS Sens. 2018;3(12):2637–2643. doi: 10.1021/acssensors.8b01047. [DOI] [PubMed] [Google Scholar]
- 159.Li J., Li Y., Chen S., Duan W., Kong X., Wang Y., Zhou L., Li P., Zhang C., Du L., Wang C. Highly sensitive exosome detection for early diagnosis of pancreatic cancer using immunoassay based on hierarchical surface-enhanced Raman scattering substrate. Small Methods. 2022 doi: 10.1002/smtd.202200154. n/a(n/a. [DOI] [PubMed] [Google Scholar]
- 160.Dong S., Wang Y., Liu Z., Zhang W., Yi K., Zhang X., Zhang X., Jiang C., Yang S., Wang F., Xiao X. Beehive-inspired macroporous SERS probe for cancer detection through capturing and analyzing exosomes in plasma. ACS Appl. Mater. Interfaces. 2020;12(4):5136–5146. doi: 10.1021/acsami.9b21333. [DOI] [PubMed] [Google Scholar]
- 161.Wang Z., Zong S., Wang Y., Li N., Li L., Lu J., Wang Z., Chen B., Cui Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale. 2018;10(19):9053–9062. doi: 10.1039/c7nr09162a. [DOI] [PubMed] [Google Scholar]
- 162.Lee J.U., Kim W.H., Lee H.S., Park K.H., Sim S.J. Quantitative and specific detection of exosomal miRNAs for accurate diagnosis of breast cancer using a surface-enhanced Raman scattering sensor based on plasmonic head-flocked gold nanopillars. Small. 2019;15(17) doi: 10.1002/smll.201804968. [DOI] [PubMed] [Google Scholar]
- 163.Kang T., Zhu J., Luo X., Jia W., Wu P., Cai C. Controlled self-assembly of a close-packed gold octahedra array for SERS sensing exosomal MicroRNAs. Anal. Chem. 2021;93(4):2519–2526. doi: 10.1021/acs.analchem.0c04561. [DOI] [PubMed] [Google Scholar]
- 164.Zhang X., Liu C., Pei Y., Song W., Zhang S. Preparation of a novel Raman probe and its application in the detection of circulating tumor cells and exosomes. ACS Appl. Mater. Interfaces. 2019;11(32):28671–28680. doi: 10.1021/acsami.9b09465. [DOI] [PubMed] [Google Scholar]
- 165.Fraire J.C., Stremersch S., Bouckaert D., Monteyne T., De Beer T., Wuytens P., De Rycke R., Skirtach A.G., Raemdonck K., De Smedt S., Braeckmans K. Improved label-free identification of individual exosome-like vesicles with Au@Ag nanoparticles as SERS substrate. ACS Appl. Mater. Interfaces. 2019;11(43):39424–39435. doi: 10.1021/acsami.9b11473. [DOI] [PubMed] [Google Scholar]
- 166.Yan Z., Dutta S., Liu Z., Yu X., Mesgarzadeh N., Ji F., Bitan G., Xie Y.-H. A label-free platform for identification of exosomes from different sources. ACS Sens. 2019;4(2):488–497. doi: 10.1021/acssensors.8b01564. [DOI] [PubMed] [Google Scholar]
- 167.Pramanik A., Mayer J., Patibandla S., Gates K., Gao Y., Davis D., Seshadri R., Ray P.C. Mixed-dimensional heterostructure material-based SERS for trace level identification of breast cancer-derived exosomes. ACS Omega. 2020;5(27):16602–16611. doi: 10.1021/acsomega.0c01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen H., Luo C., Yang M., Li J., Ma P., Zhang X. Intracellular uptake of and sensing with SERS-active hybrid exosomes: insight into a role of metal nanoparticles. Nanomedicine. 2020;15(9):913–926. doi: 10.2217/nnm-2019-0419. [DOI] [PubMed] [Google Scholar]
- 169.Kwizera E.A., O'Connor R., Vinduska V., Williams M., Butch E.R., Snyder S.E., Chen X., Huang X. Molecular detection and analysis of exosomes using surface-enhanced Raman scattering gold nanorods and a miniaturized device. Theranostics. 2018;8(10):2722–2738. doi: 10.7150/thno.21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang Y., Li Q., Shi H., Tang K., Qiao L., Yu G., Ding C., Yu S. Microfluidic Raman biochip detection of exosomes: a promising tool for prostate cancer diagnosis. Lab Chip. 2020;20(24):4632–4637. doi: 10.1039/d0lc00677g. [DOI] [PubMed] [Google Scholar]
- 171.Wang J., Kao Y.-C., Zhou Q., Wuethrich A., Stark M.S., Schaider H., Soyer H.P., Lin L.L., Trau M. An integrated microfluidic-SERS platform enables sensitive phenotyping of serum extracellular vesicles in early stage melanomas. Adv. Funct. Mater. 2022;32(3) [Google Scholar]
- 172.Ning C.-F., Wang L., Tian Y.-F., Yin B.-C., Ye B.-C. Multiple and sensitive SERS detection of cancer-related exosomes based on gold–silver bimetallic nanotrepangs. Analyst. 2020;145(7):2795–2804. doi: 10.1039/c9an02180a. [DOI] [PubMed] [Google Scholar]
- 173.Yu H., Peng Y., Yang Y., Li Z.-Y. Plasmon-enhanced light–matter interactions and applications. npj Computat. Mater. 2019;5(1):45. [Google Scholar]
- 174.Gellé A., Jin T., de la Garza L., Price G.D., Besteiro L.V., Moores A. Applications of plasmon-enhanced nanocatalysis to organic transformations. Chem. Rev. 2020;120(2):986–1041. doi: 10.1021/acs.chemrev.9b00187. [DOI] [PubMed] [Google Scholar]
- 175.Zhao Y., Tong R.-j., Xia F., Peng Y. Current status of optical fiber biosensor based on surface plasmon resonance. Biosens. Bioelectron. 2019;142 doi: 10.1016/j.bios.2019.111505. [DOI] [PubMed] [Google Scholar]
- 176.Wang Q., Zou L., Yang X., Liu X., Nie W., Zheng Y., Cheng Q., Wang K. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens. Bioelectron. 2019;135:129–136. doi: 10.1016/j.bios.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 177.Fan Y., Duan X., Zhao M., Wei X., Wu J., Chen W., Liu P., Cheng W., Cheng Q., Ding S. High-sensitive and multiplex biosensing assay of NSCLC-derived exosomes via different recognition sites based on SPRi array. Biosens. Bioelectron. 2020;154 doi: 10.1016/j.bios.2020.112066. [DOI] [PubMed] [Google Scholar]
- 178.Mao Z., Zhao J., Chen J., Hu X., Koh K., Chen H. A simple and direct SPR platform combining three-in-one multifunctional peptides for ultra-sensitive detection of PD-L1 exosomes. Sensor. Actuator. B Chem. 2021;346 [Google Scholar]
- 179.Liu C., Zeng X., An Z., Yang Y., Eisenbaum M., Gu X., Jornet J.M., Dy G.K., Reid M.E., Gan Q., Wu Y. Sensitive detection of exosomal proteins via a compact surface plasmon resonance biosensor for cancer diagnosis. ACS Sens. 2018;3(8):1471–1479. doi: 10.1021/acssensors.8b00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.VahidMohammadi A., Rosen J., Gogotsi Y. The world of two-dimensional carbides and nitrides (MXenes) Science. 2021;372(6547) doi: 10.1126/science.abf1581. [DOI] [PubMed] [Google Scholar]
- 181.Sinha A., Dhanjai, Zhao H., Huang Y., Lu X., Chen J., Jain R. MXene: an emerging material for sensing and biosensing. TrAC, Trends Anal. Chem. 2018;105:424–435. [Google Scholar]
- 182.Verger L., Xu C., Natu V., Cheng H.-M., Ren W., Barsoum M.W. Overview of the synthesis of MXenes and other ultrathin 2D transition metal carbides and nitrides. Curr. Opin. Solid State Mater. Sci. 2019;23(3):149–163. [Google Scholar]
- 183.Qiu G., Thakur A., Xu C., Ng S.-P., Lee Y., Wu C.-M.L. Detection of glioma-derived exosomes with the biotinylated antibody-functionalized titanium nitride plasmonic biosensor. Adv. Funct. Mater. 2019;29(9) [Google Scholar]
- 184.Sun X., Huang L., Zhang R., Xu W., Huang J., Gurav D.D., Vedarethinam V., Chen R., Lou J., Wang Q., Wan J., Qian K. Metabolic fingerprinting on a plasmonic gold chip for mass spectrometry based in vitro diagnostics. ACS Cent. Sci. 2018;4(2):223–229. doi: 10.1021/acscentsci.7b00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Xia C., Jiao F., Gao F., Wang H., Lv Y., Shen Y., Zhang Y., Qian X. Two-dimensional MoS2-based zwitterionic hydrophilic interaction liquid chromatography material for the specific enrichment of glycopeptides. Anal. Chem. 2018;90(11):6651–6659. doi: 10.1021/acs.analchem.8b00461. [DOI] [PubMed] [Google Scholar]
- 186.Chen H., Zhang N., Wu Y., Yang C., Xie Q., Deng C., Sun N. Investigation of urinary exosome metabolic patterns in membranous nephropathy by titania-assisted intact exosome mass spectrometry. Small Sci. 2022;2(5) [Google Scholar]
- 187.Han Z., Peng C., Yi J., Wang Y., Liu Q., Yang Y., Long S., Qiao L., Shen Y. Matrix-assisted laser desorption ionization mass spectrometry profiling of plasma exosomes evaluates osteosarcoma metastasis. iScience. 2021;24(8) doi: 10.1016/j.isci.2021.102906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang Y., Zhang K., Huang X., Qiao L., Liu B. Mass spectrometry imaging of mass tag immunoassay enables the quantitative profiling of biomarkers from dozens of exosomes. Anal. Chem. 2021;93(2):709–714. doi: 10.1021/acs.analchem.0c03904. [DOI] [PubMed] [Google Scholar]
- 189.Fang X., Duan Y., Adkins G.B., Pan S., Wang H., Liu Y., Zhong W. Highly efficient exosome isolation and protein analysis by an integrated nanomaterial-based platform. Anal. Chem. 2018;90(4):2787–2795. doi: 10.1021/acs.analchem.7b04861. [DOI] [PMC free article] [PubMed] [Google Scholar]