Highlights
-
•
Dice score and Hausdorff distance do not correlate with dose-volume-based results.
-
•
Auto-contours close to the tumor or in entry/exit beams should be checked.
-
•
Heart and esophagus must be checked for locally advanced non-small cell lung cancer.
-
•
Bronchi must be checked for peripheral early-stage non-small cell lung cancer.
-
•
Every treatment plan still passed the clinical goals for the manual organs at risk.
Keywords: Lung cancer, Radiotherapy, Treatment planning, Dose, Volume, Dice
Abstract
Background and purpose
The geometrical accuracy of auto-segmentation using convolutional neural networks (CNNs) has been demonstrated. This study aimed to investigate the dose-volume impact of differences between automatic and manual OARs for locally advanced (LA) and peripherally located early-stage (ES) non-small cell lung cancer (NSCLC).
Material and methods
A single CNN was created for automatic delineation of the heart, lungs, main left and right bronchus, esophagus, spinal cord and trachea using 55/10/40 patients for training/validation/testing. Dice score coefficient (DSC) and 95th percentile Hausdorff distance (HD95) were used for geometrical analysis. A new treatment plan based on the auto-segmented OARs was created for each test patient using 3D for ES-NSCLC (SBRT, 3–8 fractions) and IMRT for LA-NSCLC (24–35 fractions). The correlation between geometrical metrics and dose-volume differences was investigated.
Results
The average (±1 SD) DSC and HD95 were 0.82 ± 0.07 and 16.2 ± 22.4 mm, while the average dose-volume differences were 0.5 ± 1.5 Gy (ES) and 1.5 ± 2.8 Gy (LA). The geometrical metrics did not correlate with the observed dose-volume differences (average Pearson for DSC: −0.27 ± 0.18 (ES) and −0.09 ± 0.12 (LA); HD95: 0.1 ± 0.3 mm (ES) and 0.2 ± 0.2 mm (LA)).
Conclusions
After post-processing, manual adjustments of automatic contours are only needed for clinically relevant OARs situated close to the tumor or within an entry or exit beam e.g., the heart and the esophagus for LA-NSCLC and the bronchi for ES-NSCLC. The lungs do not need to be checked further in detail.
1. Introduction
Lung cancer is the second most frequent cancer worldwide, with a global incidence of over 2.2 million cases in 2020, representing 11.4 % of all cancer diagnoses, only just overtaken by breast cancer (11.7 %). It is the first cause of cancer death [1]. Surgery remains the treatment of choice for early-stage non-small cell lung cancer (ES-NSCLC) without lymph node invasion (stage I and IIA disease), but patients with unresectable tumors or unwilling to undergo surgery are candidates for stereotactic body radiation therapy (SBRT), especially if the tumor is peripherally located. In case of stage III disease, also referred to as locally-advanced NSCLC (LA-NSCLC), the loco-regional treatment strategy can consist of a surgical or a non-surgical multi-modality approach [2]. In the latter case, radiotherapy will be delivered, all or not in combination with chemotherapy and/or immunotherapy.
As such, radiotherapy is a very important part of lung cancer treatment, where errors in target volume (TV) and organ at risk (OAR) delineation may lead to suboptimal tumor control and/or increased toxicity. Model-based segmentation [3], [4], atlas-based segmentation [5], [6] and deep learning [7], [8], [9] methods have been investigated in the hope to enhance the quality of the radiation treatments delivered by improving delineation accuracy and decreasing inter-observer variability. Currently the most investigated method for auto-segmentation of medical images is the use of convolutional neural network (CNN)-based deep learning algorithms [7]. Automatic contours are increasingly used in the clinic, but time-consuming manual adjustments are often still made. Vaassen et al. have drawn up recommendations for manual checks of automatically contoured OARs in NSCLC, based on the dose-volume effect of contour variations for patients receiving 60 Gy in 30 fractions [10]. The clinical impact of (atlas-based or deep-learning based) automatic contours for thoracic OARs has been assessed by means of observers’ rating [8], [11], [12], concordance indices [13], geometrical metrics [14], [15] and dose statistics [16], [10].
This study aimed to assess the dose-volume impact of CNN-based auto-segmentation of OARs in a cohort of lung cancer patients, where ES-NSCLC was treated with SBRT and LA-NSCLC with standard or moderately hypo-fractionated schedules. The correlation between geometrical metrics (Dice score coefficients and 95th percentile Hausdorff distances) and dose-volume histogram (DVH) values was investigated, and any outliers were further examined. The impact of auto-segmentation on treatment plan acceptance based on clinical goals was also evaluated.
2. Material and methods
2.1. Patient data
CT images and manual OAR delineations of the heart, lungs, left and right main bronchus, esophagus, spinal cord and trachea were retrospectively collected from 105 lung cancer patients with LA-NSCLC or peripherally located ES-NSCLC treated at our institution between February 2019 and March 2021. All patient data were processed and analyzed anonymously according to the guidelines of Ghent University Hospital’s Ethics Committee. Detailed patient characteristics can be found in Supplementary table S1.
All OARs were manually delineated by two experienced dosimetrists and subsequently checked by an experienced radiation oncologist (RO). Manual segmentation of the lungs was performed using the automatic region growing tool in RayStation 6 (RaySearch Laboratories, Stockholm, Sweden) after which the dosimetrists manually corrected the contours. The gross target volumes (GTVs) of all patients were delineated on a thoracic CT scan with a slice thickness of 2 mm by a RO. For ES-NSCLC the delineations were aided by the maximum intensity projection (MIP) scan, derived from the 4D-CT scan. For LA-NSCLC the delineation was supported using a contrast agent administered during CT-simulation. The clinical target volume (CTV, for LA-NSCLC), planning target volume (PTV) and any planning risk volumes (PRVs) were derived from the GTV and OARs according to in-house standardized protocols.
Forty out of the 105 patients were used for CNN testing through geometrical analysis and for the subsequent dose-volume-based evaluation. Twenty patients had an early-stage peripheral lesion (ES-group), while 20 patients suffered from locally advanced disease (LA-group). In the ES-group, all patients had T1a-T3N0 NSCLC except for one patient that had a stage IV NSCLC with oligoprogression on immunotherapy in the original primary tumor. All received 3 to 8 fractions with SBRT up to a dose of 60 Gy to the PTV-D95 using a 3D conformal treatment technique because of robustness. In the LA-group, all patients were treated with step-and-shoot intensity-modulated radiotherapy (IMRT). Half of them had concurrent chemo-radiotherapy and received between 30 and 35 fractions of 2 Gy to the PTV-D50, while the other 10 patients received radiotherapy alone or sequentially to chemotherapy and were treated with hypofractionated radiotherapy (24 fractions of 2.75 Gy to the PTV-D50).
All patients were treated on an Elekta Synergy Agility or Varian Clinac iX linear accelerator using 6 and/or 15 MV photons. Clinical goals in terms of dose-volume statistics were defined for each patient separately by the RO, depending on the dose prescription and the location of the tumor. Six to 9 beams were used for each patient based on a standard 9-beam template with the following gantry/collimator/table angles: 155°/0°/0°, 0°/0°/0°, 205°/0°/0°, 315°/45°/45°, 30°/45°/45°, 60°/45°/45°, 45°/45°/315°, 330°/315°/45°, 300°/45°/315°, adjusted according to the individual patient anatomy by an experienced dosimetrist.
2.2. Convolutional neural network
A CNN for auto-segmentation of OARs was developed using data from 55 patients (14 599 images) for training, 10 patients (2 705 images) for validation and 40 patients (10 615 images) for testing. The CNN was trained on individual CT images with a batch size of 8.
Pre-processing of the training and validation data for the CNN was performed by generating organ masks based on the Digital Imaging and Communications in Medicine (DICOM) CT images and DICOM RT Struct files for each patient. Organ masks were created by activating the image voxels included within each contour for the OAR on each CT image slice. For the CT images, data normalization was performed through a linear projection of the HU from the [-1000, 3095] interval to [0, 1] [17]. Each image was cropped from 512x512 voxels to the most central 256x256 voxels as all OARs were present in this region. This reduced the input into the CNN fourfold, speeding up the training process. The body outline was not always entirely present in the cropped images. Data augmentation was performed by rotation of the CT images over a range of [-5, 5] degrees and scaling over a factor [0.9, 1.1] [18]. Because the model was trained for 100 epochs, this effectively created 1 459 900 different training images.
A U-Net model [19] was trained on the High-Performance Computing Infrastructure at Ghent University using two 2.8 GHz cores, two 32 GB GPUs and 275 GB usable memory. Pre-trained encoder weights were used from ImageNet [20] as proven useful in previous similar studies [21]. The U-Net based model was provided by the python module “segmentation models” [22]. During training the following loss metric was used: 1–2(|X||Y|)/(|X|+|Y|), where |X| and |Y| represent the number of voxels within the manual and auto-segmented areas. To classify multiple OARs in a single CNN model, an averaged loss was used for training [23]. The input format of the CNN model was a 256x256x1 matrix corresponding to the cropped CT image, while the output format was a 256x256x7 matrix containing the voxel-wise probability p̂ for each OAR (7 in total). A voxel was assigned to a certain OAR if p̂ ≥ 0.5.
The geometrical performance of the CNN was assessed using both the Dice score coefficient (DSC) and the 95th percentile Hausdorff distance (HD95). DSC was defined as 2(|X||Y|)/(|X|+|Y|), while HD95 was defined as the 95th percentile of the ordered distance measures for the maximum distance to agreement between the manual and the automatic OAR. The correlation between DSC and HD95 was assessed by means of the Pearson correlation coefficient.
Further details regarding the CNN model, in line with the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) guidelines [24], can be found in Supplementary material.
2.3. Dose-volume-based evaluation
For each of the 40 patients in the test set, a new treatment plan was created based on the auto-segmented OARs without any further manual adjustments, while keeping the target volumes from the manual delineations. Each new plan was optimized according to our standard clinical procedure, using the same beam setup and the same clinical goals as for the original plan but re-adapting the optimization objectives when necessary. Finally, each new plan was evaluated on both the auto-segmented (“auto”) and the manual OARs (“manual”).
The dose-volume statistics listed in Table 1 were compared and absolute dose-volume differences were calculated for each patient as follows: |Dauto – Dmanual| or |Vauto – Vmanual|.
Table 1.
Pearson correlation coefficients and Wilcoxon rank sum test p-values for the difference in dose-volume statistics between the auto-segmented and manually delineated OARs for each patient group. Statistically significant differences (p < 0.05) are written in bold and marked with an *. D2 corresponds to the dose that the OAR receives on 2 % of its volume and V20 and V5 correspond to the percentage volume of the organ receiving at least 20 Gy and 5 Gy, respectively. Dmean corresponds to the mean dose.
| OAR | Dose-volume statistic | Early-stage |
Locally advanced |
||
|---|---|---|---|---|---|
| P-value | Pearson | P-value | Pearson | ||
| Lungs | Dmean | 0.001* | 0.999 | 0.509 | 0.999 |
| V20 | 0.019* | 0.998 | 0.944 | 0.999 | |
| V5 | 0.254 | 0.998 | 0.529 | 1.000 | |
| Heart | Dmean | 0.865 | 0.977 | 0.529 | 0.969 |
| D2 | 0.689 | 0.966 | 0.503 | 0.913 | |
| Esophagus | Dmean | 0.889 | 0.775 | 0.682 | 0.986 |
| D2 | 0.313 | 0.994 | 0.857 | 0.990 | |
| Main left bronchus | D2 | 0.749 | 0.987 | 0.453 | 0.995 |
| Main right bronchus | D2 | 0.575 | 0.932 | 0.412 | 0.998 |
| Spinal cord | D2 | 0.267 | 0.998 | 0.857 | 0.999 |
| Trachea | D2 | 1.000 | 0.998 | 0.944 | 0.990 |
A Wilcoxon rank sum test () was performed to assess the statistical significance of the dose-volume differences between the automatic and manual delineations. The Pearson correlation coefficient was also calculated for the dose-volume differences between both contour sets, as well as between each geometrical metric (DSC and HD95) and the dose difference.
3. Results
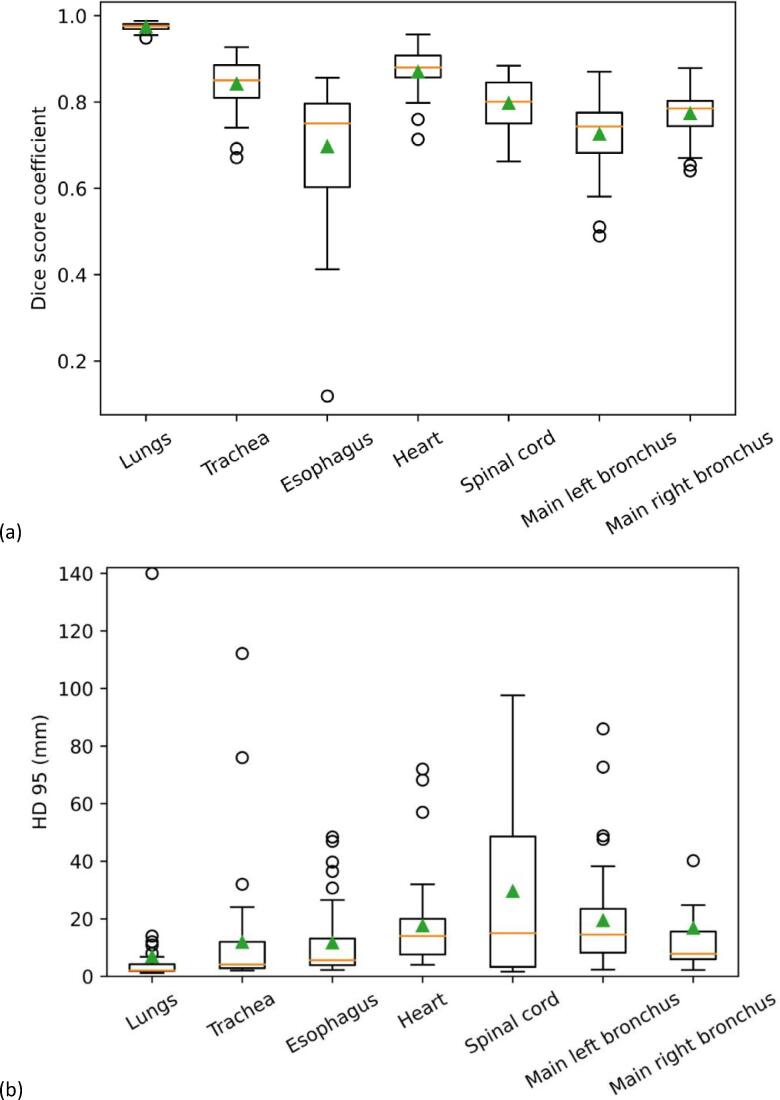
On average, the highest DSC (0.98 ± 0.01) and lowest HD95 (6.9 ± 21.8 mm) were found for the lungs, while the lowest DSC (0.72 ± 0.15) was found for the esophagus and the largest HD95 (29.6 ± 29.6 mm) for the spinal cord (Table 2 and Supplementary table S3). The mean Pearson correlation coefficient between DSC and HD95 over all OARs was –0.35, with the highest correlation found for the lungs (-0.51) and the lowest for the heart (0.09). The similarity of the DSC values for different thresholds of p̂ indicated the robustness of the CNN model for threshold variations (Supplementary table S2).
Table 2.
OAR volume and Dice score coefficients between the manual and automatic contours and a summary of average Dice score coefficients for automatic segmentation methods (including CNN methods) of thoracic OARs found in literature. If multiple values are reported in a single reference, they are mentioned between square brackets.
| OAR | Volume of the manual delineation (mean ± 1 SD) [cm3] | Volume of the automatic delineation (mean ± 1 SD) [cm3] | Average DSC ± 1 SD | Average DSC from previous studies on auto-segmentation | Average DSC from previous studies on inter-observer variability |
|---|---|---|---|---|---|
| Lungs | 3780 ± 1004 | 3708 ± 1000 | 0.98 ± 0.01 | 0.97 [40] | 0.97 [26] |
| 0.95 [39] | [0.98,0.97] [38] | ||||
| 0.99 [43] | 0.95 [39] | ||||
| 0.97 [16] | 0.98 [44] | ||||
| Trachea | 36 ± 13 | 36 ± 12 | 0.84 ± 0.06 | 0.93 [23] | 0.97 [26] |
| 0.91 [42] | |||||
| Esophagus | 46 ± 40 | 31 ± 9 | 0.72 ± 0.15 | 0.73 [40] | 0.64 [26] |
| 0.86 [23] | [0.77,0.76] [38] | ||||
| 0.64 [39] | 0.83 [39] | ||||
| 0.82 [43] | |||||
| 0.75 [16] | |||||
| Heart | 691 ± 150 | 694 ± 142 | 0.91 ± 0.06 | 0.85 [40] | 0.92 [26] |
| 0.94 [23] | [0.86,0.87] [38] | ||||
| [0.87,0.88] [41] | 0.94 [39] | ||||
| 0.91 [39] | 0.91 [44] | ||||
| 0.94 [43] | |||||
| 0.87 [16] | |||||
| Spinal cord | 56 ± 11 | 51 ± 9 | 0.80 ± 0.06 | 0.88 [40] | 0.74 [26] |
| [0.69,0.81] [41] | [0.70,0.80] [43] | ||||
| 0.76 [39] | [0.81, 0.76] [38] | ||||
| 0.90 [16] | 0.80 [39] | ||||
| 0.81 [44] | |||||
| Main left bronchus | 9 ± 4 | 8 ± 4 | 0.75 ± 0.08 | ||
| Main right bronchus | 10 ± 4 | 9 ± 3 | 0.78 ± 0.05 |
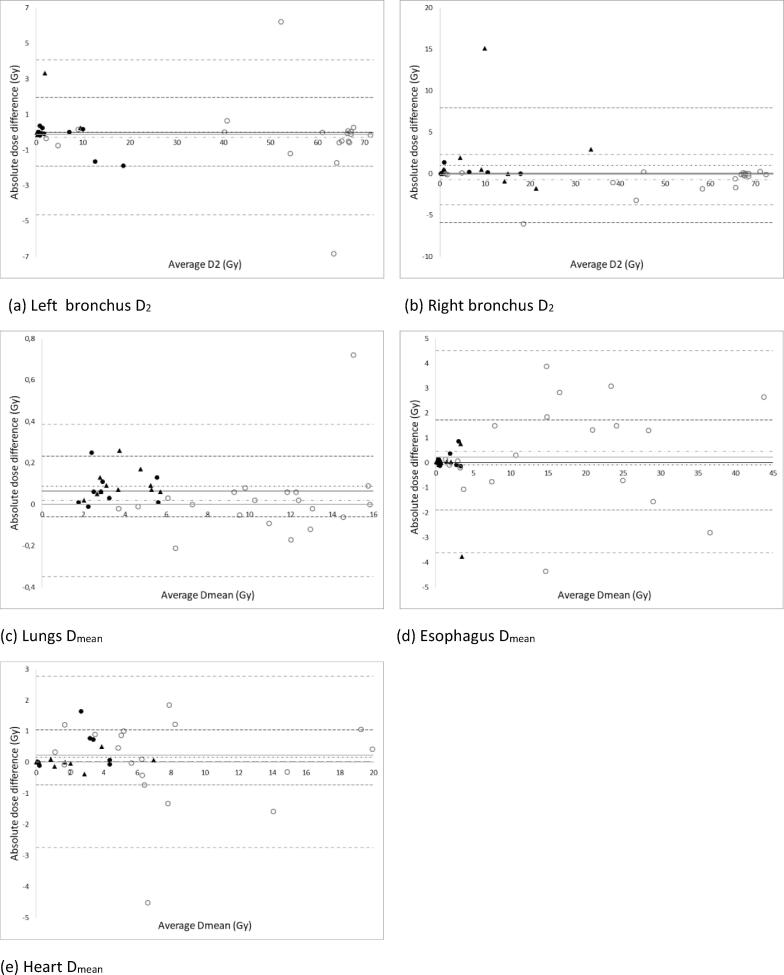
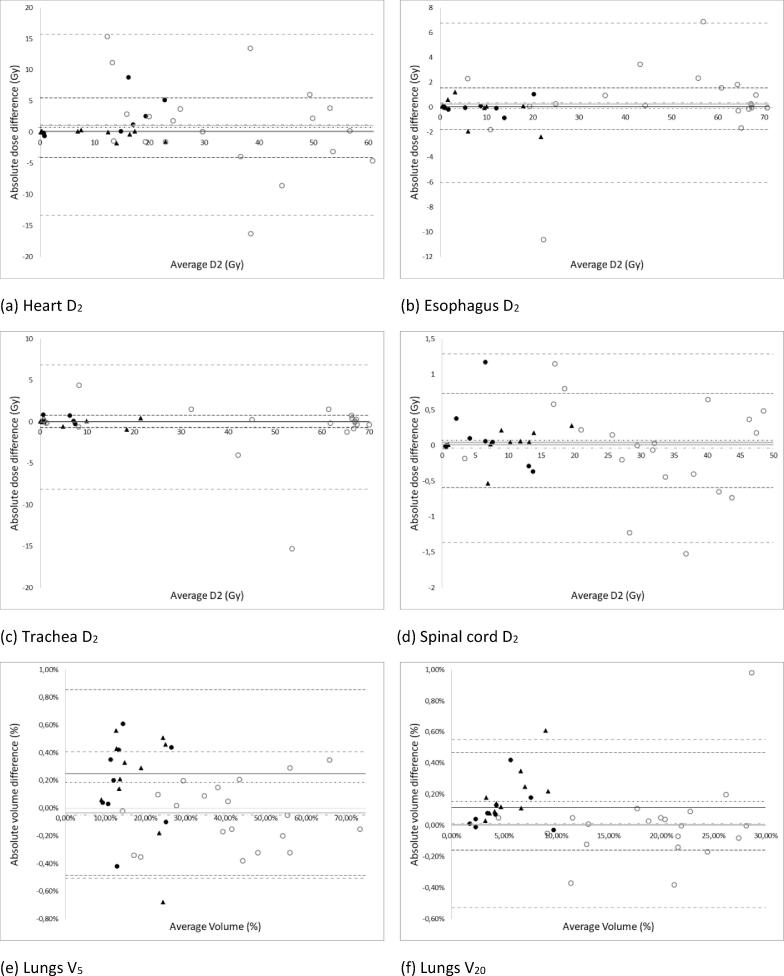
Bland-Altman plots for the absolute dose-volume differences are shown in Fig. 2, Fig. 3. The largest absolute difference was noted for ES – main right bronchus D2 (15.09 Gy), while the smallest absolute difference could be seen for the mean lung dose (ES) (0.01 Gy). The only statistically significant differences were observed in the ES group for the lungs Dmean (p = 0.001) and V20 (p = 0.019) (Table 1). However, as the absolute differences were very low (<0.3 Gy and 0.61 %) this was not clinically relevant. Apart from the mean dose to the esophagus in the ES group, all Pearson correlation coefficients between the manual and automatic contour-based dose-volume statistics were higher than 0.9. The lower value for the mean esophagus dose (0.775) was due to an extreme outlier (-3.77 Gy) (Fig. 2d). After visual inspection of the 3D dose distribution in the treatment planning system, all outliers were found to be related to delineation differences within high dose gradient regions and/or passing an entry or exit beam (see Fig. 4).
Fig. 2.
Absolute difference in dose values between the automatic and manual OAR (on the Y-axis) against the average of the dose between the automatic and manual delineation (on the X-axis). Each data point represents a patient from the test set (n = 40). The empty grey circles correspond to LA tumors and the full black discs and triangles to ES tumors. The triangles refer to patients with a right-sided tumor while the dots refer to patients with a left-sided tumor. The median (full line), the mean (dash-dotted line) and the mean ± 2 SD (dashed lines) of the absolute difference are indicated on each graph (LA = grey; ES = black). Any datapoints outside of the 2 SD limits were considered as outliers.
Fig. 3.
(a-d) Absolute difference in dose values between the automatic and manual OAR (on the Y-axis) against the average of the dose between the automatic and manual delineation (on the X-axis). (e,f) Absolute difference in volume values between the automatic and manual OAR (on the Y-axis) against the average of the volume between the automatic and manual delineation (on the X-axis). Each data point represents a patient from the test set (n = 40). The empty grey circles correspond to LA tumors and the full black discs and triangles to ES tumors. The triangles refer to patients with a right-sided tumor while the dots refer to patients with a left-sided tumor. The median (full line), the mean (dash-dotted line) and the mean ± 2 SD (dashed lines) of the absolute difference are indicated on each graph (LA = grey; ES = black). Any data points outside of the 2 SD limits were considered as outliers.
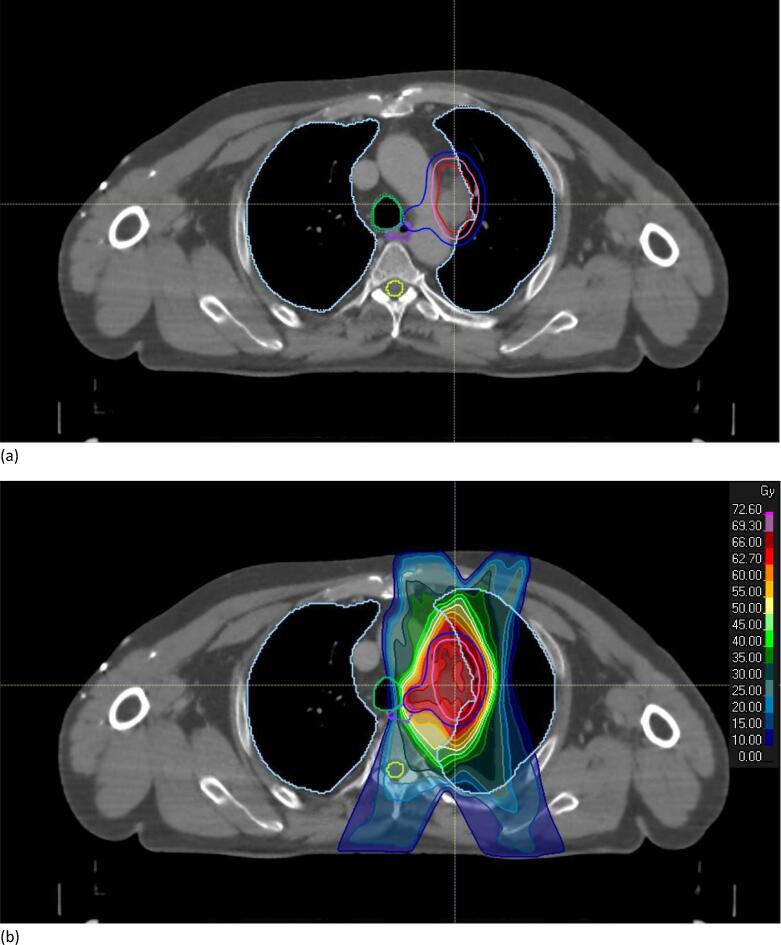
Fig. 4.
Example of a transverse CT image slice through the isocenter for a LA-NSCLC patient. (a) Comparison between manual (dotted lines) and auto-segmented OARs (full lines). (b) Dose distribution of the new treatment plan created based on the auto-segmented OARs (66 Gy in 33 fractions prescribed to the PTV-D50). GTV: red; CTV: pink; PTV: dark blue; trachea: green; spinal cord: yellow; lungs: light blue.
The Pearson correlation coefficient between HD95 and the absolute dose differences varied between 0.004 (ES, esophagus D2) and 0.69 (LA, esophagus D2), the average correlation coefficient over all OARs was 0.19 ± 0.22 for the LA group and 0.07 ± 0.25 for the ES group. The average correlation coefficient between the DSC and the absolute dose-volume difference was –0.09 ± 0.22 for the LA group and –0.27 ± 0.18 for the ES group.
All clinical goals continued to be achieved for the new treatment plans when evaluating the dose distributions on the manually segmented OARs.
4. Discussion
To assess the dose-volume impact of auto-segmentation of OARs for lung cancer patients, a U-Net CNN model was trained and new treatment plans were generated based on the automatic contours. DSC and HD95 were calculated for geometrical analysis, while the dose-volume impact was evaluated by comparing dose-volume differences. Overall, dose-volume differences were small but poor correlation was observed with the geometrical metrics. Visual inspection of the outliers revealed differences between both contour sets in regions with a high dose gradient i.e., close to the tumor or at entry or exit beams.
With regard to the geometrical analysis, the CNN model was very consistent for the lungs (Fig. 1). The other OARs showed decreasing DSC in the following order: heart, trachea, spinal cord, main right bronchus, main left bronchus and esophagus (Table 2). The location of the heart in the human body influences the position and shape of the left bronchus, therefore making the left bronchus more complex and somewhat less consistent over all patients than the right one. The esophagus showed the least contrast with the surrounding tissues on CT images and varied substantially in shape and location, making it harder to train a model for this OAR. The DSC results from the present study were in line with the available literature on auto-segmentation of OARs for lung cancer as well as published data on inter-observer variability (Table 2).
Fig. 1.
Boxplots of (a) DSC and (b) HD 95 between the manual and automatic CNN delineations. The horizontal line is the median value, the triangle is the mean value, the borders of the box are the 1st and 3rd quartiles. The whiskers are the lowest and highest value within 1.5 times the interquartile range measured from the lower, respectively upper quartile. Datapoints outside the range of the whiskers were defined as outliers. For better visualization one outlier of the right bronchus (HD95 = 248.5 mm) was taken out.
The ascending order of OARs for HD95 (Table S3) differed slightly from the DSC results. The largest values were observed for the spinal cord due to differences in the length of the contour (number of contoured CT images). All outliers in terms of HD95 were due to either a misdelineation by the CNN of a small fragment far away from the anatomic location of the OAR, or due to a delineation extending above and/or below the manual delineation. This could be solved by fast, (semi-)automatic post-processing of the auto-segmented OARs. The HD95 results from the present study were not as good as the available literature, as we did not post-process the automatic contours (Table S3).
Looking at individual patients, the model made a few non-physical predictions such as “missing” slices (i.e., CT images without a predicted contour) for the spinal cord or abrupt transitions from heart to non-heart slices. A 3D CNN model that considers the CT image slice above and below each segmented slice might learn to avoid such aberrations [25].
Manual delineations, although performed by experts, are not necessarily perfect nor consistent between clinicians [26]. This affected the training as well as the achievable evaluation accuracy of the model. It is advisable to have a diverse set of manual delineations for CNN training, to account for the inter-observer variability. Common international guidelines on OAR delineation for lung cancer may help to lower the inter-observer variability and to create better and larger data sets for deep learning [27].
Although geometrical differences between automatic and manual contours occur for all auto-segmentation methods, the impact on treatment plan evaluation has not been widely investigated yet. Dose-volume-based evaluations have been performed for inter-observer variability [28], [29] and automated planning [30], [31], while the impact of contouring variations on treatment planning for lung cancer has been investigated by Vaassen et al. [10] and Dong et al. [16]. In the present study, dose-volume differences between the automatic and manual contours were compared and individual outlier inspection was performed for both LA-NSCLC and ES-NSCLC. These outliers were also compared to the outliers of the geometrical results (DSC and HD95), but no correlation was observed.
The mean dose difference for all OARs was <1.2 Gy in both groups, while the mean volume difference was <1 %. These results are in line with the available literature: Vaassen et al. [10] found average dose-volume differences <1 Gy/1%, whereas the mean dose results of Dong et al [16] were <1.5 Gy. For different tumor locations, similar studies have reported mean dose differences <2.2 Gy [32] and <1 Gy/ 1 % [33]. None of these studies included the bronchi nor the trachea.
Overall, larger dose-volume differences were observed for OARs within a region with a high dose gradient, rather than when the delineation was less accurate. For example, the dose-volume results were the best for the mean heart dose for right-sided tumors (<1.76 Gy) in the ES-group (triangles in Fig. 2e). This corresponds to the cases where the tumor is the furthest away from the heart. The spinal cord was always in a region with a low dose gradient, while for the lungs the geometrical accuracy was high. Moreover, (near-)maximum dose values are more sensitive to delineation differences than e.g., mean dose values, as was also observed by Vaassen et al. [10].
On the other hand, for centrally located lung tumors the mediastinal organs such as the esophagus and the heart are generally located at important dose gradients and any misdelineation might lead to a wrongful dose to organ assessment. Moreover, for these tumors it has been shown that increased dose and related toxicity have a major impact on outcome [34]. Similarly, the LungTECH [35] and HILUS [36] trials on SBRT for centrally located ES-NSCLC have emphasized how critical the dose to the main bronchi is. Increasing attention is also being paid to cardiac toxicity.
As a limitation, no centrally located ES-NSCLC patients were represented in the current study. For such patients the tumor would be much closer to the heart and the esophagus, resulting in a potentially larger dose-volume impact. Including more patients could further improve the CNN model and allow for more robust clinical evaluations. Moreover, the clinical impact of dose-volume differences also depends on the fraction size, and it is well-recognized that errors in dose calculation and dose delivery are less forgiving in SBRT than in more protracted schedules [37]. As such, another limitation of the present study was that the effect of the observed dose differences on tumor response and normal tissue toxicity could not be investigated from a radiobiological perspective.
In conclusion, DSC and HD95 did not correlate with the observed dose-volume differences. Based on our observations, we recommend the following practical guidelines. All auto-segmentation contours should first be post-processed to insert missing slices and remove erroneous small fragments. This can be done quickly using standard tools available in the treatment planning system. Next, only the OARs for the most relevant dose-volume parameters should be critically assessed and manually adjusted, and only when they are situated close to the tumor or within an entry or exit beam. For the present study, this included the heart and the esophagus for LA-NSCLC and the bronchi for ES-NSCLC. Due to the performance of the CNN, no adaptation to the lungs is needed. Overall, most of the encountered dose-volume differences during the dose-volume-based evaluation were small and not clinically relevant. In the future, the need for manual corrections of auto-segmented structures could be further identified depending on the tumor stage and location as well as a combination of different metrics to better translate the geometric inaccuracies into the actual dose-volume and clinical impact.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2022.07.004.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Global cancer observatory: Cancer today [Internet]. International agency for research on Cancer.c2020 - [cited 2021 May 18]. Available from: https://gco.iarc.fr/today/data/factsheets/cancers/15-Lung-fact-sheet.pdf.
- 2.Postmus P.E., Kerr K.M., Oudkerk M., Senan S., Waller D.A., Vansteenkiste J., et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017 doi: 10.1093/annonc/mdx222. [DOI] [PubMed] [Google Scholar]
- 3.Freedman D., Radke R.J., Zhang T., Jeong Y., Lovelock D.M., Chen G.T.Y. Model-based segmentation of medical imagery by matching distributions. IEEE Trans Med Imaging. 2005;24:281–292. doi: 10.1109/tmi.2004.841228. [DOI] [PubMed] [Google Scholar]
- 4.Pekar V., McNutt T.R., Kaus M.R. Automated model-based organ delineation for radiotherapy planning in prostatic region. Int J Radiat Oncol Biol Phys. 2004;60:973–980. doi: 10.1016/j.ijrobp.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Ciardo D., Gerardi M.A., Vigorito S., Morra A., Dell’acqua V., Diaz F.J., et al. Atlas-based segmentation in breast cancer radiotherapy: Evaluation of specific and generic-purpose atlases. The Breast. 2017;32:44–52. doi: 10.1016/j.breast.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Isambert A., Dhermain F., Bidault F., Commowick O., Bondiau P.-Y., Malandain G., et al. Evaluation of an atlas-based automatic segmentation software for the delineation of brain organs at risk in a radiation therapy clinical context. Radiother Oncol. 2008;87:93–99. doi: 10.1016/j.radonc.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 7.Meyer P., Noblet V., Mazzara C., Lallement A. Survey on deep learning for radiotherapy. Comput Biol Med. 2018;98:126–146. doi: 10.1016/j.compbiomed.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Lustberg T., van Soest J., Gooding M., Peressutti D., Aljabar P., van der Stoep J., et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol. 2018;126:312–317. doi: 10.1016/j.radonc.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Brunenberg E.J.L., Steinseifer I.K., van den Bosch S., Kaanders J.H.A.M., Brouwer C.L., Gooding M.J., et al. External validation of deep learning-based contouring of head and neck organs at risk. Phys Imaging Radiat Oncol. 2020;15:8–15. doi: 10.1016/j.phro.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaassen F., Hazelaar C., Canters R., Peeters S., Petit S., van Elmpt W. The impact of organ-at-risk contour variations on automatically generated treatment plans for NSCLC. Radiother Oncol. 2021;163:136–142. doi: 10.1016/j.radonc.2021.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Gooding M.J., Smith A.J., Tariq M., Aljabar P., Peressutti D., van der Stoep J., et al. Comparative evaluation of autocontouring in clinical practice: a practical method using the Turing test. Med Phys. 2018;45:5105–5115. doi: 10.1002/mp.13200. [DOI] [PubMed] [Google Scholar]
- 12.Willigenburg T., Zachiu C., Lagendijk J.J.W., van der Voort van Zyp J.R.N., de Boer H.C.J., Raaymakers B.W. Fast and accurate deformable contour propagation for intra-fraction adaptive magnetic resonance-guided prostate radiotherapy. Phys Imaging Radiat Oncol. 2022;21:62–65. doi: 10.1016/j.phro.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Baardwijk A., Bosmans G., Boersma L., Buijsen J., Wanders S., Hochstebag M., et al. PET-CT–based auto-contouring in non–small-cell lung cancer correlates with pathology and reduces interobserver variability in the delineation of the primary tumor and involved nodal volume. Int J Radiat Oncol Biol Phys. 2007;68:771–778. doi: 10.1016/j.ijrobp.2006.12. [DOI] [PubMed] [Google Scholar]
- 14.Kim J., Han J., Ailawadi S., Baker J., Hsia A., Xu Z., et al. SU-F-J-113: Multi-atlas based automatic organ segmentation for lung radiotherapy planning. Med Phys. 2016;43:3433. doi: 10.1118/1.4956021. [DOI] [Google Scholar]
- 15.Yang J., Veeraraghavan H., Armato S.G., III, Farahani K., Kirby J.S., Kalpathy-Kramer J., et al. Autosegmentation for thoracic radiation treatment planning: A grand challenge at AAPM 2017. Med Phys. 2018;45:4568–4581. doi: 10.1002/mp.13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong X., Lei Y., Wang T., Thomas M., Tang L., Curran W.J., et al. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys. 2019;46:2157–2168. doi: 10.1002/mp.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vos BD, Wolterink JM, de Jong PA, Viergever MA, Išgum I. 2D image classification for 3D anatomy localization: employing deep convolutional neural networks. Proceedings SPIE 9784, Med Imaging 2016: Image Processing 2016 Mar 21;97841Y:517–23. doi: 10.1117/12.2216971.
- 18.Trullo R, Petitjean C, Nie D, Shen D, Ruan S. Joint segmentation of multiple thoracic organs in CT images with two collaborative deep architectures. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. DLMIA ML-CDS 2017 2017. Lecture Notes in Computer Science, vol. 10553. Cham: Springer; 2017 Sep 9. p. 21–9. doi: 10.1007/978-3-319-67558-9_3. [DOI] [PMC free article] [PubMed]
- 19.arxiv.org [Internet]. Ronneberger O, Fischer P and Brox T. U-net: Convolutional networks for biomedical image segmentation. ArXiv, c2015 [cited 2021 March 17]. Available from: doi: 10.48550/arXiv.1505.04597.
- 20.Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. ImageNet: A large-scale hierarchical image database. Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition; 2009 June 20-25, Miami, FL, USA. IEEE 2009 p.248-55. doi: 10.1109/CVPR.2009.5206848.
- 21.Zhou X., Takayama R., Wang S., Hara T., Fujita H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med Phys. 2017;44:5221–5233. doi: 10.1002/mp.12480. [DOI] [PubMed] [Google Scholar]
- 22.arxiv.org [Internet]. Yakubovskiy P. Segmentation Models. GitHub repository 2019 [cited 2020 February 17]. Available from: https://github.com/qubvel/segmentation_models.
- 23.arxiv.org [Internet]. Vesal S, Ravikumar N and Maier A. A 2D dilated residual U-Net for multi-organ segmentation in thoracic CT. ArXiv, c2019 [cited 2021 March 17]. Available from: https://doi.org/10.48550/arXiv.1905.07710.
- 24.Mongan J., Moi L., Kahn C. Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiol: Artif Intell. 2020;2 doi: 10.1148/ryai.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milletari F, Navab N and Ahmadi S. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. Proceedings of the 4th Int Conf on 3D Vis (3DV); 2016 Oct 25-26, Stanford, CA, USA. IEEE, 2016 p. 565-71. doi: 10.1109/3DV.2016.79.
- 26.Tsang Y., Hoskin P., Spezi E., Landau D., Lester J., Miles E., et al. Assessment of contour variability in target volumes and organs at risk in lung cancer radiotherapy. Tech Innov Patient Support Radiat Oncol. 2019;10:8–12. doi: 10.1016/j.tipsro.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindberg K., Grozman V., Karlsson K., Lindberg S., Lax I., Wersäll P., et al. The HILUS-Trial-a prospective nordic multicenter phase 2 study of ultracentral lung tumors treated with stereotactic body radiotherapy. J Thorac Oncol. 2021;16:1200–1210. doi: 10.1016/j.jtho.2021.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Aliotta E., Nourzadeh H., Siebers J. Quantifying the dosimetric impact of organ-at-risk delineation variability in head and neck radiation therapy in the context of patient setup uncertainty. Phys Med Biol. 2019;64 doi: 10.1088/1361-6560/ab205c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo A.C., Liu M., Chan E., Lund C., Truong P.T., Loewen S., et al. The impact of peer review of volume delineation in stereotactic body radiation therapy planning for primary lung cancer: a multicenter quality assurance study. J Thorac Oncol. 2014;9:527–533. doi: 10.1097/JTO.0000000000000119. [DOI] [PubMed] [Google Scholar]
- 30.Vanderstraeten B., Goddeeris B., Vandecasteele K., van Eijkeren M., De Wagter C., Lievens Y. Automated instead of manual treatment planning? A plan comparison based on dose-volume statistics and clinical preference. Int J Radiat Oncol Biol Phys. 2018;102:443–450. doi: 10.1016/j.ijrobp.2018.05.063. [DOI] [PubMed] [Google Scholar]
- 31.Bijman R., Sharfo A.W., Rossi L., Breedveld S., Heijmen B. Pre-clinical validation of a novel system for fully-automated treatment planning. Radiother Oncol. 2021;158:253–261. doi: 10.1016/j.radonc.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 32.van Rooij W., Dahele M., Ribeiro Brandao H., Delaney A.R., Slotman B.J., Verbakel W.F. Deep learning-based delineation of head and neck organs at risk: Geometric and dosimetric evaluation. Int J Radiat Oncol Biol Phys. 2019;104:677–684. doi: 10.1016/j.ijrobp.2019.02.040. [DOI] [PubMed] [Google Scholar]
- 33.Ji Z., Xinyuan C., Bining Y., Nan B., Tao Z., Kuo M., et al. Evaluation of automatic segmentation model with dosimetric metrics for radiotherapy of esophageal cancer. Front Oncol. 2020;10:1–9. doi: 10.3389/fonc.2020.564737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradley J.D., Paulus R., Komaki R., Masters G., Blumenschein G., Schild S., et al. Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage IIIA or IIIB non-small-cell lung cancer (RTOG 0617): a randomised, two-by-two factorial phase 3 study. Lancet Oncol. 2015;16:187–199. doi: 10.1016/S1470-2045(14)71207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adebahr S., Collette S., Shash E., Lambrecht M., Le Pechoux C., Faivre-Finn C., et al. Lungtech, an EORTC phase II trial of stereotactic body radiotherapy for centrally located lung tumours: a clinical perspective. Br J Radiol. 2015;88 doi: 10.1259/bjr.20150036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bland J.M., Altman D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;327:307–310. doi: 10.1016/S0140-6736(86)90837-8. [DOI] [PubMed] [Google Scholar]
- 37.Fowler J.F., Tomé W.A., Fenwick J.D., Mehta M.P. A challenge to traditional radiation oncology. Int J Radiat Oncol Biol. 2004;60:1241–1256. doi: 10.1016/j.ijrobp.2004.07.691. [DOI] [PubMed] [Google Scholar]
- 38.Cui Y., Chen W., Kong F.M., Olsen L.A., Beatty R.E., Maxim P.G., et al. Contouring variations and the role of atlas in non-small cell lung cancer radiation therapy: Analysis of a multi-institutional preclinical trial planning study. Pract Radiat Oncol. 2015;5:e67–e75. doi: 10.1016/j.prro.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J., Zhang J., Qiu B., Liu Y., Liu X., Chen L. Comparison of the automatic segmentation of multiple organs at risk in ct images of lung cancer between deep convolutional neural network-based and atlas-based techniques. Acta Oncol. 2019;58:257–264. doi: 10.1080/0284186X.2018.1529421. [DOI] [PubMed] [Google Scholar]
- 40.Lei Y, Liu Y, Dong X, Tian S, Wang T, Jiang X, et al. Automatic multi-organ segmentation in thorax CT images using U-Net-GAN. Proceedings of SPIE 10950, Med Imaging. 2019: Computer-Aided Diagnosis; 2019 Mar 13, San Diego, Cal, USA. SPIE, 2019;10950:262–7. doi: 10.1117/12.2512552.
- 41.Fellin F., Amichetti M., La Macchia M., Cia,chetti M., Gianolini S., Paola V., et al. Systematic evaluation of three different commercial software solutions for automatic segmentation for adaptive therapy in head-and-neck, prostate and pleural cancer. Radiat Oncol. 2012 doi: 10.1186/1748-717X-7-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Harten LD, Noothout JMH, Verhoeff JJC, Wolterink JM, Išgum I. Automatic segmentation of organs at risk in thoracic CT scans by combining 2D and 3D convolutional neural networks. In Petitjean C, Ruan S, Lamber Z, Dubray B, editors, SegTHOR 2019: Proceedings of the 2019 Challenge on Segmentation of THoracic Organs at Risk in CT Images (SegTHOR2019). CEUR. 2019. (CEUR workshop proceedings). http://ceur-ws.org/Vol-2349/SegTHOR2019_paper_12.pdf.
- 43.Chen P.H., Huang C.H., Chiu W.T., Liao C.M., Lin Y.R., Hung S.K., et al. A multiple organ segmentation system for CT image series using Attention-LSTM fused U-Net. Multimed Tools Appl. 2022;81:11881–11895. doi: 10.1007/s11042-021-11889-7. [DOI] [Google Scholar]
- 44.Lappas G., Staut N., Lieuwes N.G., Biemans R., Wolfs C.J.A., van Hoof S.J., et al. Inter-observer variability of organ contouring for preclinical studies with cone beam Computed Tomography imaging. Phys Imaging Radiat Oncol. 2022;21:11–17. doi: 10.1016/j.phro.2022.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.