Abstract
Bladder cancer (BC), as a genitourinary system tumor, is a highly prevalent tumor type. Ferroptosis is an iron-dependent oxidative cell death mechanism that is becoming increasingly recognized as a promising avenue for cancer therapy. However, further determination of the prospective prognostic value of ferroptosis for BC and investigation of the underlying mechanisms is required. The mRNA expression profiles and associated clinical data were downloaded from public databases such as The Cancer Genome Atlas, Gene Expression Omnibus and the IMvigor210 database. To construct a predictive formula, the least absolute shrinkage and selection operator Cox regression algorithm was used. In addition, a prognostic multigene signature was constructed using previously selected ferroptosis-related genes (FRGs). A total of 28 FRGs were differentially expressed between tumor and normal samples with |log2 fold change| >1 and adjusted P<0.05. A prognostic model was then established and it was validated in the GEO cohort using six genes: Glutamate-cysteine ligase modifier subunit, crystallin α-B, transferrin receptor, zinc finger E-box binding homeobox 1, squalene epoxidase and glucose-6-phosphate dehydrogenase (G6PD). Numerous important pathways involved in the development of the immune system and cancer were indicated to be significantly different between the two risk groups. In addition, it was discovered that G6PD expression subgroups that were associated with immunotherapy response in patients with BC had similar prognostic features to risk score subgroups. In the present study, a gene signature with a prognostic value for ferroptosis in BC was successfully developed and the potential value of G6PD was identified for future research.
Keywords: ferroptosis, TCGA, bladder cancer, GEO, glucose-6-phosphate dehydrogenase
Introduction
BC is one of the most prevalent malignant tumor types. It affects ~550,000 females each year (1,2). Approximately 90% of BC cases originate from urothelial cells (1,3). Smoking has long been recognized as the most significant risk factor for BC (4). BC is classified into two subtypes based on the depth of invasion: Non-muscle-invasive BC (NMIBC) and muscle-invasive BC (MIBC). This classification helps guide treatment decisions (1). Currently, NMIBC is frequently treated by adjuvant intravesical chemotherapy, bacillus Calmette-Guérin (BCG) instillations (3) and cystectomy in cases of BCG-unresponsive tumors or high risk of progression and recurrence (5). Furthermore, patients with MIBC are routinely treated with radical cystectomy and neoadjuvant/adjuvant chemotherapy (6). BC is associated with a high risk of morbidity and mortality worldwide if it is not treated properly (7). In a population-based study of 321 patients with BC in Iran's Kurdistan Province (2013–2018), the 5-year survival rate was 54% (8). In Europe, the 5-year age-standardized relative survival rate was ~70%, varying amongst specific countries by an average of 60-80% (2). Given the disease limitations in terms of treatment options and drug resistance, it is critical to investigate more effective therapeutic targets.
Ferroptosis is a type of cell death with distinct properties and functions associated with physical conditions or numerous diseases, including cancers (9). Compared with normal cells, cancer cells require more iron to support their increased rate of proliferation. This iron dependency makes cancer cells more vulnerable to iron-catalyzed necrosis (10). Du et al (11) identified that calumenin may affect the prognosis of BC through ferroptosis based on the analysis of a The Cancer Genome Atlas (TCGA) dataset of 408 patients with BC using multi-omics bioinformatics. According to Chen et al (12), compound 7j, a quinazolinyl-arylurea derivative, triggered ferroptosis in BC cells when used at high concentrations and over an extended incubation time by effectively regulating the SystemXc-/glutathione peroxidase 4 (GPX4)/reactive oxygen species (ROS) and PI3K/Akt/mTOR/ unc-51 like autophagy activating kinase 1 pathways. Guo et al (13) developed a triple therapy for inducing ferroptosis in BC cells, which may possess significant potential for clinical translation. As accumulating evidence indicates, triggering ferroptosis for cancer therapy may be a viable option, particularly for the eradication of aggressive malignancies that are resistant to traditional therapies (14). Previous research has established a critical role of ferroptosis in BC and four prognostic targets have been identified (15). However, additional research is necessary.
In the present study, mRNA expression profiles and corresponding clinical data of patients with BC were downloaded from public databases. A multigene signature associated with prognosis was constructed using genes that were related to ferroptosis and differentially expressed in the TCGA cohort. In addition, the results were validated in cohorts from the Gene Expression Omnibus (GEO) database. Next, the underlying mechanisms were explored using functional enrichment and phenotypic analysis. Finally, an analysis of glucose-6-phosphate dehydrogenase (G6PD), a ferroptosis-related gene (FRG) was performed, which deserves additional research in BC.
Materials and methods
Data collection and sources
The mRNA expression data and clinical follow-up data of patients with BC were retrieved from the TCGA database up to March 2021 (TCGA-BLCA; http://portal.gdc.cancer.gov/) (16). The validation cohort comprised 165 tumor samples obtained from the GEO database (GEO; http://www.ncbi.nlm.nih.gov/geo/) (dataset no. GSE13507) (17). In addition, the immunotherapy dataset was downloaded from IMvigor210 (http://research-pub.gene.com/IMvigor210CoreBiologies), a publicly available data package. The ‘limma’ R package was used to normalize the gene expression profiles. As all data were acquired following the relevant guidelines and policies for data access of the TCGA and GEO, no local ethical approval was required for this study. The datasets GSE76211 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76211) and GSE130598 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130598) were used to evaluate the mRNA level of G6PD in BC and adjacent normal tissue (18,19).
FRGs were identified from previous studies (10,20–22). All of the FRGs are listed in Table SI. The immunohistochemistry (IHC) staining of normal and BC samples was obtained from the human protein atlas (HPA; http://www.proteinatlas.org/), an open-access database containing multiple protein expression images.
Construction and external validation of the 6-gene signature
Using the ‘limma’ package in the R software, differential gene expression between BC and normal tissues was analyzed using P<0.05 and false discovery rate (FDR)<0.05 as the screening criteria. Cox-proportional hazard regression was used to perform univariate survival analysis to identify prognostic FRGs. The protein-protein interaction network of differentially expressed genes (DEGs) was analyzed using the Search Tool for the Retrieval of Interacting Genes and proteins (STRING) database (http://string-db.org/) (23). The R package ‘glmnet’ was used to develop a gene-related prognostic model for the least absolute shrinkage and selection operator (LASSO) Cox regression analysis (24,25). The normalized expression matrix of the prognostic DEGs served as independent variables, while the overall survival (OS) and survival status (alive or dead) of patients served as response variables in the regression. The optimal value of the penalty parameter (λ) was determined according to the minimum 10-fold cross-validations and used to select significant features for the model. The risk score for each patient consisted of the normalized expression values of each gene and the corresponding regression coefficients. It was calculated using the following formula: risk score=esum (each gene's normalized expression × each gene's corresponding coefficient). To analyze the distribution of different groups, principal component analysis (PCA) was performed using the R package ‘prcomp’ and t-distributed stochastic neighbor embedding (t-SNE) was performed using the ‘Rtsne’ R package. For the evaluation of the impact of various factors on OS, Kaplan-Meier curves were drawn and differences were assessed using the log-rank test. The optimal cut-off value for the survival analysis was determined using the R package ‘survminer’. The R package ‘survivalROC’ was used to depict time-dependent receiver operating characteristic (ROC) curves.
Functional and gene set enrichment analysis (GSEA)
Gene Ontology (GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment was performed using the ‘clusterProfiler’ R package. Gene set variant analysis (GSVA) was performed using the ‘gsva’ R package. GSEA was performed based on groups according to their risk score or G6PD expression (26). Both analyses used the Hallmark gene set with FDR<0.25 and nominal (NOM) P<0.05 as the criterion. Single sample (ss)GSEA was carried out using the ‘gsva’ package (27).
Cell lines
A total of three BC cell lines (5637, J82 and T24) and human normal bladder epithelial cells (SV-HUC-1) were obtained from the Cell Bank of the Chinese Academy of Sciences. Cells were cultured under conventional cell culture conditions of 5% CO2 at 37°C in RPMI 1640 medium (MilliporeSigma) with fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted using an RNA extraction kit (cat. no. 74004; Qiagen GmbH) and then reverse-transcribed into cDNA using the FastLane Cell cDNA Kit (cat. no. 215011; Qiagen GmbH) according to the manufacturer's instructions. The primers (G6PD forward, 5′-CGAGGCCGTCACCAAGAAC-3′ and reverse, 5′-GTAGTGGTCGATGCGGTAGA-3′; GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′) were designed and synthesized in Tsingke. qPCR was performed using the SYBR Green PCR system (cat. no. A25776; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The PCR cycling program included a 30 sec incubation step at 95°C and then 40 cycles of 5 sec at 95°C and 30 sec at 60°C. A MyCycler Thermal Cycler (cat. no. 170-9703; Bio-Rad Laboratories, Inc.) was used for qPCR. GAPDH was used for normalization and the relative expression levels were calculated using the 2−∆∆Cq method (28).
Statistical analysis
The Wilcoxon signed-rank test was used to compare gene expression levels in BC tissues to those in adjacent non-tumorous tissues. The χ2 test was used to compare proportions between groups. The ssGSEA scores of different groups were compared using the Mann-Whitney U test. All reported P-values were adjusted for multiple comparisons using the Benjamini-Hochberg method. Univariate and multivariate were performed to determine the independent prognostic factors. Association analysis was performed using the R packages ‘ggplot2’ and ‘ggpubr’ and the tests used were the unpaired t-test (between two groups) and analysis of variance (among multiple groups). Tukey's post hoc test was applied to analyze the statistical significance between each of the cancer cell lines and the normal bladder cell line. All analyses were conducted using R software (v.3.6.3), Excel (Microsoft; 2016) or GSEA (v.4.0.1). All P-values were two-tailed and P<0.05 was considered to indicate statistical significance, unless otherwise specified.
Results
Data sourcing
Fig. 1 depicts a flow chart of the present study. A total of 400 BC samples and 19 adjacent non-tumor samples from the TCGA database, as well as 165 tumor samples from the GEO database, were included in the analysis. Table I summarizes the detailed clinical characteristics of the patients with BC. In the TCGA cohort, the average participant age was 68 (34–89) years and 74.2% of the patients were male. In the GEO cohort, the mean age was 65 (24–88) years and the proportion of male patients was 81.8%.
Figure 1.
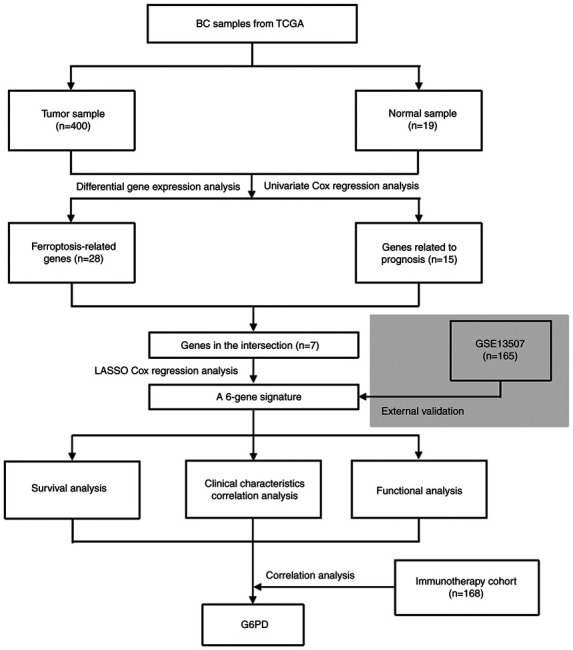
Flow chart of the present study. BC, bladder cancer; TCGA, The Cancer Genome Atlas; G6PD, glucose-6-phosphate dehydrogenase; LASSO, least absolute shrinkage and selection operator.
Table I.
Characteristics of patients used in this study.
| Item | TCGA cohort (n=400) | GEO cohort (n=165) |
|---|---|---|
| Age, years | 68 (34–89) | 65 (24–88) |
| Sex | ||
| Female | 103 (25.8) | 30 (18.2) |
| Male | 297 (74.2) | 135 (81.8) |
| Grade | ||
| Low | 20 (5.0) | 105 (63.6) |
| High | 377 (94.3) | 60 (36.4) |
| Unknown | 3 (0.7) | 0 |
| Vascular invasion | ||
| Yes | - | 62 (37.6) |
| No | - | 103 (62.4) |
| Stage | ||
| I | 2 (0.5) | - |
| II | 127 (31.8) | - |
| III | 138 (34.5) | - |
| IV | 131 (32.7) | - |
| Unknown | 2 (0.5) | - |
| Overall survival, months | 48.4 (1.03-136.97) | - |
| Risk, based on the 6-gene signature | ||
| Low | 200 (50.0) | 83 (50.3) |
| High | 200 (50.0) | 82 (49.7) |
Values are expressed as n (%) or the median (range). GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas; OS, overall survival.
Identification of FRGs in the TCGA cohort
When tumor and normal samples were compared, differential expression analysis revealed 28 (46%) significant DEGs. Univariate Cox regression analysis showed that 15 genes were associated with prognosis, 7 of which were FRGs (Fig. 2A). The genes in the intersection included glutamate-cysteine ligase (GCL), crystallin α-B (CRYAB), transferrin receptor (TFRC), zinc finger E-box binding homeobox 1 (ZEB1), squalene epoxidase (SQLE), G6PD and acyl-CoA synthetase family member 2 (ACSF2) (ACSF2 was excluded due to the deficiency of gene expression data in the GEO cohort). Their expression in normal and tumor tissue is presented in Fig. 2B and C. The interaction network and correlation are also presented in Fig. 2D and E.
Figure 2.
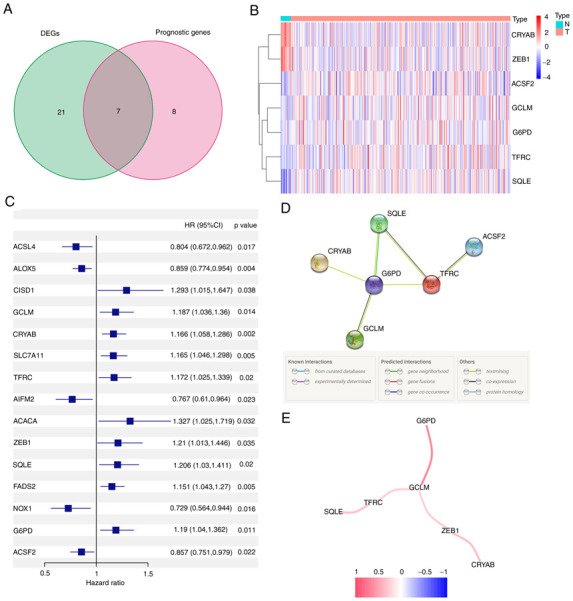
Identification of the candidate FRGs in The Cancer Genome Atlas cohort. (A) Venn diagram of DEGs. (B) Heat map of DEGs between tumor and adjacent non-tumor patient tissues. (C) Forest plot demonstrating the results of a univariate Cox regression analysis between FRGs expression and overall survival. (D) Protein-protein interaction network of candidate genes. (E) Correlation network of candidate genes. DEG, differentially expressed gene; HR, hazard ratio; G6PD, glucose-6-phosphate dehydrogenase; FRG, ferroptosis-related gene.
Prior to conducting regression modeling, GSVA was used to estimate the enrichment of hallmark gene sets in the TCGA cohort. The results are presented in Fig. S1. The volcano plot and heatmap displayed the significant enrichment of DNA repair, glycolysis, unfolded protein response, mammalian target of rapamycin complex 1 (mTORC1) signaling, E2 factor targets and MYC targets in BC tumors.
Construction of a 6-gene signature and validation in GEO cohort
To build a model for the prognosis of patients with BC, the 6 genes listed above were identified using LASSO Cox regression. A 6-gene signature was successfully established using the optimal value of λ. This model was formulated as follows: Risk score = e[0.066 × GCL modifier (GCLM) expression level + 0.157 × CRYAB expression level + 0.109 × TFRC expression level + 0.074 × ZEB1 expression level + 0.135 × SQLE expression level + 0.118 × G6PD expression level].
After calculating the median cut-off value in the TCGA cohort, all the patients were divided into two groups: The high-risk (n=200) and low-risk (n=200) group (Fig. 3A). Table II presents the characteristics of patients from the different groups. The high-risk group was associated with increased age (P<0.01), a higher tumor grade (P<0.01) and an advanced TNM stage (P<0.01). Fig. 3B and C present the results of the PCA and t-SNE analysis. The analysis indicated that different risk groups were distributed in two directions and exhibited a certain separation. As indicated in Fig. 3D, patients in the high-risk group had a higher probability of death. Kaplan-Meier survival analysis consistently demonstrated that patients in the high-risk group had unfavorable OS as compared with those in the low-risk group (Fig. 3E). The predictive performance of the model was evaluated using time-dependent ROC curves and the area under the curve (AUC) reached 0.606 at 1 year, 0.648 at 3 years and 0.669 at 5 years (Fig. 3F), which was significantly better than the predictive value of the TNM stage (Fig. S2A).
Figure 3.
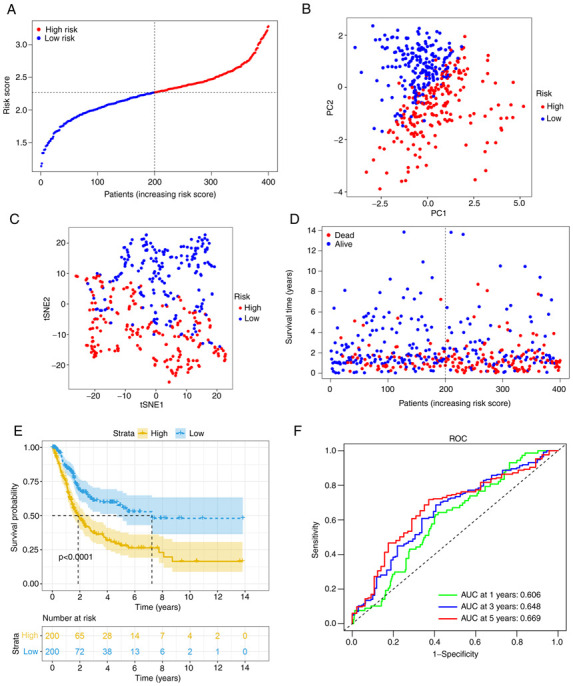
Analysis of the 6-gene signature model in The Cancer Genome Atlas cohort for potential prognostic value. (A) Risk score distribution and median value. (B) PC analysis plot. (C) t-SNE analysis. (D) Survival status distribution. (E) Kaplan-Meier curves for patients in different risk groups. (F) The AUC of time-dependent ROC curves was determined to confirm the prognostic performance of the risk score. ROC, receiver operating characteristic; AUC, area under the ROC curve; PC, principal component; t-SNE, t-distributed stochastic neighbor embedding.
Table II.
Distribution of characteristics between different risk groups.
| TCGA cohort | GEO cohort | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Characteristic | High risk | Low risk | P-value | High risk | Low risk | P-value |
| Age, years | <0.01a | <0.01a | ||||
| <60 | 29 | 57 | 12 | 30 | ||
| ≥60 | 171 | 143 | 70 | 53 | ||
| Sex | 0.30a | 0.97a | ||||
| Female | 56 | 47 | 15 | 15 | ||
| Male | 144 | 153 | 67 | 68 | ||
| Grade | <0.01b | <0.01a | ||||
| High | 197 | 180 | 48 | 12 | ||
| Low | 1 | 19 | 34 | 71 | ||
| Unknown | 2 | 1 | ||||
| TMN stage | <0.01b | - | ||||
| I+II | 37 | 92 | - | - | ||
| III+IV | 162 | 107 | - | - | ||
| Unknown | 1 | 1 | - | - | ||
| Muscle invasion | - | <0.01a | ||||
| Yes | - | - | 47 | 15 | ||
| No | - | - | 35 | 68 | ||
χ2 test;
Fisher's exact test. GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas.
Using the same formula, the risk score of patients in the GEO cohort was calculated. Similar to the TCGA cohort, they were divided into high- (n=82) or low-risk (n=83) groups (Fig. 4A). Consistent with the TCGA cohort data presented above, the high-risk group also demonstrated a significantly advanced age (P<0.01) and grade (P<0.01). In addition, the high-risk group had a higher risk of muscle-invasive disease (Table II). PCA and t-SNE analysis also indicated that when patients were divided into 2 groups, there was a certain separation between the groups of high and low risk (Fig. 4B and C). Again, it appeared that patients assigned to the high-risk group succumbed earlier and had a shorter OS than those in the low-risk group (Fig. 4D and E). Furthermore, the AUC value obtained from the ROC analysis was 0.671 at 1 year, 0.655 at 3 years and 0.621 at 5 years (Fig. 4F).
Figure 4.
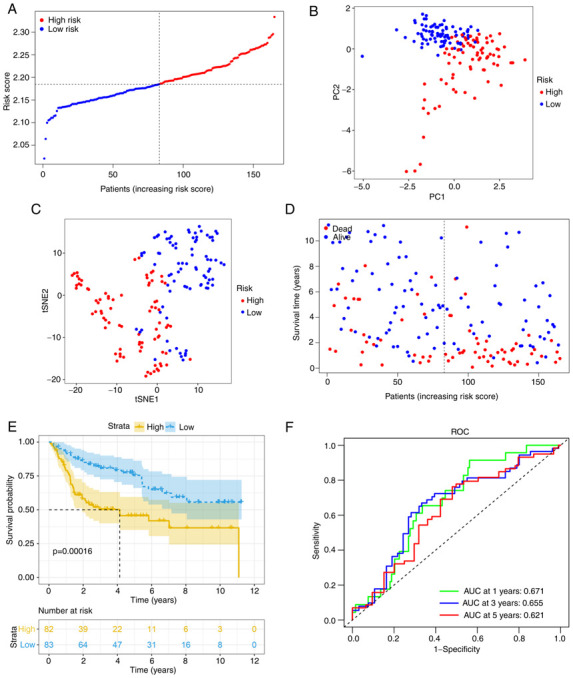
Validation in the Gene Expression Omnibus cohort. (A) Risk score distribution and median value. (B) PCA plot. (C) t-SNE analysis. (D) Survival status distribution. (E) Kaplan-Meier curves for patients in different risk groups. (F) The AUC of time-dependent ROC curves was determined to confirm the prognostic performance of the risk score. ROC, receiver operating characteristic; AUC, area under the ROC curve; PC, principal component; t-SNE, t-distributed stochastic neighbor embedding.
Prognostic value of the risk score and clinical characteristics
Univariate and multivariate Cox regression analyses were performed to determine the independent prognostic value of the model. In the univariate Cox regression analysis, age, stage and risk score were indicated to have a significant influence on OS (Fig. 5A). Furthermore, using multivariate Cox regression analysis, stage and risk score were identified to have an independent predictive value in OS (Fig. 5B). Furthermore, association analysis revealed that having a high risk score was associated with advanced age (>60 years), male sex, tumor infiltration, lymph node metastasis and distant metastasis in both the TCGA and GEO cohorts (P<0.05; Fig. 5C-G).
Figure 5.
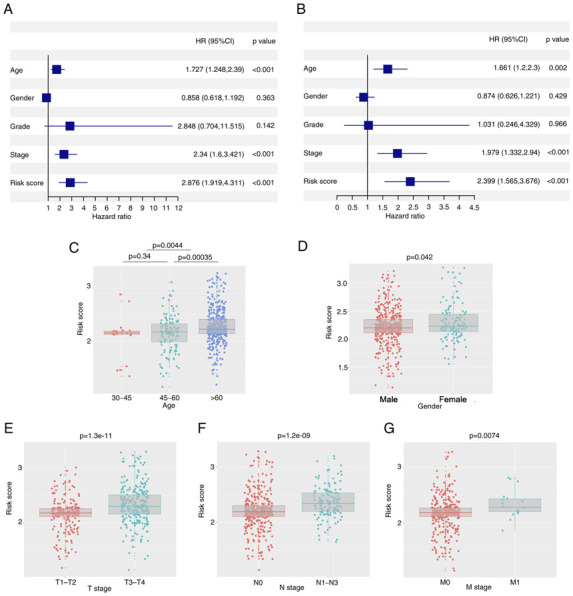
Association between the risk score and clinical factors in the TCGA cohort. (A) The results of univariate Cox regression analysis in the TCGA cohort. (B) The risk score was an independent prognostic factor in the multivariate Cox regression analysis. (C-G) The boxplots indicated a correlation between the risk score and (C) age, (D) sex, (E) T stage, (F) N stage and (G) M stage. TCGA, The Cancer Genome Atlas; HR, hazard ratio (risk of death).
Functional analyses in TCGA
To further elucidate the biological functions and pathways associated with differential FEGs, GO enrichment and KEGG pathway analyses were performed. The findings of the GO enrichment analysis indicated that the differential genes were enriched in extracellular structure organization and extracellular matrix organization in the category Biological Process. In addition, collagen-containing extracellular matrix was markedly enriched in the category Cellular Component. In the category Molecular Function, glycosaminoglycan binding, extracellular matrix structural constituent and antigen-binding were significantly enriched terms (adjusted P<0.05). Meanwhile, the KEGG pathway analysis revealed that the PI3K-Akt signaling pathway was significantly enriched. In addition, certain pathways associated with immunity were revealed to be enriched (Fig. 6A-D).
Figure 6.
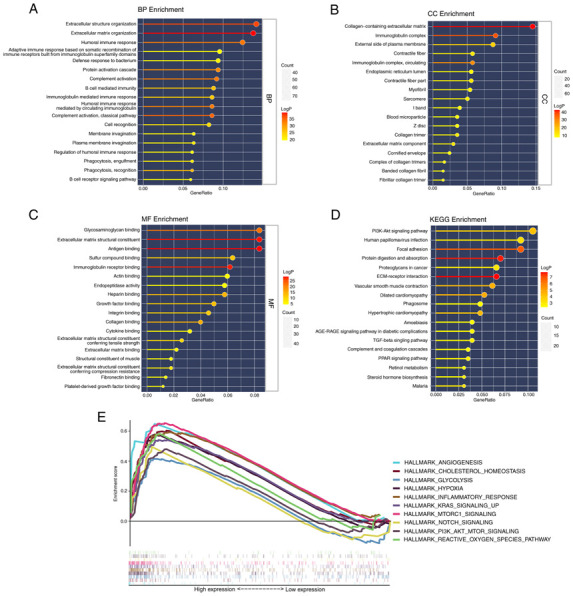
Functional enrichment analysis in The Cancer Genome Atlas cohort. (A-C) Gene Ontology enrichment analysis included (A) BP analysis, (B) CC analysis and (C) MF analysis. (D) KEGG analysis results. (E) GSEA results of hallmark gene sets for the high-risk group. BP, Biological Process; CC, Cellular Component; MF, Molecular Function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
The results of the two risk groups using GSEA on the cancer hallmarks gene sets are presented in Fig. 6E. The pathways with FDR <0.25 and NOM P<0.05 were selected and they are associated with ferroptosis or bladder cancer risk.
Analysis of immune-related characteristics in patients with BC
ssGSEA was further used to examine the differences in immune status between the two risk groups. In the TCGA cohort, B cells, dendritic cells (DCs), macrophages, neutrophils, follicular helper T cells (Tfh), Th1 cells, tumor-infiltrating lymphocytes (TIL) and regulatory T cells (Treg) scored significantly higher in the high-risk group (Fig. 7A). The immune function analysis revealed that the high-risk group had stronger antigen-presenting cell (APC) stimulation, chemokine receptors (CCR), immune checkpoint, parainflammation, T-cell inhibition and T-cell stimulation (Fig. 7B). The cells and pathways in antigen presentation-related processes significantly differed between the two risk groups. In comparison, in the GEO cohort, DCs, macrophages, neutrophils, Tfh, Th1 cells, TIL, CCR, immune checkpoint, parainflammation, T-cell inhibition and T-cell stimulation demonstrated a similar change (adjusted P<0.01; Fig. 7C and D).
Figure 7.
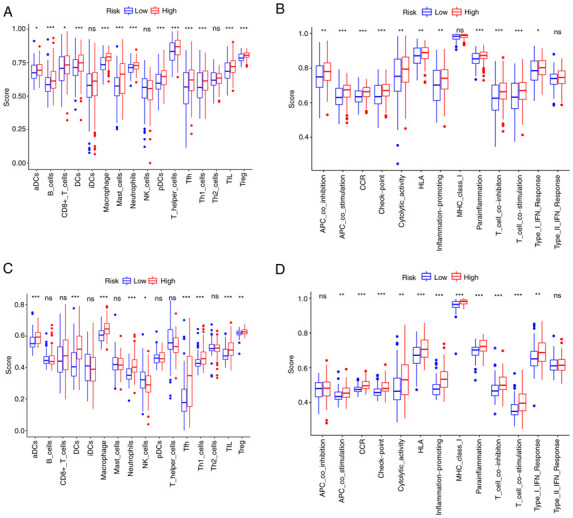
Single-sample gene set enrichment analysis results of different risk groups in the (A and B) The Cancer Genome Atlas cohort: (A) Scores of the 16 immune cells and (B) 13 immune-related functions. (C and D) Gene Expression Omnibus cohort: (C) Scores of the 16 immune cells and (D) 13 immune-related functions. ns, not significant; *P<0.05; **P<0.01; ***P<0.001. DC, dendritic cells; NK, natural killer; Tfh, follicular helper T cells; Th1, type I T-helper cells; TIL, tumor-infiltrating lymphocytes; Treg, regulatory T cells; HLA, human leukocyte antigen; APC, antigen-presenting cell; MHC, major histocompatibility complex; CCR, chemokine receptors; pDCs, plasmacytoid DCs; aDCs, activated DCs; iDCs, immature DCs.
Association between G6PD and BC-related characteristics
Association analysis between gene expression and demographic characteristics revealed that G6PD expression was significantly different between gender-defined subgroups (P<0.05; Fig. S2B). The survival analysis of single-gene expression (formula=single gene expression × corresponding coefficient) indicated that there were significant differences between two risk groups when CRYAB, G6PD, GCLM and TFRC expression levels were used to predict patient prognosis (P<0.05; Fig. S2C-H). Furthermore, the relationship between each FRG and patient prognosis was investigated. G6PD was indicated to have a significant association with prognosis in the univariate Cox regression analysis (P<0.05; Table SII).
To further identify the prognostic potential, genes related to ferroptosis were analyzed in the BC immunotherapy cohort using IMvigor210 and a significant correlation between G6PD gene expression and the patients' immunotherapy response was discovered (Fig. S3A). The ssGSEA of patients divided by G6PD gene expression in the TCGA cohort suggested that the immune cell infiltration score was higher and immune-related processes were stronger in the high-expression group. However, the type II IFN response of the high-expression group scored much lower in comparison (Fig. 8A and B). The increase in G6PD expression followed a similar trend toward advanced age, tumor infiltration, lymph node metastasis and distant metastasis (Fig. S4A-F). Further analysis revealed that immune response genes, including cytotoxic T-lymphocyte associated protein 4, programmed cell death 1 and programmed cell death 1 ligand 2 (PD-L2), had higher expression levels in the high G6PD expression group than in the low G6PD expression group (Fig. S5).
Figure 8.
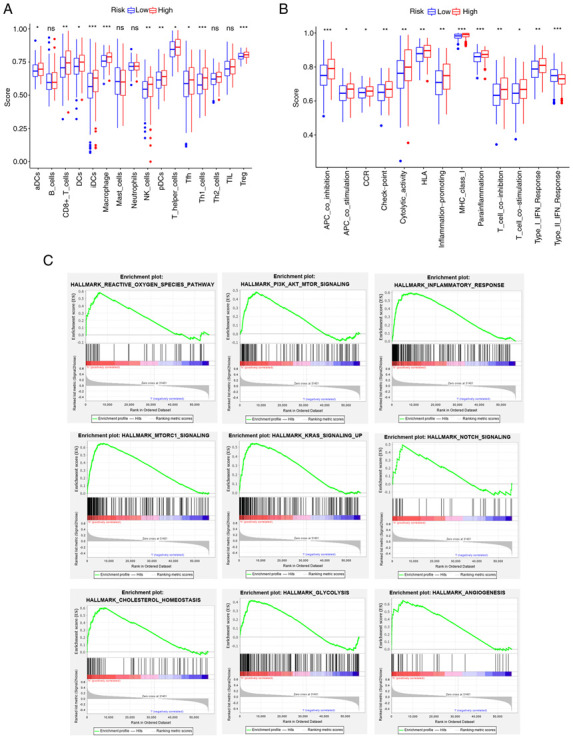
Results of single-sample GSEA and GSEA analyses for the high- and low-G6PD groups in The Cancer Genome Atlas cohort. (A) Differences in immune cell infiltrating score. (B) Differences in infiltrating score in immune-related pathways. (C) GSEA results of hallmark gene sets for the high G6PD expression group. ns, not significant; *P<0.05; **P<0.01; ***P<0.001. GSEA, gene set enrichment analysis; G6PD, glucose-6-phosphate dehydrogenase. DC, dendritic cells; NK, natural killer; Tfh, follicular helper T cells; Th1, type I T-helper cells; TIL, tumor-infiltrating lymphocytes; Treg, regulatory T cells; HLA, human leukocyte antigen; APC, antigen-presenting cell; MHC, major histocompatibility complex; CCR, chemokine receptors; pDCs, plasmacytoid DCs; aDCs, activated DCs; iDCs, immature DCs.
Furthermore, the GSEA results of the G6PD expression subgroups in the TCGA cohort are displayed in Fig. 8C. The pathways enriched in the risk groups were also enriched in the G6PD expression groups (FDR<0.25, NOM P<0.05).
Validation of G6PD expression level
RT-qPCR was performed to determine the level of G6PD mRNA expression in BC cell lines and tissue (Fig. 9A and B). The results of the cell line analysis revealed that BC cell lines (5637, J82 and T24) expressed more G6PD than the normal cell line SV-HUC-1 (Fig. 9A). Furthermore, it was indicated that G6PD expression was much higher in BC tissues than in adjacent tissues using tissue data from GSE76211, GSE130598 and TCGA-BLCA (Fig. 9B-D; GSE76211: Three paired BC and normal tissues; GSE76211: 24 paired BC and normal tissues, TCGA-BLCA: 19 paired BC and normal tissues). In addition, IHC images obtained from the HPA database indicated that the G6PD protein level in BC tissue was relatively higher than that in normal bladder tissue (Fig. 9E).
Figure 9.
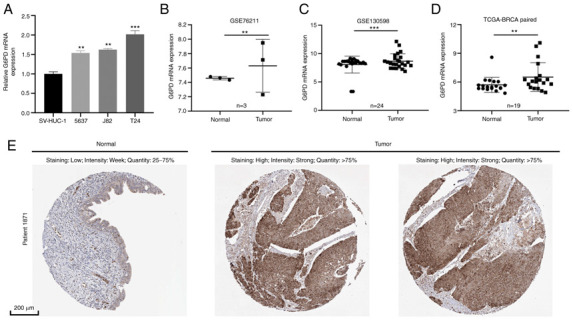
Reverse transcription-quantitative PCR and immunohistochemistry results. (A) mRNA expression level of G6PD in BC cell lines. Data were analyzed by ANOVA followed by Tukey's post-hoc test. (B-D) mRNA expression level of G6PD in BC tissue and normal tissue in (B) GSE76211, (C) GSE130598 and (D) The Cancer Genome Atlas-BLCA paired. (E) Representative immunohistochemistry images of cancerous and adjacent noncancerous tissues obtained from the Human Protein Atlas database (scale bar, 200 µm). **P<0.01; ***P<0.001. vs. SV-HUC-1/normal. BC/BLCA, bladder cancer; G6PD, glucose-6-phosphate dehydrogenase.
Discussion
BC is a complex disease associated with a high rate of morbidity and mortality (7). In the Kurdistan province of Iran, a population-based study of 321 patients with BC indicated a 5-year survival rate of 54% (2013–2018) (8). Numerous studies have characterized DNA and RNA alterations in NMIBC, which is one of the most frequently mutated human cancer types in terms of mutation rates, third to lung and skin cancer (29). The identification of common mutations has resulted in the development of novel therapeutic approaches, as well as urine and blood surveillance for both early detection and disease monitoring following treatment (30). In recent years, accumulating evidence has indicated that the tumor microenvironment has an important role in BC progression. A study suggested that radiation caused immunogenic tumor cell death through ferroptosis and polarized microenvironmental M2 tumor-associated macrophages (M2-TAMs) to M1-TAMs (31). In BC, the polarized M2 phenotype of tumor-associated macrophages is related to tumor grade and stage (32). In the terms of BC treatment, one study suggested that methyl-2-cyano-3,11-dioxo-18b-olean-1,12-dien-30-oate exerted its anti-tumor activity by inducing ROS (33). Sequential administration of gemcitabine→tamoxifen amplified the cytotoxicity of gemcitabine in BC cells (34). These studies provided new directions for BC therapy.
Ferroptosis has a unique role in anti-cancer therapeutic strategies. Ferroptosis is more lethal to tumor cells that rely on the suppression of ferroptosis to survive than apoptosis. At the same time, ferroptosis is also sensitive in certain special cases, such as tumor cells that are drug-resistant, aggressive or undergo epithelial-mesenchymal transition (35,36). In addition, a study conducted by Luan et al (37) identified nine drugs with the potential to treat BC.
In the present study, 60 FRGs that were selected from previous studies, 28 DEGs and 15 prognosis-associated genes were identified. GSVA analysis was used to investigate the phenotypic characteristics of TCGA samples. The enrichment of hallmark gene sets in tumor samples was indicated to be strongly associated with BC oncogenesis and progression. Among them, DNA repair was previously associated with the occurrence of prostate cancer (38), while mTORC1 was related to metastasis in colorectal cancer (39). Subsequently, six genes were selected to build the prognostic model based on their intersection with other gene sets. They were all associated with poor prognosis (hazard ratio >1, P<0.05).
GCLM is a component of the first rate-limiting enzyme of glutathione synthesis. GCL, which is composed of the catalytic (C) subunit GCLC and modifier subunit GCLM, is one of the major determinants of glutathione (GSH) synthesis (40). When GSH-dependent lipid peroxide repair systems are compromised, the lethal accumulation of lipid-based ROS results in ferroptosis, which is induced by the inhibition of cysteine uptake, decreased GSH levels or inactivation of the lipid repair enzyme GPX4 (41). Genetic loss of GCLM impairs a tumor's ability to drive malignant transformation. A study by Zhu et al (42) suggested that GCLM is an oncogene in esophageal adenocarcinoma. In addition, it is established that Andrographis treatment is effective in colorectal cancer and gastric cancer through dysregulating the expression of genes such as GCLM within the ferroptosis pathway (43,44). These findings provide evidence to support the use of GCLM as an adjuvant target treatment in cancers. Although GCLM has not been described in BC, the present findings indicate that it may also be a suitable target in BC.
CRYAB is a type of small heat shock protein (HSP), also known as HspB5. It was first discovered in the lens of the eye and has since been demonstrated to be ubiquitously expressed (45). Previous studies have concentrated on its role in apoptosis, where it acts as a negative regulator of Bax and caspase-3 (46). HspB1 has been identified as a negative regulator of ferroptosis-associated cancer cell death (47). In addition, HspB5 is associated with redox activity and inflammation. When CRYAB is expressed, it is possible to observe a decrease in the mitochondrial membrane potential and GSH. Subsequently, it may gradually inhibit iron uptake (48,49). It may also have a significant impact on ferroptosis-mediated cancer therapy. Increasing evidence suggests that CRYAB is associated with tumors. Overexpression of CRYAB inhibits BC cell migration and invasion through the PI3K/AKT and ERK pathways (50).
TFRC contributes to the ferroptosis process by collecting extracellular iron and replenishing the intracellular iron pool (51). This receptor is a promising target for cancer therapy due to its high expression on malignant cells, ability to internalize iron and requirement for cell proliferation. Targeting the TFRC has been demonstrated to be effective at delivering a wide variety of therapeutic agents and inducing cytotoxicity in cancer cells both in vitro and in vivo (52).
SQLE is a rate-limiting enzyme in cholesterol synthesis (53). Ferroptosis is a condition caused by the accumulation of iron-dependent lipid peroxides during cholesterol metabolism. Intracellular cholesterol promotes tumor formation or growth (54). In the present study, through GSEA analysis, enrichment of the cholesterol homeostasis pathway was identified in both high risk score groups and high G6PD expression groups. SQLE reduction caused by cholesterol accumulation aggravates colorectal cancer progression by activating the β-catenin oncogenic pathway and deactivating the p53 tumor suppressor pathway (55). Increased SQLE expression promotes cholesteryl ester biosynthesis, which induces hepatocellular carcinoma cell growth (56).
ZEB1, a central transcriptional component of fat cell differentiation, directly targets and modulates the expression of the majority of early and late adipogenic regulators (57). The sensitivity of GPX4 inhibitors has been reported to be associated with ZEB1 (36). GPX4 has a critical role in the regulation of ferroptotic cancer cell death (58).
G6PD is a rate-limiting enzyme of the pentose phosphate pathway, which promotes progression in a variety of cancer types (59–61). According to a previous study, patients with BC expressing a high level of G6PD may have a poor prognosis (62). In addition, inhibiting G6PD may decrease ROS accumulation and block the AKT pathway, affecting BC cell proliferation (62).
Ferroptosis in tumors has received increasing attention in recent years, although associated molecular changes and mechanisms in BC remain largely elusive. GO and KEGG enrichment analysis revealed potential mechanisms linking ferroptosis and BC, including immune-related, redox-related and stromal-related pathways. The KEGG pathway enrichment analysis revealed a significant enrichment of the PI3K/AKT/mTOR pathway, which has been implicated in helping cancer cells evade ferroptosis (63). A previous study on colorectal cancer suggested that the benzopyran derivative 2-imino-6-methoxy-2H-chromene-3-carbothioamide regulated the activity of the AMPK/mTOR/p70S6k signaling pathway, which is related to ferroptosis (64). However, this pathway warrants further investigation in BC. In addition, enrichment of numerous immune-related pathways indicated an association between ferroptosis and tumor immunity. A recent study by Wang et al (65) identified that tumor-specific ferroptosis regulators cysteinyl-tRNA synthetase exhibited a strong association with the expression of immune checkpoint genes, particularly PD-L1, demonstrating the influence of ferroptosis on the tumor microenvironment. Simultaneously, serum PD-L1 levels and the binding of CD68 and PD-L1 were associated with poor prognosis in patients with BC (66,67). Thus, ssGSEA was used for further study. While it has been established that a high infiltration of certain immune cells, such as macrophages, is associated with a poor prognosis in patients with BC, the role of others, such as Tregs, remains controversial (68). In addition, it was observed that the antigen presentation processes were more pronounced in the high-risk group. These points warrant further investigation.
G6PD was chosen for further study based on clinical characteristics analysis and the results of the immunotherapy cohort analysis. G6PD encodes glucose-6-phosphate dehydrogenase, which provides NADPH for fatty acid and nucleic acid synthesis. It has been suggested that G6PD is one of the genes involved in ferroptosis (69). High levels of G6PD expression were associated with unfavorable overall survival in patients with BC (62). Gu et al (70) reported that the G6PD-NADPH redox system was related to T cell metabolism. Given the association between high G6PD expression and immunotherapy response, as well as the characteristics of the immune microenvironment, it is indicated that follow-up studies on G6PD in patients with BC are necessary, while the majority of previous research has focused on G6PD in malaria.
Simultaneously, it was observed that several ferroptosis-related pathways, such as angiogenesis, glycolysis and mTORC1 signaling, were active in both the high-risk and high G6PD expression groups. A recent study indicated that the Rag/mTORC1/eukaryotic initiation factor 4E-binding proteins axis promoted the synthesis of GPX4 protein, which inhibits ferroptosis (71). As previously reported, PI3K/AKT/mTOR signaling may also suppress ferroptosis via lipogenesis (63). Chen et al (72) demonstrated that targeting activating transcription factor 4 increased ferroptosis sensitivity of tumor cells by suppressing angiogenesis.
There are still several limitations to this study. First, the sample size in the present study was small, and therefore, the data used for analysis are not comprehensive. Additional prospective clinical data are required to validate the reliability of the present data. Finally, it should be emphasized that the relationship between ferroptosis and BC requires additional basic experimental validations.
In conclusion, the present study successfully constructed an FRG signature with prognostic value for BC and further identified the potential value of G6PD for future research. This opens up new avenues for targeted therapies in BC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
This work is supported by the Innovation and Entrepreneurship Training Program for college students, Education Department of Jiangsu Province (grant no. KY102J2020058).
Availability of data and materials
The data generated in the present study are included in the figures and/or tables of this article. The data generated in the present study may be found under the following URLs: TCGA-BLCA: https://portal.gdc.cancer.gov/; GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13507.
Authors' contributions
YTW, HXH and MJJ designed and managed the entire study. YTW collected all the data. YTW, WCS and YQF analysed the results. JZT, DZ and QCW performed the experiments. YTW, WCS, QCW and YQF wrote the manuscript. YTW, JZT and DZ checked and confirmed the authenticity of the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lenis AT, Lec PM, Chamie K, Mshs MD. Bladder cancer: A review. JAMA. 2020;324:1980–1991. doi: 10.1001/jama.2020.17598. [DOI] [PubMed] [Google Scholar]
- 2.Richters A, Aben KKH, Kiemeney L. The global burden of urinary bladder cancer: An update. World J Urol. 2020;38:1895–1904. doi: 10.1007/s00345-019-02984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeGeorge KC, Holt HR, Hodges SC. Bladder cancer: Diagnosis and Treatment. Am Fam Physician. 2017;96:507–514. [PubMed] [Google Scholar]
- 4.Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: A global overview and recent trends. Eur Urol. 2017;71:96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Babjuk M, Burger M, Compérat EM, Gontero P, Mostafid AH, Palou J, van Rhijn BWG, Rouprêt M, Shariat SF, Sylvester R, et al. European association of urology guidelines on non-muscle-invasive bladder cancer (TaT1 and carcinoma in situ)-2019 update. Eur Urol. 2019;76:639–657. doi: 10.1016/j.eururo.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 6.Chang SS, Bochner BH, Chou R, Dreicer R, Kamat AM, Lerner SP, Lotan Y, Meeks JJ, Michalski JM, Morgan TM, et al. Treatment of non-metastatic muscle-invasive bladder cancer: AUA/ASCO/ASTRO/SUO guideline. J Urol. 2017;198:552–559. doi: 10.1016/j.juro.2017.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamat AM, Hahn NM, Efstathiou JA, Lerner SP, Malmström PU, Choi W, Guo CC, Lotan Y, Kassouf W. Bladder cancer. Lancet. 2016;388:2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- 8.Amiri M, Heshmatollah S, Esmaeilnasab N, Khoubi J, Ghaderi E, Roshani D. Survival rate of patients with bladder cancer and its related factors in Kurdistan province (2013–2018): A population-based study. BMC Urol. 2020;20:195. doi: 10.1186/s12894-020-00769-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mou Y, Wang J, Wu J, He D, Zhang C, Duan C, Li B. Ferroptosis, a new form of cell death: Opportunities and challenges in cancer. J Hematol Oncol. 2019;12:34. doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Du Y, Miao W, Jiang X, Cao J, Wang B, Wang Y, Yu J, Wang X, Liu H. The epithelial to mesenchymal transition related gene calumenin is an adverse prognostic factor of bladder cancer correlated with tumor microenvironment remodeling, gene mutation, and ferroptosis. Front Oncol. 2021;11:683951. doi: 10.3389/fonc.2021.683951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JN, Li T, Cheng L, Qin TS, Sun YX, Chen CT, He YZ, Liu G, Yao D, Wei Y, et al. Synthesis and in vitro anti-bladder cancer activity evaluation of quinazolinyl-arylurea derivatives. Eur J Med Chem. 2020;205:112661. doi: 10.1016/j.ejmech.2020.112661. [DOI] [PubMed] [Google Scholar]
- 13.Guo P, Wang L, Shang W, Chen J, Chen Z, Xiong F, Wang Z, Tong Z, Wang K, Yang L, et al. Intravesical in situ immunostimulatory gel for triple therapy of bladder cancer. ACS Appl Mater Interfaces. 2020;12:54367–54377. doi: 10.1021/acsami.0c15176. [DOI] [PubMed] [Google Scholar]
- 14.Liang C, Zhang X, Yang M, Dong X. Recent progress in ferroptosis inducers for cancer therapy. Adv Mater. 2019;31:e1904197. doi: 10.1002/adma.201904197. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Ma H, Meng L, Liu X, Lv Z, Zhang Y, Wang J. Construction and external validation of a ferroptosis-related gene signature of predictive value for the overall survival in bladder cancer. Front Mol Biosci. 2021;8:675651. doi: 10.3389/fmolb.2021.675651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Jensen MA, Zenklusen JC. A practical guide to the cancer genome atlas (TCGA) Methods Mol Biol. 2016;1418:111–141. doi: 10.1007/978-1-4939-3578-9_6. [DOI] [PubMed] [Google Scholar]
- 17.Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS, Jeong P, Kim MJ, Yun SJ, Lee KM, Moon SK, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer. 2010;9:3. doi: 10.1186/1476-4598-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandrashekar DS, Chakravarthi B, Robinson AD, Anderson JC, Agarwal S, Balasubramanya SAH, Eich ML, Bajpai AK, Davuluri S, Guru MS, et al. Therapeutically actionable PAK4 is amplified, overexpressed, and involved in bladder cancer progression. Oncogene. 2020;39:4077–4091. doi: 10.1038/s41388-020-1275-7. [DOI] [PubMed] [Google Scholar]
- 19.Lu M, Ge Q, Wang G, Luo Y, Wang X, Jiang W, Liu X, Wu CL, Xiao Y, Wang X. CIRBP is a novel oncogene in human bladder cancer inducing expression of HIF-1α. Cell Death Dis. 2018;9:1046. doi: 10.1038/s41419-018-1109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, et al. Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, Roberts MA, Tong B, Maimone TJ, Zoncu R, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doll S, Freitas FP, Shah R, Aldrovandi M, da Silva MC, Ingold I, Goya Grocin A, Xavier da Silva TN, Panzilius E, Scheel CH, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox's proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–395. doi: 10.1002/(SICI)1097-0258(19970228)16:4<385::AID-SIM380>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran L, Xiao JF, Agarwal N, Duex JE, Theodorescu D. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21:104–121. doi: 10.1038/s41568-020-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan C, Sun Y, Tian Y, Lu L, Dai X, Meng J, Huang J, He Q, Wu B, Zhang Z, et al. Irradiated tumor cell-derived microparticles mediate tumor eradication via cell killing and immune reprogramming. Sci Adv. 2020;6:eaay9789. doi: 10.1126/sciadv.aay9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi H, Tanaka M, Tanaka A, Tsunemi A, Yamamoto H. Predominance of M2-polarized macrophages in bladder cancer affects angiogenesis, tumor grade and invasiveness. Oncol Lett. 2016;11:3403–3408. doi: 10.3892/ol.2016.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi H, Taoka R, Mmeje CO, Jinesh GG, Safe S, Kamat AM. CDODA-Me decreases specificity protein transcription factors and induces apoptosis in bladder cancer cells through induction of reactive oxygen species. Urol Oncol. 2016;34:337.e11–8. doi: 10.1016/j.urolonc.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi H, Mmeje CO, Jinesh GG, Taoka R, Kamat AM. Sequential gemcitabine and tamoxifen treatment enhances apoptosis and blocks transformation in bladder cancer cells. Oncol Rep. 2015;34:2738–2744. doi: 10.3892/or.2015.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, Galeas J, Dhruv HD, Berens ME, Schreiber SL, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–250. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, Kaffenberger SD, Eaton JK, Shimada K, Aguirre AJ, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luan JC, Zeng TY, Zhang QJ, Xia DR, Cong R, Yao LY, Song LB, Zhou X, Zhou X, Chen X, et al. A novel signature constructed by ferroptosis-associated genes (FAGs) for the prediction of prognosis in bladder urothelial carcinoma (BLCA) and associated with immune infiltration. Cancer Cell Int. 2021;21:414. doi: 10.1186/s12935-021-02096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu B, Lu X, Shen H, Yuan X, Wang X, Yin N, Sun L, Shen P, Hu C, Jiang H, Wang D. Intratumoral heterogeneity and genetic characteristics of prostate cancer. Int J Cancer. 2020;146:3369–3378. doi: 10.1002/ijc.32961. [DOI] [PubMed] [Google Scholar]
- 39.Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M, Lee EY, Weiss HL, O'Connor KL, Gao T, Evers BM. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011;71:3246–3256. doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu SC. Regulation of glutathione synthesis. Mol Aspects Med. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kajarabille N, Latunde-Dada GO. Programmed cell-death by ferroptosis: Antioxidants as mitigators. Int J Mol Sci. 2019;20:4968. doi: 10.3390/ijms20194968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu L, Yang F, Wang L, Dong L, Huang Z, Wang G, Chen G, Li Q. Identification the ferroptosis-related gene signature in patients with esophageal adenocarcinoma. Cancer Cell Int. 2021;21:124. doi: 10.1186/s12935-021-01821-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma P, Shimura T, Banwait JK, Goel A. Andrographis-mediated chemosensitization through activation of ferroptosis and suppression of β-catenin/Wnt-signaling pathways in colorectal cancer. Carcinogenesis. 2020;41:1385–1394. doi: 10.1093/carcin/bgaa090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma R, Shimura T, Yin C, Okugawa Y, Kitajima T, Koike Y, Okita Y, Ohi M, Uchida K, Goel A, et al. Antitumor effects of Andrographis via ferroptosis-associated genes in gastric cancer. Oncol Lett. 2021;22:523. doi: 10.3892/ol.2021.12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Liu J, Wu J, Li W, Chen Z, Yang L. Progression of the role of CRYAB in signaling pathways and cancers. Onco Targets Ther. 2019;12:4129–4139. doi: 10.2147/OTT.S201799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu WF, Gong L, Cao Z, Ma H, Ji W, Deng M, Liu M, Hu XH, Chen P, Yan Q, et al. αA- and αB-crystallins interact with caspase-3 and Bax to guard mouse lens development. Curr Mol Med. 2012;12:177–187. doi: 10.2174/156652412798889036. [DOI] [PubMed] [Google Scholar]
- 47.Sun X, Ou Z, Xie M, Kang R, Fan Y, Niu X, Wang H, Cao L, Tang D. HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene. 2015;34:5617–5625. doi: 10.1038/onc.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christopher KL, Pedler MG, Shieh B, Ammar DA, Petrash JM, Mueller NH. Alpha-crystallin-mediated protection of lens cells against heat and oxidative stress-induced cell death. Biochim Biophys Acta. 2014;1843:309–315. doi: 10.1016/j.bbamcr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prabhu S, Srinivas V, Ramakrishna T, Raman B, Rao Ch M. Inhibition of Cu2+-mediated generation of reactive oxygen species by the small heat shock protein αB-crystallin: The relative contributions of the N- and C-terminal domains. Free Radic Biol Med. 2011;51:755–762. doi: 10.1016/j.freeradbiomed.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 50.Ruan H, Li Y, Wang X, Sun B, Fang W, Jiang S, Liang C. CRYAB inhibits migration and invasion of bladder cancer cells through the PI3K/AKT and ERK pathways. Jpn J Clin Oncol. 2020;50:254–260. doi: 10.1093/jjco/hyz172. [DOI] [PubMed] [Google Scholar]
- 51.Feng H, Schorpp K, Jin J, Yozwiak CE, Hoffstrom BG, Decker AM, Rajbhandari P, Stokes ME, Bender HG, Csuka JM, et al. Transferrin receptor is a specific ferroptosis marker. Cell Rep. 2020;30:3411–3423.e7. doi: 10.1016/j.celrep.2020.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daniels TR, Bernabeu E, Rodríguez JA, Patel S, Kozman M, Chiappetta DA, Holler E, Ljubimova JY, Helguera G, Penichet ML. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim Biophys Acta. 2012;1820:291–317. doi: 10.1016/j.bbagen.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto S, Bloch K. Studies on squalene epoxidase of rat liver. J Biol Chem. 1970;245:1670–1674. doi: 10.1016/S0021-9258(19)77144-0. [DOI] [PubMed] [Google Scholar]
- 54.Xu H, Zhou S, Tang Q, Xia H, Bi F. Cholesterol metabolism: New functions and therapeutic approaches in cancer. Biochim Biophys Acta Rev Cancer. 2020;1874:188394. doi: 10.1016/j.bbcan.2020.188394. [DOI] [PubMed] [Google Scholar]
- 55.Jun SY, Brown AJ, Chua NK, Yoon JY, Lee JJ, Yang JO, Jang I, Jeon SJ, Choi TI, Kim CH, Kim NS. Reduction of squalene epoxidase by cholesterol accumulation accelerates colorectal cancer progression and metastasis. Gastroenterology. 2021;160:1194–1207.e28. doi: 10.1053/j.gastro.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 56.Liu D, Wong CC, Fu L, Chen H, Zhao L, Li C, Zhou Y, Zhang Y, Xu W, Yang Y, et al. Squalene epoxidase drives NAFLD-induced hepatocellular carcinoma and is a pharmaceutical target. Sci Transl Med. 2018;10:eaap9840. doi: 10.1126/scitranslmed.aap9840. [DOI] [PubMed] [Google Scholar]
- 57.Gubelmann C, Schwalie PC, Raghav SK, Röder E, Delessa T, Kiehlmann E, Waszak SM, Corsinotti A, Udin G, Holcombe W, et al. Identification of the transcription factor ZEB1 as a central component of the adipogenic gene regulatory network. Elife. 2014;3:e03346. doi: 10.7554/eLife.03346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu T, Zhang C, Tang Q, Su Y, Li B, Chen L, Zhang Z, Cai T, Zhu Y. Variant G6PD levels promote tumor cell proliferation or apoptosis via the STAT3/5 pathway in the human melanoma xenograft mouse model. BMC Cancer. 2013;13:251. doi: 10.1186/1471-2407-13-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang C, Zhang Z, Zhu Y, Qin S. Glucose-6-phosphate dehydrogenase: A biomarker and potential therapeutic target for cancer. Anticancer Agents Med Chem. 2014;14:280–289. doi: 10.2174/18715206113136660337. [DOI] [PubMed] [Google Scholar]
- 61.Lu M, Lu L, Dong Q, Yu G, Chen J, Qin L, Wang L, Zhu W, Jia H. Elevated G6PD expression contributes to migration and invasion of hepatocellular carcinoma cells by inducing epithelial-mesenchymal transition. Acta Biochim Biophys Sin (Shanghai) 2018;50:370–380. doi: 10.1093/abbs/gmy009. [DOI] [PubMed] [Google Scholar]
- 62.Chen X, Xu Z, Zhu Z, Chen A, Fu G, Wang Y, Pan H, Jin B. Modulation of G6PD affects bladder cancer via ROS accumulation and the AKT pathway in vitro. Int J Oncol. 2018;53:1703–1712. doi: 10.3892/ijo.2018.4501. [DOI] [PubMed] [Google Scholar]
- 63.Yi J, Zhu J, Wu J, Thompson C, Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc Natl Acad Sci USA. 2020;117:31189–31197. doi: 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang L, Liu W, Liu F, Wang Q, Song M, Yu Q, Tang K, Teng T, Wu D, Wang X, et al. IMCA induces ferroptosis mediated by SLC7A11 through the AMPK/mTOR pathway in colorectal cancer. Oxid Med Cell Longev. 2020;2020:1675613. doi: 10.1155/2020/6901472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang S, Chen S, Ying Y, Ma X, Shen H, Li J, Wang X, Lin Y, Liu B, Zheng X, Xie L. Comprehensive analysis of ferroptosis regulators with regard to PD-L1 and immune infiltration in clear cell renal cell carcinoma. Front Cell Dev Biol. 2021;9:676142. doi: 10.3389/fcell.2021.676142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang LR, Zhang N, Chen ST, He J, Liu YH, Han YQ, Shi XQ, Yang JJ, Mu DY, Fu GH, Gao F. PD-1-positive tumor-associated macrophages define poor clinical outcomes in patients with muscle invasive bladder cancer through potential CD68/PD-1 complex interactions. Front Oncol. 2021;11:679928. doi: 10.3389/fonc.2021.679928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krafft U, Olah C, Reis H, Kesch C, Darr C, Grünwald V, Tschirdewahn S, Hadaschik B, Horvath O, Kenessey I, et al. High serum PD-L1 levels are associated with poor survival in urothelial cancer patients treated with chemotherapy and immune checkpoint inhibitor therapy. Cancers (Basel) 2021;13:2548. doi: 10.3390/cancers13112548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Wilpe S, Gerretsen E, van der Heijden A, de Vries I, Gerritsen W, Mehra N. Prognostic and predictive value of tumor-infiltrating immune cells in urothelial cancer of the bladder. Cancers (Basel) 2020;12:2692. doi: 10.3390/cancers12092692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu M, Zhou X, Sohn JH, Zhu L, Jie Z, Yang JY, Zheng X, Xie X, Yang J, Shi Y, et al. NF-κB-inducing kinase maintains T cell metabolic fitness in antitumor immunity. Nat Immunol. 2021;22:193–204. doi: 10.1038/s41590-020-00829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Swanda RV, Nie L, Liu X, Wang C, Lee H, Lei G, Mao C, Koppula P, Cheng W, et al. mTORC1 couples cyst(e)ine availability with GPX4 protein synthesis and ferroptosis regulation. Nat Commun. 2021;12:1589. doi: 10.1038/s41467-021-21841-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen D, Fan Z, Rauh M, Buchfelder M, Eyupoglu IY, Savaskan N. ATF4 promotes angiogenesis and neuronal cell death and confers ferroptosis in a xCT-dependent manner. Oncogene. 2017;36:5593–5608. doi: 10.1038/onc.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in the present study are included in the figures and/or tables of this article. The data generated in the present study may be found under the following URLs: TCGA-BLCA: https://portal.gdc.cancer.gov/; GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13507.


